Abstract
A platform of discrete microscopic magnetic elements patterned on a surface offers dynamic control over the motion of fluid-borne cells by reprogramming the magnetization within the magnetic bits. T-lymphocyte cells tethered to magnetic microspheres and untethered leukemia cells are remotely manipulated and guided along desired trajectories on a silicon surface by directed forces with average speeds up to 20 μm/s. In addition to navigating cells, the microspheres can be operated from a distance to push biological and inert entities and act as local probes in fluidic environments.
Introduction
The manipulation of biological cells and micro- and nanoparticles is important in fields such as biological research, engineering, and colloid science. Several techniques have been applied to control the movement of small particles that underlies the functions of addressing, transport, sorting, and assembly. These approaches include the use of optical (1–4) and magnetic tweezers (5,6), electrophoresis (7), acoustic force (8), and hydrodynamics-based schemes (9), and each approach exhibits advantages and distinct features. Simple surface patterns of micromagnetic structures are also emerging as manipulation templates, and such guides have been used in conjunction with fluid dispersion of magnetic microparticles (10), cantilever tips (11,12) and external magnetic fields (13,14) to manipulate biologically inert particles. Because constant magnetic fields have far fewer adverse effects on cells than do optical or electric fields, surface magnetic forces would be especially useful for manipulating and sorting biological cells. For instance, a microelectromagnetic template that relies on Amperian fields originating from programmed electric currents to wires in a matrix has been proposed for manipulating magnetically tethered cells (15). Whereas one drawback of magnetic-field-based approaches for cell manipulation is the need to tether magnetic particles to the cell surface, methods such as cell dielectrophoresis do not require tethering (16,17). Other disadvantages include the need to functionalize each cell type with specific biomarkers and the potential effects of such surface attachments on intrinsic cell activity.
In this study, we employ reprogrammable magnetization profiles created through lithographically patterned discrete ferromagnetic disks as a template for producing highly localized trapping fields. The resulting tunable magnetic field gradients in the vicinity of the disk periphery enable directed forces to be applied to 1), immunomagnetically labeled biological cells, and 2), magnetic microspheres that act as magnetically actuated miniature force-transmitting probes to navigate fluid-borne unlabeled cells with micrometer precision. The principal features of this study are demonstrated by remotely transporting and arranging multiple or individual T-lymphocyte and leukemia cells using programmed routines (à la joystick). Without producing damage, the forces transport the cells with speeds up to 20 μm/s across a silicon platform to predetermined sites.
Methods
The experimental layout is illustrated in Fig. 1. Central to the study are the circular permalloy disks (5–10 μm in diameter and 40 nm thick) lithographically patterned on a silicon surface (see Appendix A). The externally applied tuning magnetic fields are provided by two pairs of orthogonal miniature electromagnets to create rotating in-plane fields Hx and Hy, whereas a circular coil (solenoid) generates the out-of-plane field Hz. Relatively uniform in- and out-of-plane magnetic fields up to 100 Oe are produced with this setup. The electromagnets were each connected to independent current channels programmed in LabVIEW software (National Instruments, Austin, TX) to produce controlled planar magnetic fields; the direction of Hz was also reversed through programmed routines.
Figure 1.
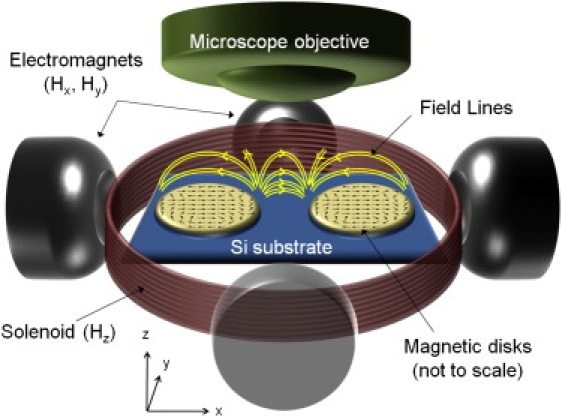
Schematic representation of experimental setup. Circular ferromagnetic permalloy disks are lithographically patterned on a silicon surface. Also shown are the vectorial maps from OOMMF simulations of the magnetic configurations using a cell size of 50 nm over disks 5 μm in diameter by 40 nm thick in the presence of Hx = 50 Oe are shown along with the disks. The magnetization profiles are tuned by four remotely controlled electromagnets that produce rotating in-plane fields Hx and Hy. Reversible perpendicular fields, Hz, are produced by the circular solenoid coil. Movements of cells are recorded by an optical microscope (Reichert) with a 20× objective and high-speed camera.
