Abstract
Despite much work, sub-cellular neurons of Caenorhabditis elegans have not been studied at nanometer resolution with millisecond time resolution. Nor has there been an effective way to immobilize C. elegans. Here we show that, without using anesthetic or paralyzing agents, Fluorescence Imaging with One Nanometer Accuracy (FIONA) can be successfully applied to fluorescently labeled molecules within C. elegans nerves. GFP- and DENDRA2-labeled ELKS punctae can be localized with sub-10 nm accuracy in ~5 milliseconds. Our results show that the protein ELKS is occasionally transferred by microtubule-based motors. This is the first example of FIONA applied to a living organism.
Because of its transparent body and overall simplicity, Caenorhabditis elegans have been used as a model organism in medicine, physiology and neuroscience (1). For example, the nervous system is just 302 neurons and approximately 7000 synapses. But the ability to localize various proteins with nanometer accuracy and millisecond time resolution, as was done previously, in vitro(2) and in vivo, i.e., in cultured mammalian cells(3), has not been done in situ, i.e. in a living organism.
We have chosen to isolate the protein ELKS, which is found in the six mechanosensory touch neurons. ELKS (which is named for its amino acid composition) plays a role in regulating synaptic development and co-localizes with other synaptic active proteins at synapses (4). This can be seen when other synaptogenic pathways are compromised(5), although many of the features of ELKS are not well established. For our use, ELKS was fused with either green fluorescent protein (GFP) or DENDRA2 (a photoconvertible fluorescent protein)(6). (See supplemental material for details about constructs.)
The three criteria for being able to perform in situ FIONA are met with GFP::ELKS. First, previous studies have shown that ELKS fused with GFP and its derivatives can be observed as very bright puncta in C. elegans neurons both in processes and at synapses(5). The ELKS process puncta may represent an invertebrate counterpart to active zone precursor vesicles that numerous vertebrate active zone proteins have been shown associate with during axonal transport to the synapses (7). Second, all of the C. elegans touch receptor neurons are in very close proximity to the cuticle (~100 nm)(8) -- the neuronal processes are aligned directly adjacent to the coverslip. This makes it possible to excite them with regular TIR microscopy and thus achieve high signal-to-noise ratio. Third, we have come up with a way to immobilize C. elegans.
The existing procedure for immobilization is to glue the worms to glass. Unfortunately, this takes a considerable amount of time (around two minutes per worm) since the worms have to be glued one at a time with cyanoacrylate glue(9). Another technique is to anesthetize the worms with sodium azide(10). However, this drastically alters the metabolism of worms, especially in the presence of ATP (11, 12). Other drugs such as levimasole and muscimol are also used for immobilizing C elegans(13). They stabilize the worms to a great extent, but there are still some spasmodic movements that make it impossible to perform nanoscale recordings.
We have used a straightforward technique to stabilize multiple worms simultaneously in the absence of any paralyzing drugs or microfluidic devices (14, 15). In our experiments the worms are pipetted away from the NGM (Nematode Growth Media) plates into Eppendorf tubes with M9 buffer (3 g of KH2PO4, 6 g of, Na2HPO4, 5 g of NaCl, 1 ml MgSO4 (1M), 1 L deionized water). 5–10 µliters of buffer (containing the worms) is put on a 2” glass bottom dish (WillCo Wells BV, GWST 5040, Amsterdam). A coverslip (Fisher Sci., 12-548-5A, 30 × 22 mm) is put on top of the drop in order to sandwich the worms between the two surfaces as the buffer disperses due to the capillary effect. Around 10 minutes later, as the water evaporates from the edges, the distance between the two surfaces gets narrower, immobilizing ~90 % of the worms (thicker worms; adults and L4s, can be stabilized instantly). After this point, evaporation has to be slowed down by closing the lid of the dish since excessive pressure turns out to be lethal for adult and L4 stage worms. When the pressure is released by carefully adding buffer to the dish, the worms start moving again, verifying their vitality.
