Abstract
During the past decade, we have experienced a revolution in the biological sciences resulting from the flux of information generated by genome-sequencing efforts. Our understanding of living organisms, the metabolic processes they catalyze, the genetic systems encoding cellular protein and stable RNA constituents, and the pathological conditions caused by some of these organisms has greatly benefited from the availability of complete genomic sequences and the establishment of comprehensive databases. Many research institutes around the world are now devoting their efforts largely to genome sequencing, data collection and data analysis. In this review, we summarize tools that are in routine use in our laboratory for characterizing transmembrane transport systems. Applications of these tools to specific transporter families are presented. Many of the computational approaches described should be applicable to virtually all classes of proteins and RNA molecules.
Key Words: Transport proteins, Phylogeny, Superfamily, Computer program, Evolution
Introduction
Reflecting on the great scientific advances of the 20th century, Albert Einstein stated that ‘the bound power of the atom has changed everything, but not our thought processes. We need a totally new way of thinking if humankind is to survive’. Genomics may prove to be the greatest advance in the biological sciences of the past two decades [Saier, 1998]. If so, the equivalent statement for biologists might be: ‘Genomics has changed everything but not our thought processes. We need a totally new way of thinking if humankind is to extract the information made available by genomics.’
E. coli is the best understood organism on Earth. A majority of its gene products have been functionally identified [Riley et al., 2006; Rudd, 2000; Serres et al., 2004]. However, we would maintain that less than 1% of the information encoded within its fully sequenced genome is understood. For example, protein sequences must contain information about protein-protein interactions and the formation of stable and transient metabolons, but this information is difficult to discern from genomic data [Amar et al., 2008; Norris et al., 2007a, b; Skrabanek et al., 2008].
In fact, every detail, or at least every programmed detail, of every living organism is encoded within its genome. It is the immense task of bioinformatics to decipher that information. And it is the even greater task of biosystematics to render that information intelligible to the feeble human mind [Busch and Saier, 2002; Rehm, 2001].
So what perspective should be used in designing systematic approaches to genomics? As we now recognize, ‘Nothing in biology makes sense except in the light of evolution’ [Dobzhansky, 1964]. Thus, any biosystematic approach to the classification of biological entities must take cognizance of evolution. Molecular phylogeny reflects the evolutionary processes, and is therefore the most suitable guide to structure, function, mechanism, metabolism, physiology, pathology and ecology – in short, everything that is of interest to the biologist [Chang et al., 2004; Doolittle, 1992; Saier, 1998; Weiss and Bachanan, 2003].
We can think of the genome as a book! Bioinformatics is the tool that allows us to read it, and evolution is the framework upon which it all makes sense. The genomes of all living organisms comprise the library of life. Unfortunately, few of us have the time, energy or ingenuity to read all the books in the library. We therefore need as much help as possible in order to understand the molecular bases by which genomes provide the instructions of life. However, slowly, they are revealing, among other things, how protein structural complexity arose over evolutionary time [Doolittle, 1989; Saier, 2003].
Dozens of eukaryotic genomes and hundreds of prokaryotic genomes have now been fully sequenced, and the rates of completion of genome sequencing projects continue to increase exponentially due to technological advances [Mulder et al., 2008]. Unfortunately, the work of bioinformaticists lags behind sequencing efforts, and consequently, valuable information, potentially available through genome sequencing, has not yet come to light. In this article we summarize the approaches that we have developed and routinely use for genome-derived protein sequence analysis. This work has led to a much clearer understanding of molecular evolution and its many ramifications [Barabote et al., 2006; Bush and Saier, 2002; Serres and Riley, 2005].
The Essentiality of Transmembrane Transport
Transport systems are essential to every living cell [Hollenstein et al., 2007; Papanikou et al., 2007; Saier, 2000]. They (1) allow the entry to all essential nutrients into the cell and its compartments at rates sufficient to support life, (2) regulate the cytoplasmic concentrations of metabolites by both uptake and excretion mechanisms, (3) provide physiologically relevant cellular concentrations of ions that can differ by several orders of magnitude from those in the external medium, (4) export macromolecules such as complex carbohydrates, proteins, lipids and DNA, (5) catalyze export and uptake of signaling molecules that mediate intercellular communication, (6) prevent toxic effects of drugs and toxins by catalyzing their active efflux, (7) promote the generation of ion electrochemical gradients, and (8) participate in biological warfare by exporting biologically active agents that insert into or permeate the membranes of target cells. Thus, transport is an essential aspect of all life-endowing processes: metabolism, communication, biosynthesis, reproduction, and both cooperative and antagonistic interorganismal behaviors [Hollenstein et al., 2007; Saier, 2000].
The Transporter Classification Database (TCDB) (www.tcdb.org/)
The transporter classification (TC) system [Busch and Saier, 2002; Saier, 2000], formally adopted by the International Union of Biochemistry and Molecular Biology (IUBMB) in June 2001, provides a guide to the known types of transport proteins present in living organisms on earth. The development of a classification system for transport proteins has allowed us to comprehensively view transport systems from structural, functional, and evolutionary standpoints, and to trace pathways taken for their evolution [Busch and Saier, 2004; Saier, 2003]. This development has been strongly influenced by recent progress in computational biology and genome sequencing. Since our last two comprehensive descriptions of the TC system [Busch and Saier, 2002; Saier, 2000], we have expanded the transporter classification system by (1) introducing new families and classes of transporters, (2) expanding the memberships of pre-existing families, (3) providing more detailed annotations of these families and proteins, (4) updating reference citations relevant to proteins described in the TC system, and (5) creating a more interactive database (TCDB). The results of our analyses, made possible by these updates, are summarized here as are some of the most important software tools developed to support it [for more detailed but less current accounts of these efforts, see Busch and Saier, 2002 and Saier et al., 2006, 2009].
More than 500 protein families are currently in the TC system (see TCDB). Affiliation with a family requires satisfying rigorous statistical criteria of homology (see next section). Whereas the classes and subclasses distinguish functionally distinct types of transporters, the families and subfamilies provide a phylogenetic basis for classification (table 1). The TC system is thus a functional/phylogenetic system of classification. Families sometimes, but rarely, cross class or subclass lines [Busch and Saier, 2002; Saier, 1998]. Hyperlinks have been constructed to define superfamilies, identify disease-related transporters, and identify sources of high-resolution 3-D structural data. Several types of search tools facilitate rapid protein identification and characterization.
Table 1.
