Summary
Fluorescent proteins have become valuable tools for biomedical research as protein tags, reporters of gene expression, biosensor components, and cell lineage tracers. However, applications of fluorescent proteins for deep tissue imaging in whole mammals have been constrained by the opacity of tissues to excitation light below 600 nm, due to absorbance by hemoglobin. Fluorescent proteins that excite efficiently in the “optical window” above 600 nm are therefore highly desirable. We report here the evolution of far-red fluorescent proteins with peak excitation at 600 nm or above. The brightest one of these, Neptune, performs well in imaging deep tissues in living mice. The crystal structure of Neptune reveals a novel mechanism for red-shifting involving the acquisition of a new hydrogen bond with the acylimine region of the chromophore.
Introduction
Mammalian tissues are opaque to most wavelengths of visible light due to the strong absorption by hemoglobin of wavelengths below 600 nm (Stamatas et al., 2006). Between 600 nm and 1200 nm, where infrared absorption by water molecules begins, there exists an “optical window” in which tissue is relatively transparent to light (Tromberg et al., 2000). While many fluorescent proteins (FPs) emit in the optical window, none have been found or developed to excite maximally in the optical window (Shcherbo et al., 2009). The fluorescent protein with the most red-shifted emission spectrum is the dsRed derivative mPlum, with excitation and emission peaks at 590 nm and 649 nm (Wang et al., 2004). The protein with the most red-shifted excitation peak wavelength is the closely related mRaspberry (Wang et al., 2004), with excitation and emission peaks at 598 nm and 625 nm, but mRaspberry excitation drops off quickly with increasing wavelength, reaching 10% of peak excitation at 630 nm. The eqFP578 derivatives Katushka, mKate, and mKate2 have been proposed for use in whole-animal imaging based on their intrinsic brightness (Shcherbo et al., 2007; Shcherbo et al., 2009), but they are neither the most red-shifted in excitation (at 588 nm) nor in emission (at 635 nm). An ideal fluorescent protein for imaging in mammals would combine the best characteristics of the above proteins and be further red-shifted to allow excitation in the optical window.
Excitation wavelength tuning in fluorescent proteins is determined by the covalent structure of the chromophore and influenced by hydrogen bonding and electronic interactions of the chromophore with nearby chemical groups (Remington, 2006). The chromophore of autocatalytic fluorescent proteins such as jellyfish green fluorescent protein (GFP) is composed of phenol and imidazolinone rings derived from autocatalytic modifications of a Tyr residue and the backbone atoms of adjacent residues. The two aromatic rings are linked by an methylidene bridge to create an extended conjugated p orbital system (Ormo et al., 1996). In most red fluorescent proteins (RFPs), including dsRed from the corallimorph Discosoma and eqFP611 from the sea anenome Entacmaea, oxidation of the N-Cα bond immediately N-terminal to the chromophore allows conjugation to extend through the acylimine group formed by the resulting imine and the carbonyl oxygen of the preceding amino acid (Gross et al., 2000; Petersen et al., 2003), leading to the dramatic bathochromic shift of RFPs relative to GFP. Notably, the RFP variants mOrange and zFP538 contain covalent modifications of this common RFP chromophore that remove the terminal carbonyl group, and correspondingly exhibit blue-shifted excitation and emission spectra. In mOrange, a threonine hydroxyl group attacks the carbonyl to create a hemi-acetal (Shu et al., 2006), while in zFP538, a reaction of a lysine amino group with the acylimine results in the formation of a new imine-containing ring (Remington et al., 2005).
Side chains near but not inherent to the chromophore also influence fluorescent protein spectra. This was first demonstrated in GFP with the mutation of Thr-203, located above the phenol ring, to Tyr to create a π–π stacking interaction. The resulting yellow fluorescent protein (YFP) exhibited red-shifted excitation and emission spectra, as would be expected from the greater polarizability of the stacked π orbital system (Ormo et al., 1996). A cation-π interaction between a histidine side chain and the chromophore has the opposite effect of creating a hypsochromic shift in teal fluorescent protein (TFP). This can be explained by preferential stabilization of the ground state due to stabilization of negative charge over the chromophore phenolate (Henderson et al., 2007). Preferential stabilization of the ground state would increase the energy difference between ground and excitation states, requiring the absorption of higher-energy photons for excitation.
We set out to create fluorescent proteins that can be excited in the optical window at 600 nm and above. We carried out a directed evolution strategy starting from Discosoma- and Entacmaea-family RFPs involving site-directed and random mutagenesis. One of the resulting proteins, Neptune, has excitation and emission peaks at 600 and 650 nm, and performs well in deep-tissue imaging in live mammals using excitation light in the optical window. Our analysis of the structure of Neptune revealed multiple mechanisms contributing to the long wavelengths of Neptune, including enhanced chromophore coplanarity and the addition of a novel water-chromophore hydrogen bond to the acylimine oxygen of the chromophore.
Results
Evolution of far-red fluorescent proteins with excitation in the optical window
We first attempted to create red-shifted fluorescent proteins based on mRFP1 by introducing a tyrosine side chain near the chromophore to create a π-π stacking interaction similar to the one responsible for the red-shifted excitation and emission spectra of YFP relative to GFP. We introduced a I197Y mutation (197 in RFPs is homologous to 203 of GFP) into mRFP1.1 (Shaner et al., 2004) and then evolved the resulting protein in bacteria for brightness and red-shifted absorbance by screening for blue-appearing colonies. One clone, named mGrape1 for its blue color, showed maximal absorbance at 595 nm, 11 nm red-shifted relative to mRFP1.1 (Supplemental Data, Table S1). Further evolution yielded two proteins with dual absorbance peaks: 470 and 605 nm for mGrape2 and 470 and 608 nm for mGrape3 (Figure S1, Table 1). Illumination by 465-495 nm light converts the 470 nm-absorbing species into the longer wavelength-absorbing species (complete in 15 minutes at power density 0.1 W/cm2), while the reverse process occurs slowly in the dark (half-life of 70 minutes at 37° C). These observations are most likely attributed to cis-trans isomerization of the chromophore, as seen previously in other fluorescent proteins (see Supplementary Data). Although the mGrapes exhibit the most red-shifted absorbance peaks of all autocatalytic fluorescent proteins, in light of their complex photophysics and low fluorescence quantum yields of 0.03 (Table S1), we pursued alternative approaches to red-shifted fluorescent proteins.
Table 1. Characteristics of far-red fluorescent proteins.
| Protein | Excitation peaka | Emission peaka | εb | φc | Brightness excited at peakd | Brightness excited at 633nme | Photostabilityf | pKa |
|---|---|---|---|---|---|---|---|---|
| dTomatog | 554 | 581 | 69000 | 0.69 | 48 | 0 | 64 | 4.7 |
| mCherryg | 587 | 610 | 72000 | 0.22 | 16 | 0.084 | 68 | <4.5 |
| mPlumg | 590 | 649 | 41000 | 0.10 | 4.1 | 0.26 | 53 | <4.5 |
| mRaspberryg | 598 | 625 | 86000 | 0.15 | 13 | 0.45 | 15 | <4.5 |
| mGrape3 | 608 | 646 | 40000h | 0.03 | 1.2 | 0.29 | 5 | 7.0 |
| mKate | 585 | 635 | 42000 | 0.30 | 13 | 0.43 | 82 | 6.2i |
| mKate S158A | 585 | 630 | 75000 | 0.30 | 23 | 0.48 | 327 | 5.4i |
| mKate2 | 586 | 630 | 50000 | 0.36 | 18 | 0.42 | ND | 5.4i |
| mKate S158C | 586 | 630 | 63000 | 0.33 | 21 | 0.56 | 220j | ND |
| mKate M41G S158C | 593 | 648 | 73000 | 0.22 | 16 | 1.2 | ND | ND |
| Neptune (mKate M41G S61C S158C Y197F) | 600 | 650 | 72000 | 0.18 | 13 | 1.7 | 185 | 5.8 |
| mNeptune (Neptune M146T) | 600 | 650 | 67000 | 0.20 | 13 | 1.8 | 160 | 5.4 |
Excitation and emission maxima in nm.
