Summary
Microscale techniques have been applied to biological assays for nearly two decades, but haven’t been widely integrated as common tools in biological laboratories. The significant differences between several physical phenomena at the microscale versus the macroscale have been exploited to provide a variety of new types of assays (such as gradient production or spatial cell patterning). However, the use of these devices by biologists seems to be limited by issues regarding biological validation, ease of use, and the limited available readouts for assays done using microtechnology. Critical validation work has been done recently that highlights the current challenges for microfluidic methods and suggest ways in which future devices might be improved to better integrate with biological assays. With more validation and improved designs, microscale techniques hold immense promise as a platform to study aspects of cell biology that are not possible using current macroscale techniques.
An introduction to microfluidics for cell biology assays
Various influences determine the phenotype of cells in vivo and contribute to their coordinated responses to stimuli. These influences include interactions with neighboring cells (e.g. epithelia–stromal), interactions with the extracellular matrix (ECM), and systemic factors (e.g. hormones). Yet, these interactions are not easily replicated or controlled in traditional formats. Current methods (Petri dish, microtiter plates, which are in general macroscale techniques, with dimensions in centimeters and larger) afford a limited degree of microenvironmental control. Approaches that aim to recapitulate aspects of in vivo microenvironments are often laborious (e.g. Dunn chamber for soluble gradients and chemotaxis), expensive (e.g. three-dimensional gel culture) or both (trans-well membrane inserts for migration, co-culture or invasion assays). Microfluidic technologies for cell-based assays have the potential to increase the biological relevance of cell culture models while maintaining or increasing the throughput of current methods.
Microscale techniques for cell biology (i.e., those using devices that have dimensions ranging from micrometers to millimeters), range from single-cell analyses and flow cytometry-like techniques,(1) to treating fields of cells in gradient generating devices,(2) patterned three-dimensional cultures,(3,4) to microscale versions of more traditional assay types such as cell culture (via perfusion,(5,6) or static cultures).(3,7–9) These microfluidic devices typically provide unique functionalities beyond traditional techniques either by controlling the cellular microenvironment in ways not previously possible, by allowing existing assays to be performed on significantly smaller samples (down even to the single cell level) or by using reagents that are many-fold less costly. Microfluidic systems enable spatial patterning of molecules and cells(10) as well as both passive(11) and active cell handling and environmental control. Temporal and spatial control on the micrometer scale (0.1–100 μm has been used in fundamental studies from the subcellular(12) to the organismal(13) level in studies of cell division axis orientation(14) and geometric influence on cell survival.(15) Thus it is clear that, at its core, microfluidics has the potential to have a great impact in cell biology as many of the leading questions in cell biology are well suited to study using these functionalities.
Although the applications of microtechnology to cell biology have been considered for nearly two decades,(16–18) the field continues to progress via a plethora of demonstrations that provide glimpses of the potential impact of microtechnology on the methods used for cell biology and the types of data that can be obtained. To date, however, microfluidics and “lab on a chip” techniques have not made a large impact on cell biology either academically or commercially. The relative lack of integration of microtechnology in biological laboratories could be due in part to a disconnect between the engineers who design and fabricate the devices and the biologists who would ultimately use them. This disconnect has resulted in devices that, while functional and potentially useful, are often technically challenging to use and obtain reliable, biologically meaningful data from. Meanwhile, the lack of biological validation of data obtained in microscale devices has potentially hindered the process as well. Finally, when new types of data are obtained using these new technologies, the challenge of interpreting the data in the context of what is currently known (but obtained using traditional techniques) can create yet another hurdle to presenting data to either field.
In this essay, we will illustrate how some existing microfluidic methods have been applied to biological assays and begun to be validated. These examples highlight the steps required to move from demonstration to utility and to more closely integrate microtechnology with traditional cell biology techniques. By understanding how physics affects the microenvironments found in microfluidic devices, we can better predict and understand on a general level the strengths and limitations of doing biological assays with microfluidic devices. We will briefly review some of the critical physical phenomena that will or in some cases have already been shown to affect the biological outcome of an assay performed in microfluidic devices. Ideally, this will provide some insight into how to interpret data obtained using these methods and how experiments can be designed to maximize the unique capabilities of microtechnology.
Current cell biological applications of microfluidics
To date, much of the work in microfluidics for cell-biology-based applications has focused on assays of cell behavior in the presence or absence of specific soluble factors. The application of controlled gradients of soluble factors has highlighted microfluidics’ potential for expanding current techniques to include new assays, or providing a platform for simplifying and improving current techniques.
Example: microfluidic gradient generation devices
Stimulating a field of cells with a controlled gradient of a soluble factor is a unique type of microfluidic assay that can effectively produce different microenvironments in a single device.(2) Few traditional techniques for gradient production, such as the Zigmond chamber,(19) have been able to produce as defined, controlled and repeatable gradients as those produced using microfluidic techniques. Precisely defined chemical gradients in microfluidic devices have been applied to many biological systems, such as to stimulate migratory cells (e.g. neutrophils, bacteria, sperm cells) using chemoattractants,(20–26) investigate cancer cells responding to a drug or growth factor,(27–29) or to stimulate the differentiation of embryonic stem cells.(30) This class of microfluidic devices has the potential to improve the sensitivity and complexity of experiments studying cellular responses to gradients beyond what is currently possible via traditional techniques.
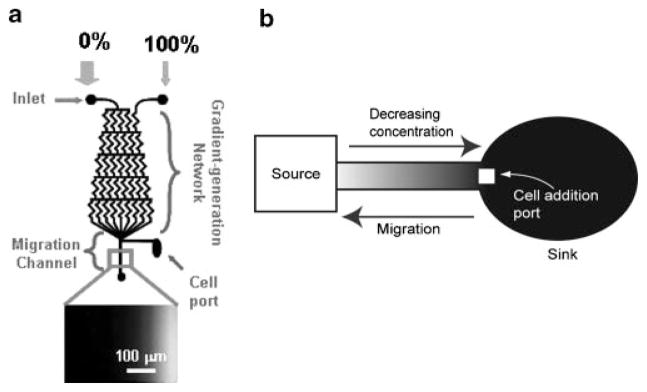
One general class of gradient-producing devices is based on the mixing of chemical species between two streams of fluid in laminar flow solely due to diffusion rather than convection (Fig. 1a). Flowing streams allow gradients of species created by diffusion to be formed at their interfaces, which then can be flowed over cells of interest to expose them to the gradient formed. Often microfluidic chemotaxis assays include a gradient of a single chemoattractant or growth factor but some devices have incorporated more complex combinations of factors.(22,31) The temporal and spatial control over defined gradients of soluble factors or immobilized factors (on surfaces) provided by flow-based microfluidic devices are a significant improvement over the widely available methods. Laminar flow-based systems facilitate quantitative correlations between environmental cues and observed cellular behavior, which may provide insight into the mechanisms that affect signaling cascades and expression.
