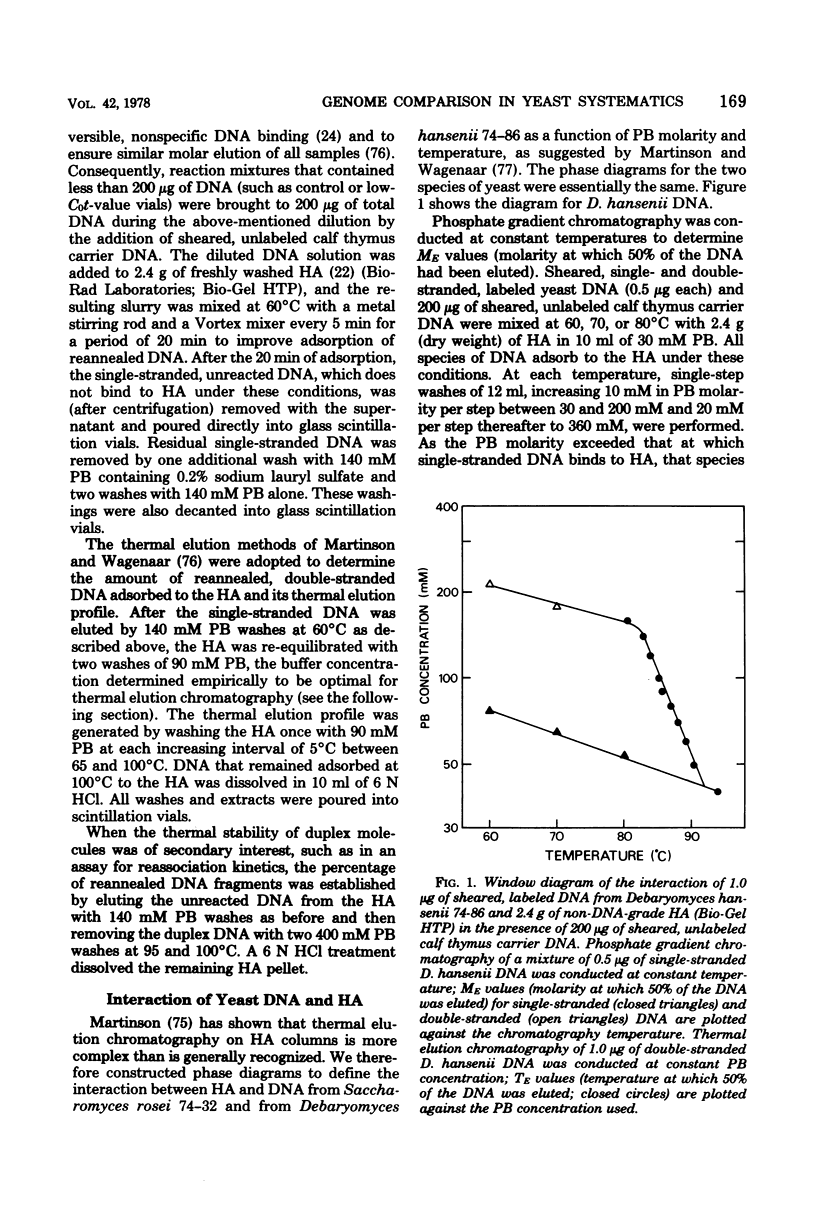
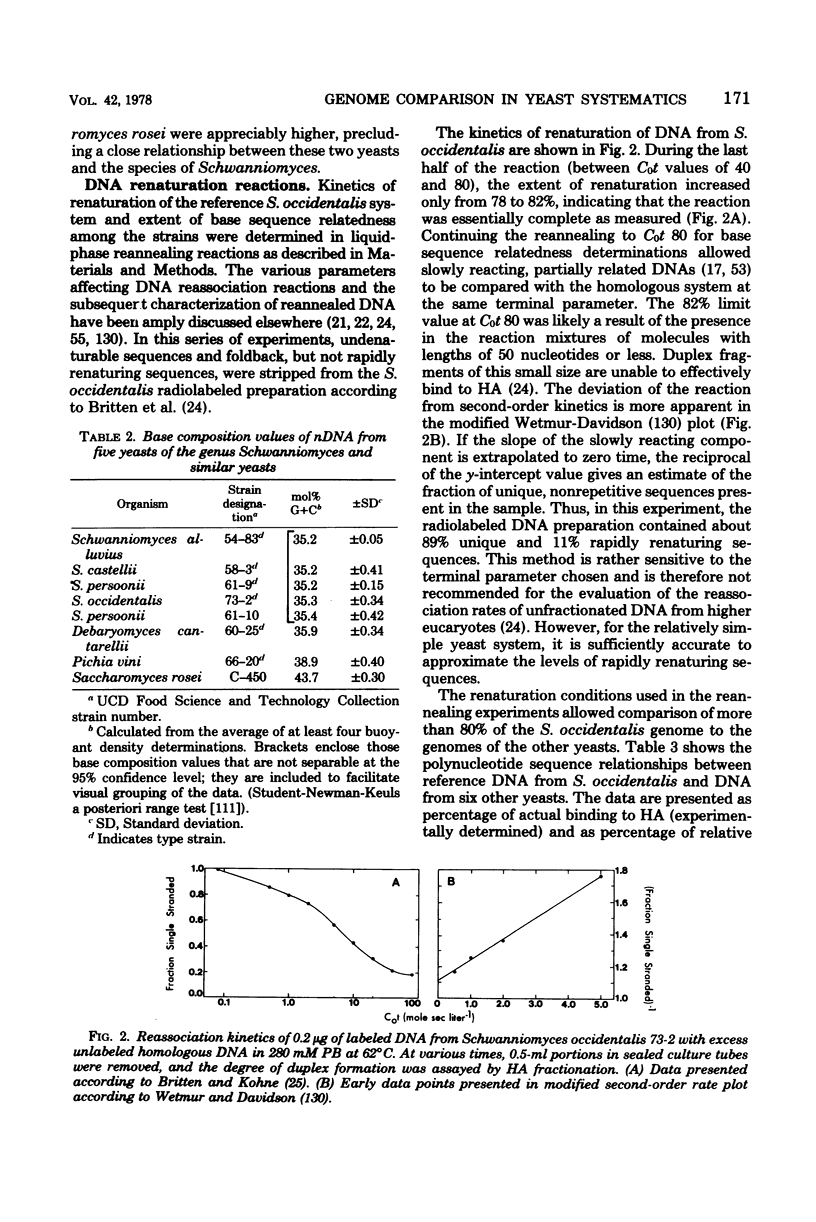
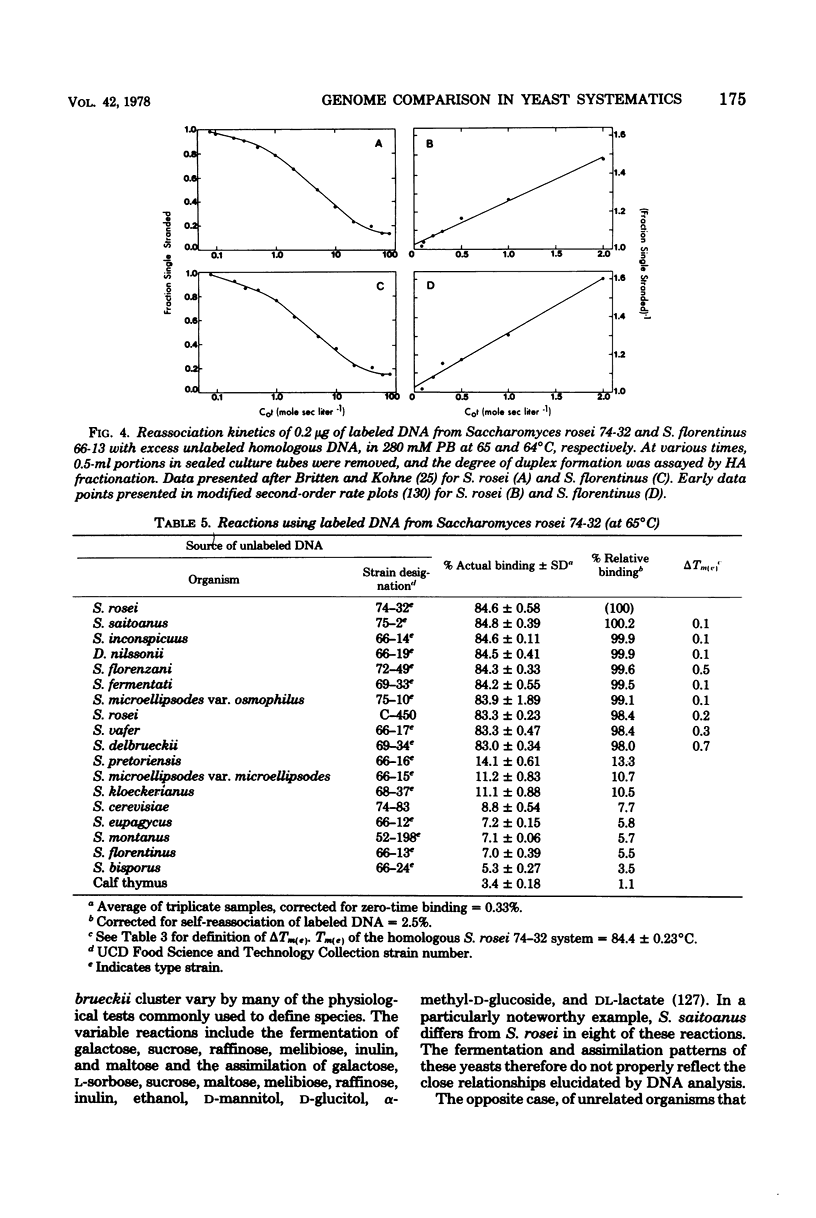
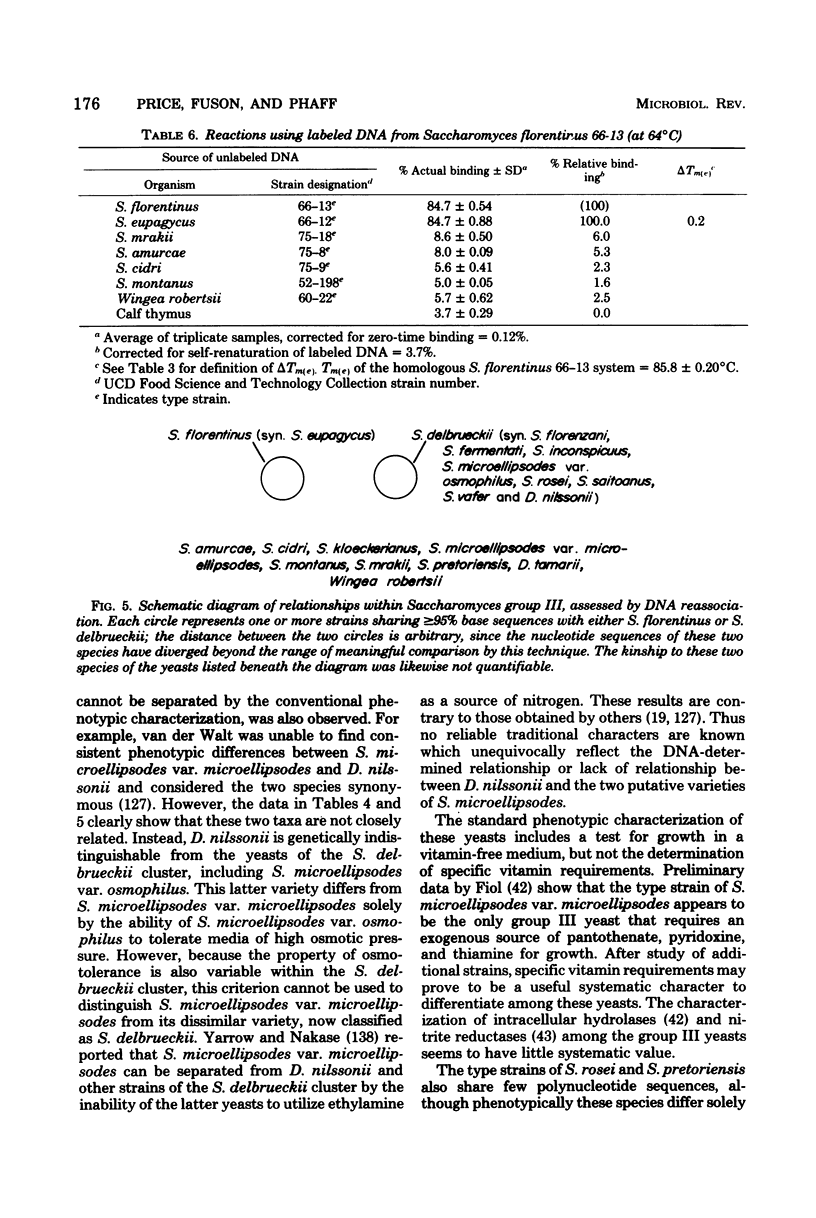
Full text
PDF



























Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BOLTON E. T., McCARTHY B. J. A general method for the isolation of RNA complementary to DNA. Proc Natl Acad Sci U S A. 1962 Aug;48:1390–1397. doi: 10.1073/pnas.48.8.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRADY B. L. UTILIZATION OF AMINO COMPOUNDS BY YEASTS OF THE GENUS SACCHAROMYCES. Antonie Van Leeuwenhoek. 1965;31:95–102. doi: 10.1007/BF02045879. [DOI] [PubMed] [Google Scholar]
- BURTON K. A study of the conditions and mechanism of the diphenylamine reaction for the colorimetric estimation of deoxyribonucleic acid. Biochem J. 1956 Feb;62(2):315–323. doi: 10.1042/bj0620315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bak A. L., Christiansen C., Christiansen G. Circular, repetitive DNA in yeast. Biochim Biophys Acta. 1972 May 29;269(3):527–530. doi: 10.1016/0005-2787(72)90144-x. [DOI] [PubMed] [Google Scholar]
- Bak A. L. DNA base composition in mycoplasma, bacteria and yeast. Curr Top Microbiol Immunol. 1973;61:89–149. doi: 10.1007/978-3-642-65531-9_3. [DOI] [PubMed] [Google Scholar]
- Bak A. L., Stenderup A. Deoxyribonucleic acid homology in yeasts. Genetic relatedness within the genus Candida. J Gen Microbiol. 1969 Nov;59(1):21–30. doi: 10.1099/00221287-59-1-21. [DOI] [PubMed] [Google Scholar]
- Ballou C. E. Some aspects of the structure, immunochemistry, and genetic control of yeast mannans. Adv Enzymol Relat Areas Mol Biol. 1974;40(0):239–270. doi: 10.1002/9780470122853.ch6. [DOI] [PubMed] [Google Scholar]
- Baptist J. N., Kurtzman C. P. Comparative enzyme patterns in Cryptococcus laurentii and its taxonomic varieties. Mycologia. 1976 Nov-Dec;68(6):1195–1203. [PubMed] [Google Scholar]
- Baptist J. N., Shaw C. R., Mandel M. Comparative zone electrophoresis of enzymes of Pseudomonas solanacearum and Pseudomonas cepacia. J Bacteriol. 1971 Nov;108(2):799–803. doi: 10.1128/jb.108.2.799-803.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baptist J. N., Shaw C. R., Mandel M. Zone electrophoresis of enzymes in bacterial taxonomy. J Bacteriol. 1969 Jul;99(1):180–188. doi: 10.1128/jb.99.1.180-188.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnet J. A., Bascomb S., Gower J. C. A maximal predictive classification of Klebsielleae and of the yeasts. J Gen Microbiol. 1975 Jan;86(1):93–102. doi: 10.1099/00221287-86-1-93. [DOI] [PubMed] [Google Scholar]