Maximum extinction coefficient per chain in M-1cm-1 measured by the alkali denaturation method (Chalfie and Kain, 2005; Gross et al., 2000).
Quantum yield of fluorescence.
Calculated as (peak extinction coefficient per chain • quantum yield)/(1000 M-1cm-1).
Calculated as (extinction coefficient per chain at 633 nm • quantum yield • emission fraction above 633 nm)/(1000 M-1cm-1).
Time in s to photobleach 50% from an initial rate of 1000 emitted photons s-1 molecule-1.
Published values for dTomato, mCherry, and mPlum (Shaner et al., 2005).
After photoactivation by 470nm light.
Published values (Pletnev et al., 2008).
Monoexponential decay.
We selected mKate as another starting point for directed evolution because of its high quantum yield and large Stokes shift. Saturation mutagenesis of amino acids Ser-143 and Ser-158, residues predicted to be located near the chromophore based on the structure of mKate (Pletnev et al., 2008), was first performed to improve mKate characteristics. Mutation of Ser-158 to Cys or Ala improved the peak extinction coefficient of mKate (Figure 1A, Table 1). In the course of this work, mKate S158A (Pletnev et al., 2008) and its improved folding variant mKate2 (Shcherbo et al., 2009) were described. mKate and its S158A and S158C mutants all exhibit residual green fluorescent components (Figure S2A). While TagRFP (Shaner et al., 2008), TagRFP-T (Shaner et al., 2008), mKate, and mKate S158A (Figure S2A) bleached with complex kinetics, mKate S158C bleached in a monoexponential manner with a normalized half-life of 220 s, making it the most photostable fluorescent protein with monoexponential photobleaching. This rare attribute makes mKate S158C well suited for quantitative time-lapse experiments.
Figure 1.

Neptune spectra and sequence. (A) Absorbance spectra of oxyhemoglobin (red), deoxyhemoglobin (dotted red), mKate (green dashed), mKate S158C (orange), mKate2 (dotted black), Neptune (dashed blue), and mNeptune (light blue). Units are M-1cm-1 versus nm. Hemoglobin spectra are previously published (Stamatas et al., 2006). Inset: Purified mCherry, mKate, S158C, and Neptune proteins at 0.5 mg/mL in visible light. (B) Normalized excitation (left) and emission (right) spectra of mKate (green dashed), mKate S158C (orange), Neptune (dashed blue), and mNeptune (light blue). mKate2 excitation and emission spectra are identical to mKate, as described (Shcherbo et al., 2009). (C) Sequence of mNeptune aligned with its parent, mKate. Changes responsible for the red shift are highlighted in red. The additional monomerization mutation to create mNeptune from Neptune is highlighted in green.
We next attempted to further red-shift mKate S158C by adopting the mechanism of emission red-shifting in mPlum. In mPlum, Glu-13 hydrogen bonds to the terminal carbonyl of the conjugated bond system of the chromophore, but the interaction is dependent on chromophore excitation and therefore primarily affects the emission spectrum (Abbyad et al., 2007; Shu et al., 2009). We hypothesized that a stable hydrogen bonding interaction with the terminal carbonyl in the ground state could cause a red-shift in excitation as well. Screening of libraries with all possible substitutions at position 13 and nearby positions 28, 41, and 65, which are not conserved between mKate and mPlum, did not recover an obvious hydrogen bond donor at one of these positions. However we found that mutation of Met-41 to Gly alone resulted in red-shifted excitation and emission (Table 1). Two rounds of random mutagenesis by error-prone PCR then yielded Y194F and S61C mutations, resulting in a protein with 600 nm excitation and 650 nm emission (Figure 1B, Table 1). Because this protein (mKate M41G S61C S158C Y194F, Figure 1C) looks blue in ambient lighting (Figure 1A) and has fluorescence wavelengths furthest away from our starting point among mKate derivatives, we named it Neptune.
Neptune is the first bright fluorescent protein (extinction coefficient 72,000 M-1cm-1, quantum yield 0.18) with an excitation peak reaching 600 nm. Compared to its parent mKate, its absorption and excitation peaks are 18 nm redder (600 vs 582 nm) and its peak extinction coefficient 71% larger (72000 vs 42000 M-1cm-1), resulting in more efficient excitation beyond 600 nm (Figure 1A, Table 1). Neptune exhibits a photobleaching half-life longer than EGFP (Figure S2A), a pKa of 5.8, and fast maturation (half-maximal in 35 minutes, Figure S2C). Like mKate and the brighter mKate S158A and S158C variants, Neptune shows a residual green component upon excitation at 470 nm (Figure S2C).
Visualization of Neptune in cells and deep tissues of living mammals
Compared to previous FPs, Neptune has the largest extinction coefficient at 633 nm, a wavelength well within the optical window and a common laser line used for exciting the organic fluorophore Cy5 (Figure 1A, Table 1). We tested the ability to detect Neptune or mKate in cells with 633 nm excitation. Neptune was well detected in liver sections following adenovirus-mediated gene transfer in mice (Figure 2A) with 532 or 633 nm laser excitation, while mKate was only detected above background with the 532 nm laser (Figure 2B). Thus the bathochromic shift of Neptune relative to its parent mKate has conferred improved excitability at 633 nm, well within the optical window.
Figure 2.