Figure 1.
Examples of microfluidic gradient production devices. Flow based gradients like that shown in (a) are based on diffusional mixing solely at the interface between fluid streams. Here two solutions with different concentrations of the solute of interest (0% and 100% of the desired final concentration in this case) are introduced to the inputs of a gradient generation network. Diffusional mixing occurs at the interfaces of the fluid streams and creates a gradient of a defined profile (dependent on input concentrations) at the point labeled Migration channel where cells are treated with the flowing gradient of interest. Static gradient systems like that shown in (b) can be used to create stable gradients in a static fluid, by addition of fluid of the maximum concentration at the Source and allowing the solute to diffuse to the sink, thus exposing cells in the channel to a gradient of the factor. Adapted from: (a) Biomedical Microdevices, A parallel-gradient microfluidic chamber for quantitative analysis of breast cancer cell chemotaxis, 8, 2006, page 109–118, Saadi W, Wang SJ, Lin F, Jeon NL, Figure 1 with kind permission from Springer Science +Business Media: and (b) from Abhyankar VV, Lokuta MA, Huttenlocher A, Beebe DJ. 2006. Characterization of a membrane-based gradient generator for use in cell-signaling studies. Lab Chip 6:389393. Reproduced by permission of The Royal Society of Chemistry, http://dx.doi.org/10.1039/b514133h.
The limitations of these systems are primarily due to practicality issues, cost and potential biological artifacts. From a practical point of view, these systems require very stable fluid flow and therefore complicated fluid handling setups, which rely on specialized pumps and tubing not typically found in most laboratories. Also, to maintain a stable gradient in a continuously flowing system, relatively large total volumes of reagents must be used and experiments become increasingly costly the longer the time course for the response of interest. In these systems, continuous flow constantly renews nutrients and chemoattractants, while also depleting waste products and intercellular signaling molecules, resulting in temporally uniform concentrations of media and experimental components. Although this uniformity is beneficial in that the response of cells over time to a uniform stimulus or the effects of rapid changes in a defined stimulus can be determined, the contribution to the response of any soluble cell–cell communication is obscured. By disrupting cell–cell communication, the location and migration behavior of nearby cells may not influence a cell’s response to the stimulus as it might in vivo. The effects of flow alone on neutrophils has been addressed and mechanical activation by shear from laminar flow in microchannels was demonstrated.(32) Walker and colleagues have also shown that the flow rate used to create gradients can affect and therefore bias the migratory behavior of these cells.(33)
In situations where cell–cell communication plays an important role in modulating cellular response, the continuously flowing streams necessary to maintain the chemical gradients make laminar flow-based methods unsuitable for probing cellular responses. Generation of gradients in static fluid preserves paracrine signaling, while still providing gradients of factors defined in both time and location (Fig. 1b).(20) By allowing diffusion between a source and a sink along a thin channel, passively generated gradients can be formed and kept intact for long periods of time (over 24 hours). The source–sink concept can be used to create stable or temporally varying gradients along length of a channel. The gradient profile can then be controlled by adjusting the input concentration, distance from source to sink, or by changing the geometry of the channel (e.g. uniform width versus expanding or contracting), allowing for a range of linear or non-linear gradient profiles which may more accurately mimic in vivo gradients.
While flow based gradient generation devices often rely on more complicated designs and fluid-handling systems, many static gradient devices are much simpler to use. Because these devices typically require no additional equipment beyond common laboratory supplies (pipettes, microscopes, etc), they have the potential to be integrated more easily than more complicated designs. Also, with no fluid flow required for gradient maintenance, a more coordinated response of a population of cells can be observed by allowing paracrine signaling, and minimal total reagent is consumed for even long experiments. However, without constant flow, steps must be taken to minimize evaporation, which can be a significant factor for microfluidic devices with small volumes, and the time required to set up the gradients is often rather long (on the order of hours rather than seconds). Additionally, if the cell population requires more media renewal than that occurring via diffusional mixing between the channel and the source–sink (which both serve as a source for other media components), then artifacts may occur.
By treating cells with gradients of soluble components in microfluidic devices, a wide range of new assays can be performed that are more challenging or impossible to perform as accurately with traditional methods. However, the potential artifacts introduced by these devices will be important to establish for further integration of these techniques into biological research. Also, close collaboration between engineers and biologists will aid in developing devices that are more user friendly, a critical step in enabling these devices to become more accessible methods for experimentation. While microfluidics could provide a wealth of new information, it is unclear how data from microfluidics assay might fit in with data obtained using traditional methods without a baseline for comparison.
Example: cell culture in microfluidic devices
Another application of microfluidics that, while seemingly simple, holds immense promise is cell culture.(34,35) Microfluidic devices for cell culture provide a platform for higher throughput analyses of cellular responses to soluble stimuli with a variety of cost and resource benefits. Because each assay can be performed on a smaller total number of cells, more different assays can be performed with the same sample size when done in microfluidic cultures. This is particularly beneficial for rare or expensive cell types such as stem cells, or flow sorted cell populations. Additionally, typical microfluidic cultures require far less media and potentially costly inhibitors, growth factors or other reagents than even 96-well plates.
Many dominant phenomena in microfluidic devices are unique to the microscale and, by leveraging the scalability (or lack thereof) of specific designs, more control and flexibility of the microenvironments that the cells are exposed to can be obtained allowing new and different assays to be performed. Stimulating cultures of cells with drugs,(27,36,37) or other components that induce differentiation(38) in microfluidic platforms allow the replication of traditional tissue culture analyses in smaller volumes with fewer cells allowing expensive assays to be performed using minimal resources.