Families within the APC superfamily
| TC No. | Family name | Family abbreviation | Number of proteins in TCDB | Organismal type |
|---|---|---|---|---|
| 2.A.3.1 | amino acid transporter family | AAT | 13 | B |
| 2.A.3.2 | basic amino acid/polyamine antiporter family | APA | 6 | B |
| 2.A.3.3 | cationic amino acid transporter family | CAT | 5 | E |
| 2.A.3.4 | amino acid/choline transporter family | ACT | 6 | E |
| 2.A.3.5 | ethanolamine transporter family | EAT | 2 | A, B |
| 2.A.3.6 | archaeal/bacterial transporter family | ABT | 1 | A, B |
| 2.A.3.7 | glutamate: GABA antiporter family | GGA | 1 | B |
| 2.A.3.8 | L-type amino acid transporter family | LAT | 15 | B, E |
| 2.A.3.9 | spore germination protein family | SGP | 3 | B |
| 2.A.3.10 | yeast amino acid transporter family | YAT | 20 | E |
| 2.A.3.11 | aspartate/glutamate transporter family | AGT | 1 | A, B |
| 2.A.3.12 | polyamine: H+ symporter family | PHS | 1 | E |
| 2.A.3.13 | amino acid efflux family | AAE | 1 | B |
| 2.A.18 | amino acid/auxin permease family | AAAP | 26 | E |
| 2.A.25 | alanine or glycine:cation symporter family | AGCS | 3 | A, B |
| 2.A.30 | cation-chloride cotransporter family | CCC | 10 | E |
| 2.A.42 | hydroxy/aromatic amino acid permease family | HAAAP | 6 | B |
APC = Amino acid/polyamine/organocation.
Family transporter classification (TC) number for the 17 current members of the APC superfamily. In the current study, only the AGCS (TC 2.A.25) family was not included, as it was not known to be a member of the APC superfamily when these studies were initiated. The last four entries are established TC families that were later shown to be members of the APC superfamily.
Their superfamily status is indicated in TCDB by a hyperlink. The original family TC numbers were retained because of the stipulation by the International Union of Biochemistry and Molecular Biology (IUBMB) that the TC system must be a static system of classification.
Organismal type from which the proteins derive: B = bacteria; A = archaea; E = eukaryotes.
Recognition of a phylogenetic relationship based on sequence similarity allows certain conclusions to be drawn regarding three-dimensional structural features. Any two proteins that can be shown to be homologous (i.e. that exhibit sufficient primary and/or secondary structural similarity to establish that they arose from a common evolutionary ancestor) can be expected to exhibit strikingly similar topological features and 3-D structures, although a few exceptions have been noted [Emanuelsson et al., 2007; Saier, 2003]. Therefore, extrapolation from one member of a family of known structure to other members becomes justifiable, and the degree of confidence in such an extrapolation process is inversely related to the degree of sequence divergence. However, extrapolation of structural data to other proteins is never justified if homology has not been established.
Similar arguments apply to mechanistic considerations. Thus, the mechanism of solute transport is likely to be similar for all members of a permease family, with variations on a specific mechanistic theme being greatest when the sequence divergence is greatest [Pollock, 2002; Yen et al., 2002]. By contrast, for members of any two independently evolving permease families, the transport mechanisms may be entirely different. Extensive experimental work has established that phylogenetic data can also be used to predict substrate specificity, polarity of transport, and even intracellular localization, depending on the family and the degree of sequence divergence observed within that family [Busch and Saier, 2002; Emanuelsson et al., 2007].
Transport system families included in the current TC system are described in database format in TCDB [Saier et al., 2006]. TCDB provides detailed descriptions of and reference citations for (1) TC classes, (2) subclasses, (3) families, (4) subfamilies, and (5) individual proteins. Additionally, relevant software tools can be found on the TC website, facilitating examination and analysis of the world of transport proteins. The current list of transporter classes and subclasses is provided in table 1. TCDB is equipped with search tools that allow the user to search by key word, gene name, family, or protein sequence. Most proteins demonstrably homologous to a TC family member can be identified using TC-BLAST. Finally, TCDB is interconnected with other useful databases and websites [Saier et al., 2006; 2009].
Establishing Homology between Proteins
Statistical algorithms are used to establish homology between two proteins, two families of proteins, or two repeat sequences within the proteins of a single family [Zhai and Saier, 2002; Zhou et al. 2003]. In general, these depend on the ‘superfamily principle’ [Doolittle, 1981; 1986; Saier, 1994]. This principle simply states that if A is homologous to B, and B is homologous to C, then A is homologous to C. Care must be taken, however, that in establishing homology, corresponding domains or regions of the protein are being compared [Barabote et al., 2006; Serres and Riley, 2005]. Moreover, a reliable program must take into account unusual residue compositions as, for example, occur with membrane proteins that have a disproportionate percentage of hydrophobic residues, or proteins with multiple short repeat sequences that comprise a substantial fraction of the proteins or protein segments compared [Serres and Riley, 2005; Zhai and Saier, 2002].
An average protein domain is roughly 60 residues long. Therefore, we have arbitrarily set the minimal length of sequences to be compared for purposes of establishing homology as 60 residues [Saier, 1994]. Further, if the two proteins are expected to be homologous throughout their lengths, these homologous segments should occur in comparable regions of the two proteins unless it can be shown that repeat sequences exist or domain shuffling has occurred [Barabote et al., 2005; 2006]. Then the two segments must occur in comparable regions of the repeats or domains. When protein domain order changes as sequences change through the evolutionary process, these changes must be taken into account [Barabote and Saier, 2005].
In 1994, we set up somewhat arbitrary, but very rigorous criteria for the purpose of establishing common evolutionary ancestry [Saier, 1994]. In the past 15 years, these procedures have never yielded false-positives. To be considered homologous, two proteins, when correctly aligned to maximize identities and similarities and minimize gaps, must give a comparison score of 9 SD. This value corresponds to a probability of 10–19 that this degree of sequence similarity could have occurred by chance [Dayhoff et al., 1983]. As noted above, the segments to be compared must align over a stretch of a least 60 residues. This requirement eliminates the possibility that convergent sequence evolution could account for the degree of similarity observed [Saier, 1994; 2000].
It is useful, not merely to establish homology when possible, but also to delineate independent origins for distinct families showing no significant sequence similarity. This is only possible if it can be shown that members of these two families arose by two different evolutionary pathways. Thus, two families with members possessing 6 transmembrane α-helical spanners (TMSs) must have arisen independently if one displays 3 repeat units of 2 TMSs while the other displays 2 repeats of 3 TMSs. The first arose by intragenic triplication of a 2 TMS-encoding genetic segment, while the other arose by intragenic duplication of a 3 TMS-encoding genetic unit. Of course, there still remains the possibility that the primordial 3 TMS unit arose from the 2 TMS unit by addition of an extra TMS, or vice versa.
When we began our bioinformatics studies 22 years ago, there were only about 3,000 nonredundant protein sequences in the NCBI GenBank database, and half of these sequences had been determined by protein rather than DNA sequencing [Saier and McCaldon, 1988]. The probability of finding homologous sequences was meager, partly because search tools were not well developed, but also because so few sequences were available for comparative analyses. By May 2008, the number had increased to 6.6 million. The probability of finding a homologue today is over 2000 fold what it was in 1988. Similarly, the probability of identifying two sequences with a specified degree of sequence similarity by chance has increased proportionally to the numbers of sequences available. Consequently, the rigor of the criteria used to establish homology should be increased as the number of sequences increases. Since the probability has increased over 103-fold, we should require a corresponding increase in rigor. We suggest that 10 SD (probability of 10–24 that the similarity of two sequences arose by chance) should replace 9 SD (probability of 10–19). A value of 10 SD should allow a 100× increase in database size over the present value without loss of rigor [Dayhoff et al., 1983]. It is likely that this degree of expansion will be achieved within the next few years.