Dynabeads 2.8 μm in diameter (M-280, Invitrogen, Carlsbad, CA) were used to demonstrate magnetically induced movement of the system. For immunomagnetically labeled cell studies, we utilized previously separated (<24 h old) T-lymphocyte cells (CD3-positive) from human blood cells according to a previously published protocol (see Appendix B). These T-lymphocytes were labeled with tetrameric antibody complex. This complex targeted the CD3 receptor on the T-lymphocytes and simultaneously dextran-coated 1-μm magnetic particles. Unlabeled leukemia cells were directly manipulated using magnetic particles with no biomarkers. Movements of the cells are observed by an optical microscope (Leica Microsystems, Wetlzar, Germany) equipped with a 20× objective and a high-speed camera.
Modeling
To model the magnetic forces on a microsphere in the lateral and vertical directions, we considered magnetization profiles within permalloy disks 5 μm in diameter and 40 nm thick. As discussed in Appendix C, computer simulations based on the 2D version of the object-oriented micromagnetic framework (OOMMF) program (18) yielded the micromagnetic structure and its response to the external fields Hx, Hy, and Hz. The vectorial maps of the magnetic configurations using a cell size of 50 nm over the permalloy disks (saturation magnetization 8.6 × 105 A/m) in the presence of Hx = 50 Oe, , are illustrated in Fig. 1. The results reveal the magnetization at the disk center to be largely parallel to Hx and curling around the periphery. The vector data from the OOMMF simulation provides for the disk magnetic scalar potential, , and field, HDisk (ÑFM):
| (1) |
where is the magnetization/unit volume as a function of position derived from the OOMMF simulation and is the normal to the disk (19). The volume integral is performed as a summation over all OOMMF cells. Due to strong shape anisotropy, the magnetization remains largely in-plane for the modest out-of-plane fields (Hz < 75 Oe) utilized in this study. When the superparamagnetic particles are in the regime where they are linearly magnetizable, the net magnetic force on the sphere is F = (1/2)∇(m · B) = (1/2)χV∇B2/μo, where μo, V, and χ are the free space permeability, volume, and linear magnetic susceptibility, respectively, of the microsphere.
Figs. 2 and 3 illustrate the energy, trapping fields, and forces associated with a permalloy disk 5 μm diameter by 40 nm thick by external fields Hx and Hy, as well as their tunability with Hz. Fig. 2, a–c, shows the potential energy profiles for different in-plane field directions for Hz = 50 Oe. The sharp potential energy minimum moves along the outer disk periphery tracking synchronously with the direction of the in-plane field. Fig. 2, d–f, shows real images of a magnetic microsphere trapped and transported along with the energy minimum around the disk.
Figure 2.
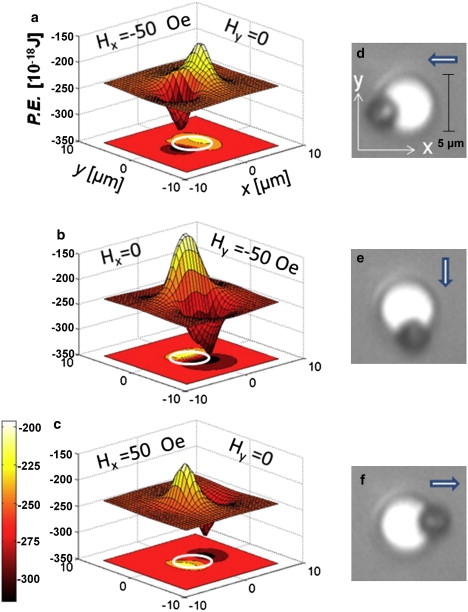
Calculated energy and force profiles from a permalloy disk 5 μm in diameter and 40 μm thick magnetized with an in-plane field |Hx + Hy| = 50 Oe and perpendicular field Hz = + 50 Oe. (a–c) Potential energy (P.E.) profiles of a 2.8-μm magnetic particle (χ = 0.85) and contour plot of the energy profile in the x-y plane. The trap strengths are tunable with Hz. The trap strengths are tunable with Hz. (d–f) Real images of a microsphere moved along the periphery of the disk. Arrows indicate orientation of in-plane field Hx + Hy.
Figure 3.