- Baumann L., Baumann P. Regulation of aspartokinase activity in the genus Beneckea and marine, luminous bacteria. Arch Mikrobiol. 1973;90(3):171–188. doi: 10.1007/BF00424970. [DOI] [PubMed] [Google Scholar]
- Baumann P., Baumann L. Biology of the marine enterobacteria: genera Beneckea and Photobacterium. Annu Rev Microbiol. 1977;31:39–61. doi: 10.1146/annurev.mi.31.100177.000351. [DOI] [PubMed] [Google Scholar]
- Bernardi G., Faures M., Piperno G., Slonimski P. P. Mitochondrial DNA's from respiratory-sufficient and cytoplasmic respiratory-deficient mutant yeast. J Mol Biol. 1970 Feb 28;48(1):23–42. doi: 10.1016/0022-2836(70)90216-0. [DOI] [PubMed] [Google Scholar]
- Bicknell J. N., Douglas H. C. Nucleic acid homologies among species of Saccharomyces. J Bacteriol. 1970 Feb;101(2):505–512. doi: 10.1128/jb.101.2.505-512.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blamire J., Cryer D. R., Finkelstein D. B., Marmur J. Sedimentation properties of yeast nuclear and mitochondrial DNA. J Mol Biol. 1972 Jun 14;67(1):11–24. doi: 10.1016/0022-2836(72)90382-8. [DOI] [PubMed] [Google Scholar]
- Bonner T. I., Brenner D. J., Neufeld B. R., Britten R. J. Reduction in the rate of DNA reassociation by sequence divergence. J Mol Biol. 1973 Dec 5;81(2):123–135. doi: 10.1016/0022-2836(73)90184-8. [DOI] [PubMed] [Google Scholar]
- Bos P., de Bruyn J. C. The significance of hydrocarbon assimilation in yeast identification. Antonie Van Leeuwenhoek. 1973;39(1):99–107. doi: 10.1007/BF02578845. [DOI] [PubMed] [Google Scholar]
- Brenner D. J., Fanning G. R., Rake A. V., Johnson K. E. Batch procedure for thermal elution of DNA from hydroxyapatite. Anal Biochem. 1969 Apr 4;28(1):447–459. doi: 10.1016/0003-2697(69)90199-7. [DOI] [PubMed] [Google Scholar]
- Britten R. J., Graham D. E., Neufeld B. R. Analysis of repeating DNA sequences by reassociation. Methods Enzymol. 1974;29:363–418. doi: 10.1016/0076-6879(74)29033-5. [DOI] [PubMed] [Google Scholar]
