Abstract
A novel microwave-assisted synthetic method has been used to synthesise a series of mixed ligand ruthenium(ii) compounds containing diimine as well as bidentate thiosemicarbazone ligands. The compounds contain the diimine 1,10-phenanthroline (phen) or 2,2′-bipyridine (bpy) and the thiosemicarbazone is derived from 9-anthraldehyde. Based on elemental analyses and spectroscopic data, the compounds are best formulated as [(phen)2Ru(thiosemicarbazone)](PF6)2 and [(phen)2Ru(thiosemicarbazone)](PF6)2 where thiosemicarbazone = 9-anthraldehydethiosemicarbazone, 9-anthraldehyde-N(4)-methylthiosemicarbazone, and 9-anthraldehyde-N(4)-ethylthiosemicarbazone. Fluorescence competition studies with ethidium bromide, along with viscometric measurements suggests that the complexes bind calf thymus DNA (CTDNA) relatively strongly via an intercalative mode possibly involving the aromatic rings of the diimine ligands. The complexes show good cytotoxic profiles against MCF-7 and MDA-MB-231 (breast adenocarcinoma) as well as HCT 116 and HT-29 (colorectal carcinoma) cell lines.
Introduction
Thiosemicarbazones are of considerable pharmacological interest since a number of derivatives have shown a broad spectrum of chemotherapeutic properties. The wide range of biological activities possessed by substituted thiosemicarbazones includes cytotoxic, anti-tumour,1 anti-bacterial,2 and anti-viral3 properties. The biological properties of the ligands can be modified and in fact enhanced, by the linkage to metal ions.4–6
Ruthenium complexes of various types are actively studied as metallodrugs as they are believed to have low toxicity and good selectivity for tumours.7 Very recently, two ruthenium(iii) complexes have also successfully completed phase I clinical trials, namely, NAMI-A8–10 (NAMI-A = (ImH)[trans-Ru(iii)Cl4Im(Me2SO)]; Im=imidazole), and KP1019 indazolium trans-[tetrachlorobis(1H-indazole)ruthenate(iii)].11,12 Organometallic compounds exhibit different ligand exchange kinetics in solution to coordination complexes, as well as novel structural motifs and organometallic ruthenium(ii)-arene complexes (of the type [(η6-arene)Ru(LL)Cl]+, LL = ligand) are also currently attracting increasing interest as anticancer compounds.13,14
For many years, research into the interaction of ruthenium(ii)-containing complexes with DNA has been the primary focus for many researchers. During the last few decades, a number of transition-metal complexes have been utilized to probe nucleic acid structures,15,16 DNA photoprobes,17,18 and DNA-molecular light switches.19,20 It has also been documented that metal complexes can bind to DNA covalently as well as non-covalently.21–23 Transition metal complexes of diimine ligands such as 2,2′-bipyridine (bpy) and 1,10-phenanthroline (phen) are widely used in bioinorganic chemistry particularly as a probe for DNA. These types of compounds possess interesting anticancer properties.24 In this paper, we report on a study (synthesis, characterization and cytotoxicity) of a series of mixed-ligand diimine ruthenium complexes of the type [(bipy)2Ru(TSC)](PF6)2 and [(phen)2Ru(TSC)](PF6)2 where TSC is a chelating thiosemicarbazone ligand. We also report on their interaction with CTDNA.
Results and discussion
Syntheses and characterization
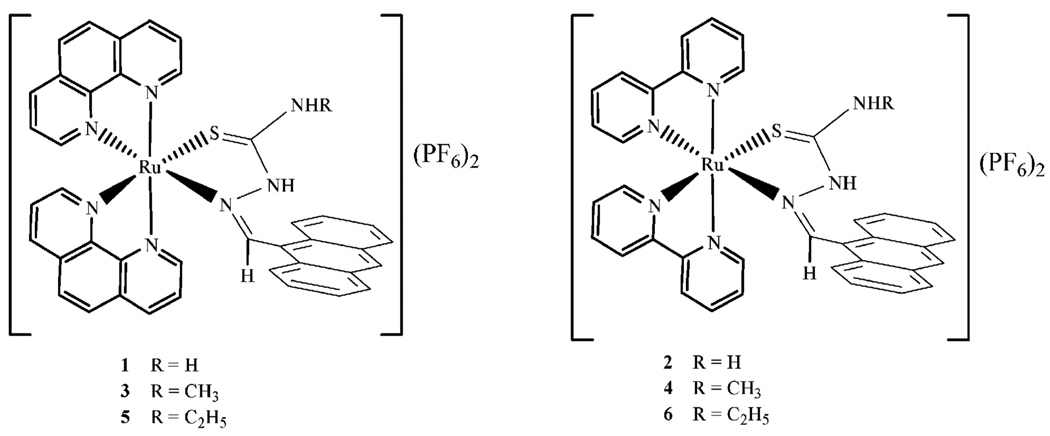
The ligands were synthesized by the acid catalyzed condensation of 9-anthraldehyde with the corresponding N-alkyl substituted thiosemicarbazide in ethanol. The reaction produced orange or yellow-orange microcrystalline solids. The complexes were made using a microwave-assisted thermal reaction of the dichlorobis(diimine)ruthenium(ii) starting material with the ligands. [Ru(diimine)2Cl2] and ligand were suspended in the ethylene glycol solvent and the reaction mixture saturated with argon. The mixture was then heated using a dynamic method that was developed as follows: time = 5 min; temperature = 150 °C; power = variable; stirring = max; cooling = on. The reaction produced red solids that are insoluble in alcohols and water but are very soluble in acetone, CH2Cl2 and DMSO. From microanalytical and spectroscopic data we propose that the complexes can be formulated as [(diimine)2Ru(TSC)](PF6)2 (Fig. 1).
Fig. 1.
Proposed structures of the compounds.
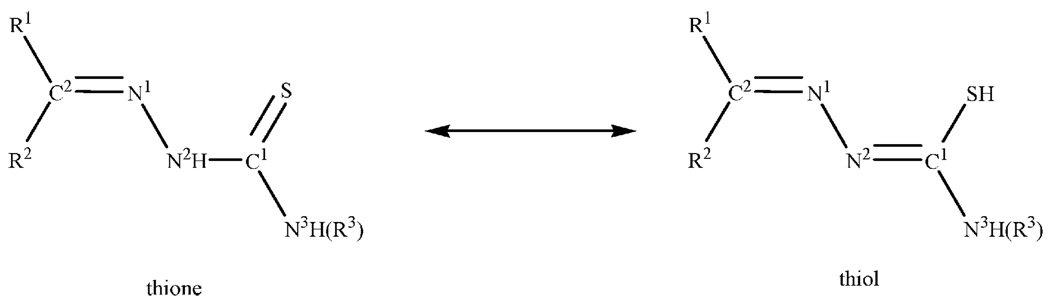
We attempted to further purify the compounds by chromatography on alumina using a 3 : 1 mixture of acetonitrile and toluene but those attempts were unsuccessful. The process produced dark red-black solids that have similar absorption spectra as the starting material. However, based on elemental analysis of these compounds, we can suggest that a tautomerization reaction occurred on the column resulting in the formation of the complex [(phen)2Ru(TSC)]PF6 or [(phen)2Ru(TSC)]PF6. These complexes contain the anionic form of the thiosemicarbazone ligand (Fig. 2).
Fig. 2.
Tautomerism in thiosemicarbazones.