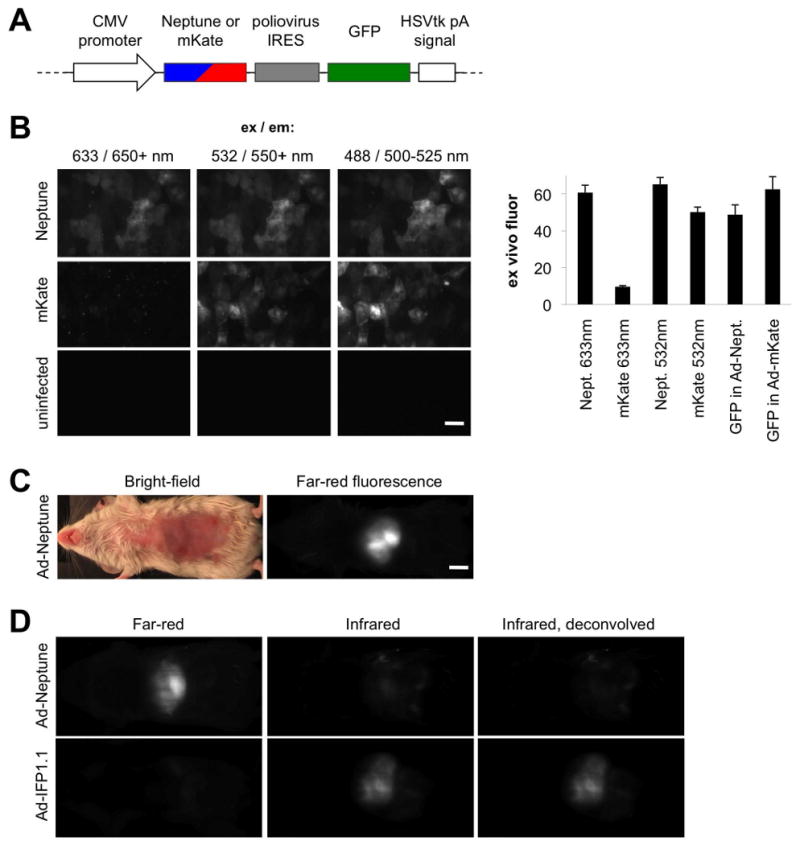
Performance of Neptune in cells and animals. (A) Design of a monocistronic adenovirus vector for coexpression of GFP and Neptune (Ad-Neptune) or (Ad-mKate) in hepatocytes. (B) Sections of liver were removed from a mouse injected with Ad-Neptune or Ad-mKate and imaged at red or far-red wavelengths on a confocal microscope. GFP was also imaged to confirm expression of the cistron. Fluorescence intensities are shown in the chart. Scale bar, 50 μm. (C) Epifluorescence of Neptune in the liver of a living mouse acquired on a Maestro imaging system (CRI) with excitation and emission at 610-630 nm and 660-700 nm, respectively. Scale bar, 5 mm. (D) Dual fluorescence imaging in the optical window is possible with Neptune and infrared fluorescent protein (IFP). Images of live mice expressing Neptune or IFP1.1 in liver were acquired with Cy5 and Cy5.5 channels. Excitation/emission wavelengths used were 610-630/660-700 nm for Cy5 and 625-675/700-730 nm for Cy5.5. In the right column, spectral unmixing was used to correct for Neptune cross-excitation and -detection in the Cy5.5 channel. Restricting IFP excitation to above 650 nm with custom filters would produce similar results.
The ability to be excited at ∼630 nm makes Neptune potentially suitable for fluorescence imaging in living mammals using existing Cy5 filter sets. The liver in particular is a heavily vascularized organ where hemoglobin absorbance hinders imaging (Colin et al., 2000). Using an available Cy5 filter set (610-630 nm excitation, 660-700 nm emission), Neptune fluorescence in livers of living mice following adenovirus-mediated gene transfer was easily visible, with the brightest areas displaying 44-fold contrast over uninfected tissue autofluorescence (Figure 2C). We recently engineered a bacterial phytochrome into a biliverdin-dependent infrared fluorescent protein, IFP1.1, with excitation and emission maxima at 660 nm and 700 nm (Shu et al., 2009). To see whether Neptune could be visualized orthogonally to IFP1.1, we imaged living mice expressing either Neptune or IFP1.1, using filter sets separately optimized for Cy5 (far-red emission) and Cy5.5 (infrared emission), respectively. Indeed liver-expressed Neptune was selectively imaged in the Cy5 channel while IFP1.1 was preferentially imaged in the Cy5.5 channel (Figure 2D). A small amount of cross-detection of Neptune occurred using our Cy5.5 excitation filter due to its broad excitation range (625-675 nm). This signal can be subtracted by linear unmixing (Figure 2D) or preferably eliminated by the future design of an appropriately narrow excitation filter for IFP1.1. We also confirmed that excitation wavelengths in the optical window detected liver fluorescence more efficiently than shorter wavelengths for both Neptune and its parent mKate (Figure S3 and Table S2).
Quaternary structure of Neptune and further monomerization
We studied the quaternary structure of Neptune, mKate, and mKate derivatives in solution by size-exclusion chromatography (SEC) with in-line multi-angle laser light-scattering. At low micromolar concentrations, Neptune and mKate (Figure 3A), as well as mKate S158A and mKate S158C (Figure S4A), primarily co-migrate with the dimeric standard dTomato. However, the elution profiles are not monodisperse, but rather exhibit a trailing tail that overlaps with the elution profile of the monomeric standard mCherry. By contrast, mGrape3, EGFP, and mCherry all co-migrate in a sharp monodisperse peak (Figure S4A). Light scattering reveals average molecular weights for Neptune and mKate of 62 and 58 kD at the elution peak, consistent with dimers (Figure 3A). Average measured molecular weight in the tail is consistent with the late-eluting subpopulation of mKate proteins being predominantly monomers (34 kD), while late-eluting Neptune proteins exhibit an average molecular weight of 40 kD, suggesting a mix of monomers and dimers. Finally, at similar concentrations, Neptune and mKate migrate as dimers in native gel electrophoresis but as monomers in SDS, a behavior between that of mCherry, which migrates as a monomer in both conditions, and dTomato, which migrates as a dimer in both conditions (Figure S4B and S4C). Together, these data suggest that mKate and Neptune exist in a monomer-dimer equilibrium at low micromolar concentrations.
Figure 3.

A functionally monomeric variant of Neptune for protein fusions. (A) Neptune exhibits more dimerization relative to mKate, while mNeptune exhibits less. Normalized elution profiles on size-exclusion HPLC (thin traces, axis on left) and estimated molecular weights upon elution by in-line light scattering (thick traces, axis on right) are shown for dTomato (orange solid), Neptune (dark blue dashed), mNeptune (light blue solid), mKate (green dashed), mKate2 (red dashed), and mCherry (maroon solid). Dots show estimated molecular weights upon elution by in-line light scattering. Proteins were analyzed sequentially in the same day at 5 μM in PBS. mCherry and dTomato serve as monomeric and dimeric controls, respectively. (B) mNeptune-actin and mNeptune-tubulin are correctly localized upon expression in HeLa cells, similarly to mKate2 fusions. Scale bar, 10 μm.
To improve its performance in fusion proteins, we further monomerized Neptune. The Y194F mutation in Neptune alters an external side chain located adjacent to Met-146, which participates in a hydrophobic intersubunit interaction. Removal of a polar hydroxyl group by the Y194F mutation in Neptune might have resulted in increased dimerization tendency. To restore a polar group near position 194, we performed saturation mutagenesis of Met-146 and found a M146T mutant that retained the brightness and spectral profile of Neptune (Table 1, Figure 1), while exhibiting enhanced dissociation into monomers compared to mKate (Figure 3A). We performed equilibrium analytical ultracentrifugation to measure dissociation constants for Neptune and Neptune M146T. Sedimentation profiles were best fit to a monomer-dimer equilibrium model with a dissociation constant of 2.1 μM for the M146T mutant compared with 0.5 μM for Neptune (Table S3), confirming the increased monomeric character of the mutant. Fusions of Neptune 146T protein, which we refer to as mNeptune, to actin and tubulin localized correctly in HeLa cells (Figure 3b), similarly to mKate2 (Figure 3B), demonstrating its usefulness as a monomeric fusion tag. Hydrolysis of the chromophore acylimine in mNeptune, which occurs upon exposure of the chromophore to water during protein denaturation, was observed in less than 7% of the protein in samples aged for 4 months (Figure S4D), indicating that mNeptune is structurally stable.