- Britten R. J., Kohne D. E. Repeated sequences in DNA. Hundreds of thousands of copies of DNA sequences have been incorporated into the genomes of higher organisms. Science. 1968 Aug 9;161(3841):529–540. doi: 10.1126/science.161.3841.529. [DOI] [PubMed] [Google Scholar]
- Campbell I. Methods of numerical taxonomy for various genera of yeasts. Adv Appl Microbiol. 1974;17(0):135–156. doi: 10.1016/s0065-2164(08)70557-4. [DOI] [PubMed] [Google Scholar]
- Campbell I. Numerical analysis of the genera Saccharomyces and Kluyveromyces. J Gen Microbiol. 1972 Nov;73(2):279–301. doi: 10.1099/00221287-73-2-279. [DOI] [PubMed] [Google Scholar]
- Christiansen C., Bak A. L., Stenderup A., Christiansen G. Repetitive DNA in yeasts. Nat New Biol. 1971 Jun 9;231(23):176–177. doi: 10.1038/newbio231176a0. [DOI] [PubMed] [Google Scholar]
- Clark-Walker G. D. Isolation of circular DNA from a mitochondrial fraction from yeast. Proc Natl Acad Sci U S A. 1972 Feb;69(2):388–392. doi: 10.1073/pnas.69.2.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen T. Measurement of 32P activity in a liquid scintillation counter without the use of scintillator. Anal Biochem. 1968 Jan;22(1):70–73. doi: 10.1016/0003-2697(68)90260-1. [DOI] [PubMed] [Google Scholar]
- Crombach W. H. Deep-freezing of bacterial DNA for thermal denaturation and hybridization experiments. Antonie Van Leeuwenhoek. 1973;39(2):249–255. doi: 10.1007/BF02578857. [DOI] [PubMed] [Google Scholar]
- Davidson E. H., Hough B. R., Amenson C. S., Britten R. J. General interspersion of repetitive with non-repetitive sequence elements in the DNA of Xenopus. J Mol Biol. 1973 Jun 15;77(1):1–23. doi: 10.1016/0022-2836(73)90359-8. [DOI] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Dutta S. K. DNA homologies among heterothallic species of Neurospora. Mycologia. 1976 Mar-Apr;68(2):388–401. [PubMed] [Google Scholar]
- Dutta S. K., Ojha M. Relatedness between major taxonomic groups of fungi based on the measurement of DNA nucleotide sequence homology. Mol Gen Genet. 1972;114(3):232–240. doi: 10.1007/BF01788892. [DOI] [PubMed] [Google Scholar]
- Dutta S. K., Sheikh I., Choppala J., Aulakh G. S., Nelson W. H. DNA homologies among homothallic, pseudo-homothallic and heterothallic species of Neurospora. Mol Gen Genet. 1976 Sep 23;147(3):325–330. doi: 10.1007/BF00582884. [DOI] [PubMed] [Google Scholar]