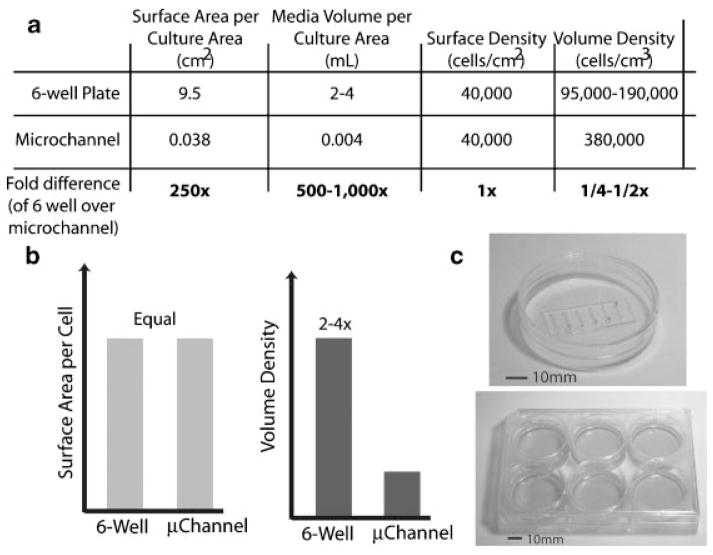
Typical volume densities in cultures using traditional techniques can be quite small (large media volumes for very few cells), and often are much higher in static microfluidic culture devices (Fig. 2). The effects on cellular behavior of volume density alone can be significant(39,40) but have not been addressed for many microfluidic cell culture systems and seem to be cell-type dependent. The lack of flow in static cultures results in less disturbance of the soluble cellular microenvironment than traditional macroscale cultures where bulk fluid flows result in convective mass transport being dominant (and thus eliminate local concentration gradients set up via diffusion to or from a cell). These devices generally do not require any equipment beyond pipettes for fluid handling and microscopes for visualization and analysis, both common laboratory equipment. The ease of use of these types of devices and the integration with existing automated fluid handling (or manual pipeting) has allowed this type of microfluidic device to be easily integrated into biological laboratories without the need for additional specialized equipment.(3)
Figure 2.
A comparison between volume densities of culture conditions in traditional, macroscale culture in 6-well plates and in microscale, microchannel culture. Given the same cell surface density, even a rather large microchannel (750 μm wide, 5 mm long, and 250 μm tall) can provide a volume density 2–4 times that of a traditional well in a 6-well plate, use 250 times fewer cells, and 500–1000 times less media and costly reagents.
Microfluidic devices also have optical benefits over smaller multiwell plates such as 96-well plates. Visualizing cells via either phase or fluorescence in a 96-well plate is nearly impossible due to the meniscus of fluid in the well (interferes with phase contrast) and thickness of the plastic bottom (not suitable for high magnification fluorescence images). Microfluidic devices for culture keep fluid menisci at the ends of a channel only, allowing easy visualization via phase contrast along the length of the channel. Additionally, most devices can be placed on any substrate, allowing glass to be used when necessary for fluorescent detection and analysis.
Meanwhile, continuously perfused microfluidic cultures can provide continuous supply of nutrients and allow for longer-term cultures as compared to static cultures, but at the expense of effective volume density and soluble cell–cell communication.(6) These culture conditions employ laminar flow for continuous transport of solutes and thus are convection based devices that limit diffusive transport of solutes to and from cells in a culture.
Perfusion cultures of murine embryonic stem cells in microchannels at flow rates orders of magnitude apart showed improved morphology after several days with higher flow.(41) Experiments that benefit from or require flow such as studying how cells respond to shear stresses (such as endothelial cells(42)) are potentially well suited to microfluidic assay because very precise control over flow rates and channel dimensions can provide accurate shear stresses at the surfaces. However, perfusion systems have the potential to impart a range of artifacts to the culture, due to aspects such as continuous flow (e.g. elimination of soluble cell–cell communication, constant concentration of all components in the media) and low effective volume densities (e.g. the total volume perfused rather than the channel volume, divided by the number of cells in the culture area). Just as flow-based gradient systems do, these perfusion culture devices also require additional fluid handling setups, not commonly found in biological laboratories.
Finally, membrane-based culture devices employ a membrane to allow only diffusional mass transport between the static fluid in a channel, with flowing media in source channels (convective flow), which allow longer term cultures to be performed without sacrificing soluble cell–cell communication. Nutrient exchange via convection at the boundaries of hydrogels seeded with cells has also been shown to effectively culture cells in microchannels.(43) Conceivably, these types of devices could also produce an intermediate effective volume density but are more challenging to fabricate and use than typical static culture devices.
While many cell types have been shown to be compatible with a wide variety of microdevices, proliferation kinetics are not always the same in microculture versus macrocultures.(7,9,40) Differences in the responses of cells to the engineered microenvironments of microfluidic devices to those in macroscale techniques not only has been reflected in proliferation, but has also been assayed via microarray. A notable study done to analyze the artifacts imparted by a microfluidic culture chamber via the analysis of cellular expression profiles by DNA microarray(44,45) showed significant differences between the profiles of macroscale and microscale cultures, though most were less than 3-fold induction or reduction. Comparisons between macroscale and microscale cultures on a variety of engineered surfaces were performed to study any differences in baseline expression of cells in microfluidic assays. This work is the most-comprehensive analysis of the differences in cellular behavior (in this case expression) in microfluidic devices to date.
Example: a unique device
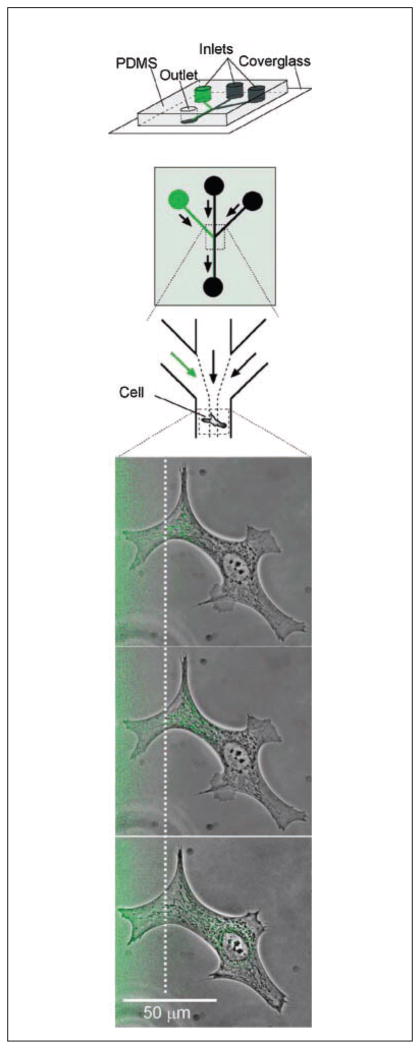
In contrast with these more widely applied areas of microfluidics for cell-based assays, a notable example of a device with great potential that was not as widely applied is Takayama et al’s device for subcellular domain treatments.(46) This device took advantage of diffusion-based mass transport between streams of liquid in laminar flow to create regions within a device that could be treated with a compound independently from the rest of the device. The resolution was so fine that portions of single cells could be stimulated without altering the microenvironment of the rest of the cell (see Fig. 3).
Figure 3.