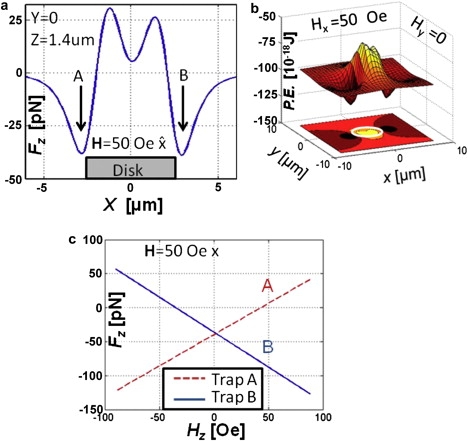
(a) Fz, the z component of the force, on a 2.8-μm magnetic particle with Hx = 50 Oe and Hz = 0. With Hz = 0 the two traps (A and B) are formed at opposite ends of the disk along the x-axis. The lack of complete symmetry of the energy profile of traps A and B arises from the magnetization of the disk being not perfectly symmetric in the OOMMF simulation. (b) Potential energy profiles of traps A and B for Hz = 0. (c) The attractive force from trap A decreases with increasing Hz, with the reverse response for trap B.
With Hz = 0 (Fig. 3 a), two traps, A and B, are located near diametrically opposite ends parallel to the in-plane field Hx. Fig. 3, a and b, shows that these two traps are approximately symmetric for Hz = 0. The introduction of an axial field, Hz (+50 Oe), directed away from the disk, renders trap B more attractive, at the same time weakening trap A (Fig. 3 c). Reversing Hz (−50 Oe) inverts the character of traps A and B (Fig. 3 c), which illustrates how trapping forces are tunable to hundreds of piconewtons.
Results
Fig. 4 and Movies S1, S2, S3, and S4 in the Supporting Material show transporting examples of labeled T-lymphocyte cells or untethered leukemia cells. The directional forces are regulated by the clockwise or counterclockwise rotation in 90° steps of the in-plane field (60 Oe) and timed reversal of Hz. The tethered cells hop from one disk to the next by reversing Hz to create an attractive trapping center on the targeted disk, and the trajectory is reversed by reordering the sequence of the planar (Hx + Hy) and Hz fields. The controlled navigation of single (Fig. 4 a) and multiple (Fig. 4 b) tethered T-lymphocyte cells traveling at an average speed of 20 μm s−1 illustrates the labeling sphere and cell locked in with the magnetic potential well (Fig. 2, a–c), which is micromanipulated along the surface. The Reynolds number of a cell traveling at this speed is ∼2.4 × 10−4. The effect of the resulting hydrodynamic forces arising from the induced movement of the cells is negligible. For example, Gregoriades et al. demonstrated that it took flow through a flow contraction with a corresponding Reynolds number of 323 to remove Chinese hamster ovary cells attached to 200-μm microcarriers (21). Later, Mollet et al. experimentally demonstrated that flow through a 225-μm contraction at 21.7 m s−1, corresponding to a Reynolds number of 8146, was required before detectable damage to suspended Chinese hamster ovary cells is observed (22).
Figure 4.
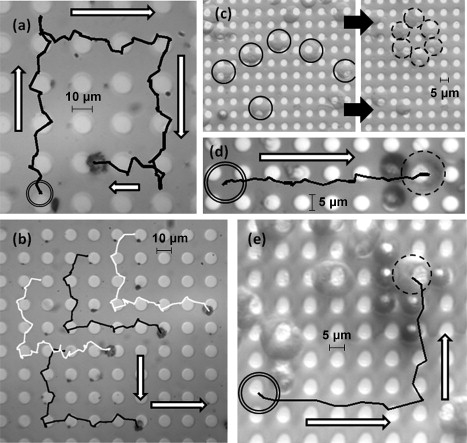
(a) Sequential applications of planar and perpendicular fields transport a labeled T-lymphocyte cell on a Si platform along the periphery of a 10-μm-diameter disk before hopping to a neighboring disk. The initial position of the cell (double circle) is shown and the trajectory of the cell indicated by connected lines at discrete intervals. (b) Simultaneous transport of four labeled fluid-borne T-cells traveling from top to bottom and then to the right in unison along the platform. Lines identify trajectories of the four cells. (c) Six unlabeled leukemia cells (left, solid circles) being sequentially manipulated by a single microsphere to create a hexagonal lattice (right, dashed circles). The magnetic disks are 5 μm in diameter. (d) Two magnetic microspheres navigating a single leukemia cell on the surface from initial position (double circle) to final location (dashed circle). (e) Several microspheres surrounding and transporting a single unlabeled leukemia cell. Successive center positions of remotely controlled movement of the cell in solution are shown by the solid lines. White arrows indicate direction of travel of the cells.