- EATON N. R. New press for disruption of microorganisms. J Bacteriol. 1962 Jun;83:1359–1360. doi: 10.1128/jb.83.6.1359-1360.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erke K. H., Schneidau J. D., Jr Relationship of some Cryptococcus neoformans hypha-forming strains to standard strains and to other species of yeasts as determined by deoxyribonucleic acid base ratios and homologies. Infect Immun. 1973 Jun;7(6):941–948. doi: 10.1128/iai.7.6.941-948.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiol J. B. A critical study of the taxonomic value of some tests of assimilation used for the classification of the sporogenous yeasts. Mycopathologia. 1975 Dec 23;57(2):79–88. doi: 10.1007/BF01365708. [DOI] [PubMed] [Google Scholar]
- Fiol J. B. Systématique des Saccharomyces: osidases et besoins vitaminiques. Mycopathologia. 1976 Jun 4;58(1):49–58. doi: 10.1007/BF00493593. [DOI] [PubMed] [Google Scholar]
- Gillespie D., Spiegelman S. A quantitative assay for DNA-RNA hybrids with DNA immobilized on a membrane. J Mol Biol. 1965 Jul;12(3):829–842. doi: 10.1016/s0022-2836(65)80331-x. [DOI] [PubMed] [Google Scholar]
- Gruenwedel D. W., Hsu C. H., Lu D. S. The effects of aqueous neutral-salt solutions on the melting temperatures of deoxyribonucleic acids. Biopolymers. 1971;10(1):47–68. doi: 10.1002/bip.360100106. [DOI] [PubMed] [Google Scholar]
- Hartwell L. H. Saccharomyces cerevisiae cell cycle. Bacteriol Rev. 1974 Jun;38(2):164–198. doi: 10.1128/br.38.2.164-198.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenberg C. P., Borst P., van Bruggen E. F. Mitochondrial DNA. V. A 25 micron closed circular duplex DNA molecule in wild-type yeast mitochondria. Stucture and genetic complexity. Biochim Biophys Acta. 1970 May 21;209(1):1–15. [PubMed] [Google Scholar]
- Hough B. R., Davidson E. H. Studies on the repetitive sequence transcripts of Xenopus oocytes. J Mol Biol. 1972 Oct 14;70(3):491–509. doi: 10.1016/0022-2836(72)90555-4. [DOI] [PubMed] [Google Scholar]
- Hutton J. R., Wetmur J. G. Effect of chemical modification on the rate of renaturation of deoxyribonucleic acid. Deaminated and glyoxalated deoxyribonucleic acid. Biochemistry. 1973 Jan 30;12(3):558–563. doi: 10.1021/bi00727a032. [DOI] [PubMed] [Google Scholar]
- Kohne D. E. Evolution of higher-organism DNA. Q Rev Biophys. 1970 Aug;3(3):327–375. doi: 10.1017/s0033583500004765. [DOI] [PubMed] [Google Scholar]