Illustration of a microfluidic device capable of treating subcellular domains with specific reagents while leaving the rest of a cell or region unaffected. Schematics of the device are shown at the top in which the green channels represent the channel in which dye is included. Fluid from the three inputs flows alongside one another and only mix via diffusion, allowing part of the cell shown below to be stained prior to significant mixing of the reagent. In this case, BCE cells are shown in the lower panels after being labeled with MitoTracker Green for 5, 11 and 35 minutes of exposure to the dye, from top to bottom respectively. Reprinted from Chemistry and Biology, 10/2, Takayama S, Ostuni E, LeDuc P, Naruse K, Ingber DE, Whitesides GM, Selective chemical treatment of cellular microdomains using multiple laminar streams., 123–130, Copyright 2003, with permission from Elsevier.
This device illustrates the incredible potential that microtechnology holds. No other traditional technique could control both temporally and spatially the stimulation of a cell with soluble factors as precisely as this microfluidic device. Thus, the data produced by the device could provide access to previously inaccessible cellular responses. Paradoxically, in cases like this the methods are so novel and thus the data so unique, the challenge arises of interpreting the data. No other method can be used to verify the data as a check for the validity of the assay. Unfortunately, this device likely also suffered from not being simple to operate and integrate into biology research labs. Because of the difficulties associated with obtaining data and having that data widely accepted, often very novel and potentially useful devices go unused.
Understanding how the physics affect the microenvironment
When the scale of the culture device is reduced, the dominant physical phenomena that define how materials behave and how fluid and molecules move change as well. Surface effects and material interactions can substantially change microenvironment composition due to increases in surface area to volume ratios as the scale of the culture is reduced. Purcell provided a very useful account of what environments are like when dominated by diffusion and laminar flow (low Reynolds number), such as those found in typical microfluidic devices.(47) Since then, engineers have identified many of the major physical differences between macroscale and microscale environments,(48) some of which will be discussed briefly here in new contexts, along with other phenomena that are beginning to be more thoroughly examined.
Materials
Most macroscale cultures are performed in polysytrene (or glass-bottomed) tissue-culture flasks, dishes and plates. While many microfluidic cultures are performed with similar substrates as macroscale cultures by adding micropatterned channel materials(49) onto tissue culture substrates, new materials are used to fabricate the body of the devices. Understanding how the materials and processes used to fabricate the devices impact cellular behavior and the readouts of assays will be important in order to analyze the data produced by microfluidic culture systems.
While many new materials are being integrated into microfluidic devices for cell-based assays, the limitations of these materials are also being evaluated. Often the materials that cells interact with are considered to be “inert” with respect to their effects on cellular behavior and are largely ignored unless they are designed specifically to be bioactive. However, many reagents used during common processes in cellular assays, such as fixation and permeabilization or staining and labeling, do interact with these polymers.
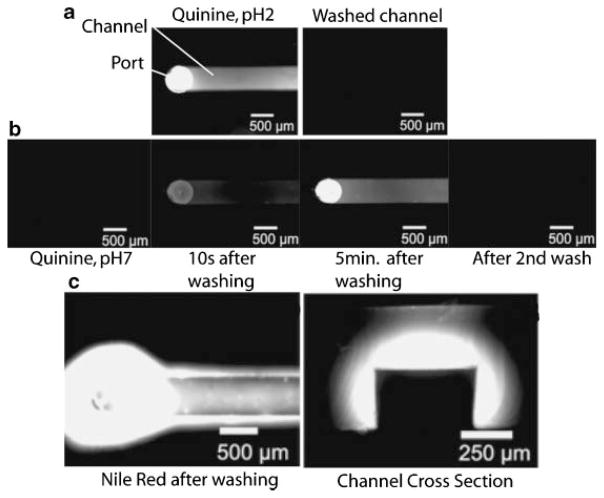
Recent work has shown for a common polymer used for microfabrication, poly(dimethyl siloxane), or PDMS, that the partitioning of hydrophobic molecules into the polymer bulk can result in significant changes in the solution concentrations (Fig. 4).(50) This issue becomes particularly important when compounds used to stimulate or block cellular processes or pathways are both small and hydrophobic such as many small molecule inhibitors or other compounds used in drug screening. When basic procedures such as fixation and staining are performed in microfluidic devices, the possibility for fluorescent reagents to leach into the bulk must be addressed via very simple, no-cell reagent only controls. Additionally, titrations of compounds used for screening or controls that may potentially interact with the materials used can be done to determine whether or not this might be a significant issue for the molecules/materials of interest.
Figure 4.
Microfluidic channels fabricated from poly(dimethyl siloxane), (PDMS) have been shown to absorb small hydrophobic molecules. a: Quinine (fluoresceces at pH2, but not at pH7) was put into a channel and then washed out with pH2 water and fluorescence images of the channel taken. b: If quinine is incubated for 5 minutes in pH7 water in the channel no fluorescence is seen, but after the channel is washed with pH2 water, quinine begins to leach back into solution from the PDMS channel walls and remains until it is washed again. c: A similar phenomenon was shown for Nile Red, as even after the channel is washed with detergent and water, significant fluorescence indicates that the Nile Red was absorbed into the walls of the channels. Adapted from Toepke MW, Beebe DJ. 2006. PDMS absorption of small molecules and consequences in microfluidic applications. Lab On A Chip 6:1484–1486. Reproduced by permission of The Royal Society of Chemistry http://dx.doi.org/10.1039/b612140c.
Evaporation
Recent work has brought to light many important phenomena whose effects can become quite influential as the scale of the cultures are reduced, a prime example is evaporation. Evaporation dynamics of fluids is a very complex phenomenon, as it depends on many environmental variables that often change even as fluid is evaporating. Because of thermodynamic factors involved in the phase change between a liquid and a gas, evaporation is very temperature, pressure and humidity dependent.
Most materials used in traditional culture, such as polysytrene, are relatively impermeable to water and air and only suffer evaporation from between the lids and the substrates. Many commercially available multiwell plates now have “low evaporation” lids to further minimize the surface area in which water can escape the cell culture chamber. For macroscale cultures where media volumes often range from 20mL in a large flask to 200μL in the wells of a 96-well plate, the level of humidification in a typical incubator (approximately 70–90% relative humidity) is sufficient to prevent appreciable evaporation.
However, when microfluidic cultures are performed, typical volumes can range from 5–10μL down to nanoliter volumes and additional means are required to prevent significant evaporation and subsequent concentration of the media.(51,52) However, evaporation is often a limitation of microfluidic cell culture platforms, and many ways have been proposed to combat the loss of water from culture areas. The most common of these include using continuously perfused chambers, employing additional local sacrificial water reservoirs beyond those typically found in standard incubators,(42,53) covering exposed media with oil,(54–56) or submerging the entire chamber in water,(57) though how well these methods limit evaporation is generally unknown.