The GAP program [Devereaux et al., 1984] randomly shuffles two sequences being compared 100 times and compares the actual aligned sequences with alignments of the shuffled sequences. This method eliminates artifacts due to unusual amino acid compositions. However, our empirical experience with this program has revealed that 500 random shuffles are required to obtain reliable results. Therefore, we designed a modified program (the Intra/InterCompare program; IC) [Zhai and Saier, 2002; unpubl. modifications], which has three principal advantages over GAP: (1) It automatically conducts five 100-shuffle runs and averages the results. (2) It can take any number of sequences known to be homologous to a protein or protein domain A, and compares them to any number of sequences known to be homologous to protein or domain B. If protein/domain C (homologous to A) shows over 10 SD with protein/domain D (homologous to B), then since A is homologous to C, C is homologous to D, and D is homologous to B, by the superfamily principle [Doolittle, 1981, 1986], A must be homologous to B. The IC program can, for example, compare 100 homologues of A with 100 homologues of B to give 10,000 comparison scores. (3) The IC program presents the results as specified by the user. It is most useful to arrange the results according to the value of the average comparison score with the best values at the top. This allows the investigator to quickly identify the best comparisons for further examination [Chang et al., 2004].
The IC program can take a few hours (e.g. overnight) to compare 100 sequences with another 100 sequences, yielding 10,000 comparisons expressed in SD. Consequently, the number of proteins that can be compared is limited. If BLAST searches of proteins A and B yield 500 sequences each, this number must be reduced. This becomes possible due to the availability of the CD-Hit program [Li and Godzik, 2006]. This program eliminates all redundancies and all sequences with a percent identity greater than some specified value. The default setting is 90%. Thus, with the default setting, only one protein of all the retrieved sequences with greater than 90% identity will be retained. If too many sequences are still retained, a lower cut off value (80% or 70% or 60% identity) can be used. In this way, the desired number of sequences can be fed into the IC program.
A problem with the CD-Hit program is that the retained sequences may be fragments of complete protein sequences (rather than the full-length sequences). We have therefore modified CD-Hit so that only sequences of ‘normal’ length are retained. The program works as follows: the script summarizes the sizes of all the proteins obtained by a BLAST search. A decision is then made to exclude presumed fragmentary sequences. This is done by selecting a size range. All sequences of less than a specified value are then eliminated. Similarly, the program can eliminate sequences that are greatly in excess of the average. Thus, in addition to redundancies, fragments and abnormally long sequences can be eliminated if desired. The product, a list of ‘normal’ sized homologues, is tabulated. The result is a table made by the MakeTable program (unpublished program available at our TCDB website). The table summarizes (1) the protein abbreviation, (2) a description of the protein from the NCBI database, (3) its organismal source, (4) the size of the protein in numbers of amino acyl residues (aas), (5) the GI number of the sequence, (6) the organismal type, and (7) the organismal domain (bacteria, eukaryotes or archaea). It allows easy access to information such as organismal distribution of homologues and size distribution.
Extra large homologues are often ‘fusion’ proteins. Their identity can reveal functional aspects of a transporter of unknown substrate specificity as demonstrated in several publications [Barabote et al., 2005; 2006; Felce and Saier, 2004; Harvat et al., 2005].
The two sets of proteins are then compared with each other. Two programs are available for this purpose: IC and GS (Get Score). The IC program is described above [Zhai and Saier, 2002]. The GS program functions as follows. The two lists of proteins can be compared by: (1) BLAST [Altschul et al., 1997; Yu et al., 2006] or (2) SSearch [Pearson, 1998]. In the latter program, for any binary comparison, the two bit scores are averaged, and based on a standard curve, they are converted to a comparison score expressed in SDs. Because SSearch compares the binary alignment with 500 randomly shuffled sequences, this program, like GAP and IC, corrects for abnormal amino acid compositions. An advantage of GS over IC is that it takes only about 1% as much computer time. Thus, comparing 100 sequences with 100 sequences takes several hours with IC, but only a few minutes with GS.
Establishing Homology between Internal Protein Repeats
The IC or GS program can also be used to compare internal regions within a set of homologous sequences. However, one needs first to identify the boundaries between putative repeat sequences, and then to cut them into the segments to be compared. [Zhou et al. 2003] developed novel programs for displaying and analyzing the α-helical transmembrane segments (TMSs) in the aligned sequences of homologous integral membrane proteins. TMS_ALIGN predicts the positions of putative TMSs in multiply aligned protein sequences and graphically shows the TMSs in the alignment. TMS_SPLIT (1) predicts the positions of TMSs for each sequence, (2) allows a user to select proteins with a specified number of TMSs, and (3) splits the sequences into groups of TMSs of equal numbers. TMS_CUT works like TMS_SPLIT, but it can cut sequences with any combination of TMSs as specified. The BASS program [Zhou et al., 2003] similarly allows comparison of protein repeat elements, equivalent to TMS_SPLIT plus IC, but it provides the comparison data expressed in BLAST e values instead of S.D. values. These programs, together with the IC program, facilitate the identification of repeat sequences in integral membrane proteins. They also facilitate the bioinformatic determination of integral membrane protein topology and the prediction of evolutionary origin pathways. Theoretically, these programs can be used to establish homology for internal segments within any type of protein or nucleic acid.
Estimating Topologies of Transmembrane Proteins
We have also designed programs for displaying the topological features of individual proteins. One such program is the Web-based Hydropathy, Amphipathicity and Topology (WHAT) program [Zhai and Saier, 2001a]. This program uses a sliding window (default setting of 19 residues for α-helices or 9 residues for β-strands) to determine and plot the hydropathy, amphipathicity, secondary structure and predicted transmembrane topology along the length of any protein sequence. This method is based on programs designed by us, but also on preexisting programs including the hydrophobic moment program [Eisenberg et al., 1982], the Tree program [Feng and Doolittle, 1990; 1996], JNET [Cuff et al., 1998], MEMSAT [Jones et al., 1994], and HMMTOP [Tusnady and Simon, 2001], programs for secondary structure and transmembrane topology predictions. The WHAT program has a user-friendly interface and uses a convenient input format.
Recently, we have modified this program in two principle respects. In contrast to the original WHAT program, WHAT2 does not store the sequences analyzed (for security reasons), and it is written in JAVA instead of C (because of ease of use and increased flexibility). It gives results that are very similar to those obtained with WHAT. To predict orientation in the membrane, we now use the HMMTOP program, which is a combined transmembrane topology and signal peptide predictor [Emanuelsson et al., 2007; Melen et al., 2003; Sonnhammer et al., 1998; Tusnady and Simon, 2001].