- Kowalski S., Fresco J. R. Preparation of highly labeled (32P)nucleic acids from yeasts; isolation of "denaturable" leucine acceptor transfer RNA. Science. 1971 Apr 23;172(3981):384–385. doi: 10.1126/science.172.3981.384. [DOI] [PubMed] [Google Scholar]
- Kreger van Rij N. J., Veenhuis M. Electron microscopy of ascus formation in the yeast debaryomyces hansenii. J Gen Microbiol. 1975 Aug;89(2):256–264. doi: 10.1099/00221287-89-2-256. [DOI] [PubMed] [Google Scholar]
- Kreger-van Rij N. J. Electron microscopy of sporulation in Schwanniomyces alluvius. Antonie Van Leeuwenhoek. 1977;43(1):55–64. doi: 10.1007/BF02316210. [DOI] [PubMed] [Google Scholar]
- Kreger-van Rij N. J., Veenhuis M. Ultrastructure of the ascospores of some species of the Torulaspora group. Antonie Van Leeuwenhoek. 1976;42(4):445–455. doi: 10.1007/BF00410175. [DOI] [PubMed] [Google Scholar]
- Kurtzman C. P., Smiley M. J., Baker F. L. Scanning electron microscopy of ascospores of Debaryomyces and Saccharomyces. Mycopathologia. 1975 Feb 28;55(1):29–34. doi: 10.1007/BF00467088. [DOI] [PubMed] [Google Scholar]
- Kurtzman C. P., Smiley M. J., Baker F. L. Scanning electron microscopy of ascospores of Schwanniomyces. J Bacteriol. 1972 Dec;112(3):1380–1382. doi: 10.1128/jb.112.3.1380-1382.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- LaRue T. A., Spencer J. F. Utilization of organic nitrogen compounds by yeasts of the genus Saccharomyces. Antonie Van Leeuwenhoek. 1968;34(2):153–158. doi: 10.1007/BF02046425. [DOI] [PubMed] [Google Scholar]
- Lam K. B., Marmur J. Isolation and characterization of Saccharomyces cerevisiae glycolytic pathway mutants. J Bacteriol. 1977 May;130(2):746–749. doi: 10.1128/jb.130.2.746-749.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- London J., Kline K. Aldolase of lactic acid bacteria: a case history in the use of an enzyme as an evolutionary marker. Bacteriol Rev. 1973 Dec;37(4):453–478. doi: 10.1128/br.37.4.453-478.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARMUR J., DOTY P. Determination of the base composition of deoxyribonucleic acid from its thermal denaturation temperature. J Mol Biol. 1962 Jul;5:109–118. doi: 10.1016/s0022-2836(62)80066-7. [DOI] [PubMed] [Google Scholar]