One group has recently analyzed how evaporation through PDMS affects the osmolarity of the cellular microenvironment.(54) Heo et al. tested the osmolarity of 50 μL of media in a well with a 200 μm thick PDMS membrane under it (and mineral oil on top) over time. The osmolarity of the media increased by ~18% over 48 hours while the control culture conditions (an organ culture dish), showed very little increase. The authors also show distinct effects of this increase in osmolarity on the maturation of embryos and the survival of an endothelial cell line (HDMECs). In these examples, definite phenotypic changes result from shifts in osmolarity although, in other systems, the sensitivity of the cells of interest may not be so easily observed. Thus, evaluation of the means of preventing evaporation used for each specific device will be critical to ensure that, during the assays performed, cells are not simultaneously undergoing osmotic shock, potentially altering the results.
Fluid flow and mass transport
The most-striking difference between the physical environment in microfluidic culture devices and traditional macroscale culture and also the most well understood is the dominance of diffusional mass transport and laminar flow.(48) Briefly, at very small length scales (milli- or micrometers typically) and low flow rates such as those found in most microfluidic culture devices, fluid flow becomes laminar (smooth, streamlined) rather than turbulent. With little unsteady fluid flow to mix the contents of the fluid, diffusion can become a significant mechanism for soluble components to move through the culture volume (e.g. diffusion between flowing streams as in flow-based gradient devices).
Alternatively, in traditional macroscale cultures, the larger volumes and longer length scales allow for more chaotic flow, obscuring any mixing due to diffusion, resulting in more rapid homogenizing of the media due to improved mixing efficiencies. With better control of fluid flow and mass transport mechanisms, microfluidic techniques can provide temporal and spatial patterning of soluble factors or cells not otherwise possible. These capabilities enable a wide variety of new functionalities such as gradient generation discussed previously to be integrated with cell-based assays using microfluidic-based devices.
Next steps for microfluidic cell biology assays
On the topic of validation
The results of biological assays are very sensitive to variations in both intrinsic cellular factors and extrinsic environmental factors. Sources of extrinsic variation common to currently accepted tissue culture techniques range from factors such as lot-to-lot variation of reagents (e.g. fetal bovine serum components, or the degree of hydrophilicity of plastic tissue culture-ware), to pipetting error or other experimenter errors (e.g. differences in reagent concentrations, exposure times or temperatures such as during fixation and staining), to environmental differences (e.g. the temperature and humidity fluctuations in the incubator). Thus, positive and negative controls, and multiple replicates are crucial for verification that results seen in an experiment are truly due to the variables of interest and not the artifacts from any of the multitude of experimental factors that are inadvertently being altered each time an experiment is run.
For example, it is well known that proteins spontaneously adsorb onto polystyrene, the most-common tissue-culture ware component. For a culture in which soluble secreted factors are important for the cellular response of interest, this adsorption results in loss of functional, active protein. Because protein adsorption per unit volume is dependent on the surface-area-to-volume ratio in a culture, while a 75cm2 flask, 6-well and 96-well plates all share the same bulk material and are generally used interchangeably, 6-well plates have 6% higher surface area to volume ratio than the 75cm2 flask, and 96-well plates have 30% more for typical media volumes. While these different formats are similar because the cell culture surfaces and materials are the same, each type of culture ware may impart its own artifacts to a culture.
Though this simple difference between these culture options could alter a sensitive assay, it is rare for the format of the culture from which the data are obtained to be discussed in the literature. This is because re-validation of existing laboratory techniques and materials is being done constantly. Titrations of relevant reagents to determine the best dilution for the cells, assay, or materials used are performed as a first step in any new assay. Positive and negative controls give the experimenter a way to troubleshoot experiments gone wrong and provide a comparison for the data of interest to existing data. Multiple replicates are done with different stock cell suspensions in case differences in the numerous sources of variation present in any biological assay significantly affect the outcome. All of these measures are standard measures that serve the purpose of validation of all of the culture conditions.
Understanding how the microenvironments found in microdevices for biological assays affect both the cellular baseline and responses to stimuli will be key to better understanding the context of any future assays. The challenge posed to novel methods for cell-based assays is substantial. For instance, when a Western blot is presented in a paper, the limitations and caveats inherent in the assay are widely understood: proteins are not in the native conformation (for non-native, SDS–PAGE), antibodies may not be completely specific, loading controls of housekeeping proteins or total amounts of a protein of interest control for variations in loading, etc. However, when data are obtained using new techniques, the understanding of its limitations and strengths is simply not there and, in some cases, not even known at all. If the specific caveats of an assay or technique are not well understood, it can be challenging for readers and reviewers (and experimenters) to accurately interpret the results. To overcome this challenge, it is important for authors to include additional background data to support the novel data to make it more accessible to the reader.
Thus, while new culture techniques such as microfluidic methods and materials do require some degree of validation before they can truly be integrated as a tool in a biological laboratory, this isn’t a particularly new process. Performing the typical validation steps described above in microfluidic devices is critical for initial validation. Also, understanding the unique limitations and benefits of the microfluidic systems in use for biological assays will provide insight into what controls will be necessary to more fully validate the results in context of current techniques. As the platforms are validated, a better understanding of how to interpret data produced by them will result.
Ease of use and appropriate design
Clearly, microfluidic devices have shown utility in cell biology research by allowing for new ways of controlling the cellular microenvironment both spatially and temporally, with a variety of potential cost and resource benefits. By using the unique physical phenomena that dominate in these devices, we can expand the types of assays and cellular responses that we can study in vitro beyond what is currently available. While the demonstrated utility is clear, ease of use and integration with existing laboratory techniques, resources and equipment is an often overlooked issue that can present a significant barrier to use by biologists. Because microfluidic devices have often been relatively complex to fabricate and use (compared to using Petri dishes or multiwell plates), the potential gain from the additional functionalities have not always outweighed the extra work required to use the devices. Thus, an ongoing challenge is to design devices and methods of using the devices that are well suited for cell biology. Close collaborations between the potential end users and the engineers who design the devices can address this challenge.
By constraining designs to operate with commonly available (biological) laboratory tools and analysis equipment (e.g. pipettors, microscopes, plate readers), devices will be significantly more accessible. Focusing microfluidic device designs to provide their unique capabilities without excess complexity will be crucial for them to be easy to use and to become common tools in biological laboratories.