Mass spectral analysis
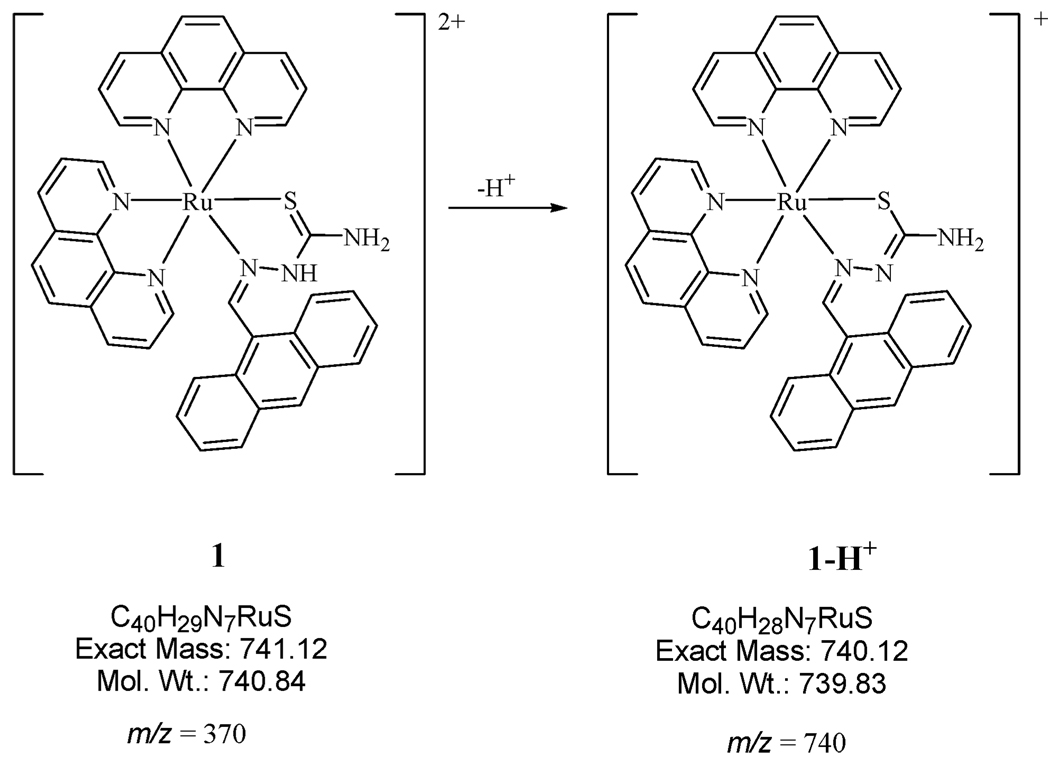
The m/z values (see ESI, Fig. S1 and S2†) detected suggest that during ionization, in addition to the loss of the two PF6− counterions, deprotonation of the hydrazinic N(2)-H proton also occurs. This leads to the formation of the anionic thiolate form of the thiosemicarbazone ligand coordinated to the metal. Hence, the species detected are singly charged (Scheme 1).
Scheme 1.
Formation of species detected by mass spectrometry.
NMR spectral studies
The NMR spectra of the ligand and its metal complexes were acquired in DMSO-d6 as they are very soluble in this solvent. Fig. 2 shows the atom numbering used for assignment of protons in the ligand. The 1H NMR spectrum of all the ligands shows a singlet at approximately δ = 11.70 ppm. On the basis of spectroscopic data available in the literature, it is suggested that this is due to the N(2)-H proton.25 In general, the hydrazinic protons (N(2)-H) of free ligands appear as single broad peaks in a fairly wide range, δ = 8.7–15 ppm, depending on the nature of the substituents at C2 carbon.26–29 This signal is also diagnostic for isomer identification. According to Afrasiabi,30 it falls between δ = 13–15 ppm for the E form and δ = 9–12 ppm for the Z form. Using that analysis the ligands under our conditions exists as the Z isomer. Coupled with the lack of a resonance signal at ca. δ = 4.0 ppm attributable to a –SH proton resonance, we can say that the ligand also exist in the thione form in solution (of even a polar solvent as DMSO). In the spectrum of the ATSC ligand two resonances at δ = 9.32 and δ = 8.17 ppm were observed, which we assign to the geminal N(3)-H2 protons. This is not uncommon31 and indicates hindered rotation due to the SC–N(3)H2 bond containing some double bond character.32 It was possible to assign almost all the resonance signals in the 13C NMR spectra. The primary assignments are for the two low-field signals at δ = 178 ppm assigned to the C1=S and δ = 142 ppm assigned to the C2=N group. The aromatic signals show up in the usual place (δ = 120–130 ppm). For MeATSC methyl protons are at δ = 3.02 ppm and the carbon signal is at δ = 31 ppm. In the case of EtATSC, the methylene protons of the ethyl group are at δ = 3.59 ppm and the methyl protons are at δ = 1.25 ppm. The corresponding carbon signals are observed at δ = 39.05 ppm and δ = 15.05 ppm, respectively. A simple comparison of the NMR spectra of the ligand and the metal complexes reveals that the presence of the N(2)H signal in the spectra of the complexes is indicative of the non-deprotonation of the ligand confirming the neutrality of the coordinated thiosemicarbazone. In all the complexes there is a general upfield shift (of nearly 2 ppm), which reflects coordination through the azomethine nitrogen. The signal ascribed to the N(3)H proton in the free ligands generally move downfield (by as much as 0.5 ppm). This is indicative of the binding of the thiocarbonyl group and is a result of a decrease in the electron density caused by electron withdrawal by the metal ions from the thione sulfur.
Infrared spectra
Thiosemicarbazones exhibit characteristic bands corresponding to various groups in specific energy regions. It can be seen that the characteristic absorption peaks of all complexes are similar. The absence of a ν(S–H) absorption in the region 2600–2500 cm−1 is considered as evidence that the thione form of the ligands exist in the solid state (Fig 2).33,6 There are two or three bands in the ν(N–H) region and these signals play an important role in evaluating the nature of the bonding in thiosemicarbazone complexes. The presence of a band corresponding to N(2)-H group, suggests the coordination of a thiosemicarbazone to the metal centre in a neutral form, while its absence, is suggestive of deprotonation of hydrazinic N(2)-H proton in the complexes. The band at ~3200–3150 cm−1, which is assigned to the N(2)-H group, support the thione formulation of the ligand in the complexes. The other band(s) (at ~3450–3350 cm−1) are the stretching vibrations of the terminal N(3)-H group and do not shift significantly on complexation. The ligands show a medium intensity band at 1621 cm−1 that we ascribe to C=N; these are shifted slightly to higher or lower energy upon complexation. Considering the two principal bands, the C=N band, shifts by 19–40 cm−1 to lower wavenumbers. This negative shift indicates that the azomethine nitrogen (N(1)) coordinates to the metal.34,25 That the N(2)-H stretching frequency also shifts (due to change in the electron density upon complexation of the thiocarbonyl sulfur) supports this theorization. The involvement of the thiocarbonyl group can similarly be inferred from the wavenumber shifts that occur on binding. The band in the free ligand at ~840 cm−1, which we attribute to the C=S group shifts to lower frequencies by 13–19 cm−1. The size of the shifts suggest that the ligand coordinates as a neutral, bidentate (through the azomethine nitrogen and thiocarbonyl sulfur) ligand in all the complexes. This is supported by the absence of all the tell-tale signs of thiolate formation particularly the presence of the N(2)-H in all the complexes.