- Mandel M. New approaches to bacterial taxonomy: perspective and prospects. Annu Rev Microbiol. 1969;23:239–274. doi: 10.1146/annurev.mi.23.100169.001323. [DOI] [PubMed] [Google Scholar]
- Martini A., Phaff H. J., Douglass S. A. Deoxyribonucleic acid base composition of species in the yeast genus Kluyveromyces van der Walt emend. van der Walt. J Bacteriol. 1972 Aug;111(2):481–487. doi: 10.1128/jb.111.2.481-487.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinson H. G. The nucleic acid-hydroxylapatite interaction. II. Phase transitions in the deoxyribonucleic acid-hydroxylapatite system. Biochemistry. 1973 Jan 2;12(1):145–150. doi: 10.1021/bi00725a024. [DOI] [PubMed] [Google Scholar]
- Martinson H. G., Wagenaar E. B. The effect of the amount of nucleic acid load on hydroxylapatite chromatography. Can J Biochem. 1974 Mar;52(3):267–271. doi: 10.1139/o74-041. [DOI] [PubMed] [Google Scholar]
- Martinson H. G., Wagenaar E. B. Thermal elution chromatography and the resolution of nucleic acids on hydroxylapatite. Anal Biochem. 1974 Sep;61(1):144–154. doi: 10.1016/0003-2697(74)90341-8. [DOI] [PubMed] [Google Scholar]
- Martinson H. G., Wagenaar E. B. Thermal elution chromatography of nucleic acids on hydroxyapatite. Biochim Biophys Acta. 1977 Feb 3;474(3):445–455. doi: 10.1016/0005-2787(77)90273-8. [DOI] [PubMed] [Google Scholar]
- Messenguy F., Wiame J. -M. The control of ornithinetranscarbamylase activity by arginase in Saccharomyces cerevisiae. FEBS Lett. 1969 Apr;3(1):47–49. doi: 10.1016/0014-5793(69)80093-1. [DOI] [PubMed] [Google Scholar]
- Meyer S. A., Anderson K., Brown R. E., Smith M. T., Yarrow D., Mitchell G., Ahearn D. G. Physiological and DNA characterization of Candida maltosa, a hydrocarbon-utilizing yeast. Arch Microbiol. 1975 Aug 28;104(3):225–231. doi: 10.1007/BF00447328. [DOI] [PubMed] [Google Scholar]
- Moore R. L. Nucleic acid reassociation as a guide to genetic relatedness among bacteria. Curr Top Microbiol Immunol. 1974;64(0):105–128. doi: 10.1007/978-3-642-65848-8_4. [DOI] [PubMed] [Google Scholar]
- O'Connor R. M., McArthur C. R., Clark-Walker G. D. Respiratory-deficient mutants of Torulopsis glabrata, a yeast with circular mitochondrial deoxyribonucleic acid of 6 mu m. J Bacteriol. 1976 May;126(2):959–968. doi: 10.1128/jb.126.2.959-968.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojha M., Dutta S. K., Turian G. DNA nucleotide sequence homologies between some zoosporic fungi. Mol Gen Genet. 1975;136(2):151–165. doi: 10.1007/BF00272036. [DOI] [PubMed] [Google Scholar]