In order to test how effectively this procedure can immobilize the worms, we have used a strain which is cultured in NGM plates that were seeded with 1 µm diameter fluorescent beads (FluoSpheres F8819, Molecular Probes/ 1000× diluted, 1 mL for a 9 cm diameter plate). In Fig. 1 the beads swallowed by the worms decorate the intestinal lumen and can be used as stable markers for determining the positions of the worms with high accuracy. After sandwiching the worms we have imaged the fluorescent beads that are trapped in the lumen with epifluorescence. We observed that the beads move extremely slow (~300 nm over a minute) suggesting that the immobilization is very strong. The fact that the intestinal lumen is situated microns above the coverslip shows that the whole animal is immobilized. After the recording, we have released the pressure to verify the vitality (see the supplementary movie). Also, this technique can immobilize the worms that were previously treated by levamisole or muscimol, enabling high-resolution recordings in the absence of spasmodic movements. Previously, a similar approach was used to take pictures of formaldehyde-fixed (dead) C. elegans at different orientations where a few Sephadex G-100 particles between the two surfaces were used to glide the coverglass gently to roll the animals to the desired position(16).
Figure 1.
Movement of fluorescent beads in immobilized C. elegans (at the L2 stage). A) Beads at the anterior side are shown in the inset. The beads move ~300 nm at maximum in 65 seconds. B) Beads at the posterior side are shown in the inset. The beads move ~100 nm at maximum in 65 seconds. Beads move in different directions and different amounts showing that the movement is not due to a stage drift but the movement of the worm itself. See the supplementary movie showing how worms start moving when the pressure is released.
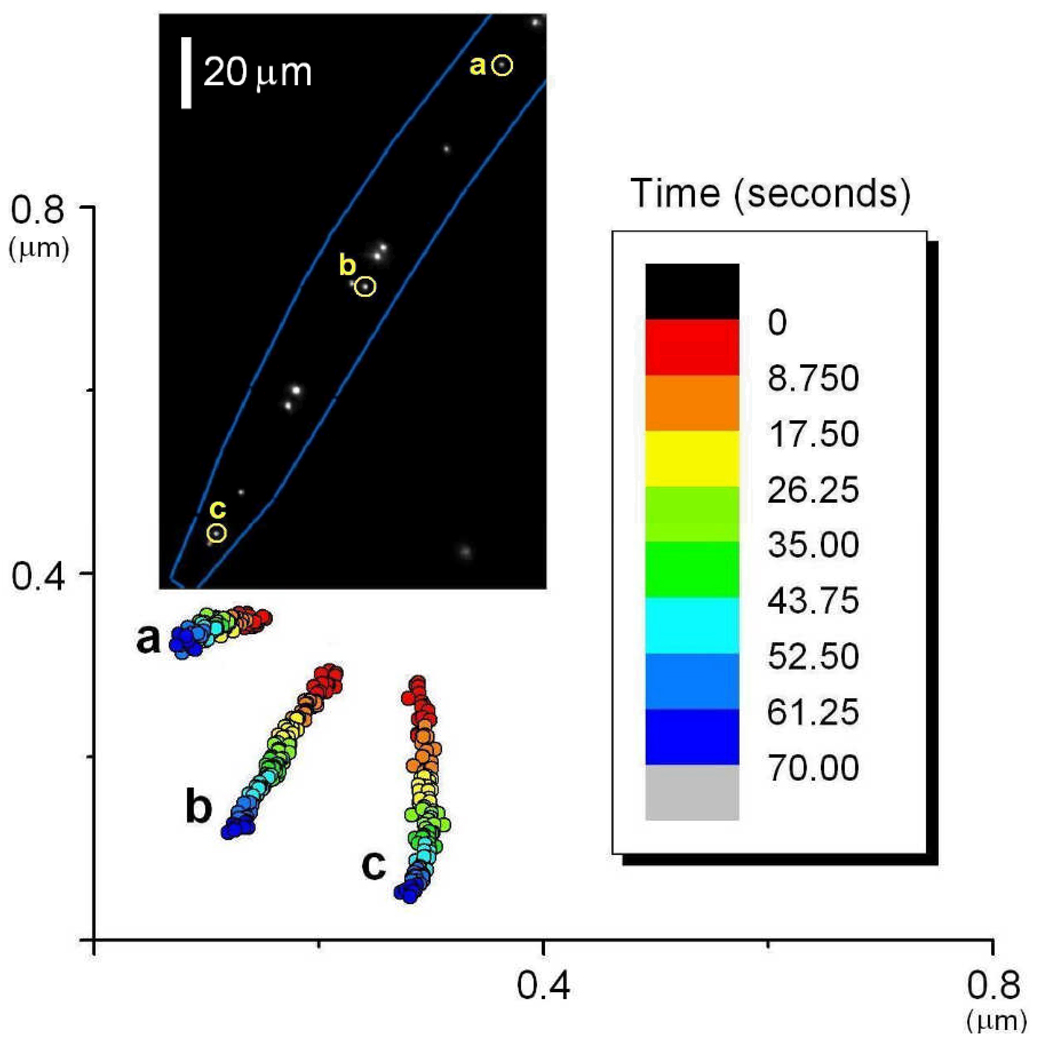
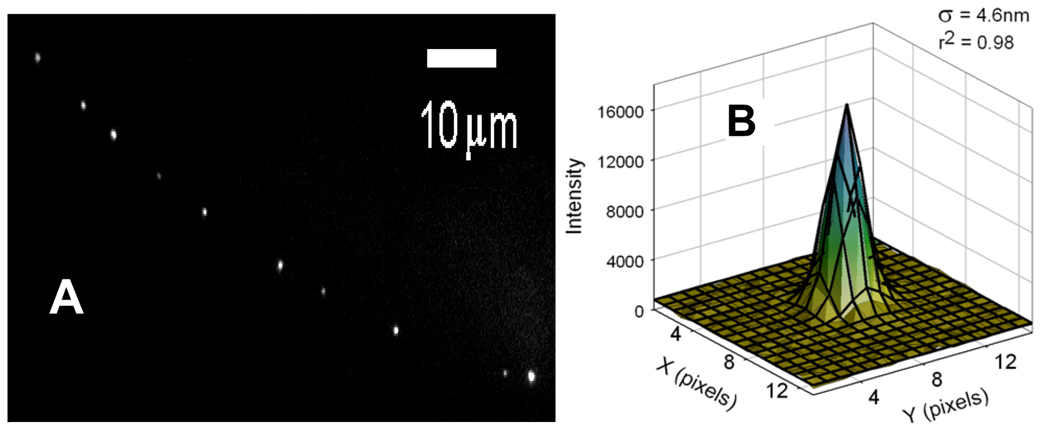
We find that fluorescently-labeled ELKS punctae are found in the form of bright clumps that are distributed with a distance of ~5 microns between them within the neuronal processes of the touch neurons (Fig. 2a). We also find that fluorescent punctae found in touch neurons of live C. elegans can be tracked by FIONA with sub-10 nm spatial resolution within ~5 milliseconds (Fig. 2b). This is similar, though not quite as good, as FIONA detected on GFP-labeled organelles in cultured cells, where we got 1.5 nm and 1 msec resolution.
Figure 2.
A) Stable DENDRA2::ELKS spots that are distributed in a punctual way along the touch neuron process. B) Two-dimensional Gaussian fit to the emission pattern of a single DENDRA2::ELKS puncta. B) The peak can be localized within 4.6 nm in 4 msecs.