Because the WHAT program analyzes a single sequence, it has limited reliability. Much greater accuracy results when the plots for several correctly aligned homologous sequences are used. The more sequences analyzed, the more reliable the prediction. The program that allows this to be accomplished is the AveHas program [Zhai and Saier, 2001b]. It has been used in many applications [Lee et al., 2007; Yamaguchi and Saier, 2007; Yen and Saier, 2007].
To align homologous sequences, numerous programs are available. Each is based on a different set of assumptions. These programs include CLUSTAL X (neighbor joining), ProtPars (parsimony) and PAUP (maximum likelihood). All of the programs function reliably when the sequence similarity between homologues is sufficient to insure correct alignment. The CLUSTAL X program [Thompson et al., 1994] has been used for the generation of phylogenetic trees [Zhai et al., 2002] and for producing average hydropathy, amphipathicity, and similarity plots [AveHAS; Zhai and Saier, 2001b]. This method is based on the TREEMOMENT and Hydro programs [Le et al., 1999]. It has a user-friendly interface, a convenient input format and an improved algorithm. We have modified this program so it is written in JAVA rather than C, and so it provides predictions of the transmembrane segments as well as orientation in the membrane. All of these programs can be found on our Biotools Server (http://saier-144–37.ucsd.edu) associated with TCDB.
An Example of the Use of These Programs to Characterize the 4-Toluene Sulfonate Uptake Permease (TSUP) Family (9.A.29)
The putative 4-toluene sulfonate uptake permease (TSUP) family (TC# 9.A.29; also called the DUF81 family) is large (over 500 members) and diverse in sequence. These proteins are present in Gram-negative and Gram-positive bacteria as well as archaea and eukaryotes. The eukaryotic proteins can be similar in size to the prokaryotic homologues or else about twice as large. One bacterial member, TsaS (239 aas), has been proposed to be a secondary carrier for 4-toluene sulfonate uptake in Comamonas testosteroni T2 [Locher et al., 1993; Mampel et al., 2004; Tralau et al., 2001]. A homologue, TanE in Cupriavidus necator, may be a sulfite exporter, involved in the metabolism of C2 sulfonates [Weinitschke et al., 2007]. However, the phylogenetic relationships of TSUP family members, the evolutionary origins of these proteins, the mechanism(s) of transport and the energy coupling mechanism(s) had not been investigated.
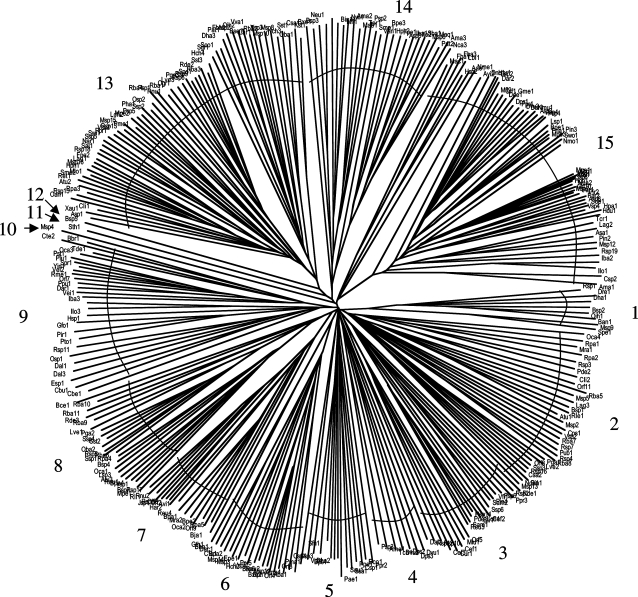
Using the putative 4-toluene sulfonate uptake permease of Comamonas testosteroni (TC# 9.A.29.1.1), hundreds of bacterial DUF81 homologues were retrieved in PSI-BLAST searches with a single iteration. The modified CD-Hit program (65% identity cutoff) was used to eliminate redundancies and closely related sequences. 318 sequences remained as listed in online supplementary table S1 (www.karger.com/doi/10.1159/000239667), which also presents protein designations and abbreviations, genbank index (GI) numbers and organismal sources. This table, generated with the MakeTable program, presents these homologues according to phylogenetic cluster as revealed in the tree shown in figure 1. The multiple alignment, upon which this tree was based, is shown in online supplementary figure S1.
Fig. 1.
Phylogenetic tree of 309 prokaryotic members of the TSUP family. The tree is based on a CLUSTAL X (neighbor joining) multiple alignment (online suppl. fig. S1).
A brief analysis of online supplementary table S1 reveals that almost all proteins exhibit 230–280 aas per polypeptide chain and an estimated 7–9 transmembrane segments (TMSs). A few larger proteins have C-terminal hydrophilic extensions. These proteins were analyzed according to phylogenetic cluster, and in all 15 clusters, the average sizes of the proteins varied from 242 to 266 aas (average overall size of 254 aas) with estimated numbers of TMSs = 6.5–8.1 (average value of 7.7). It is clear that these proteins exhibit a surprisingly uniform size and topology.
Organisms represented that possess homologues of the TSUP family include many bacteria, archaea and eukaryotes, but only prokaryotic proteins were included in this study. The bacterial homologues derive from α-, β-, γ- and δ-proteobacteria, firmicutes, actinobacteria, cyanobacteria, bacteroides, and a few other bacterial phyla including Planktomycetes, Verrucomicrobia, Aquificae and Lentisphaerae (online suppl. table S1).
The phylogenetic tree (fig. 1 and online suppl. table S1) reveals that surprisingly, each of the 15 clusters includes proteins from diverse bacterial kingdoms, almost without exception. Thus, for example, cluster 1 includes proteins from α- and γ-proteobacteria as well as firmicutes; clusters 2 and 3 include proteins from α-, β- and γ-proteobacteria as well as actinobacteria, and cluster 4 includes proteins from δ-proteobacteria, firmicutes and a crenarchaeon. It is clear that phylogenetic clustering does not correlate with organismal source, suggesting that extensive horizontal transfer of genes encoding these homologues has occurred over evolutionary time.
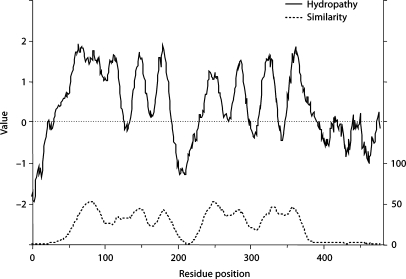
The average hydropathy and similarity plots for members of the TSUP family were derived using the modified AveHAS program described above. TMS predictions, based on all 318 homologues included in this study, are shown in figure 2. The plot reveals two sets of four putative TMSs, one in the N-terminal halves of these proteins, the other within the C-terminal halves. All four peaks are about equally well conserved, although peaks 1 and 5 are clearly better conserved than the others.
Fig. 2.
Average hydropathy (top, solid line) and similarity (bottom, dotted line) plots for the TSUP family of transporters. A modified AveHAS program was used to derive the plots as described in this review article.