- Palleroni N. J., Ballard R. W., Ralston E., Doudoroff M. Deoxyribonucleic acid homologies among some Pseudomonas species. J Bacteriol. 1972 Apr;110(1):1–11. doi: 10.1128/jb.110.1.1-11.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petes T. D., Byers B., Fangman W. L. Size and structure of yeast chromosomal DNA. Proc Natl Acad Sci U S A. 1973 Nov;70(11):3072–3076. doi: 10.1073/pnas.70.11.3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle J. R. Methods for avoiding proteolytic artefacts in studies of enzymes and other proteins from yeasts. Methods Cell Biol. 1975;12:149–184. doi: 10.1016/s0091-679x(08)60956-5. [DOI] [PubMed] [Google Scholar]
- Richards M. Serology and yeast classification. Antonie Van Leeuwenhoek. 1972;38(2):177–192. doi: 10.1007/BF02328090. [DOI] [PubMed] [Google Scholar]
- STUDIER F. W. SEDIMENTATION STUDIES OF THE SIZE AND SHAPE OF DNA. J Mol Biol. 1965 Feb;11:373–390. doi: 10.1016/s0022-2836(65)80064-x. [DOI] [PubMed] [Google Scholar]
- Seidler R. J., Mandel M. Quantitative aspects of deoxyribonucleic acid renaturation: base composition, state of chromosome replication, and polynucleotide homologies. J Bacteriol. 1971 May;106(2):608–614. doi: 10.1128/jb.106.2.608-614.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. T., Simione F. P., Jr, Meyer S. A. Kloeckera apis st. nov.; the imperfect state of Hanseniaspora guilliermondii Pijper. Antonie Van Leeuwenhoek. 1977;43(2):219–223. doi: 10.1007/BF00395676. [DOI] [PubMed] [Google Scholar]
- Spencer J. F., Gorin P. A., Rank G. H. The genetic control of the two types of mannan produced by Saccharomyces cerevisiae. Can J Microbiol. 1971 Nov;17(11):1451–1454. doi: 10.1139/m71-230. [DOI] [PubMed] [Google Scholar]
- Stanier R. Y., Palleroni N. J., Doudoroff M. The aerobic pseudomonads: a taxonomic study. J Gen Microbiol. 1966 May;43(2):159–271. doi: 10.1099/00221287-43-2-159. [DOI] [PubMed] [Google Scholar]