Structural basis of the bathochromic shift in Neptune
Of the mutations differentiating Neptune from mKate S158A, no single one is necessary for the majority of the bathochromic shift (Table 2), implying multiple additive mechanisms for red-shifting. To explore the biophysical basis of wavelength tuning in Neptune, we determined the crystal structure of Neptune at pH 7 to 1.6 Å resolution (Figure 4). Neptune crystallized in tetragonal space group P42212 with unit cell dimensions of a = b = 92.1 Å, c = 53.2 Å, α = β = γ = 90°, and with one subunit per unit cell (Table 3). Symmetry operations generate a tetramer with similar organization as mKate. Despite being characterized in a different space group, the structure of Neptune is well superimposed upon the pH 7 structure of mKate (minimum r.m.s. of 0.24 Å over the Cα atoms of residues 1-220). Not surprisingly, side chains atoms show more structural divergence from mKate than backbone atoms (minimum r.m.s. of 0.38 Å vs. 0.24 Å). As in mKate at pH 7.0, the chromophore of Neptune exists entirely in the cis, or Z, state, with no electron density observed in the region expected for a trans-state chromophore even at a sigma value of 0.2 (Fig. S5). The covalent structure of the chromophore is identical to that of dsRed, with primarily sp2 geometry at the Cα atom of Met-63 (observed bond angles of 110, 117, and 126°), consistent with the presence of an acylimine group.
Table 2. Contributions of individual mutations to red-shifting in dNeptune.
| Reversion from Neptune | Excitation peaka | Emission peaka | εb | φc |
|---|---|---|---|---|
| none (Neptune) | 600 | 650 | 72000 | 0.18 |
| C61S | 597 | 649 | 80000 | 0.18 |
| F197Y | 596 | 649 | 56000 | 0.16 |
| G41M | 595 | 637 | 60000 | 0.21 |
| C158A | 593 | 644 | 97000 | 0.19 |
| C61S F197Y G41M C158A (mKate S158A) | 585 | 630 | 75000 | 0.30 |
| C61S F197Y G41M (mKate S158C) | 586 | 630 | 63000 | 0.33 |
Excitation and emission maxima in nm.
Maximum extinction coefficient in M-1cm-1 measured by the alkali denaturation method (Chalfie and Kain, 2005; Gross et al., 2000).
Quantum yield of fluorescence.
Figure 4.

Structural basis for red-shifting in Neptune. (A) Crystal structures of mKate at pH 7 (Pletnev et al., 2008) and Neptune at pH 7 were aligned, with mKate colored pink and Neptune light blue. The conjugated π system of the chromophore and side chain changes involved in red-shifting are shown in stick representation with nitrogen in blue, oxygen in red, and sulfur in yellow. Thin lines depict other side chains. (B) Neptune contains an additional hydrogen bond to the chromophore. Structures of mKate and Neptune cut away to the level of the chromophore are shown. The conjugated system of the chromophore and the Met-41 side chain are shown in stick representation colored as in (A), and the van der Waals surfaces of water oxygen atoms depicted as dotted spheres colored pink in mKate and light blue in Neptune. In Neptune, a water molecule occupies the space created by the M41G mutation and is in hydrogen bond distance of Ser-28 and the chromophore acylimine. Ser-28 is located beneath Met-41 in mKate and is not visible.
Table 3. Crystallographic data.
| Space group | P42212 |
| Cell dimensions: a, b (Å) | 92.144 |
| Cell dimensions: c (Å) | 53.216 |
| Wavelength (eV) | 11111 |
| Resolution (Å) | 10.4-1.60 (1.69-1.60)* |
| Total reflections | 260383 |
| Unique reflections | 30735 |
| Completeness (%) | 99.9 (99.7)* |
| Rsym | 0.055 (0.778)* |
| <I/σ> | 23.8 (2.2)* |
| R/Rfree | 0.171/0.202 |
| rmsd bond lengths (Å) | 0.011 |
| rmsd bond angles (°) | 1.43 |
Highest resolution shell
Details of the structure reveal multiple possible mechanisms for red-shifting conferred by side chain changes. The first is the addition of a novel hydrogen bond to the terminal oxygen of the conjugated π system of the chromophore. The cavity created by the M41G mutation in Neptune is filled by a water molecule simultaneously positioned to hydrogen bond to the hydroxyl of Ser-28 (inter-oxygen distance of 3.0 Å) and the chromophore acylimine oxygen (inter-oxygen distance of 2.7 Å) (Figure 4A and 4B). Photon absorption by dsRed-like chromophores is believed to excite an electron from the highest occupied molecular orbital (HOMO), with charge distribution concentrated over the phenolate group, into the lowest unoccupied molecular orbital (LUMO) with a more delocalized charge distribution (Taguchi et al., 2009). Hydrogen bond donation to this terminal carbonyl oxygen of the chromophore π system would be expected to produce an absorbance red-shift by preferentially stabilizing the excited state and thus decreasing the HOMO-LUMO energy difference.
Another possible mechanism of red-shifting we observed in Neptune is increased coplanarity of the chromophore rings. In mKate, the Arg-197 and Ser-158 side chains extend into a space adjacent to the chromophore (Figure 4A and 5A). In Neptune, steric clash with the larger side chain of Cys-158 as well as the loss of hydrogen bonding to Ser-158 may prevent Arg-197 from assuming a similar conformation. Instead the Arg-197 side chain extends parallel to the long axis of the chromophore, and the phenolate ring of the chromophore and Cys-158 shift toward each other by 1.0 Å (5.3 Å in Neptune versus 6.3 Å in mKate) to partially fill the space previously occupied by Arg-197 (Figure 4A and 5A). The Arg-197 guanidinium group is located 3.9 Å above the methylidene bridge of the chromophore (when the molecule is viewed with N and C termini upwards), where it can participate in a cation-π interaction that would be expected to stabilize the extended Arg-197 conformation. The Arg-197 movement away from its previous location to the side of the phenolate rings also allows a reduction in the twist angle between the imidazolinone and phenolate rings of the chromophore (Figure 5A, S5), improving coplanarity and electron delocalization, which would also tend to increase wavelength, extinction coefficient, and quantum yield. The conformational flexibility of Arg-197 is made possible by the presence of an accomodating chromophore cavity. In mKate, the space above the methylidene bridge is not occupied by amino acid side chains but rather by a water molecule. In Neptune, Arg-197 projects into this space, while a water molecule occupies the portion of the chromophore cavity adjacent to the β-barrel vacated by the Arg-197 side chain movement (Figure 5A).
Figure 5.