Fig. 4, c–e, illustrates the manipulation of living leukemia cells without attachment of a magnetic microsphere. This feature overcomes a limitation of standard magnetic-tweezers-based cell manipulation schemes—namely, attachment of magnetic beads that may affect the cells' intrinsic properties. Fig. 4 c shows six leukemia cells sequentially maneuvered by a microsphere into a hexagonal pattern on the surface. To have greater regulation of the cell movement, Fig. 4 d illustrates two magnetic spheres acting as a pair of handles to navigate a cell in a controlled manner. Fig. 4 e illustrates several microspheres around a leukemia cell, which is then navigated by the multiple spheres. Direct measurements of the microsphere speed show the transmitted forces that overcome drag on the cells to be on the order of several tens of piconewtons.
The platform could also find applications related to propulsion and navigation in aqueous environments. Common micromanipulation approaches based on optical tweezers, or atomic force cantilevers, are not suitable for work at long ranges, as they require proximity to a microscope objective or an atomic force microscope tip, whereas the magnetic microspheres on the platform are readily controlled from a distance through external fields. In addition to untethered cells, the magnetic spheres can also push inert entities and act as local delivery agents to targeted sites on surfaces for realizing functional microelectromechanical systems. For instance, it should be possible to assemble dielectric microspheres to create two-dimensional photonic band lattices (23).
Development of microfabricated magnetic traps and their tunability by external programmable fields makes it possible to readily integrate them into microfluidic devices (24,25). For example, embedded microdisk arrays in one or multiple microfluid channels can capture and transport magnetic labeled species in the sample flowing perpendicular to the array channels. The arrays can then be initialized with programmed routines (Fig. 4, a and b) to move the labeled species to a different cross-channel, where it can be chemically detached and detected. The ability to manipulate multiple objects at the individual cell level coupled within existing microfluidic technology could form the basis for realizing low-cost, lab-on-a-chip analytical tools for detection of living microorganisms. Our current design enables the transport of one given functional marker at a time. In the case of sorting a heterogeneous cell population, this would be achieved with different markers transporting different cells in a sequential approach that can be readily integrated into a chip. Alternatively, we can use a “cocktail” of antibodies that target several specific markers on the cell surface at the same time and move all of these cells at once. It is envisioned that this technique will be used primarily on cells that are either grown in suspension, or previously suspended by some method. The suspended cells can come from blood, digested tissue, or cell culture.
In addition to the simplicity, reliability, and tunability of the traps, there are other advantages of the planar platform. These include ease of standard lithography fabrication, a single focal plane that allows for real-time image acquisition of the cells in a parallel manner that is greatly increased compared to point or line scanning, and ready scale up to high array densities, enabling concurrent measurement of single-particle and ensemble average responses. In addition, the array of magnetic disks can be fabricated on other biocompatible platforms, such as glass slides, thus providing low-cost platforms for manipulating cells. Moreover, the remotely controlled external fields allow for manipulation of cells by programmed routines that will not require dynamic refocusing or out-of-focus calibrations.
Conclusions
In conclusion, patterned templates of magnetic disks enable remotely controlled (joystick) directional transport of fluid borne individual or multiple labeled cells on a silicon surface. The template also offers the capability to maneuver untethered cells to precise surface locations, thereby offering an important advantage over approaches that rely on the need to attach magnetic beads to cells. The platform may also be readily utilized in combination with tip based probes to study single cell responses, and is amenable to replication for wide integration with other biological, microfluidic and nanoscale measurement devices.
Appendix A: Platform Fabrication
Two layers of e-beam resist, first methyl methacrylate (8.5) MAA E 9, then 950 polymethyl methacrylate C 4, both from MicroChem (Newton, MA), were spin-coated, then baked onto a silicon/silicon oxide substrate. A Helios NanoLab 600 focused ion beam (FEI, Hillsboro, OR) was used to pattern circle arrays into the resist.
A 40-nm layer of permalloy was deposited onto the entire silicon surface by magnetron sputtering. The sample was then submerged in warm acetone (50°C) for ∼30 min to lift off the e-beam resist and excess magnetic material.