The previous examples of applications of microfluidics to cell biology illustrate why simply designed devices are more likely to be incorporated into laboratories and widely used than their more complex counterparts. Many flow-based gradient generation devices require complex fluid-handling systems, (including extensive tubing, syringe pumps and associated electronics). But if simpler passive gradient generation systems (such as that in Fig. 1b)(20) can provide a suitable gradient for studying the response of interest but only require a few pipeting steps, they are much more likely to be used. Similarly, the application of plate readers for microfluidic proliferation assays(8) provides a simple, fast, accessible readout for a microfluidic culture rather than devices that rely upon frame-by-frame microscopy analysis, or manual cell counting. The introduction of microfluidic cultures to automated liquid handling systems has also highlighted the potential for a high-throughput microfluidic platform for cell culture, which could be easily integrated and widely used and can provide throughput beyond manual pipette-loaded microfluidic channels for cell-based assays.(3) In all of these cases, a strength of the devices, beyond the benefits provided by the microdevices, is the smaller hurdle from the user’s perspective because the devices are simpler to use or employ methods that are both familiar and more widely available. Ultimately, microfluidics will only have a significant impact on current experimental methods if they are widely accepted and used by the end users, cell biologists.
Expanding the available assays
Much of the initial work in integrating cell culture into microfluidics focused on cell survival alone, as many of the original fabrication components and processes were cytotoxic or not compatible with mammalian cell culture. Thus, with the development of more biocompatible polymers, improved methods of spatially patterning proteins and cells and better understanding of the microenvironments that the cells are exposed to in these devices, additional assays have been incorporated.(58)
Cell proliferation is a common readout from a microfluidic culture, as often entire culture areas can be imaged and analyzed via imaging software and total adherent or non-adherent cell numbers per channel can be obtained and tracked over time.(7,40) Recent work has integrated microfluidic cultures into a format that can be analyzed by a standard plate reader for cell enumeration purposes.(8)
Integration of current microfluidic culture techniques with existing biological analysis technology (such as DNA arrays), will allow us to further study the effects of microscale cultures on cellular behavior at the molecular level and widen the available range of cellular readouts for microfluidic biological applications. The integration of the research being performed in both engineering and biology has the potential to provide new methods and technologies that may allow biology to be studied in different ways. New assays and new ways of researching biological phenomena will come from the use of technologies that provide novel functionalities. Future studies integrating cell biological assays with microfluidic cultures will rely upon well-designed studies with correct and thorough positive and negative controls for validation purposes.
While some traditional techniques can already be easily integrated into microfluidic devices, some of their characteristics can provide access to new cellular assays, previously inaccessible via traditional methods (e.g. due to improved control over fluid flow and reagent delivery). These will need new methods for quantitation of phenomena that previously weren’t an issue (e.g. quantifying cell migration in complex gradients). Also, other characteristics of these devices can result in different results from the same assay performed in a macroscale culture (e.g. differences in cell proliferation due to volume density differences). It will be important to understand these factors when interpreting the data produced in these devices and to address any differences in results as they too may provide insights into the mechanism being studied. These differences might then be leveraged to provide new ways to assay cellular responses by comparing macroscale and microscale assays.
More types of assays and readouts need to be adapted into microfluidic cultures. Currently, traditional assays that require significantly more cell numbers or cell lysate than a typical microfluidic culture device would produce cannot be easily integrated into microfluidic assays. Commonly used methods such as performing cell separations/flow cytometric analyses or gel electrophoresis based techniques (Westerns, Northerns, Southerns) have yet to be well integrated into microfluidic culture devices in a user friendly manner. Improving and altering the protocols for these types of readouts to compliment the techniques used for microfluidic assays will be important to better provide accessible and accurate microfluidic versions of existing technology or enable new assays to be performed due to the unique capabilities of microfluidic devices.
Conclusion
Microfluidic devices for cell-based assays have provided new types of microenvironments and new methods for controlling and observing the cellular responses to them. The field has begun to analyze the biological effects of the physical differences of microfluidic devices for cell-based assays, ranging from evaporation in static microfluidic cultures to flow-induced artifacts in gradient generation devices. Nonetheless, the relative lack of quantitative biological analysis techniques that have been interfaced with microfluidic devices has prevented more facets of cellular function beyond viability or proliferation to be analyzed in them. Without a better understanding of the effects of the micro-environments present in microdevices from a cellular perspective, it will continue to be challenging to integrate work done in microdevices with biological data obtained via traditional methods. As more microfluidic devices for cell biology are developed and implemented that address the current roadblocks such as ease of use, biological validity, and limitations in readouts, the unique strengths of these devices will become more accessible to the general biology community as common laboratory tools.