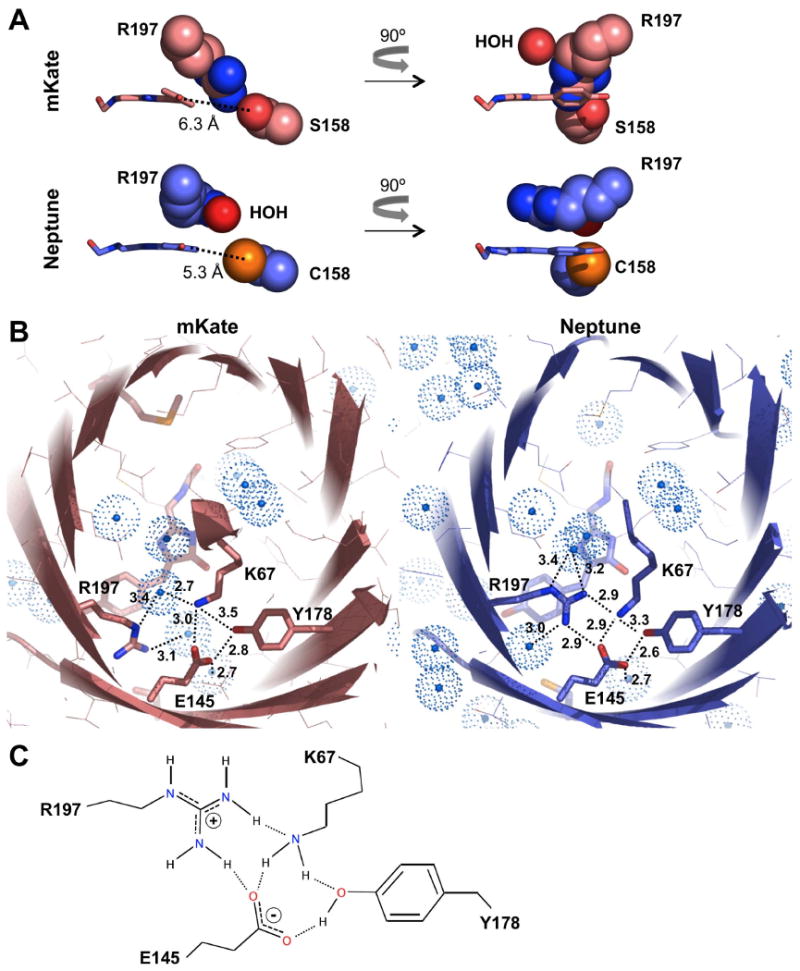
Changes in Arg-197 conformation and interactions in Neptune. (A) In mKate (upper), the Arg-197 and Ser-158 side chains (in space-fill representation) fill a space adjacent to the chromophore (stick), and the two rings of the chromophore are non-coplanar. A water molecule occupies a space above the methylidene bridge. Atoms are colored as in Figure 4A. In Neptune (lower), only Cys-158 is adjacent to the chromophore, while Arg-197 extends along the chromophore axis with its guanidinium group above the methylidene bridge. A water molecule occupies the former location of the guanidinium group. (B) In mKate (left), Arg-197 is hydrogen-bonded to water molecules. In Neptune (right), it also participates in a hydrogen-bond network involving Lys-67, Tyr-178, and Glu-145, including an unusual Arg-Lys hydrogen bond. Possible hydrogen bonds (interatomic distances < 3.5 Å) are shown as dotted lines. Colors are as in Figure 4A, except water molecules are in turquoise. (C) Detailed model of the hydrogen-bonding network with proposed donor-acceptor relationships. Hydrogen atoms are attached to donors with solid lines and to acceptors with dotted lines. Water molecules are omitted for clarity.
In conjuction with the repositioning of Arg-197, Neptune contains an unusual hydrogen bond between Arg and Lys sidechains. In mKate, the nitrogen atoms of Arg-197 are involved in hydrogen bond interactions with water molecules. In Neptune, the Nη atoms have acquired new hydrogen-bond partners in Lys-67 and Glu-145. Specifically, one Nη atom is positioned 2.9 Å from the Lys-67 nitrogen while the other is positioned 2.9 Å from an oxygen atom of Glu-145. (Figure 5B). The short distance observed between the Arg-197 and Lys-67 nitrogen atoms is compatible with hydrogen bonding and also argues strongly against both residues carrying formal positive charges. Indeed, the program PROPKA (Li et al., 2005) predicts Lys-67 to be predominantly uncharged (pKa of 5.97 for the conjugate acid). The retention of positive charge primarily on the Arg-197 is also consistent with the greater ability of Arg to stabilize charge through resonance delocalization and its higher pKa relative to Lys. The addition of Glu-145, which is rotated to become more coplanar with Arg-197 in Neptune (Figure 5B), as an additional hydrogen bond acceptor further stabilizes the positive charge on Arg-197. By acquiring these hydrogen-bonding interactions with Lys-67 and Glu-145, Arg-197 extends a hydrogen-bond network already present in mKate comprising Lys-67, Glu-145, and Tyr-178. We propose a model for this hydrogen-bond network consistent with the structural data (Figure 5C).
The cation-π interaction of Arg-197 could conceivably function in stabilizing the anionic state of the chromophore, in color tuning by influencing the electron distribution of the chromophore, and/or in catalysis of chromophore formation. To determine the function of Arg-197, we performed saturation mutagenesis at position 197 and obtained fluorescent variants with Val, Thr, and Tyr side chains at this position. Neptune R197V and R197T exhibit excitation peaks and shoulders shaped identically to Neptune (Fig. S6A). This observation implies that the positive charge of Arg-197 surprisingly does not have a function in wavelength tuning. On the other hand, both R197V and R197T show increased absorbance at 400 nm (Fig. S6B), suggesting that Arg-197 stabilizes the anionic state (with 600 nm peak absorption) of the chromophore in Neptune.
The R197Y mutations causes a blue-shift of the peak excitation to 594 nm (Fig. S6A). This suggests that Tyr-197 may counteract the bathochromic effects of hydrogen-bonding to the acylimine oxygen. One possible mechanism may be π–π stacking between Tyr-197 and the phenolate ring preferentially stabilizing electron distribution away from the acylimine group. The absorption spectrum of Neptune R197Y also shows a distinct hump at 557 nm (Fig. S6B). This likely reflects the presence of a subpopulation with a trans chromophore conformation. The Neptune precursor mKate contains both cis and trans chromophores in a pH-dependent manner, and the closely related protein eqFP611, which is spectrally similar to the mKate precursor TagRFP, contains primarily a trans chromophore at neutral pH with an absorption peak at 559 nm. As in the R197V and R197T mutants, protonated species absorbing at 400 nm are also present in Neptune R197Y, confirming the importance of Arg-197 in stabilizing the anionic state of the chromophore in Neptune.
Notably, red chromophore maturation still occurs in the R197V, R197T, and R197Y variants, implying that a positive charge at position 197 is not required for chromophore maturation. Indeed, positive charges that are known to promote chromophore maturation in other RFPs, namely Arg-92, which catalyzes ring cyclization (Sniegowski et al., 2005), and Lys-67, which promotes acylimine formation (Baird et al., 2000), are already present in Neptune, further arguing against an unique requirement for a positive charge at 197 for chromophore maturation. Taken together, the results of mutational analysis at position 197 indicate that Arg-197 functions to stabilize an anionic cis chromophore state in Neptune.
Interestingly, although critical for the bathochromic shift in Neptune (Table 2), the S158C mutation alone in mKate has a negligible effect on wavelength, whereas the combination of S158C and M41G produces a 7 nm red-shift. The repositioning of Arg-197 seen in Neptune may therefore be energetically favorable only upon loss of Met-41 due to coordinated conformational changes. One possibility may be a Met-41-dependent change in barrel ellipticity. The barrel cross-section in the plane of the chromophore is lengthened along the axis parallel to the chromophore in Neptune relative to mKate (20.0 Å versus 19.7 Å measured between the Cα atoms of residues 197 and 117), while it is narrower along the axis perpendicular to the chromophore (21.0 Å versus 21.5 Å between residues 41 and 175). A change in barrel ellipticity may be required for Arg-197 to assume its conformation in Neptune but may be energetically disfavored in the presence of the Met-41 side chain due to incompatible packing interactions.
The mechanisms of red-shifting by Y194F and S62C are less clear, although together they account for another 7 nm of the 14 nm difference between mKate and Neptune excitation peaks (Table 2). Within the Neptune structure, these residues are located near residues involved in red-shifting, namely Cys-158, Arg-197, and Gly-41. Y194F resides at the dimeric interface, but its 4-nm contribution to red-shifting does not depend on dimerization, as Neptune spectra and quantum yield were independent of concentration from 0.25 μM to 16 μM (Table S4). Furthermore, the red-shift is maintained even after the introduction of the monomerizing mutation M146T (Figure 1, Table 1). Instead, we hypothesize that the Y194F may lead to conformational changes at the beta barrel surface that are propagated to the nearby Cys-158 and Arg-197 side chains. S62C is also likely to function via subtle conformational changes. Through steric interactions, S62C may alter the positioning of the internal alpha helix to which the chromophore is linked at its acylimine group. This repositioning may influence the strength of the hydrogen bond between the water molecule and the acylimine oxygen.