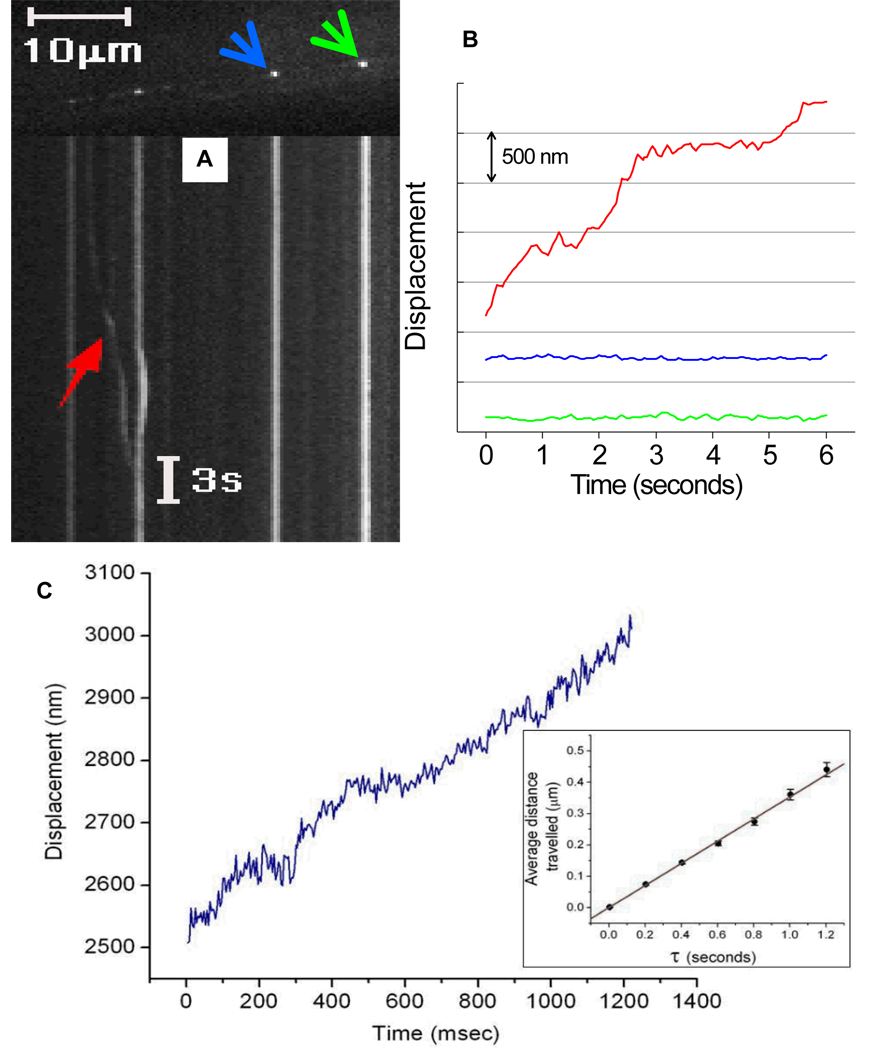
We have observed that GFP::ELKS punctae are motionless most of the time. However we have recorded some rare, long-range directed movements (Fig. 3a). This suggests that, at times, GFP::ELKS punctae are intact and can move as a whole within the axon. The high-resolution tracking of moving GFP::ELKS spots shows that the average travelled distance increases linearly over time, with an average velocity of 0.35 µm/sec (Fig. 3b). This shows that the spots are not diffusing but are being transported by molecular motors.
Figure 3.
A) Kymograph showing the movement of a GFP::ELKS puncta (shown with red arrow). Nonmoving punctae (shown with blue and green arrows) found in the same process confirm that the touch neuron is motionless during the movie. B) Traces of puncta shown in A. Red, blue and green traces correspond to the kymograph shown in red arrow and the spots shown with blue and green arrows, respectively. C) Trace of a GFP::ELKS spot imaged with 4 msec temporal resolution. Average distance travelled shown in inset.