UV-vis spectra
In the absorption spectra of the compounds, essentially identical absorption bands are observed in related complexes (see ESI, Fig. S3).† For the phen analogs (1, 3, 5), four major absorption features are observed: 441 nm, 395 nm (very broad), 290 nm and 235 nm. Consistent with the data in the literature, the spectral feature at 441 nm may be assigned as a MLCT, t2g(Ru) → π*(phen), transition. This band shows up as a lower energy shoulder on the main MLCT band at centred at 395 nm, which we ascribe to a transition to a π* orbital on the thiosemicarbazone ligand and is a stronger σ donor and weaker π acceptor than the diimine. Similar arguments can be made for the bpy analogs (2, 4, 6). For this set of complexes, the MLCT to the thiosemicarbazone is observed at higher energies (370 nm) indicating that is a much weaker π acceptor than bpy compared to phen. In both sets of complexes the commonality of the other two high energy bands are most reasonably assigned to π → π* transitions localized on the diimine ligand. Given the constancy of the bands within each set of complexes, it appears that the alkyl group on N(3) of the thiosemicarbazone is an innocent substituent with no effect on the electronic properties of the complexes.
Electrochemistry
The electrochemical (cyclic voltammetry) behaviour of the have been studied in dichloromethane. (See ESI.)† The complexes showed similar features in the investigated sweep range. For complexes with 1 and 2 there is a quasi-reversible redox couple at +1.12 V for 1 and +1.16 for 2. This is a metal-based couple and is due to the RuIII/II redox process. For a classic reversible reaction the ratio of ip,a to ip,c (ip,a, ip,c are the anodic and cathodic currents, respectively) should be close to one. None of the compounds in this study had this ideal ratio. It is known however that chemical reactions can modify the electrode process leading to significant variation in the ratio of peak currents. The quasi-reversible nature of the redox couple is also seen in the large peak separations: ΔEp averages 206 mV (range 151–302 mV) which is much higher than the theoretical value of 59 mV for a reversible 1-electron transfer. When N(3) is alkylated the RuIII/II couple occurs at approximately +1.10 V in all cases. All the potentials are vs. the Ag/AgCl system.
Interaction of complexes with ct-DNA
Competitive binding between EB and complexes for ct-DNA
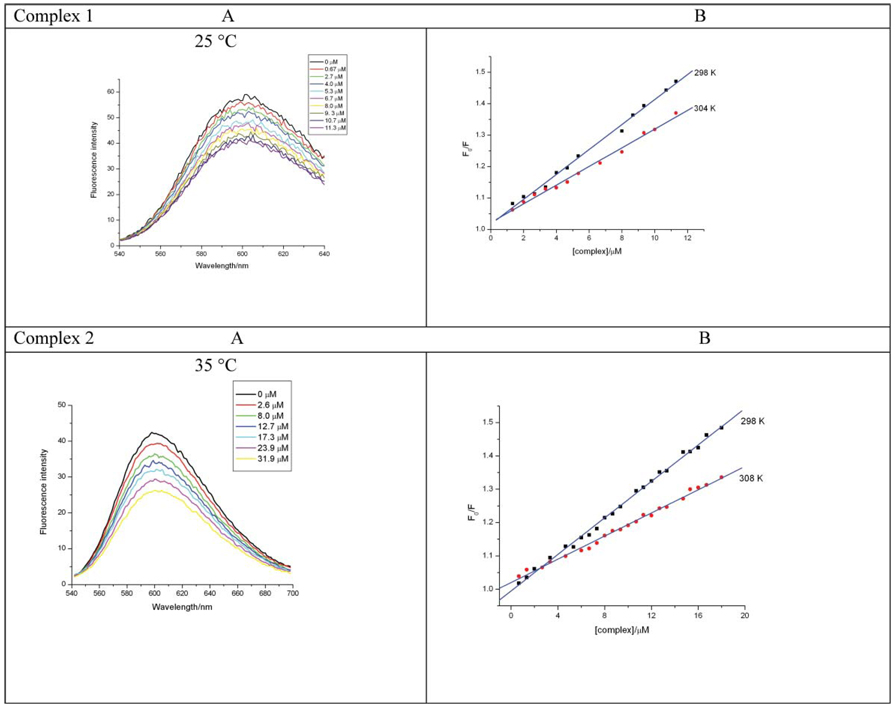
In order to investigate the interaction mode between the complexes and ct-DNA, the ethidium bromide (EB) fluorescence displacement experiment was also employed. EB is a planar cationic dye that is widely used as a sensitive fluorescence probe for native DNA. EB emits intense fluorescent light in the presence of DNA due to its strong intercalation between the adjacent DNA base pairs. The displacement technique is based on the decrease of this fluorescence resulting from the displacement of EB from a DNA sequence by a quencher.35 The quenching is due to the reduction of the number of binding sites on the DNA that is available to the EB. The method therefore provides indirect evidence for an intercalative binding mode. The extent of fluorescence quenching may also be used to determine the extent of binding between the quencher and DNA. Fig 3 shows the emission spectra of the EB-DNA solution on titration with two selected complexes. The spectra show no significant changes in shape or wavelength. It is obvious that there is a reduction in the fluorescence intensity at λem ≈ 600 nm as the concentration of the ruthenium(ii) complex increases. This clearly indicates that some EB molecules are displaced from their DNA binding sites and replaced by the complexes.
Fig. 3.
(A) Fluorescence emission spectra of EB-DNA in the presence of increasing amounts of 1 and 2. [EB] and [DNA] are 0.33 µM and 10 µM, respectively. [1] = (a) 0, (b) 0.667, (c) 2.67, (d) 4.00, (e) 5.34, (f) 6.67, (g) 8.00, (h) 9.34, (i) 10.7, (j) 11.3 µM. [2] = (a) 0, (b) 2.66, (c) 8.00, (d) 12.7, (e) 17.3, (f) 23.9 (g) 31.9 µM. (B) Stern–Volmer curves for 1 and 2.
Fluorescence quenching may result from a variety of processes including ground state complex formation (static quenching) and collisional processes (dynamic quenching). A quantitative estimation of quenching can be obtained from a Stern–Volmer analysis of the data. According to the Stern–Volmer eqn (1)36 the relative fluorescence is directly proportional to the concentration of the quencher:
| (1) |
where F0 and F are the fluorescence intensities of the EB-DNA system before and after the addition of the complexes, KSV is the Stern–Volmer quenching constant and [Q] is the concentration of the quencher (the complexes). The quenching constants were calculated from the slope of the Stern–Volmer plot (F0/F vs. [Q]) shown in Fig 3 and the results listed in Table 1.
Table 1.