Discussion
Hemoglobin effectively absorbs light at wavelengths below 600 nm and impedes imaging of traditional autocatalytic fluorescent proteins in deep tissues of mammals. The development of fluorescent proteins that excite effectively in the optical window at 600 nm and above is therefore highly desirable (Ntziachristos, 2006). We have engineered fluorescent proteins with excitation peaks at 600 nm and above through a strategy of random and site-directed mutagenesis. The brightest of these, Neptune, can be detected using excitation wavelengths commonly used for the organic fluorophore Cy5, and performs well in intravital imaging in mammals. As a “traditional” Cnidarian autocatalytic fluorescent protein, Neptune encodes its own chromophore, in contrast to phytochrome-based fluorescent proteins such as IFP. Thus its brightness is not dependent on the availability of cofactors such as the biliverdin. Table 4 summarizes current relative advantages of Neptune versus IFPs.
Table 4. Characteristics of mNeptune versus IFP1.4.
| Protein | mNeptune | IFP1.4 |
|---|---|---|
| Peak excitation (nm) | 600 | 684 |
| Peak emission (nm) | 650 | 708 |
| Extinction coefficient (M-1cm-1) | 67000 | 92000 |
| Quantum yield | 0.20 | 0.07 |
| Overall brightness* | 13.4 | 6.4 |
| Photostability* | 160 | 8.4 |
| Green emission component | present | absent |
| Cofactor dependence | none | biliverdin |
| Molecular weight (kD) | 30 | 36 |
| Stoichiometry (at 5 μM) | monomer-dimer equilibrium | monomer |
Calculated as in Table 1.
It would be useful to further evolve FPs to produce even more red-shifted variants in the future. To this end, understanding wavelength tuning mechanisms in autocatalytic FPs informs future strategies to evolve additional color variants. The crystal structure of Neptune that we describe here reveals a novel mechanism for excitation wavelength red-shifting, specifically a new hydrogen bond between a water molecule and the acylimine oxygen of the chromophore. Hydrogen bonding also occurs between the acylimine oxygen and the side chain of an internal amino acid, Glu-16, in mPlum, but this interaction selectively causes a red-shift in the emission spectrum only. Based on ultrafast spectroscopy and structural analysis of mPlum and its mutants, an explanation for this effect has been proposed that involves two dynamic changes: First, the hydrogen bond is relatively weak in the ground state and strengthens in the excited state (Shu et al., 2009). Glu-16 side-chain rotation then occurs in the excited state, causing relaxation to a lower energy intermediate state and emission at red-shifted wavelengths (Abbyad et al., 2007; Shu et al., 2009). In the case of Neptune, hydrogen bonding between the acylimine oxygen and a water molecule causes bathochromic shifts in both the excitation and emission spectra. The conformational flexibility of water may allow strong hydrogen bonding and thereby influence the electron distribution in the ground state in Neptune.
Together with the studies on mPlum, our findings raise the possibility of inducing further red-shifts in RFPs by introducing other hydrogen bond donors or acidic groups near the acylimine oxygen. Other strategies that could be explored include enhancing chromophore co-planarity between the imidazolinone ring and acylimine group, and the introduction of electrostatic interactions near the chromophore to preferentially stabilize the excited state electron distribution. Continued evolution of monomeric autocatalytic RFPs along these lines has the potential to further expand their utility in intracellular and intravital applications.
Significance
We have developed the first autocatalytic fluorescent proteins with excitation peaks at ≥ 600 nm. Among many potential applications, Neptune should be useful for tracking movement and proliferation in vivo of specific cell populations during development, of implanted tumors during metastasis, or of implanted stem cells in therapy models. Used together with bacterial phytochrome-based fluorescent proteins such as IFP, Neptune allows for the simultaneous tracking of two protein or cell populations in the optical window.
Experimental Procedures
Mutagenesis and Screening
Random mutagenesis was performed by error-prone PCR using the GeneMorph I or GeneMorph II kit (Stratagene). Mutations at specific residues were introduced by overlapping PCR or by QuikChange (Stratagene). For all library construction methods, chemically competent or electrocompetent Escherichia coli strain JM109(DE3) (for pRSETB) or LMG194 (for pBAD) were transformed and grown overnight on LB/agar (supplemented with 0.02% (wt/vol) L-arabinose (Fluka) for pBAD constructs) at 37°C and maintained thereafter at room temperature. LB/agar plates were screened for transmitted color by eye and for fluorescence using a Gel-DocIt imaging system (UVP). JM109(DE3) colonies of interest were cultured overnight in 2 ml LB supplemented with ampicillin. For secondary screening, colonies of interest were cultured in LB/ampicillin with 0.2% (wt/vol) arabinose overnight. Lysates were extracted with B-PER II (Pierce), and spectra were obtained on a Safire 96-well fluorescence plate reader (TECAN).
Fluorescence Activated Cell Sorting
LMG194 bacteria were electroporated with a modified pBAD vector containing the gene library and the transformed cells were grown in 30 ml RM media (M9 media with 0.2% glucose, 2% casamino acids, 1mM MgCl2) with ampicillin. After 8 hours, RFP expression was induced by adding L-arabinose to a final concentration of 0.2% (wt/vol). Overnight induced cultures were diluted 1:100 into PBS supplemented with ampicillin prior to FACS sorting. Multiple rounds of cell sorting were performed on a FACSDiva (BD Biosciences) in yield mode for the first sort and purity or single cell mode for subsequent sorts of the same library. Sorted cells were grown overnight in 4 ml RM with ampicllin and 0.2% (wt/vol) D-glucose and the resulting saturated culture was diluted 1:100 into 30 ml of the same media to start the next culture to be sorted. After 3 to 4 rounds of FACS sorting, the bacteria were plated onto LB agar with 0.02% (wt/vol) L-arabinose and grown overnight, after which individual clones were screened manually as described above.