Examination of the multiple alignment upon which figures 1 and 2 were based revealed that no residue position is fully conserved for either a particular residue or a particular residue type. However, two glycine residues, almost exclusively substituted by small semipolar residues (A, S, and T) were by far the best conserved. They occur at positions 79 (beginning of TMS 1) and 246 (beginning of TMS 5). The occurrence of two sets of four TMSs with similar apparent topologies and patterns of residue conservation suggested that these 8 TMS proteins may have arisen by an intragenic duplication event.
Using the GAP and IC programs (see above) to compare the first with the second halves of these proteins, homology between them could be established. For example when the first half of Tcr1 of Thiomicrospira crunogena (cluster 15) was compared with the second half of Ama2 of Acaryochloris marina (cluster 14), a comparison score of 19 SD was obtained (fig. 3). 216 comparisons gave greater that 9 SD, thus establishing that both halves derived from a common ancestor, probably as a result of an ancient intragenic duplication event. The large comparison scores noted above clearly suggest that the presumed intragenic duplication event that generated the 8 TMS proteins occurred more recently than in most other families of integral membrane transport proteins examined [Saier, 2003]. This suggestion is in agreement with the uniform size and topological characteristics noted above.
Fig. 3.
Alignment of the first half of one TSUP family member (Tcr1) with the second half of another (Ama2). All family members are homologous to each other throughout their lengths. The modified IC and GAP programs were used with 500 random shuffles, a gap penalty of 8 and a gap extension penalty of 2. The aligned sequences gave a comparison score of 19 SDs.
Establishing Superfamily Relationships between Distantly Related Families
The programs described above are useful for identifying distant relationships between proteins [Chang et al., 2004; Kim et al., 2006; Mansour et al., 2007]. Once homology is established, phylogenetic tree construction is justified. If nonhomologous sequences are included, phylogenetic trees present meaningless data. It is, therefore, always important to establish homology before conducting phylogenetic analyses.
A major problem arises when the sequences are so divergent from each other that accurate multiple alignments cannot be generated. Incorrect alignments mean that the trees generated will similarly be inaccurate and misleading. A novel program is therefore required for detecting increasingly distant relationships.
We have developed such a procedure using programs called SuperfamilyTree 1 (SFT1) and SFT2. The former program is based on BLAST searches and the resultant bit scores. There are several steps in its use: (1) The query protein sequences (from TCDB; one for each family within the superfamily) are BLASTed against the NCBI protein database. (2) Five sequences from each set are randomly selected by the program. (3) All resultant sequences from one set are compared with all resultant sequences from another set using the Blastall program. (4) The mean score of 25 comparisons (5 × 5) is tabulated for each inter-family comparison within the superfamily. Thus, a mean score is obtained for each family comparison. (5) The resultant matrix is then used to generate a Fitch tree (PHYLIP, http://evolution.genetics.washington.edu/phylip.html). (6) This process is conducted 100 times, generating a 100 Fitch consensus tree using the program Consense (PHYLIP). (7) The tree is drawn using the TreeView (TV) program [Zhai et al., 2002]. The SFT2 program combines the query proteins from each TCDB family before integrating the data along family lines to generate a consensus tree where each family is found at the end of a distinct branch.
Application of the SuperfamilyTree Programs to the Amino Acid/Polyamine/Organocation (APC) Superfamily
The APC superfamily was described by [Jack et al. 2000]; it included 10 families. Since then, this superfamily has expanded with the inclusion of 6 more families. A seventh family (AGCS) was added after the completion of this work. See table 1 and TCDB for all families and proteins within the APC superfamily. The three or four letter abbreviations of the families are used in this analysis (table 1). Protein abbreviations (3 letters indicating the organism, e.g. Eco for E. coli followed by a number to indicate the paralogue) are used. Last, members assigned to the proteins in family 2.A.3 of TCDB are used to identify the proteins, and the last two numbers in the TC entries of the proteins in all other families (2.A.18, 2.A.25, 2.A.30 and 2.A.42) are used in figures 4 and 5.
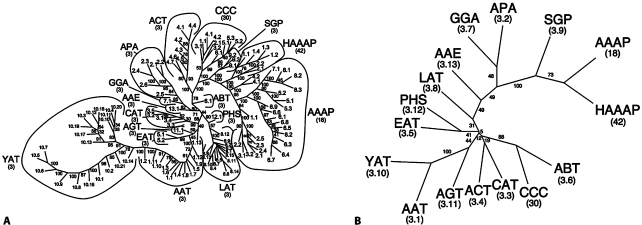
Fig. 4.
Phylogenetic (Fitch) trees for the APC superfamily [Jack et al., 2000]. The trees were generated with (A) the SFT1 program and (B) the SFT2 program. A The tree presents the relationships of all proteins of the APC superfamily as of June, 2008. These include all 16 families of the APC superfamily as indicated by their three letter abbreviations as defined in TCDB. Numbers refer to the individual TC numbers of the proteins within the various families. Bootstrap values are provided adjacent to each branch. The same convention is also used for figures 4B, 5D and 6B below. B A FITCH tree of the entire APC superfamily generated with the SFT2 program. The tree reveals the phylogenetic relationships of the 16 families relative to each other.
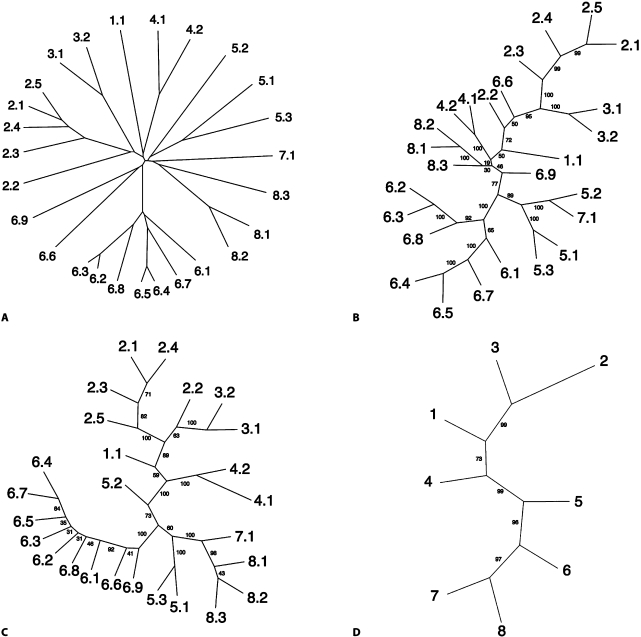
Fig. 5.
Phylogenetic trees generated with four programs for the AAAP family [Young et al., 1999] within the APC superfamily. A CLUSTAL X-TREEVIEW-neighbor joining tree. B Parsimony (Protpars) tree based on the CLUSTAL X-generated multiple alignment. C The corresponding tree generated with the SFT1 program. D The tree generated with the SFT2 program. The SFT1-based tree (C) and the SFT2-based tree (D) show the eight subfamilies of the AAAP family. Note: this tree resembles the AAAP cluster for the APC superfamily tree shown in figure 4A. All members in TCDB included in the eight AAAP subfamilies (C) were averaged to provide the positions of each of these subfamilies relative to each other (D).