Acknowledgments
DJB, NIH grants R21CA122672 and K25CA104162, and ALP, DOD/BRCP W81XWH-06-1-0487.
References
- 1.Sims CE, Allbritton NL. Analysis of single mammalian cells on-chip. Lab On A Chip. 2007;7:423–440. doi: 10.1039/b615235j. [DOI] [PubMed] [Google Scholar]
- 2.Keenan TM, Folch A. Biomolecular gradients in cell culture systems. Lab Chip. 2008;8:34–57. doi: 10.1039/b711887b. Epub 2007 Dec 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meyvantsson I, Warrick JW, Hayes S, Skoien A, Beebe DJ. Automated cell culture in high density tubeless microfluidic device arrays. Lab Chip. 2008;8(5):717–24. doi: 10.1039/b715375a. [DOI] [PubMed] [Google Scholar]
- 4.Tan W, Desai TA. Microfluidic patterning of cells in extracellular matrix biopolymers: effects of \ channel size, cell type, and matrix composition on pattern integrity. Tissue Eng. 2003;9:255–567. doi: 10.1089/107632703764664729. [DOI] [PubMed] [Google Scholar]
- 5.Fisher RJ, Peattie RA. Controlling tissue microenvironments: biomimetics, transport phenomena, and reacting systems. Adv Biochem Eng Biotechnol. 2007;103:1–73. doi: 10.1007/10_018. [DOI] [PubMed] [Google Scholar]
- 6.Kim L, Toh YC, Voldman J, Yu H. A practical guide to microfluidic perfusion culture of adherent mammalian cells. Lab on a Chip. 2007;7:681–694. doi: 10.1039/b704602b. [DOI] [PubMed] [Google Scholar]
- 7.Walker GM, Ozers MS, Beebe DJ. Insect cell culture in microfluidic channels. Biomedical Microdevices. 2002;4:161–166. [Google Scholar]
- 8.Yu H, Alexander CM, Beebe DJ. A plate reader-compatible microchannel array for cell biology assays. Lab Chip. 2007;7:388–391. doi: 10.1039/b612358a. [DOI] [PubMed] [Google Scholar]
- 9.Yu HM, Alexander CM, Beebe DJ. Understanding microchannel culture: parameters involved in soluble factor signaling. Lab On A Chip. 2007;7:726–730. doi: 10.1039/b618793e. [DOI] [PubMed] [Google Scholar]
- 10.Folch A, Toner M. Microengineering of cellular interactions. Annu Rev Biomed Eng. 2000;2:227–256. doi: 10.1146/annurev.bioeng.2.1.227. [DOI] [PubMed] [Google Scholar]
- 11.Walker GM, Beebe DJ. A passive pumping method for microfluidic devices. Lab Chip. 2002;2:131–134. doi: 10.1039/b204381e. [DOI] [PubMed] [Google Scholar]
- 12.Sawano A, Takayama S, Matsuda M, Miyawaki A. Lateral propagation of EGF signaling after local stimulation is dependent on\ receptor density. Dev Cell. 2002;3(2):245–57. doi: 10.1016/s1534-5807(02)00224-1. [DOI] [PubMed] [Google Scholar]
- 13.Lucchetta EM, Munson MS, Ismagilov RF. Characterization of the local temperature in space and time around a developing\ Drosophila embryo in a microfluidic device. Lab Chip. 2006;6:185–190. doi: 10.1039/b516119c. [DOI] [PubMed] [Google Scholar]
- 14.Thery M, Racine V, Pepin A, Piel M, Chen Y, et al. The extracellular matrix guides the orientation of the cell division axis. Nat Cell Biol. 2005;7:947–953. doi: 10.1038/ncb1307. [DOI] [PubMed] [Google Scholar]
- 15.Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Geometric control of cell life and death. Science. 1997;276:1425–1428. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- 16.Folch A, Toner M. Microengineering of Cellular Interactions. Annual Review of Biomedical Engineering. 2000;2:227–256. doi: 10.1146/annurev.bioeng.2.1.227. [DOI] [PubMed] [Google Scholar]
- 17.Masuda S, Washizu M, Nanba T. Novel methods of cell fusion and handling using fluid integrating circuit. Oxford. 1987:69–74. [Google Scholar]
- 18.Masuda S, Washizu M, Nanba T. Novel method of cell fusion in field constriction area in fluid integrated circuit. IEEE Trans Ind Appl. 1989;25:732–737. [Google Scholar]
- 19.Zigmond S. Ability of polymorphonuclear leukocytes to orient in gradients of chemotactic factors. Journal of Cell Biology. 1977;75:606–616. doi: 10.1083/jcb.75.2.606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abhyankar VV, Lokuta MA, Huttenlocher A, Beebe DJ. Characterization of a membrane-based gradient generator for use in cell-signaling studies. Lab Chip. 2006;6:389–393. doi: 10.1039/b514133h. [DOI] [PubMed] [Google Scholar]
- 21.Diao JP, Young L, Kim S, Fogarty EA, Heilman SM, et al. A three-channel microfluidic device for generating static linear gradients and its application to the quantitative analysis of bacterial chemotaxis. Lab On A Chip. 2006;6:381–388. doi: 10.1039/b511958h. [DOI] [PubMed] [Google Scholar]
- 22.Frevert CW, Boggy G, Keenan TM, Folch A. Measurement of cell migration in response to an evolving radial chemokine gradient triggered by a microvalve. Lab On A Chip. 2006;6:849–856. doi: 10.1039/b515560f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Irimia D, Liu S, Tharp W, Samadani A, Toner M, Poznansky M. Microfluidic system for measuring neutrophil migratory responses to fast switches of chemical gradients. Lab on a Chip. 2006;6:191–198. doi: 10.1039/b511877h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jeon NL, Baskaran H, Dertinger SKW, Whitesides GM, Van de Water L, Toner M. Neutrophil chemotaxis in linear and complex gradients of interleukin-8 formed in a microfabricated device. Nature Biotechnol. 2002;20:826–830. doi: 10.1038/nbt712. [DOI] [PubMed] [Google Scholar]
- 25.Koyama S, Amarie D, Soini H, Novotny M, Jacobson SC. Chemotaxis of mouse sperm on microfluidic devices. Anal Chem. 2006;78:3354–3359. doi: 10.1021/ac052087i. [DOI] [PubMed] [Google Scholar]
- 26.Lin F, Nguyan C, Wang S, Saadi W, Gross S, Jeon NL. Neutrophil migration in opposing chemoattractant gradients using microfluidic chemotaxis devices. Anals of Biomedical Engineering. 2005;33:475–482. doi: 10.1007/s10439-005-2503-6. [DOI] [PubMed] [Google Scholar]
- 27.Fujii S, Uematsu M, Yabuki S, Abo M, Yoshimura E, Sato K. Microbioassay system for an anti-cancer agent test using animal cells on a microfluidic gradient mixer. Analyt Sci. 2006;22:87–90. doi: 10.2116/analsci.22.87. [DOI] [PubMed] [Google Scholar]
- 28.Saadi W, Wang SJ, Lin F, Jeon NL. A parallel-gradient microfluidic chamber for quantitative analysis of breast cancer cell chemotaxis. Biomed Microdevices. 2006;8:109–118. doi: 10.1007/s10544-006-7706-6. [DOI] [PubMed] [Google Scholar]
- 29.Wang SJ, Saadi W, Lin F, Nguyen CMC, Jeon NL. Differential effects of EGF gradient profiles on MDA-MB-231 breast cancer cell chemotaxis. Experimental Cell Res. 2004;300:180–189. doi: 10.1016/j.yexcr.2004.06.030. [DOI] [PubMed] [Google Scholar]