Protein Production and Characterization
For spectral characterization, bacteria were lysed in B-PER (Pierce) and His-tagged proteins were purified by Ni-NTA chromatography (Qiagen). Purified proteins were exchanged into PBS pH 7.4 by dialysis or gel filtration. Absorbance measurements were performed on a Cary UV-Vis spectrophotometer. Fluorescence measurements were obtained on a Fluorolog-3 spectrofluorimeter with excitation spectra corrected for the xenon lamp spectrum and emission corrected for detector quantum yield. The excitation monochromator was calibrated to the xenon spectrum peak at 467 nm and the emission monochromator to the water Raman spectrum peak at 397 nm. Cresyl violet in methanol was used as a quantum yield standard with a correction factor applied for the refractive index difference between methanol and water (Lakowicz, 2006). Visible light photography was performed on a light table with a digital camera with white balance calibrated to the background. For maturation experiments, bacteria were grown in sealed flasks in broth deoxygenated by bubbling with nitrogen gas. Fluorescence intensities were measured at various times after lysis on the Safire fluorescence plate reader. For size-exclusion chromatography and light-scattering, 15 μg of purified purified protein in 100 μL was applied to a Shodex KW803 column and run in PBS pH 7.0 at a flow rate of 0.5 mL/min using an Agilent 1100 Series HPLC system controlled by ChemStation software. Protein elution was dually monitored with 280 nm absorbance and 568 nm absorbance for RFPs and 480 nm absorbance for EGFP. In-line light scattering was performed by a Wyatt Heleos system running ASTRA software. For native PAGE, 10 μg purified protein was run on a 4-16% Bis-Tris polyacrylamide gel (Invitrogen) in 50 mM BisTris 50 mM Tricine pH 6.8 with 0.002% Coomasssie Blue G-250. Gels were imaged for fluorescence at indicated wavelengths in a UVP iBox imaging system. For SDS PAGE, samples were loaded without boiling onto 4-12% Bis-Tris polyacrylamide gels and electrophoresed in MOPS electrophoresis buffer. Gels were then fixed and stained with 0.02% Coomassie Blue in 40% ethanol 10% acetic acid, and destained in 40% ethanol 10% acetic acid. For equilibrium analytical ultracentrifugation, purified protein samples in 10mM Tris pH 7.4 were centrifuged for 48 hours at 25° in a Beckman Optima XL-I at speeds of 16k, 20k, and 24k rpm, measuring absorbance as a function of radius. Absorbance at 220 nm was measured at 0.13, 0.38, 0.63, 1.0, 3.0, and 5.0 μM protein concentrations. Profiles were globally fitted by nonlinear least-squares analysis to single-component and monomer-dimer equilibrium models using Ultrascan version 9.9 software.
Photobleaching measurements
Aqueous droplets of purified protein in phosphate-buffered saline were formed under mineral oil in a chamber on the fluorescence microscope stage. For reproducible results it proved essential to pre-extract the oil with aqueous buffer, which would remove any traces of autoxidized or acidic contaminants. The droplets were small enough (5 – 10 μm diameter) so that all the molecules would see the same incident intensity. The absolute excitation irradiance in photons/(cm2·s·nm) as a function of wavelength was computed from the spectra of a xenon lamp, the transmission of the excitation filter, the reflectance of the dichroic mirror, the manufacturer-supplied absolute spectral sensitivity of a miniature integrating-sphere detector (SPD024 head and ILC1700 meter, International Light Corp.), and the measured detector current. The predicted rate of initial photon emission per chromophore (before any photobleaching had occurred) was calculated from the excitation irradiance and absorbance spectrum (both as functions of wavelength), and the quantum yield. The observed photobleaching time courses were normalized to a common arbitrary standard of 1000 initially emitted photons s-1 chromophore-1.
Animal imaging
Transcription units containing mKate, dNeptune, or IFP1.1 followed by the poliovirus IRES and a GFP coding sequence were constructed in pENTR1 by standard methods, then recombined into pAd-CMV-DEST by the Gateway system (Invitrogen). Linearized plasmid was used to transfect HEK293A cells for adenovirus production, followed by amplification and purification on FastTrap columns (Millipore) and elution into HBSS + 10% glycerol. Serial dilutions onto HEK293A cells and GFP visualization were performed to assess titers. Injections, imaging, and dissection were carried out in duplicate. Mice were anesthesized with ketamine/xylazine, then injected via the tail vein with 50 μL of 5×1010 infectious units per mL of purified adenoviruses. Five days later, mice were shaved on their ventral side and anesthesized with ketamine/xylazine. Images were acquired on a Maestro 1 animal fluorescence imaging system (CRI) with indicated excitation filters and a tunable liquid crystal emission filter of 40nm bandwidth centered on the indicated wavelengths. For quantification, off-animal dark counts were first subtracted, then mid-sagittal line intensities were obtained in ImageJ with the Plot Profile function. Similar results were obtained in the duplicate samples for each condition. For the charts in Figure 3, line intensities from duplicate conditions were averaged, and background was defined as the mean mid-sagittal intensity of uninfected liver, treatment-matched in terms of pre- or post-dissection condition. Defining background as the mid-thoracic region of infected animals gave similar results. For confocal imaging, three randomly oriented freshly prepared liver slices were placed on a coverslip and imaged with a 40× water objective on a Zeiss LSM 5Live microscope. For each slice, fluorescence intensities were measured from maximum intensity projections of a 10-section stack of total thickness 40μm. All animal procedures were performed according to protocols approved by the UCSD institutional animal care and use committee. For spectral unmixing, the predicted Neptune distribution in the infrared image is removed based on the Neptune distribution in the far-red channel and the relative efficiencies in the two channels in detecting Neptune, as calculated from the Neptune spectra.
Crystallization and refinement
For crystallization, the His tag on the protein was removed by TEV protease digestion. Crystals were grown by the sitting drop vapor diffusion method. 1 μL of protein at 10 mg/mL was mixed with 1 μL of 0.1 M HEPES pH 7.0, 0.2 M CaCl2, 20% PEG 6000 at 18° C. Crystals were cryoprotected with a mixture of 66.5% Paratone-N, 28.5% paraffin oil, and 5% glycerol (Sugahara and Kunishima, 2006) prior to immersion in liquid nitrogen. Data were collected at Beamline 8.3.1 at the Advanced Light Source, Lawrence Berkeley National Laboratory and processed with ELVES (Holton and Alber, 2004) and MOSFLM/SCALA (Evans, 2006; Leslie, 2006). The crystal structure was solved by molecular replacement using PHASER (McCoy et al., 2007) with mKate at pH 4.2 without the chromophore (PDB 3BXA) as the search model. The structure was refined with TLS parameters using PHENIX (Zwart et al., 2008), alternating with manual revision of the model using COOT (Emsley and Cowtan, 2004). No planarity restraints were applied to the chromophore during refinement. Crystallographic data and refinement statistics are presented in Table 3. Structure validation was performed with WHAT_CHECK (Hooft et al., 1996). Neptune and mKate pH 7.0 (PDB file 3BXB) were aligned and root mean square deviations were calculated using the align command in PyMol (http://www.pymol.org). Coordinates and structure factors have been deposited in the PDB (3IP2).