Stern–Volmer quenching parameters for the binding of 1 and 2 with CTDNA
| 1 | 2 | |||
|---|---|---|---|---|
| T/K | 10−4KSV/M−1 | 10−12kq/M−1 s−1 | 10−4KSV/M−1 | 10−12kq/M−1 s−1 |
| 298 | 3.90 | 1.77 | 2.77 | 1.26 |
| 304 | 2.91 | 1.32 | — | — |
| 308 | — | — | 1.71 | 0.78 |
The linearity of the quenching plots illustrate that the quenching is in good agreement with the Stern–Volmer equation. The linearity also indicates that only one type of quenching process is in operation. It is well known that quenching occurs through a static or dynamic process. Dynamic quenching refers to a process where the fluorophore and the quencher collide during the lifetime of the excited state. On the other hand, static or contact quenching involves the formation of quencher-fluorophore complex. Static quenching occurs when both the fluorophore and quencher are in the ground state. High temperatures tend to disrupt ground state complex formation. This fact can be used to establish which mechanism is in operation. The value of the Stern–Volmer quenching constant should decrease with an increase in temperature as the ground state complex becomes less stable. The reverse will be observed for dynamic quenching. The trend observed in the current study (Table 1) illustrates quenching by the complexes is predominantly static. A bimolecular quenching constant (Kq) can be calculated from the Stern–Volmer constant: KSV = Kqτ0, where τ0 is the lifetime of the fluorophore and is 22 ns.37 For both complexes Kq is two orders of magnitude larger than the limiting value of 1010 M−1 s−1 36 considered the largest possible value in aqueous medium. This confirms that the fluorescence quenching is not the result of dynamic quenching, but rather a consequence of static quenching. Complex 2 with phen being more hydrophobic compared to bpy, can be expected to have better hydrophobic interaction when compared to 1 and this is reflected in the quenching constants of the two complexes. This view is supported by calculating the binding constants for the reaction. Since the quenching was initiated by a static process (the observed changes in fluorescence results from the interaction between EB-DNA and the complex) the binding constant for the reaction can be calculated using the following equation:
| (2) |
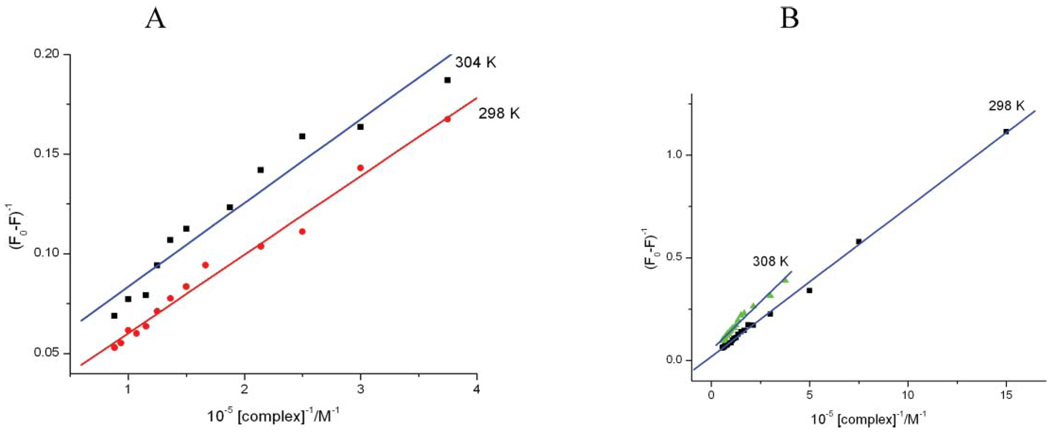
K is the binding constant and is obtained by taking the ration of the intercept to the slope of the plot of (F0 − F)−1 vs. 1/[complex] (Fig. 4). The results are given in Table 2. It can be seen from those results that the binding to ct-DNAis stronger for 1, which contains the phen ligand.
Fig. 4.
Plots 1/(Fo − F) vs. 1/[Ru] at different temperatures for 1 (A) and 2 (B).
Table 2.
Binding constants and thermodynamic parameters for interaction of 1 and 2 with the EB-DNA complex
| Compound | T/K | 10−4 K/M−1 |
R2 | ΔG°/kJ mol−1 |
ΔH°/ kJ mol−1 |
ΔS°/ J mol−1 K−1 |
|---|---|---|---|---|---|---|
| 1 | 298 | 10.1 | 0.987 | −28.5 | 43.6 | 242.3 |
| 304 | 14.3 | 0.981 | −30.0 | — | — | |
| 2 | 298 | 3.23 | 0.994 | −27.4 | 23.9 | 166.6 |
| 308 | 4.42 | 0.989 | −25.7 | — | — |
The typical thermodynamic parameters for a reaction (ΔG°, ΔH°, and ΔS°) can be calculated from the binding constants using the following standard relationships:
| (3) |
| (4) |
The values obtained are also given in Table 2. There are several intermolecular forces at play when a small molecule binds to a macromolecule. Both ΔH° and ΔS° are positive in our case and that suggest that the major forces in action are hydrophobic in nature.38 This is probably due to the water molecules around the DNA being disrupted on binding to the compounds and the whole system acquiring a more random configuration.
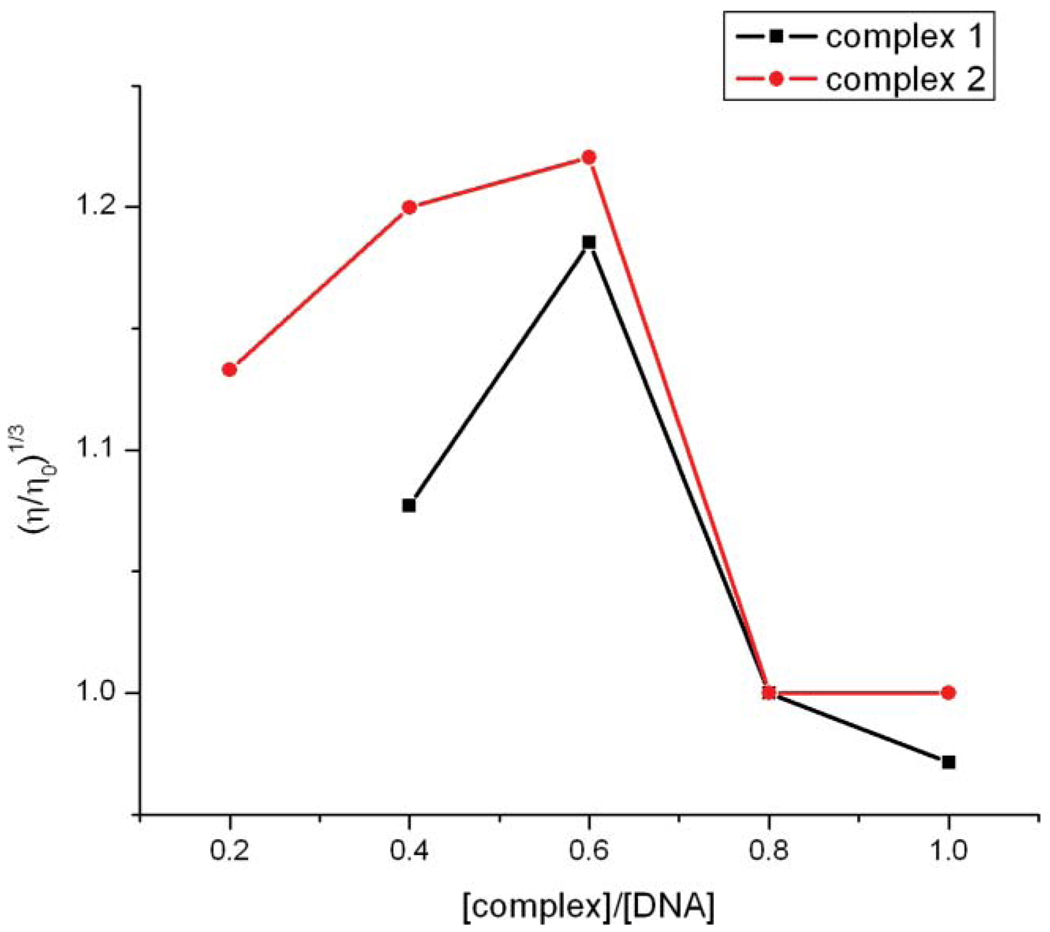
Viscometric studies
As a means for further clarifying the binding of the complexes with DNA, the viscosity of DNA solutions containing varying amount of added complex were measured. Photophysical probes such as absorption or fluorescence measurements generally provide significant but inconclusive evidence to support an intercalative binding model. Among the common methods, hydrodynamic methods (viscometry in particular) that are sensitive to DNA length changes are the most definitive tests of the classical intercalation model of binding in solution. Besides the ability to unwind DNA, a classical intercalator will cause an increase in the viscosity of a DNA solution since the DNA helix must lengthen as base pairs are separated to accommodate the binding ligand.39 In this study 1 mL of DNA solutions (10 µM in DNA) containing 0–10 µM of metal complexes were placed in the viscometer and flow times were measured after thermal equilibrium. According to theory of Cohen and Eisenberg40 viscosity data were plotted as (η/η0)1/3 vs. the binding ratio ([Ru]/[DNA]) as shown in Fig. 5. It was observed that increasing the complex concentration led to a gentle increase in the viscosity of the DNA solution at lower complex to DNA ratios. However at higher ratios there is a decrease in the viscosity of the solutions. Thus, together with the results from the EB displacement experiments, we may conclude that the complexes are only mild intercalators.