We find that GFP::ELKS spots are distributed in a surprisingly even way along the microtubule cells. Except for a few cases, we observed that these bright spots do not change their positions in the neuronal processes. We used several different mutant strains of C. elegans to try and understand why the distribution of ELKS spots does not change. The first is a mec-7 mutant, in which the 15-protofilament microtubules are replaced by 11 protofilaments. Mechanosensory neurons have microtubule bundles of 15 protofilaments rather than 11 protofilaments which are found in all other microtubules of the worm. When the 15-protofilament microtubules are disrupted, or are replaced by 11 protofilament microtubules by the mutation of mec-7 gene (MEChanosensory abnormality), the touch sensitivity is lost(17). The second strain is mec-4 mutants in which degenerin-channel formation is disrupted. Mechanosensory transduction in microtubule cells are assumed to be mediated by degenerin/epithelial Na+ channels(18), which are believed to be encoded by mechanosensory abnormality genes; mec-4, mec-10 and mec-6(19). Previous work have revealed that MEC-4 proteins are distributed in a punctuate pattern (20) indicating that the degenerin-channels are assembled heterogeneously along the touch neuron processes. This punctate distribution is dependent on the MEC-1 and MEC-5 extracellular matrix proteins which are thought to link the degenerin-channel complexes to the epidermis (cuticle). We have used the mec-1, mec-4, mec-5 mutant strain to observe if ELKS punctae are associated with degenerin-channel clusters.
We have applied in situ FIONA tracking on DENDRA2::ELKS spots in both mec-7 and mec-4 mutants. In these two strains we have observed no significant difference either of the mobility, or of the distribution of ELKS punctae. Furthermore, epi-fluorescence microscopy revealed that GFP::ELKS puncta were not differentially distributed in mec-1 or mec-5 mutants in contrast to that which is observed for the degenerin channels. These finding imply that ELKS dynamics does not depend on 15-protofilement microtubule structure or the localization of degenerin-channels.
In conclusion, our findings are an important step in extending the current techniques for high-resolution real-time imaging of cultured cells to a widely used multi-cellular model organism. We have shown that fluorescently-labeled ELKS proteins accumulated in touch neurons of live C. elegans can be tracked within sub-10 nm in a few milliseconds using the technique of FIONA. Furthermore, C. elegans can be immobilized without affecting the biochemistry of the nematode for high-resolution imaging experiments. Generally, ELKS punctae positions are stable. On the other hand, seldom long-range directed movements mediated by motor proteins imply that either the ELKS proteins are aggregated on membranous vesicles that are carried by motor proteins or are a part of some complexes which interact with molecular motors. Our results suggest that replacing the 15 protofilament microtubules with 11 protofilaments does not affect the motor activity on the ELKS punctae in touch neurons. Moreover, ELKS punctae distribution is not affected by the loss of degenerin channels or extracellular matrix components required for degenerin channel clustering, suggesting that ELKS does not interact with mechanosensory channels.
Supplementary Material
Acknowledgments
This work was support by NSF DBI 06-49779 and NIH AR44420 to PRS and NIH NS040094 to MN.
Footnotes
Supplemental materials about ELKS constructs may be accessed free of charge online at http://pubs.acs.org.
References
- 1.Riddle D, Blumenthal T, Mayer B, Priess J. C. elegans II. New York: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 2.Yildiz A, Forkey JN, McKinney SA, Ha T, Goldman YE, Selvin PR. Myosin V walks hand-over-hand: single fluorophore imaging with 1.5-nm localization. Science. 2003;300:2061–2065. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- 3.Kural C, Kim H, Syed S, Goshima G, Gelfand VI, Selvin PR. Kinesin and dynein move a peroxisome in vivo: a tug-of-war or coordinated movement? Science. 2005;308:1469–1472. doi: 10.1126/science.1108408. [DOI] [PubMed] [Google Scholar]