Supplementary Material
Acknowledgments
We thank Paul Steinbach for assisting with protein photobleaching and related analysis, and Larry Gross for performing analytical ultracentrifugation. We acknowledge the following sources of support: the Jane Coffin Childs Foundation and a Burroughs Wellcome Career Award for Medical Scientists (M.Z.L.), the UCSD Chancellor's Award for Undergraduate Research (M.R.M.), a Howard Hughes Medical Institute (HHMI) predoctoral fellowship (N.C.S.), a Canadian Institutes of Health Research postdoctoral fellowship (R.E.C.), the National Institutes of Health (NIH) Medical Scientist Training Program and U.S. Army Breast Cancer Research Program (BCRP) grant W81XWH-05-01-0183 (T.A.A.), NIH grant R01 GM048958 (H-L.N., W.M., and T.A.), and HHMI, NIH grants R01 GM086197 and R37 NS027177, and BCRP grant W81XWH-05-01-0183 (R.Y.T.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abbyad P, Childs W, Shi X, Boxer SG. Dynamic Stokes shift in green fluorescent protein variants. Proc Natl Acad Sci U S A. 2007;104:20189–20194. doi: 10.1073/pnas.0706185104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird GS, Zacharias DA, Tsien RY. Biochemistry, mutagenesis, and oligomerization of DsRed, a red fluorescent protein from coral. Proc Natl Acad Sci U S A. 2000;97:11984–11989. doi: 10.1073/pnas.97.22.11984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie M, Kain SR. Methods of Biochemical Analysis, Green Fluorescent Protein: Properties, Applications and Protocols. 2nd. Vol. 47. Wiley-Liss; 2005. [Google Scholar]
- Colin M, Moritz S, Schneider H, Capeau J, Coutelle C, Brahimi-Horn MC. Haemoglobin interferes with the ex vivo luciferase luminescence assay: consequence for detection of luciferase reporter gene expression in vivo. Gene Ther. 2000;7:1333–1336. doi: 10.1038/sj.gt.3301248. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Evans P. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- Gross LA, Baird GS, Hoffman RC, Baldridge KK, Tsien RY. The structure of the chromophore within DsRed, a red fluorescent protein from coral. Proc Natl Acad Sci U S A. 2000;97:11990–11995. doi: 10.1073/pnas.97.22.11990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson JN, Ai HW, Campbell RE, Remington SJ. Structural basis for reversible photobleaching of a green fluorescent protein homologue. Proc Natl Acad Sci U S A. 2007;104:6672–6677. doi: 10.1073/pnas.0700059104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holton J, Alber T. Automated protein crystal structure determination using ELVES. Proc Natl Acad Sci U S A. 2004;101:1537–1542. doi: 10.1073/pnas.0306241101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooft RW, Vriend G, Sander C, Abola EE. Errors in protein structures. Nature. 1996;381(6580):272. doi: 10.1038/381272a0. [DOI] [PubMed] [Google Scholar]
- Lakowicz JR. Principles of Fluorescence Spectroscopy. New York: Springer; 2006. [Google Scholar]
- Leslie AG. The integration of macromolecular diffraction data. Acta Crystallogr D Biol Crystallogr. 2006;62:48–57. doi: 10.1107/S0907444905039107. [DOI] [PubMed] [Google Scholar]
- Li H, Robertson AD, Jensen JH. Very fast empirical prediction and rationalization of protein pKa values. Proteins. 2005;61:704–721. doi: 10.1002/prot.20660. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntziachristos V. Fluorescence molecular imaging. Annu Rev Biomed Eng. 2006;8:1–33. doi: 10.1146/annurev.bioeng.8.061505.095831. [DOI] [PubMed] [Google Scholar]
- Ormo M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ. Crystal structure of the Aequorea victoria green fluorescent protein. Science. 1996;273:1392–1395. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- Petersen J, Wilmann PG, Beddoe T, Oakley AJ, Devenish RJ, Prescott M, Rossjohn J. The 2.0-A crystal structure of eqFP611, a far red fluorescent protein from the sea anemone Entacmaea quadricolor. J Biol Chem. 2003;278:44626–44631. doi: 10.1074/jbc.M307896200. [DOI] [PubMed] [Google Scholar]
- Pletnev S, Shcherbo D, Chudakov DM, Pletneva N, Merzlyak EM, Wlodawer A, Dauter Z, Pletnev V. A crystallographic study of bright far-red fluorescent protein mKate reveals pH-induced cis-trans isomerization of the chromophore. J Biol Chem. 2008;283:28980–28987. doi: 10.1074/jbc.M800599200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remington SJ. Fluorescent proteins: maturation, photochemistry and photophysics. Curr Opin Struct Biol. 2006;16:714–721. doi: 10.1016/j.sbi.2006.10.001. [DOI] [PubMed] [Google Scholar]
- Remington SJ, Wachter RM, Yarbrough DK, Branchaud B, Anderson DC, Kallio K, Lukyanov KA. zFP538, a yellow-fluorescent protein from Zoanthus, contains a novel three-ring chromophore. Biochemistry. 2005;44:202–212. doi: 10.1021/bi048383r. [DOI] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Shaner NC, Lin MZ, McKeown MR, Steinbach PA, Hazelwood KL, Davidson MW, Tsien RY. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat Methods. 2008;5:545–551. doi: 10.1038/nmeth.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- Shcherbo D, Merzlyak EM, Chepurnykh TV, Fradkov AF, Ermakova GV, Solovieva EA, Lukyanov KA, Bogdanova EA, Zaraisky AG, Lukyanov S, Chudakov DM. Bright far-red fluorescent protein for whole-body imaging. Nat Methods. 2007;4:741–746. doi: 10.1038/nmeth1083. [DOI] [PubMed] [Google Scholar]
- Shcherbo D, Murphy CS, Ermakova GV, Solovieva EA, Chepurnykh TV, Shcheglov AS, Verkhusha VV, Pletnev VZ, Hazelwood KL, Roche PM, Lukyanov S, Zaraisky AG, Davidson MW, Chudakov DM. Far-red fluorescent tags for protein imaging in living tissues. Biochem J. 2009 doi: 10.1042/BJ20081949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu X, Royant A, Lin MZ, Aguilera TA, Lev-Ram V, Steinbach PA, Tsien RY. Mammalian expression of infrared fluorescent proteins engineered from a bacterial phytochrome. Science. 2009;324:804–807. doi: 10.1126/science.1168683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu X, Shaner NC, Yarbrough CA, Tsien RY, Remington SJ. Novel chromophores and buried charges control color in mFruits. Biochemistry. 2006;45:9639–9647. doi: 10.1021/bi060773l. [DOI] [PubMed] [Google Scholar]
- Shu X, Wang L, Colip L, Kallio K, Remington SJ. Unique interactions between the chromophore and glutamate 16 lead to far-red emission in a red fluorescent protein. Protein Sci. 2009;18:460–466. doi: 10.1002/pro.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sniegowski JA, Lappe JW, Patel HN, Huffman HA, Wachter RM. Base catalysis of chromophore formation in Arg96 and Glu222 variants of green fluorescent protein. J Biol Chem. 2005;280:26248–26255. doi: 10.1074/jbc.M412327200. [DOI] [PubMed] [Google Scholar]
- Stamatas GN, Southall M, Kollias N. In vivo monitoring of cutaneous edema using spectral imaging in the visible and near infrared. J Invest Dermatol. 2006;126:1753–1760. doi: 10.1038/sj.jid.5700329. [DOI] [PubMed] [Google Scholar]
- Sugahara M, Kunishima N. Novel versatile cryoprotectants for heavy-atom derivatization of protein crystals. Acta Crystallogr D Biol Crystallogr. 2006;62:520–526. doi: 10.1107/S0907444906009012. [DOI] [PubMed] [Google Scholar]
- Taguchi N, Mochizuki Y, Nakano T, Amari S, Fukuzawa K, Ishikawa T, Sakurai M, Tanaka S. Fragment molecular orbital calculations on red fluorescent proteins (DsRed and mFruits) J Phys Chem B. 2009;113:1153–1161. doi: 10.1021/jp808151c. [DOI] [PubMed] [Google Scholar]
- Tromberg BJ, Shah N, Lanning R, Cerussi A, Espinoza J, Pham T, Svaasand L, Butler J. Non-invasive in vivo characterization of breast tumors using photon migration spectroscopy. Neoplasia. 2000;2:26–40. doi: 10.1038/sj.neo.7900082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Jackson WC, Steinbach PA, Tsien RY. Evolution of new nonantibody proteins via iterative somatic hypermutation. Proc Natl Acad Sci U S A. 2004;101:16745–16749. doi: 10.1073/pnas.0407752101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwart PH, Afonine PV, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, McKee E, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Storoni LC, Terwilliger TC, Adams PD. Automated structure solution with the PHENIX suite. Methods Mol Biol. 2008;426:419–435. doi: 10.1007/978-1-60327-058-8_28. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