To prevent antibody-coated beads and T-lymphocyte cells from adhering to the substrate, the entire substrate was coated with a 1-nm seed layer of Permalloy followed by a 2-nm layer of gold. The sample was then submerged in a solution of 1 mM triethylene glycol mono-11-mercaptoundecyl ether (from Aldrich, St. Louis, MO) in ethanol for 12 h, and rinsed with ethanol.
Appendix B: Cell Preparation
Peripheral blood buffy coats from healthy donors were purchased from the American Red Cross, Central Ohio Region. Isolation of peripheral blood leukocytes was carried out by diluting buffy coat aliquot with Hank's balanced salt solution (HyClone, Logan, UT) at a volume ratio of 1:2, and laid over a Ficoll-Hypaque (Mediatech, Manassas, VA) density gradient (1.077g ml−1) at a volume ratio of 1:1. Tubes were spun for 30 min at 350 × g with the centrifuge brake off. The recovered white blood cell layer was washed twice with phosphate-buffered saline (PBS) without calcium and magnesium (Mediatech).
The concentrations of cells in the PBS suspension were determined by hemocytometer using the Unopette microcollection system (BD Biosciences, San Jose, CA). Trypan Blue (GIBCO, Carlsbad, CA) was utilized in addition to indicate cell viability.
Raji Burkitt's lymphoma cells, obtained from American Type Culture Collection (Manassas, VA), were cultured in RPMI 1640 media supplemented with 10% heat-inactivated fetal bovine serum (Hyclone), 2 mM L-glutamine (Invitrogen, Carlsbad, CA), and penicillin (100 U/mL)/streptomycin (100 μg/ml; Sigma-Aldrich, St. Louis, MO) at 3°C in an atmosphere of 5% CO2. Cells were washed twice with PBS (pH 7.4), then fixated with 2% paraformaldehyde for 30 min.
The CD3 expressing T-lymphocytes were immunomagnetically labeled using tetrameric antibodies from Stem Cell Technologies (Vancouver, BC, Canada). 10μl tetrameric antibody complex was added to a 107 leukocyte pellet and the cell suspension was incubated for 15 min at room temperature in a shaker. After incubation, 10μl magnetic microparticles/107 leukocytes was added 10 min before separation, and the cells were separated according to the procedure of Tong et al. (2007) (26,27).
Appendix C: Simulations
OOMMF simulations were performed for disks 5 μm in diameter and 40 nm in height with cell size 50 nm × 50 nm × 40 nm (17). The parameters used for permalloy were as follows: exchange stiffness constant, 13 × 10−12 J m−1, saturation magnetization of 8.6 × 105 A/m, and zero magnetocrystalline anisotropy. The simulation provides for the three-dimensional magnetization vector within each micromagnetic cell.
Supporting Material
Five movies are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(09)01678-6.
Supporting Material
Acknowledgments
The authors acknowledge support from the National Science Foundation (EEC-0425626), the Army Research Office (W911NF-08-1-0455), and the National Cancer Institute (R01 CA97391-01A1).
References
- 1.Ashkin A., Dziedzic J.M., Chu S. Observation of a single-beam gradient force optical trap for dielectric particles. Optics Letters. 1986;11:288–290. doi: 10.1364/ol.11.000288. [DOI] [PubMed] [Google Scholar]
- 2.Ashkin A. Accelerating and trapping of particles by radiation pressure. Phys. Rev. Lett. 1970;24:156–159. [Google Scholar]
- 3.Neuman K.C., Nagy A. Single-molecule force spectroscopy: optical tweezers, magnetic tweezers and atomic force microscopy. Nat. Methods. 2008;5:491–505. doi: 10.1038/nmeth.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grier D.G. A revolution in optical manipulation. Nature. 2003;424:21–27. doi: 10.1038/nature01935. [DOI] [PubMed] [Google Scholar]
- 5.Gosse C., Croquette V. Magnetic tweezers: micromanipulation and force measurement at the molecular level. Biophys. J. 2002;82:3314–3329. doi: 10.1016/S0006-3495(02)75672-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strick T., Allemand J.-F., Bensimon D. The manipulation of single biomolecules. Phys. Today. 2001;54:46–51. [Google Scholar]