- 4.Deken SL, Vincent R, Hadwiger G, Liu Q, Wang ZW, Nonet ML. Redundant localization mechanisms of RIM and ELKS in Caenorhabditis elegans. J Neurosci. 2005;25:5975–5983. doi: 10.1523/JNEUROSCI.0804-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dai Y, Taru H, Deken SL, Grill B, Ackley B, Nonet ML, Jin Y. SYD-2 Liprin-alpha organizes presynaptic active zone formation through ELKS. Nat Neurosci. 2006;9:1479–1487. doi: 10.1038/nn1808. [DOI] [PubMed] [Google Scholar]
- 6.Chudakov DM, Lukyanov S, Lukyanov KA. Tracking intracellular protein movements using photoswitchable fluorescent proteins PS-CFP2 and Dendra2. Nat Protoc. 2007;2:2024–2032. doi: 10.1038/nprot.2007.291. [DOI] [PubMed] [Google Scholar]
- 7.Zhai RG, Vardinon-Friedman H, Cases-Langhoff C, Becker B, Gundelfinger ED, Ziv NE, Garner CC. Assembling the presynaptic active zone: a characterization of an active one precursor vesicle. Neuron. 2001;29:131–143. doi: 10.1016/s0896-6273(01)00185-4. [DOI] [PubMed] [Google Scholar]
- 8.Chalfie M. The differentiation and function of the touch receptor neurons of Caenorhabditis elegans. Prog. Brain Res. 1995;105:179–182. doi: 10.1016/s0079-6123(08)63293-8. [DOI] [PubMed] [Google Scholar]
- 9.Hilliard MA, Apicella AJ, Kerr R, Suzuki H, Bazzicalupo P, Schafer WR. In vivo imaging of C. elegans ASH neurons: cellular response and adaptation to chemical repellents. Embo J. 2005;24:63–72. doi: 10.1038/sj.emboj.7600493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sulston J, Hodgkin J. The nematode Caenorhabditis elegans. Plainview, New York: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 11.Duncan HM, Mackler B. Electron transport systems of yeast. 3. Preparation and properties of cytochrome oxidase. J Biol Chem. 1966;241:1694–1697. [PubMed] [Google Scholar]
- 12.Van der Bend RL, Duetz W, Colen AM, Van Dam K, Berden JA. Differential effects of triphenyltin and 8-azido-ATP on the ATP synthesis, ATP-Pi exchange, and ATP hydrolysis in liposomes containing ATP synthase and bacteriorhodopsin. Arch Biochem Biophys. 1985;241:461–471. doi: 10.1016/0003-9861(85)90571-5. [DOI] [PubMed] [Google Scholar]
- 13.Lewis JA, Wu CH, Berg H, Levine JH. The genetics of levamisole resistance in the nematode Caenorhabditis elegans. Genetics. 1980;95:905–928. doi: 10.1093/genetics/95.4.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rohde CB, Zeng F, Gonzalez-Rubio R, Angel M, Yanik MF. Microfluidic system for on-chip high-throughput whole-animal sorting and screening at subcellular resolution. Proc Natl Acad Sci U S A. 2007;104:13891–13895. doi: 10.1073/pnas.0706513104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hulme SE, Shevkoplyas SS, Apfeld J, Fontana W, Whitesides GM. A microfabricated array of clamps for immobilizing and imaging C. elegans. Lab Chip. 2007;7:1515–1523. doi: 10.1039/b707861g. [DOI] [PubMed] [Google Scholar]
- 16.Hedgecock EM, Culotti JG, Hall DH. The unc-5, unc-6, and unc-40 genes guide circumferential migrations of pioneer axons and mesodermal cells on the epidermis in C. elegans. Neuron. 1990;4:61–85. doi: 10.1016/0896-6273(90)90444-k. [DOI] [PubMed] [Google Scholar]
- 17.Chalfie M, Thomson JN. Structural and functional diversity in the neuronal microtubules of Caenorhabditis elegans. J Cell Biol. 1982;93:15–23. doi: 10.1083/jcb.93.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ernstrom GG, Chalfie M. Genetics of sensory mechanotransduction. Annu Rev Genet. 2002;36:411–453. doi: 10.1146/annurev.genet.36.061802.101708. [DOI] [PubMed] [Google Scholar]
- 19.Huang M, Chalfie M. Gene interactions affecting mechanosensory transduction in Caenorhabditis elegans. Nature. 1994;367:467–470. doi: 10.1038/367467a0. [DOI] [PubMed] [Google Scholar]
- 20.Chelur DS, Ernstrom GG, Goodman MB, Yao CA, Chen L, R OH, Chalfie M. The mechanosensory protein MEC-6 is a subunit of the C. elegans touch-cell degenerin channel. Nature. 2002;420:669–673. doi: 10.1038/nature01205. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.