Fig. 5.
Effect of increasing concentration of 1 and 2 on the relative viscosities of CTDNA.
Cytotoxicity studies
Compounds 1–6 were evaluated for their cytotoxicity in a panel of human tumour cell lines (MCF-7, MDA-MB-231, HCT116 and HT29) by means of a colorimetric assay (MTS assay) which measures mitochondrial dehydrogenase activity as an indication of cell viability. The effects of the compounds on the viability of these cells were evaluated after an exposure period of 72 h. All the complexes showed activity and their corresponding IC50 values, corresponding to inhibition of cancer cell growth at the 50% level, are listed in Table 3. All the complexes have very moderate cytotoxic potencies, with IC50 values generally in the low micromolar concentrations. As a general observation, the phen complexes are more active than the bpy complexes in all the tested cell lines. DNA is a major cellular target for ruthenium metal complexes and the greater hydrophobicity of the phen ancillary ligand may lead to better cellular uptake leading to a higher cytotoxicity. Between cell lines there is also another weakly discernible trend in that the compounds showed higher activities against the MDA-MB-231 cells, which are oestrogen receptor negative (ER(−)) vs. the ER(+) MCF-7 cells. There is a similar but more weakly defined trend present in the colon cell lines—activity is higher in the HCT-116 cells. A simple structure-activity relationship (SAR) analysis suggest that, for both sets of complexes, as the alkyl group on N(3) gets bigger there is an increase in the cytotoxic potency. So the alkyl group on N(3) is a part of the pharmacophore and this suggests that the cytotoxicity is mainly governed by the thiosemicarbazone ligands, which are highly cytotoxic themselves, while complexation to metal ions rather serves to modulate their mode of action and activity. This is supported by the fact that the free thiosemicarbazone ligands are barely active under these conditions.
Table 3.
Anti-proliferative activity of complexes 1–6 in panel of four human cancer cell lines
| IC50/µMa | ||||
|---|---|---|---|---|
| Compound | MDA-MB-231 | MCF-7 | HCT-116 | HT29 |
| 1 | 4.47 ± 0.70 | 8.72 ± 1.22 | 7.26 ± 1.24 | 4.26 ± 2.24 |
| 2 | 18.8 ± 9.8 | 18.3 ± 7.9 | 20.7 ± 3.6 | 42.4 |
| 3 | 1.85 ± 0.03 | 3.60 ± 0.96 | 1.79 ± 0.12 | 2.00 ± 0.13 |
| 4 | 8.53 ± 4.52 | 11.2 ± 3.3 | 6.49 ± 1.14 | 19.5 ± 13.6 |
| 5 | 3.71 ± 3.24 | 1.93 ± 0.48 | 1.78 ± 0.08 | 1.87 ± 0.32 |
| 6 | 1.95 ± 0.06 | 3.56 ± 1.02 | 3.57 ± 2.77 | 2.48 ± 0.66 |
| Cisplatin | 730 | 506 ± 86 | 3.10 | 24.3 |
50% inhibitory concentration after exposure for 72 h in the MTS assay. Values are means ± standard deviations.
Conclusions
The synthesis of mixed-ligand diimine-thiosemicarbazone complexes of ruthenium has been successfully achieved using a novel method employing microwave radiation. From biophysical experiments we conclude that the complexes interact with DNA via weak to moderate intercalation likely through the aromatic rings of the diimine ligand. The complexes show good cytotoxicity against a variety of human cancer cell lines. The cytotoxic potencies indicate that these complexes are good candidates for further development as anticancer agents. We are currently studying their biochemical reactivity with serum proteins and the results will be reported in the future.
Experimental
Materials and methods
Analytical or reagent grade chemicals were used throughout. All the chemicals, including solvents, were obtained from Sigma–Aldrich (St. Louis, MO, USA) or other commercial vendors and used as received. The metal complexes were synthesized using a Discover S-Class microwave reactor (CEM, Matthews, USA). Microanalyses (C, H, N) were performed by Desert Analytics, Tucson, USA). 1H and 13C NMR spectra were recorded in dimethylsulfoxide-d6 on a JEOL ECX-300 or a Varian 300 MHz spectrometer operating at room temperature. The residual 1H and 13C present in DMSO-d6 (2.49 and 39.7 ppm, respectively) were used as internal references. Infrared (IR) spectra in the range 4000-500 cm−1 were obtained using KBr pellets or using the ATR accessory (with a diamond crystal) on a Nicolet 6700 FTIR spectrophotometer. Cyclic voltammetric (CV) data were collected on a Bioanalytical Systems Inc. Episilon workstation on a C3 cell stand at 296 K. CH2 Cl2 solutions (1 mM) containing 0.1 M tetrabutylammonium hexafluorophosphate as supporting electrolyte, were saturated with nitrogen for 15 min prior to each run. A blanket of nitrogen gas was maintained throughout the measurements. The measurements were carried out with a three-electrode system consisting of a platinum working electrode, a platinum wire auxiliary electrode and a Ag/AgCl reference electrode. Ferrocene was used as an internal standard. The working electrode was polished before each experiment with alumina slurry. The electronic spectra were recorded using quartz cuvettes on an Agilent 8453 spectrophotometer in the range 190–1100 nm using samples dissolved in DMSO. Fluorescence spectra were recorded on a Varian Cary Eclipse spectrophotometer. Melting points (triplicate measurements) were determined in open capillaries and are uncorrected. ESI MS was carried out on an HP Agilent 1956b single-quadrupole mass spectrometer. Samples were dissolved in acetonitrile and introduced by direct injection using a syringe pump and a flow rate of 20 µL min−1, while sweeping the cone voltage from 0 to 200 V at a rate of 10 V min−1.
Synthesis of ligands
The ligands 9-anthraldehydethiosemicarbazone (ATSC), 9-anthraldehyde-N(4)-methylthiosemicarbazone (MeATSC) and 9-anthraldehyde-N(4)-ethylthiosemicarbazone (EtATSC) were prepared according to the following general method: equimolar amounts (16.4 mmol) of 9-anthraldehyde and the appropriate N4 alkyl-substituted thiosemicarbazide were suspended in 100 mL of absolute anhydrous ethanol containing a few drops of glacial acetic acid. The reaction mixture was heated at reflux for 3.5 h and after cooling the light precipitate that formed was collected by filtration and washed thoroughly with ethanol followed by ether and dried in the vacuum.