The 16 families could not be analyzed using traditional methods based on multiple alignments because no program could correctly align all of these sequences. We therefore resorted to the SuperfamilyTree programs (SFT1 and SFT2) described here. The results are presented in figures 4A (SFT1) and 4B (SFT2). In figure 4A, all proteins of the APC superfamily recognized in TCDB as of June, 2008 are included. A neighbor joining (NJ) (Fitch) tree (PHYLIP) is shown. In the convention used, all branches are the same length, so only relative positions are important. In general, all proteins within a single family of the APC superfamily (e.g. YAT or CAT or APA) cluster together with few exceptions. For example, all 21 YAT family members cluster together on the lower left hand side of the tree, while all AAAP family members currently included in TCDB cluster together on the far right hand side of the tree.
The AAAP family [Young et al., 1999] within the APC superfamily was analyzed in greater detail. All of the sequences within this one family are sufficiently similar to generate a reliable tree based on a traditional CLUSTAL X-generated multiple alignment. This allows comparison of the trees generated by traditional methods with the two SuperfamilyTree programs, SFT1 and SFT2. Compare the right hand cluster of figure 4A (just the AAAP family members) with the NJ tree shown in figure 5A, and the Parsimony tree, shown in figure 5B. The two important features are: (1) clusters 7 and 8 are most closely related, with clusters 6 and 5 showing the next closest relationships, and (2) off on a separate branch, we find two families, 2 and 3, clustering tightly together, with 1 and 4 clustering more loosely with them. Now compare this tree (fig. 5A) with the arrangement of the AAAP family members within the APC superfamily tree shown in figure 4A. We see that the relationships are almost identical. 7 and 8 cluster tightly together with 5 and 6 branching from the base of the same cluster. Similarly, off on a distinct branch, 2 and 3 cluster tightly together with 1 branching from the base of the same cluster. 4 can be found to the upper left of the two major clusters (1, 2, 3 and 5, 6, 7, 8).
The SFT1 program-generated tree, just for the AAAP family, is shown in figure 5C, while the SFT2 generated tree is presented in figure 5D. This last tree shows most clearly the relationships between the 8 subfamilies of the AAAP family. Thus, in figures 5C and D (as for fig. 5A and B), clusters 7 and 8 are closely linked, followed by 6 and 5 on the lower half of the tree, while clusters 2 and 3, on the upper half of the tree, are most closely linked to 1 and 4, appearing in the more central part of the tree. The agreement between these 4 trees is striking, thus confirming the validity of the SuperfamilyTree approach when sequences are sufficiently similar to allow all four programs to produce reliable trees.
Returning to the full tree for the APC superfamily, next to and above the AAAP family in figure 4A is the HAAP family, and proceeding counterclockwise, one finds the SGP, CCC, ACT, and ABT families in that order. Continuing counterclockwise around the tree, the AAE, GGA, and APA family members appear on a distant branch. Below this branch are two other branches, one including the PHS and LAT families, the other bearing the AGT, EAT and AAT families with only one exception: AAT13 which is on the far side of AGT1 and EAT1, 2. This one protein (on the right side of the AGT and EAT proteins) is separated from all other AAT family members (on the left side of the AGT and EAT families). These are followed by the large YAT family (far lower left) mentioned above.
Finally, the composite tree obtained with the SFT2 program (fig. 4B) reveals the relative positions of the 16 families in the APC superfamily. This tree, with only one branch per family, is far easier to read, and because it averages the results for all members of an individual family, it is also more accurate. Thus, in agreement with the figure 4A tree, the yeast YAT and bacterial AAT families (lower left on both trees) cluster tightly together, with bacterial AGT branching from the base of this cluster. Off on a separate branch, we find the cation chloride cotransporter (CCC) family (mostly eukaryotic) clustering most closely with the bacterial ABT family, with the eukaryotic ACT and CAT families branching off together at the base of the same cluster. Then, progressing further up the tree, the prokaryotic ethanolamine (EAT) and the eukaryotic polyamine (PHS) families prove to cluster together although this is not immediately apparent based on the figure 4A tree. The remainder of the tree is as expected with the eukaryotic AAAP and bacterial HAAAP families clustering together, with the amino acid signaling spore germination protein receptors of the SGP family being their closest relative. Progressing further down towards the base of the tree, the bacterial GGA and APA families cluster together in both figures 4A and 4B, with the bacterial AAE and the ubiquitous LAT families branching from points closer to the center of the tree. PHS clusters with EAT while AAE clusters loosely with GGA and APA. All but one of the families at the top of the tree (AAE, GGA, APA, SGP, and HAAP) are of prokaryotic origin. The sole exception is the AAAT family which is represented primary in eukaryotes. We conclude that several, but by no means all families cluster according to organismal type. We see no correlation with substrate type, but this may be due to the similar structures of all substrates of the APC superfamilies. These transporters function exclusively as uptake systems using solute:cation symport.
Conclusions and Perspectives
The vast amount of protein sequence data now available renders data mining essential to maximize output. Towards this purpose, our laboratory has utilized a large number of preexisting programs and designed novel software in order to refine and optimize data extraction procedures concerned primarily with the topologies, structures and evolutionary origins of transport proteins. This information is then entered into TCDB. Twenty years of bioinformatic research has resulted in the functional/phylogenetic classification and characterization of these proteins as recorded in TCDB. The approaches we have developed will undoubtedly be applicable to nucleic acid and protein bioinformaticists working in many areas of biology.
We hope that this review of our bioinformatic efforts will provide incentive for expansion of phylogeny-based data mining technologies so as to allow extraction of ever-increasing amounts of information from genome sequences. The techniques and programs described can also be used as a basis for the development of more sophisticated software. TCDB can serve as a model database for the expansion of database technology.
Acknowledgement
We thank the NIH (GM1077402) for financial support.