- Stanier R. Y., Wachter D., Gasser C., Wilson A. C. Comparative immunological studies of two Pseudomonas enzymes. J Bacteriol. 1970 May;102(2):351–362. doi: 10.1128/jb.102.2.351-362.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya T., Fukazawa Y., Taguchi M., Nakase T., Shinoda T. Serologic aspects on yeast classification. Mycopathol Mycol Appl. 1974 Aug 30;53(1):77–91. doi: 10.1007/BF02127199. [DOI] [PubMed] [Google Scholar]
- Ullman J. S., McCarthy B. J. The relationship between mismatched base pairs and the thermal stability of DNA duplexes. I. Effects of depurination and chain scission. Biochim Biophys Acta. 1973 Feb 4;294(1):405–415. doi: 10.1016/0005-2787(73)90095-6. [DOI] [PubMed] [Google Scholar]
- Urrestarazu L. A., Vissers S., Wiame J. M. Change in location of ornithine carbamoyltransferase and carbamoylphosphate synthetase among yeasts in relation to the arginase/ornithine carbamoyltransferase regulatory complex and the energy status of the cells. Eur J Biochem. 1977 Oct 3;79(2):473–481. doi: 10.1111/j.1432-1033.1977.tb11830.x. [DOI] [PubMed] [Google Scholar]
- Wetmur J. G., Davidson N. Kinetics of renaturation of DNA. J Mol Biol. 1968 Feb 14;31(3):349–370. doi: 10.1016/0022-2836(68)90414-2. [DOI] [PubMed] [Google Scholar]
- Wickerham L. J., Burton K. A. Carbon Assimilation Tests for the Classification of Yeasts. J Bacteriol. 1948 Sep;56(3):363–371. doi: 10.1128/jb.56.3.363-371.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A. C., Carlson S. S., White T. J. Biochemical evolution. Annu Rev Biochem. 1977;46:573–639. doi: 10.1146/annurev.bi.46.070177.003041. [DOI] [PubMed] [Google Scholar]
- Yamada Y., Arimoto M., Kondo K. Coenzyme Q system in the classification of some ascosporogenous yeast genera in the families Saccharomycetaceae and Spermophthoraceae. Antonie Van Leeuwenhoek. 1977;43(1):65–71. doi: 10.1007/BF02316211. [DOI] [PubMed] [Google Scholar]
- Yarrow D., Nakase T. DNA base composition of species of the genus Saccharomyces. Antonie Van Leeuwenhoek. 1975;41(1):81–88. doi: 10.1007/BF02565038. [DOI] [PubMed] [Google Scholar]
- van der WALT J. Utilization of ethylamine by yeasts. Antonie Van Leeuwenhoek. 1962;28:91–96. doi: 10.1007/BF02538727. [DOI] [PubMed] [Google Scholar]
- van der Walt J. P., Taylor M. B., Liebenberg N. V. Ploidy, ascus formation and recombination in Torulaspora (Debaryomyces) hansenii. Antonie Van Leeuwenhoek. 1977;43(2):205–218. doi: 10.1007/BF00395675. [DOI] [PubMed] [Google Scholar]