ATSC
Orange-yellow solid (6.08 g, 91%); calcd for C16H13N3S: C 68.8, H 4.7, N 15.0. Found: C 68.95, H 4.7, N 14.95; mp 208–210 °C; ν/cm−1: 3438, 3214, 3155 (NH2, NH), 1600 (C=N), 1019 (N–N) and 1282, 843 (C=S); δH (300 MHz; DMSO); 11.65 (1 H), 9.32 (1 H), 8.70 (1 H), 8.55–8.58 (2 H), 8.17 (1 H), 8.13–8.15 (2 H), 7.57–7.59 (4 H).
MeATSC
Orange-yellow solid (4.95 g, 88%); calcd for C17H15N3S: C 69.6, H 5.1, N 14.3. Found: C 69.7, H 5.1, N 14.1; mp 213–214 °C; ν/cm−1: 3399, 3202 (NH2, NH), 1621 (C=N), 1040 (N–N) and 1283 (w), 841 (C=S); δH (300 MHz; DMSO); 11.72 (1 H), 9.27 (1 H), 8.68 (1 H), 8.48–8.50 (2 H), 8.11–8.14 (2 H), 7.54–7.65 (4 H), 3.02 (3 H).
EtATSC
Yellow solid (5.21 g, 87%); calcd for C18H17N3S: C 70.3, H 5.6, N 13.7. Found: C 70.2, H 4.65, N 13.2; mp 194–196 °C; ν/cm−1: 3342, 3154 (NH2, NH), 1622 (C=N), 1047 (N–N) and 1299, 840 (C=S); δH (300 MHz; DMSO); 11.67 (1 H), 9.27 (1 H), 8.68 (1 H), 8.47–8.50 (2 H), 8.12–8.15 (2 H), 7.56–7.59 (4 H), 3.58 (2 H), 1.15 (3 H).
Synthesis of the complexes
The starting ruthenium complexes, [(phen)2RuCl2]·H2O and [(bpy)2RuCl2], were synthesized as described in the literature.41 The target complexes were synthesized by the following general method: Equimolar amounts of [(phen)2RuCl2]·H2O or [(bpy)2RuCl2] and the appropriate ligand was suspended in 8–10 mL of ethylene glycol in a 35 mL reaction vessel. The vessel was capped and the reaction mixture saturated with argon for 15 min. The reaction was then heated at 150 °C for 5 min (using a dynamic method). The dark brown suspension became a dark red solution. This solution was poured onto 5 mL of a saturated aqueous solution of KPF6, which resulted in the immediate precipitation of a red solid. The solid was collected by vacuum filtration, washed with water followed by ether and then dried at the vacuum pump. The product was recrystallized from acetone and ether.
[(phen)2Ru(ATSC)](PF6)2], 1
Orange-red solid (174 mg, 42%); calcd for C40H29F12N7 P2Ru S: C46.6, H 2.8, N 9.5. Found: C47.05, H 3.2, N 9.2; mp 206 °C; ν/cm−1: 3425 (w), 3354, 3203 (w) (NH2, NH), 1580 (C=N), 1047 (N–N) and 1263, 826 (C=S); m/z (ESI) 740 (100, [M − H − 2PF6]+); λmax (CH2Cl2)/ nm 235, 395, 433 sh and 270 infl (log ε 4.30, 3.60, 3.56 and 4.12); δH (300 MHz; DMSO); 9.88 (s), 9.79 (s), 8.95 (d), 6.40–8.50 (multiple multiplets).
[(bpy)2Ru(ATSC)](PF6)2], 2
Red-brown solid (262 mg, 66%); calcd for C36H29F12N7 P2Ru S: C 44.0, H 3.0, N 10.0. Found: C 44.9, H 2.6, N 9.6; mp 207 °C; ν/cm−1: 3466, 3345, 3223 (w) (NH2, NH), 1603 (C=N), 1047 (N–N) and 1265, 825 (C=S); m/z (ESI) 692 (100, [M − H − 2PF6]+); λmax (CH2Cl2)/ nm 200, 236, 293, 372 and 446 infl (log ε 4.01, 4.30, 4.09, 3.79 and 3.62); δH (300 MHz; DMSO); 9.76 (s), 9.40 (s), 9.33 (d), 6.11–8.40 (multiple multiplets).
[(phen)2Ru(MeATSC)](PF6)2], 3
Red solid (218 mg, 55%); calcd for C37H31F12N7 P2Ru S: C 44.6, H 3.1, N 9.9. Found: C 43.95, H 2.8, N 9.6. mp 204 °C. ν/cm−1: 3381, 3242 (NH2, NH), 1601 (C=N), 1047 (N–N) and 1271, 825 (C=S); m/z (ESI) 754 (100, [M − H − 2PF6]+); λmax (CH2Cl2)/ nm 235, 395, 440 sh and 283 infl (log ε 4.35, 3.79, 3.73 and 4.12); δH (300 MHz; DMSO); 9.89 (s), 9.79 (s), 8.96 (d), 6.20–8.20 (multiple multiplets), 3.10 (s).
[(bpy)2Ru(MeATSC)](PF6)2], 4
Dark red solid (353 mg, 85%); calcd for C41H31F12N7 P2Ru S: C 47.1, H 3.0, N 9.4. Found: C 46.6, H 3.4, N 9.05; mp 201 °C; ν/cm−1: 3375 (NH2, NH), 1585 (C=N), 824 (C=S); m/z (ESI) 706 (100, [M − H − 2PF6]+); λmax (CH2Cl2)/ nm 199, 236, 291, 376 and 449 infl (log ε 4.02, 4.33, 4.10, 3.73 and 3.59); δH (300 MHz; DMSO); 9.77 (s), 9.32 (d), 6.20–8.20 (multiple multiplets), 3.11 (s).
[(phen)2Ru(EtATSC)](PF6)2], 5
Orange-red solid (275 mg 64%); calcd for C42H33F12N7 P2Ru S: C 47.4, H 2.9, N 8.7. Found: C 47.6, H 3.1, N 9.3; mp 199 °C. ν/cm−1: 3365, 3226 (w) (NH2, NH), 1580 (C=N), 1037 (N–N) and 824 (C=S); m/z (ESI) 768 (100, [M − H − 2PF6]+); λmax (CH2Cl2)/ nm 236, 395, 440 sh and 284 infl (log ε 4.31, 3.92, 3.86 and 4.09); δH (300 MHz; DMSO); 9.91 (s), 9.77 (s), 8.91 (d), 6.26–8.50 (multiple multiplets), 2.11 (s), 1.27 (bs).
[(bpy)2Ru(EtATSC)](PF6)2], 6
Red solid (258 mg, 65%); calcd for C39H37F12N7 P2Ru S: C 45.15, H 3.3, N 9.7. Found: C 44.7, H 3.2, N 9.4; mp 203 °C; ν/cm−1: 3375, 3242 (w) (NH2, NH), 1601 (C=N), 1044 (N–N) and 1272, 827 (C=S); m/z (ESI) 720 (100, [M − H − 2PF6]+); λmax (CH2Cl2)/ nm 200, 236, 292, 376 and 447 infl (log ε 4.02, 4.32, 4.09, 3.74 and 3.60); δH (300 MHz; DMSO); 9.76 (s), 9.49 (s), 9.34 (s), 5.90–8.60 (multiple multiplets), 1.25 (s), 0.96 (bs).