References
- Amar P, Legent G, Thellier M, Ripoll C, Bernot G, Nystrom T, Saier MH, Jr, Norris V. A stochastic automaton shows how enzyme assemblies may contribute to metabolic efficiency. BMC Syst Biol. 2008;25:2–27. doi: 10.1186/1752-0509-2-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabote RD, Saier MH., Jr Comparative genomic analyses of the bacterial phosphotransferase system. Microbiol Mol Biol Rev. 2005;69:608–634. doi: 10.1128/MMBR.69.4.608-634.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabote RD, Tamang DG, Abeywardena SN, Fallah NS, Fu JYC, Lio JK, Mirhosseini P, Pezeshk R, Podell S, Salampessy ML, Thever MD, Saier MH., Jr Extra domains in secondary transport carriers. Biochim Biophys Acta. 2006;1758:1557–1579. doi: 10.1016/j.bbamem.2006.06.018. [DOI] [PubMed] [Google Scholar]
- Busch W, Saier MH., Jr The IUBMB-Endorsed transporter classification system. Mol Biotech. 2002;27:253–262. doi: 10.1385/mb:27:3:253. [DOI] [PubMed] [Google Scholar]
- Chang AB, Lin R, Keith Studley W, Tran CV, Saier MH., Jr Phylogeny as a guide to structure and function of membrane transport proteins. Mol Membrane Biol. 2004;21:171–181. doi: 10.1080/09687680410001720830. [DOI] [PubMed] [Google Scholar]
- Cuff JA, Clamp ME, Siddiqui AS, Finlay M, Barton GJ. Jpred: a consensus secondary structure prediction server. Bioinformatics. 1998;14:892–893. doi: 10.1093/bioinformatics/14.10.892. [DOI] [PubMed] [Google Scholar]
- Dayhoff MO, Barker WC, Hunt LT. Establishing homologies in protein sequences. Methods Enzymol. 1983;91:524–545. doi: 10.1016/s0076-6879(83)91049-2. [DOI] [PubMed] [Google Scholar]
- Dobzhansky T. Biology, molecular and organismic. Am Zool. 1964;4:443–452. doi: 10.1093/icb/4.4.443. [DOI] [PubMed] [Google Scholar]
- Doolittle RF. Similar amino acid sequences: chance or common ancestry? Science. 1981;214:149–150. doi: 10.1126/science.7280687. [DOI] [PubMed] [Google Scholar]
- Doolittle RF. Of URFS and ORFs: A Primer on How to Analyze Derived Amino Acid Sequences. Mill Valley: University Science Books; 1986. [Google Scholar]
- Doolittle RF. Redundancies in Protein Sequences, Prediction of Protein Structure and the Principles of Protein Conformation. New York: Plenum Publishing Corporation; 1989. pp. 599–623. [Google Scholar]
- Doolittle RF. Reconstructing history with amino acid sequences. Protein Sci. 1992;1:191–200. doi: 10.1002/pro.5560010201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg D, Weiss RM, Terwilliger TC. The helical hydrophobic moment: a measure of the amphiphilicity of a helix. Nature. 1982;299:371–374. doi: 10.1038/299371a0. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP, and related tools. Nat Protoc. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- Felce J, Saier MH., Jr Carbonic anhydrases fused to anion transporters of the SulP family: evidence for a novel type of bicarbonate transporter. J Mol Microbiol Biotechnol. 2004;8:169–176. doi: 10.1159/000085789. [DOI] [PubMed] [Google Scholar]
- Feng DF, Doolittle RF. Progressive alignment of amino acid sequences and construction of phylogenetic trees from them. Methods Enzymol. 1996;266:368–382. doi: 10.1016/s0076-6879(96)66023-6. [DOI] [PubMed] [Google Scholar]
- Feng DF, Doolittle RF. Progressive alignment and phylogenetic tree construction of protein sequences. Methods Enzymol. 1990;183:375–387. doi: 10.1016/0076-6879(90)83025-5. [DOI] [PubMed] [Google Scholar]
- Feng DF, Taylor WR, Thornton JM. A model recognition approach to the prediction of all-helical membrane protein structure and topology. Biochemistry. 1994;33:3038–3049. doi: 10.1021/bi00176a037. [DOI] [PubMed] [Google Scholar]
- Harvat EM, Zhang YM, Tran CV, Zhang Z, Frank MW, Rock CO, Saier MH., Jr Lysophospholipid flipping across the Escherichia coli inner membrane catalyzed by a transporter (LplT) belonging to the major facilitator superfamily. J Biol Chem. 2005;280:12028–12034. doi: 10.1074/jbc.M414368200. [DOI] [PubMed] [Google Scholar]
- Hollenstein K, Dawson RJ, Locher KP. Structure and mechanism of ABC transporter proteins. Curr Opin Struct Biol. 2007;17:412–418. doi: 10.1016/j.sbi.2007.07.003. [DOI] [PubMed] [Google Scholar]
- Jack DL, Paulsen IT, Saier MH., Jr The amino acid/polyamine/ogranocation (APC) superfamily of transporters specific for amino acids, polyamines and organocations. Microbiology. 2000;146:1797–1814. doi: 10.1099/00221287-146-8-1797. [DOI] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. A model recognition approach to the prediction of α-helical membrane protein structure and topology. Biochemistry. 1994;33:3038–3049. doi: 10.1021/bi00176a037. [DOI] [PubMed] [Google Scholar]
- Kim SH, Chao Y, Saier MH., Jr Protein-translocating trimeric autotransporters of Gram-negative bacteria. J Bacteriol. 2006;188:5655–5667. doi: 10.1128/JB.01596-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le T, Tseng TT, Saier MH., Jr Flexible programs for the prediction of average amphipathicity of multiply aligned homologous proteins: application to integral membrane transport proteins. Mol Membr Biol. 1999;16:173–179. doi: 10.1080/096876899294634. [DOI] [PubMed] [Google Scholar]
- Lee JH, Harvat EM, Stevens JM, Ferguson SJ, Saier MH., Jr Evolutionary origins of members of a superfamily of integral membrane of cytochrome C biogenesis proteins. Biochim Biophys Acta. 2007;1768:2164–2181. doi: 10.1016/j.bbamem.2007.04.022. [DOI] [PubMed] [Google Scholar]
- Locher HH, Poolman B, Cook AM, Konings WN. Uptake of 4-toluene sulfonate by Comamonas testosteroni T-2. J Bacteriol. 1993;175:1075–1080. doi: 10.1128/jb.175.4.1075-1080.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mampel J, Maier E, Tralau T, Ruff J, Benz R, Cook AM. A novel outer-membrane anion channel (porin) as part of a putatively two-component transport system for 4-toluenesulphonate in Comamonas testosteroni T-2. Biochem J. 2004;383:91–99. doi: 10.1042/BJ20040652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour NM, Sawhney M, Vogl C, Saier MH., Jr The bile/arsenite/riboflavin transporter (BART) superfamily. FEBS J. 2007;274:612–629. doi: 10.1111/j.1742-4658.2006.05627.x. [DOI] [PubMed] [Google Scholar]
- Melen K, Krogh A, von Heijne G, Nielsen H. Reliability measures for membrane protein topology prediction algorithms. J Mol Bioltechnol. 2003;327:735–744. doi: 10.1016/s0022-2836(03)00182-7. [DOI] [PubMed] [Google Scholar]
- Mulder NJ, Kersey P, Pruess M, Apweiler R. In silico characterization of proteins: UniPort, InterPro and Integr8. Mol Biotechnol. 2008;38:165–177. doi: 10.1007/s12033-007-9003-x. [DOI] [PubMed] [Google Scholar]