- 30.Chung BG, Flanagan LA, Rhee SW, Schwartz PH, Lee AP, et al. Human neural stem cell growth and differentiation in a gradient-generating microfluidic device. Lab Chip. 2005;5:401–406. doi: 10.1039/b417651k. [DOI] [PubMed] [Google Scholar]
- 31.Barkefors I, Le Jan S, Jakobsson L, Hejll E, Carlson G, et al. Endothelial cell migration in stable gradients of VEGFA and F. F2: Effects on chemotaxis and chemokinesis. Lab Chip. 2008;8:34–57. doi: 10.1074/jbc.M704917200. [DOI] [PubMed] [Google Scholar]
- 32.Yap B, Kamm RD. Mechanical deformation of neutrophils into narrow channels induces pseudopod projection and changes in biomechanical properties. J App Physiol. 2005 doi: 10.1152/japplphysiol.01226.2004. [DOI] [PubMed] [Google Scholar]
- 33.Walker GM, Sai JQ, Richmond A, Stremler M, Chung CY, Wikswo JP. Effects of flow and diffusion on chemotaxis studies in a microfabricated gradient generator. Lab On A Chip. 2005;5:611–618. doi: 10.1039/b417245k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen CS, Jiang XY, Whitesides GM. Microengineering the Enviroment of Mammalian Cells in Culture. MRS Bulletin. 2005;30:194–201. [Google Scholar]
- 35.El-Ali J, Sorger PK, Jensen KF. Cells on chips. Nature. 2006;442:403–411. doi: 10.1038/nature05063. [DOI] [PubMed] [Google Scholar]
- 36.Linder V, Koster S, Franks W, Kraus T, Verpoorte E, et al. Microfluidics/CMOS orthogonal capabilities for cell biology. Biomedical Microdevices. 2006;8:159–166. doi: 10.1007/s10544-006-7711-9. [DOI] [PubMed] [Google Scholar]
- 37.Schwartz MP, Derfus AM, Alvarez SD, Bhatia SN, Sailor MJ. The smart petri dish: A nanostructured photonic crystal for real-time monitoring of living cells. Langmuir. 2006;22:7084–7090. doi: 10.1021/la060420n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tourovskaia A, Figueroa-Masot X, Folch A. Differentiation-on-a-chip: A microfluidic platform for long-term cell culture studies. Lab On A Chip. 2005;5:14–19. doi: 10.1039/b405719h. [DOI] [PubMed] [Google Scholar]
- 39.Raty S, Walters EM, Davis J, Zeringue H, Beebe DJ, et al. Embryonic development in the mouse is enhanced via microchannel culture. Lab On A Chip. 2004;4:186–190. doi: 10.1039/b316437c. [DOI] [PubMed] [Google Scholar]
- 40.Yu HM, Meyvantsson I, Shkel IA, Beebe DJ. Diffusion dependent cell behavior in microenvironments. Lab on a Chip. 2005;5:1089–1095. doi: 10.1039/b504403k. [DOI] [PubMed] [Google Scholar]
- 41.Kim L, Vahey MD, Lee HY, Voldman J. Microfluidic arrays for logarithmically perfused embryonic stem cell culture. Lab On A Chip. 2006;6:394–406. doi: 10.1039/b511718f. [DOI] [PubMed] [Google Scholar]
- 42.Song JW, Gu W, Futai N, Warner KA, Nor JE, Takayama S. Computer-controlled microcirculatory support system for endothelial cell culture and shearing. Analyt Chem. 2005;77:3993–3999. doi: 10.1021/ac050131o. [DOI] [PubMed] [Google Scholar]
- 43.Choi NW, Cabodi M, Held B, Gleghorn JP, Bonassar LJ, Stroock AD. Microfluidic scaffolds for tissue engineering. Nature Materials. 2007;6:908–915. doi: 10.1038/nmat2022. [DOI] [PubMed] [Google Scholar]
- 44.Stangegaard M, Petronis S, Jorgensen AM, Christensen CBV, Dufva M. A biocompatible micro cell culture chamber (mu CCC) for the culturing and on-line monitoring of eukaryote cells. Lab On A Chip. 2006;6:1045–1051. doi: 10.1039/b603379b. [DOI] [PubMed] [Google Scholar]
- 45.Stangegaard M, Wang Z, Kutter J, Dufva M, Wolff A. Whole genome expression profiling using DNA microarray for determining biocompatibility of polymeric surfaces. Molec Biosys. 2006;2:421–428. doi: 10.1039/b608239d. [DOI] [PubMed] [Google Scholar]
- 46.Takayama S, Ostuni E, LeDuc P, Naruse K, Ingber DE, Whitesides GM. Selective chemical treatment of cellular microdomains using multiple laminar streams. Chem Biol. 2003;10:123–130. doi: 10.1016/s1074-5521(03)00019-x. [DOI] [PubMed] [Google Scholar]
- 47.Purcell EM. Life At Low Reynolds-Number. Am J Physics. 1977;45:3–11. [Google Scholar]
- 48.Walker GM, Zeringue HC, Beebe DJ. Microenvironment design considerations for cellular scale studies. Lab Chip. 2004;4:91–97. doi: 10.1039/b311214d. [DOI] [PubMed] [Google Scholar]
- 49.Whitesides GM, Ostuni E, Takayama S, Jiang X, Ingber DE. Soft Lithography in Biology and Biochemistry. Ann Rev Biomed Eng. 2001;3:335–373. doi: 10.1146/annurev.bioeng.3.1.335. [DOI] [PubMed] [Google Scholar]
- 50.Toepke MW, Beebe DJ. PDMS absorption of small molecules and consequences in microfluidic applications. Lab On A Chip. 2006;6:1484–1486. doi: 10.1039/b612140c. [DOI] [PubMed] [Google Scholar]
- 51.Berthier E, Warrick J, Beebe DJ. Managing evaporation for more robust microscale assays. Part 2: Characterization of convection and diffusion for cell biology. Lab on a Chip. 2008;8:860–864. doi: 10.1039/b717423c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Berthier E, Warrick J, Yu H, Beebe DJ. Managing evaporation for more robust microscale assays. Part 1: Volume loss in droplet based assays. Lab on a Chip. 2008;8:852–859. doi: 10.1039/b717422e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Urbanski JP, Thies W, Rhodes C, Amarasinghe S, Thorsen T. Digital microfluidics using soft lithography. Lab On A Chip. 2006;6:96–104. doi: 10.1039/b510127a. [DOI] [PubMed] [Google Scholar]
- 54.Heo YS, Cabrera LM, Song JW, Futai N, Tung YC, et al. Characterization and resolution of evaporation-mediated osmolality shifts that constrain microfluidic cell culture in poly(dimethylsiloxane) devices. Anal Chem. 2007;79:1126–1134. doi: 10.1021/ac061990v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Khandurina J, McKnight TE, Jacobson SC, Waters LC, Foote RS, Ramsey JM. Integrated system for rapid PCR-based DNA analysis in microfluidic devices. Analyt Chem. 2000;72:2995–3000. doi: 10.1021/ac991471a. [DOI] [PubMed] [Google Scholar]
- 56.Lee DS, Park SH, Yang HS, Chung KH, Yoon TH, et al. Bulk-micromachined submicroliter-volume PCR chip with very rapid thermal response and low power consumption. Lab On A Chip. 2004;4:401–407. doi: 10.1039/b313547k. [DOI] [PubMed] [Google Scholar]
- 57.Zheng B, Roach LS, Ismagilov RF. Screening of protein crystallization conditions on a microfluidic chip using nanoliter-size droplets. J Am Chem Soc. 2003;125:11170–11171. doi: 10.1021/ja037166v. [DOI] [PubMed] [Google Scholar]
- 58.Weibel DB, DiLuzio WR, Whitesides GM. Microfabrication meets microbiology. Nature Revs Microbiol. 2007;5:209–218. doi: 10.1038/nrmicro1616. [DOI] [PubMed] [Google Scholar]