DNA interaction studies
All the experiments involving the interaction of the complexes with DNA were carried out in Tris-HCl buffer (5 mM, 50 mM NaCl, pH 7.20). Stock solutions of ct-DNA were prepared by dissolving commercial nucleic acids in buffer and stored at 4 °C for more than 24 h to get homogeneity. After dilution, the DNA concentration per nucleotide phosphate was determined spectrophotometrically using the molar extinction coefficient of 6600 M−1 cm−1 at 260 nm.42 A solution of ct-DNA in the buffer gave a ratio of UV absorbance at 260 and 280 nm of ≥ 1.8 indicating that ct-DNA was sufficiently free from protein.43 The ct-DNA stock solutions were stored at −20 °C and used within 1 week after their preparation. Doubly purified water used in all experiments was from a Millipore Milli-Q (18.2 MΩ cm) system.
Viscosity measurements
Viscosity studies were done using a Cannon-Manning semi micro-dilution viscometer (type 75, Cannon Instruments Co., State College, PA, USA) immersed vertically in a thermostatted water bath maintained at 31.0 ± 0.1 °C. The viscosity for DNA was measured in the presence and absence of the metal complexes. The DNA concentration was maintained at 10 µM, while the complex concentration varied from 0–10 µM. Data are presented as (η/η0)1/3 vs. 1/R, where R = [DNA]/[complex] and η is the viscosity of DNA in the presence of the complex and η0 is the relative viscosity of DNA alone. Relative viscosity values were calculated from the observed flow time of DNA solution (t) and corrected for the flow time of buffer alone (t0), using the expression η0 = (t − t0)/ t0. Flow time was measured with a digital stopwatch and each sample was measured three times and an average flow time was used.
Ethidium bromide displacement experiments
In the ethidium bromide (EB) fluorescence displacement experiment, 3 mL of a solution that is 10 µM DNA and 0.33 µM EB (saturated binding levels),44 in Tris buffer was titrated with concentrated solutions of the complexes producing the solutions with the varied mole ratio of complex to ct-DNA. After each addition the solution was vortexed for 30 s and allowed to sit at the appropriate temperature for 5 min before measurements. The fluorescence spectra of the solution were obtained by exciting at 520 nm and measuring the emission spectra from 540–700 nm using 5 nm slits.
Cell culture
All the tumour cell lines, MCF-7 and MDA-MB-231 (human breast adenocarcinoma), HT-29 (colon adenocarcinoma) and HCT116 (colon carcinoma) were obtained from the American Type Culture Collection (ATCC, Rockville, MD, USA) and maintained at the University of Rhode Island. The cells were cultured in McCoy’s 5A medium containing glutamine supplemented with 10% heat-inactivated foetal bovine serum, the antibiotic, Penicillin-Streptomycin (10 mg mL−1) (Invitrogen, Grand Island, NY, USA), and the addition of HEPES solution to control the pH of the media. All cell lines were incubated in a humidified environment at 37 °C in 5% CO2 and maintained in the linear phase of growth.
Cytotoxicity tests
The assay was carried out as described previously45 to measure the IC50 values for samples. Briefly, the in vitro cytotoxicity of samples were assessed in tumour cells by a tetrazolium-based colorimetric assay, which takes advantage of the metabolic conversion of MTS [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfenyl)-2H-tetrazolium, inner salt] to a reduced form that absorbs light at 490 nm. Cells were counted using a hemacytometer and were plated at 3750–10 000 cells per well, depending on the cell line, in a 96-well format for 24 h prior to drug addition. Test samples and a positive control, etoposide, were solubilized in DMSO by sonication. All samples were diluted with media to the desired treatment concentration and the final DMSO concentration per well did not exceed 0.3%. Control wells were also included on all plates. Following a 72 h drug-incubation period at 37 °C with serially diluted test compounds, MTS, in combination with the electron coupling agent, phenazine methosulfate, was added to the wells. The incubation was continued for 3 h, and the absorbance of the medium was measured at 490 nm with a spectrophotometer (Spectramax M2, Molecular Devices, operated by SoftmaxPro v.4.6 software) to obtain the number of surviving cells relative to control populations. The results are expressed as the median cytotoxic concentrations (IC50 values) and were calculated from six-point dose response curves using 4-fold serial dilutions. Each point on the curve was tested in triplicate. Data are expressed as mean ± SE for three replications.
Supplementary Material
Acknowledgements
The project described was supported by NIH Grant Number P20 RR-16460 from the IDeA Networks of Biomedical Research Excellence (INBRE) Program of the National Center of Research Resources. AAH would like to thank USM for funding part of this research. AAH is also very grateful for the use of an LC/Ion Trap Mass Spectrometer, which was funded by the NSF, grant number, CHE 0639208.
Footnotes
Electronic supplementary information (ESI) available: ESI MS (Fig. S1 and S2) and UV-vis and electrochemistry (Fig. S3 and S4) data.
References
- 1.Quiroga AG, Perez JM, Lopez-Solera I, Masaguer JR, Luque A, Raman P, Edwards A, Alonso C, Navarro-Ranninger C. J. Med. Chem. 1998;41:1399–1408. doi: 10.1021/jm970520d. [DOI] [PubMed] [Google Scholar]
- 2.Offiong OE, Martelli S. Farmaco. 1994;49:513–518. [PubMed] [Google Scholar]
- 3.Hadjipavlou-Litina D. Pharmazie. 1996;51:468–470. [PubMed] [Google Scholar]
- 4.Garcia-Tojal J, Garcia-Orad A, Serra JL, Pizarro JL, Lezamma L, Arriortua MI, Rojo T. J. Inorg. Biochem. 1999;75:45–54. doi: 10.1016/S0162-0134(99)00031-8. [DOI] [PubMed] [Google Scholar]
- 5.Petering DH. Bioinorg. Chem. 1972;1:255–271. [Google Scholar]