- Norris V, den Blaauwen T, Doi RH, Harshey RM, Janniere L, Jimenez-Sanchez A, Jin DJ, Levin PA, Mileykovskaya E, Minsky A, Misevic G, Ripoll C, Saier MH, Jr, Skarstad K, Thellier M. Toward a hyperstructure taxonomy. Annu Rev Microbiol. 2007a;61:309–329. doi: 10.1146/annurev.micro.61.081606.103348. [DOI] [PubMed] [Google Scholar]
- Norris V, den Blaauwen T, Cabin-Flaman A, Doi RH, Errington J, Harshey RM, Janniere L, Jimenez-Sanchez A, Jin DJ, Levin PA, Mileykovskaya E, Minsky A, Saier MH, Jr, Skarstad K. Functional taxonomy of bacterial hyperstructures. Microbiol Mol Biol Rev. 2007b;71:230–253. doi: 10.1128/MMBR.00035-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papanikou E, Karamanou S, Economou A. Bacterial protein secretion through the translocase nanomachine. Nat Rev Microbiol. 2007;5:839–851. doi: 10.1038/nrmicro1771. [DOI] [PubMed] [Google Scholar]
- Pearson WR. Empirical statistical estimates for sequence similarity searches. J Mol Biol. 1998;276:71–84. doi: 10.1006/jmbi.1997.1525. [DOI] [PubMed] [Google Scholar]
- Pollock DD. Genomic biodiversity, phylogenetics and coevolution in proteins. Appl Bioinformatics. 2002;1:81–92. [PMC free article] [PubMed] [Google Scholar]
- Rehm BH. Bioinformatic tools for DNA/protein sequence analysis, functional assignment of genes and protein classification. Appl Microbiol Biolechnol. 2001;57:579–592. doi: 10.1007/s00253-001-0844-0. [DOI] [PubMed] [Google Scholar]
- Riley M, Abe T, Arnaud MB, Berlyn MKB, Blattner FR, Chaudhuri RR, Glasner JD, Horiuchi T, Keseler IM, Kosuge T, Mori H, Perna NR, Wishart D, Wanner BL. Escherichia coli K-12: a cooperatively developed annotation snapshot – 2005. Nucleic Acids Res. 2006;34:1–9. doi: 10.1093/nar/gkj405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd KE. EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res. 2000;28:60–64. doi: 10.1093/nar/28.1.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier MH., Jr Computer-aided analyses of transport protein sequences: gleaning evidence concerning function, structure, biogenesis, and evolution. Microbiol Rev. 1994;58:71–93. doi: 10.1128/mr.58.1.71-93.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier MH., Jr Genome sequencing and informatics: new tools for biochemical discoveries. Plant Physiol. 1998;117:1129–1133. doi: 10.1104/pp.117.4.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier MH., Jr A functional-phylogenetic classification system for transmembranec solute transporters. Microbiol Mol Biol Rev. 2000;64:354–411. doi: 10.1128/mmbr.64.2.354-411.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier MH., Jr Tracing pathways of transport protein evolution. Mol Microbiol. 2003;48:1145–1156. doi: 10.1046/j.1365-2958.2003.03499.x. [DOI] [PubMed] [Google Scholar]
- Saier MH, Jr, McCaldon P. Statistical and functional analyses of viral and cellular proteins with N-terminal amphipathic α-helices with large hydrophobic moments. Importance to macromolecular recognition and organellar targeting. J Bacteriol. 1988;170:2296–2300. doi: 10.1128/jb.170.5.2296-2300.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier MH, Jr, Tran CV, Barabote RD. TCDB: The transporter classification database for membrane transport protein analyses and information. Nucleic Acids Res. 2006;34:D181–D186. doi: 10.1093/nar/gkj001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serres MH, Goswami S, Riley M. GenProtEC: an updated and improved analysis of Escherichia coli K-12 proteins. Nucleic Acids Res. 2004;32:300–302. doi: 10.1093/nar/gkh087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skrabanek L, Saini HK, Bader GD, Enright AJ. Computational prediction of protein-protein interactions. Mol. Biotechnol. 2008;38:1–17. doi: 10.1007/s12033-007-0069-2. [DOI] [PubMed] [Google Scholar]
- Sonnhammer EL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol. 1998;6:175–182. [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL X: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tralau T, Cook AM, Ruff J. Map of the IncP1beta plasmid pTSA encoding the widespread genes (tsa) for p-toluenesulfonate degradation in Comamonas testosteroni T-2. Appl Environ Microbiol. 2001;67:1508–1516. doi: 10.1128/AEM.67.4.1508-1516.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusnady GE, Simon J. The HMMTOP transmembrane topology prediction server. Bioinformatics. 2001;17:849–850. doi: 10.1093/bioinformatics/17.9.849. [DOI] [PubMed] [Google Scholar]
- Weinitschke S, Denger K, Cook AM, Smits TH. The DUF81 protein TauE in Cupriavidus necator H16, a sulfite exporter in the metabolism of C2 sulfonates. Microbiology. 2007;153:3055–3060. doi: 10.1099/mic.0.2007/009845-0. [DOI] [PubMed] [Google Scholar]
- Weiss KM, Buchanan AV. Evolution by phenotype: a biomedical perspective. Perspect Biol Med. 2003;46:159–182. doi: 10.1353/pbm.2003.0032. [DOI] [PubMed] [Google Scholar]
- Yamaguchi A, Saier MH., Jr The bile/arsenite/riboflavin transporter (BART) superfamily. FEBS Journal. 2007;274:612–629. doi: 10.1111/j.1742-4658.2006.05627.x. [DOI] [PubMed] [Google Scholar]
- Yen MR, Saier MH., Jr Gap junctional proteins of animals: the innexin/pannexin superfamily. Prog Biophys Mol Bio. 2007;94:5–14. doi: 10.1016/j.pbiomolbio.2007.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young GB, Jack DL, Smith DW, Saier MH., Jr The amino acid/auxin:proton symport permease family. Biochim Biophys Acta. 1999;1415:306–322. doi: 10.1016/s0005-2736(98)00196-5. [DOI] [PubMed] [Google Scholar]
- Zhai Y, Saier MH., Jr A web-based program (WHAT) for the simultaneous prediction of hydropathy, amphipathicity, secondary structure and transmembrane topology for a single protein sequence. J Mol Microbiol Biotech. 2001a;3:501–502. [PubMed] [Google Scholar]
- Zhai Y, Saier MH., Jr A web-based program for the prediction of average hydropathy, average amphipathicity and average similarity of multiply aligned homologous proteins. J Mol Microbiol Biotechnol. 2001b;3:285–286. [PubMed] [Google Scholar]
- Zhai Y, Saier MH., Jr A simple sensitive program for detecting internal repeats in sets of multiply aligned homologous proteins. J Mol Microbiol Biotechnol. 2002;4:375–377. [PubMed] [Google Scholar]
- Zhai Y, Tchieu J, Saier MH., Jr A web-based Tree View (TV) program for the visualization of phylogenetic trees. J Mol Microbiol Biotechnol. 2002;4:69–70. [PubMed] [Google Scholar]
- Zhou X, Yang N, Tran C, Hvorup R, Saier MH., Jr Web-based programs for the display and analysis of transmembrane α-helices in aligned protein sequences. J Mol Microbiol Biotech. 2003;5:1–6. doi: 10.1159/000068718. [DOI] [PubMed] [Google Scholar]