- 7.Kremser L., Blaas D., Kenndler E. Capillary electrophoresis of biological particles: viruses, bacteria, and eukaryotic cells. Electrophoresis. 2004;25:2282–2291. doi: 10.1002/elps.200305868. [DOI] [PubMed] [Google Scholar]
- 8.Hertz H.M. Standing-wave acoustic trap for nonintrusive positioning of microparticles. J. Appl. Phys. 1995;78:4845–4849. [Google Scholar]
- 9.Sundararajan N., Pio M.S., Berlin A.A. Three dimensional hydrodynamic focusing in polydimethylsiloxane (PDMS) microchannels. J. Microelectromech. Syst. 2004;13:559–567. [Google Scholar]
- 10.Yellen B.B., Hovorka O., Friedman G. Arranging matter by magnetic nanoparticle assemblers. Proc. Natl. Acad. Sci. USA. 2005;102:8860–8864. doi: 10.1073/pnas.0500409102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mirowski E., Moreland J., Donahue M.J. Manipulation and sorting of magnetic particles by a magnetic force microscope on a microfluidic magnetic trap platform. Appl. Phys. Lett. 2005;86:2439011–2439013. [Google Scholar]
- 12.Mirowski E., Moreland J., Donahue M.J. Integrated microfluidic isolation platform for magnetic particle manipulation in biological systems. Appl. Phys. Lett. 2004;84:1786–1788. [Google Scholar]
- 13.Gunnarsson K., Roy P.E., Oscarsson S. Programmable motion and separation of single magnetic particles on patterned magnetic surfaces. Adv. Mater. 2005;17:1730–1734. [Google Scholar]
- 14.Tierno P., Johansen T.H., Fisher T.M. Localized and delocalized motion of colloidal particles on a magnetic bubble lattice. Phys. Rev. Lett. 2007;99:0383031–0383034. doi: 10.1103/PhysRevLett.99.038303. [DOI] [PubMed] [Google Scholar]
- 15.Lee H., Purdon A.M., Westervelt R.M. Manipulation of biological cells using a microelectromagnetic matrix. Appl. Phys. Lett. 2004;82:1063–1065. [Google Scholar]
- 16.Talary M.S., Burt J.P.H., Pethig R. Electromanipulation and separation of cells using travelling electric fields. Appl. Phys. Lett. 1996;29:2198–2203. [Google Scholar]
- 17.Yang J., Huang Y., Gascoyne P.R. Cell separation on microfabricated electrodes using dielectrophoretic/gravitational field-flow fractionation. Anal. Chem. 1999;71:911–918. doi: 10.1021/ac981250p. [DOI] [PubMed] [Google Scholar]
- 18.Donahue, M. J., and D. G. Porter. 1999 OOMMF User's Guide, Version 1.0, Interagency Report NISTIR 6376, National Institute of Standards and Technology, Gaithersburg, MD. http://math.nist.gov/oommf/.
- 19.Jackson J.D. 3rd ed. John Wiley; New York: 1999. Classical Electrodynamics. [Google Scholar]
- 20.Reference deleted in proof.
- 21.Gregoriades N., Clay J., Chalmers J.J. Cell damage of microcarrier cultures as a function of local energy dissipation created by a rapid extensional flow. Biotechnol. Bioeng. 2000;69:171–182. doi: 10.1002/(sici)1097-0290(20000720)69:2<171::aid-bit6>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 22.Mollet M., Godoy-Silva R., Chalmers J.J. Acute hydrodynamic forces and apoptosis: a complex question. Biotechnol. Bioeng. 2007;98:772–788. doi: 10.1002/bit.21476. [DOI] [PubMed] [Google Scholar]
- 23.Miyazaki H.T., Miyazaki H., Sato T. Photonic band in two-dimensional lattices of micrometer-sized spheres mechanically arranged under a scanning electron microscope. J. Appl. Phys. 2000;87:7152–7156. [Google Scholar]
- 24.Andersson H., van den Berg A. Microfluidic devices for cellomics: a review. Sensor Actuat. B Chem. 2000;92:315–325. [Google Scholar]
- 25.Pamme N. Magnetism and microfluidics. Lab Chip. 2006;6:24–38. doi: 10.1039/b513005k. (and references therein) [DOI] [PubMed] [Google Scholar]
- 26.Yang L., Lang J.C., Chalmers J.J. Optimization of an enrichment process for circulating tumor cells from the blood of head and neck cancer patients through depletion of normal cells. Biotechnol. Bioeng. 2009;102:521–534. doi: 10.1002/bit.22066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tong X., Xiong Y., Chalmers J.J. A novel high throughput immunomagnetic cell sorting system for potential clinical scale depletion of T cells for allogeneic stem cell transplantation. Exp. Hematol. 2007;35:1613–1622. doi: 10.1016/j.exphem.2007.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