- 6.West DX, Liberta AE, Padhye SB, Chikate RC, Sonawane PB, Kumbhar AS, Yerande RG. Coord. Chem. Rev. 1993;123:49–71. [Google Scholar]
- 7.Sava G, Pacor S, Bergamo A, Cocchettio M, Mestroni G, Allesio E. Chem.-Biol. Interact. 1995;95:109–126. doi: 10.1016/0009-2797(94)03350-1. [DOI] [PubMed] [Google Scholar]
- 8.Sava G, Bergamo A. Int. J. Oncol. 2000;17:353–365. doi: 10.3892/ijo.17.2.353. [DOI] [PubMed] [Google Scholar]
- 9.Rademaker-Lakhai JM, Van Den Bongard D, Pluim D, Beijnen JH, Schellens JH. Clin. Cancer Res. 2004;10:3717–3727. doi: 10.1158/1078-0432.CCR-03-0746. [DOI] [PubMed] [Google Scholar]
- 10.Sava G, Gagliardi R, Bergamo A, Alessio E, Mestroni G. Anticancer Res. 1999;19:969–972. [PubMed] [Google Scholar]; Bergamo A, Gava B, Alessio E, Mestroni G, Serli B, Cocchieto M, Zorzet S, Sava G. Int. J. Oncol. 2002;21:1331–1338. [PubMed] [Google Scholar]; Groessl M, Reisner E, Hartinger CG, Eichinger R, Semenova O, Timerbaev AR, Jakupec MA, Arion VB, Keppler BK. J. Med. Chem. 2007;50:2185–2193. doi: 10.1021/jm061081y. [DOI] [PubMed] [Google Scholar]
- 11.Hartinger CG, Zorbas-Seifried S, Jakupec MA, Kynast B, Zorbas H, Keppler BK. J. Inorg. Biochem. 2006;100:891–904. doi: 10.1016/j.jinorgbio.2006.02.013. [DOI] [PubMed] [Google Scholar]; Kapitza S, Pongratz M, Jakupec MA, Heffeter P, Berger W, Lackinger L, Keppler BK, Marian B. J. Cancer Res. Clin. Oncol. 2005;131:101–110. doi: 10.1007/s00432-004-0617-0. [DOI] [PubMed] [Google Scholar]; Keppler BK, Henn M, Juhl UM, Berger MR, Niebl R, Wagner FE. Prog. Clin. Biochem. Med. 1989;10:41–69. [Google Scholar]; Pongratz M, Schluga P, Jakupec MA, Arion VB, Hartinger CG, Allmaier G, Keppler BK. J. Anal. At. Spectrom. 2004;19:46–51. [Google Scholar]
- 12.Kreuser ED, Keppler BK, Berdel WE, Piest A, Thiel E. Semin. Oncol. 1992;19:73–81. [PubMed] [Google Scholar]
- 13.Morris RE, Aird RE, Murdoch P, del S, Chen H, Cummings J, Hughes ND, Parsons S, Parkin A, Boyd G, Jodrell DI, Sadler PJ. J. Med. Chem. 2001;44:3616–3621. doi: 10.1021/jm010051m. [DOI] [PubMed] [Google Scholar]
- 14.Novakova O, Kasparkova J, Bursova V, Hofr C, Vojtiskova M, Chen H, Sadler PJ, Brabec V. Chem. Biol. 2005;12:121–129. doi: 10.1016/j.chembiol.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 15.Erkkila KE, Odom DT, Barton JK. Chem. Rev. 1999;99:2777–2796. doi: 10.1021/cr9804341. [DOI] [PubMed] [Google Scholar]
- 16.Metcalfe C, Thomas JA. Chem. Soc. Rev. 2003;32:215–224. doi: 10.1039/b201945k. [DOI] [PubMed] [Google Scholar]
- 17.Stoeffler HD, Thornton NB, Temkin SL, Schanze KS. J. Am. Chem. Soc. 1995;117:7119–7128. [Google Scholar]
- 18.Yam VW-WV, Lo KK-W, Cheung KK, Kong RY-C. J. Chem. Soc., Dalton Trans. 1997:2067–2072. [Google Scholar]
- 19.Metcalfe C, Webb M, Thomas JA. Chem. Commun. 2002:2026–2027. doi: 10.1039/b205312h. [DOI] [PubMed] [Google Scholar]
- 20.Arounaguiri S, Maiya BG. Inorg. Chem. 1999;38:842–843. doi: 10.1021/ic981109z. [DOI] [PubMed] [Google Scholar]
- 21.Liu X-W, Li J, Li H, Zheng K-C, Chao H, Ji L-N. J. Inorg. Biochem. 2005;99:2372–2380. doi: 10.1016/j.jinorgbio.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 22.Sundquist WI, Lippard SJ. Coord. Chem. Rev. 1990;100:293–322. [Google Scholar]
- 23.Kumar CV, Barton JK, Turro NJ. J. Am. Chem. Soc. 1985;107:5518–5523. [Google Scholar]
- 24.Papadia P, Margiotta N, Bergamo A, Sava G, Natile G. J. Med. Chem. 2005;48:3364–3371. doi: 10.1021/jm0500471. [DOI] [PubMed] [Google Scholar]
- 25.West DX, Swearigen JK, Valdee-Martinez J, Hernandez-Ortegs S, El-Sawaf AK, van Meurs F, Castineiras A, Garcia I, Bermejo E. Polyhedron. 1999;18:2919–2929. [Google Scholar]
- 26.Lobana TS, Bawa G, Butcher RayJ, Liaw B-J, Liu CW. Polyhedron. 2006;25:2897–2903. [Google Scholar]
- 27.Lobana TS, Sanchez A, Casas JS, Castineiras A, Sordo J, Garcia-Tasende MS, Vazquez-Lopez EM. J. Chem. Soc., Dalton Trans. 1997:4289–4299. [Google Scholar]
- 28.Lobana TS, Rekha, Butcher RJ, Failes TW, Turner P. J. Coord. Chem. 2005;58:1369–1375. [Google Scholar]
- 29.Lobana TS, Khanna S, Butcher RayJ, Hunter AD, Zeller M. Polyhedron. 2006;25:2755–2763. [Google Scholar]
- 30.Afrasiabi Z, Sinn E, Kulkami P, Ambike V, Padhye S, Deobagakar D, Heron M, Gabbutt C, Anson C, Powell A. Inorg. Chim. Acta. 2005;358:2023–2030. [Google Scholar]
- 31.Beliochi-Ferrari M, Biscoglie F, Cavalieri C, Pelosic G, Tarasconi P. Polyhedron. 2007;26:3774. [Google Scholar]
- 32.Lobana TS, Castinieras A. Polyhedron. 2002;21:1603–1611. [Google Scholar]
- 33.Mostafa MM, El-Hammid A, Shallaby M, El-Asmay AA. Transition Met. Chem. 1981;6:303. [Google Scholar]
- 34.Beraldo H, Nacif WF, Teixeria LR, Reboucas JS. Transition Met. Chem. 2002;27:85–88. [Google Scholar]
- 35.Baguley BC, Le Bret M. Biochemistry. 1984;23:937–943. doi: 10.1021/bi00300a022. [DOI] [PubMed] [Google Scholar]
- 36.Lacowicz JR. Principles of Fluorescence Spectroscopy. 3rd Ed. New York: Springer; 2006. [Google Scholar]
- 37.Ghosh KS, Sahoo BK, Jana D, Dasgupta S. J. Inorg. Biochem. 2008;102:1711–1718. doi: 10.1016/j.jinorgbio.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 38.Ross PD, Subramanian S. Biochemistry. 1981;20:3096–3102. doi: 10.1021/bi00514a017. [DOI] [PubMed] [Google Scholar]
- 39.Long EC, Barton JK. Acc. Chem. Res. 1990;23:271–273. [Google Scholar]
- 40.Cohen G, Eisenberg H. Biopolymers. 1969;8:45–55. [Google Scholar]
- 41.Sullivan P, Salmon DJ, Meyer TJ. Inorg. Chem. 1978;17:3335–3341. [Google Scholar]
- 42.Reichmann ME, Rice SA, Thomas CA, Doty PJ. J. Am. Chem. Soc. 1954;76:3047–3053. [Google Scholar]
- 43.Vijayalakshmi R, Kanthimathi M, Subramanian V, Nair BU. Biochim. Biophys. Acta, Gen. Subj. 2000;1475:157–162. doi: 10.1016/s0304-4165(00)00063-5. [DOI] [PubMed] [Google Scholar]
- 44.Barton JK, Goldberg JM, Kumar CV, Turro NJ. J. Am. Chem. Soc. 1986;108:2081–2088. [Google Scholar]
- 45.Cory AH, Owen TC, Barltrop JA, Cory JG. Cancer Commun. 1991;3:207–212. doi: 10.3727/095535491820873191. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.