Imprinting is a form of epigenetic gene regulation whereby the expression of an allele is dictated by parental origin. This parental legacy is established in the germ-line via heritable epigenetic modifications such as DNA methylation1 that are maintained throughout the somatic development of offspring. Imprinting is important in aspects of growth and development in mammals,2 flowering plants3 and in genetic disease,4 but in the context of this review, imprinted genes provide a model for dissecting epigenetic mechanisms because the active and silent alleles are in the same cell. In the presence of an identical complement of trans-regulatory factors, allelic differences in transcription are likely to be largely attributable to epigenetic factors. However, imprinting is historically associated with influences on transcription purely at the point of transcript initiation, where maternally and paternally derived alleles of genes are differentially active, allele-specific differences in termination processes have not been previously reported. We discuss how alternative polyadenylation (poly(A)) sites at the imprinted H13 gene are utilized in an allele-specific way and how other imprinted loci behave similarly.
There are a large number of poly(A) sites in the mammalian genome5 and many human genes use more than one poly(A) site.6 Epigenetic modifications can influence transcription initiation by altering the accessibility of promoter sequences to initiation complex components.7,8 Regions of heterochromatin can subtly impede the progress of an elongating polymerase complex regulating transcriptional output.9 However, it has not been shown that epigenetic modifications downstream of a promoter can lead to variant gene products through alternative poly(A) or splicing.
To study epigenetic regulation at imprinted loci, we focused on what are termed ‘intronic-host’ imprinted gene loci. These loci are organised such that an imprinted ‘intronic’ gene with a differentially methylated region (DMR) at the promoter resides in the intron of another gene known as the ‘host’ (Fig. 1). The host generally falls victim to imprinted expression as a consequence of intronic gene imprinting, but this victimisation does not extend beyond the two genes. This is in contrast to the organisation of the more complex imprinted domains, within which multiple mechanisms can act.10-14 The systematic dissection of multi-layered imprinting mechanisms at complex loci is important for understanding regulatory mechanisms genome-wide, however the study of small, less-complex intronic-host imprinted gene pairs can provide insight into the minimum features necessary and sufficient for imprinting, and facilitate the study of epigenetic regulatory mechanisms.
Figure 1.
Genes in an intronic (green)—host (blue) configuration.
How might a gene come to be resident in the intron of another functionally unrelated gene in an intronic-host configuration? Probably the most common method is by retrotransposition of the intronic gene from elsewhere in the genome. This can occur through proteins encoded by transposable elements such as LINEs acting upon an endogenous mRNA followed by reverse transcription and integration into the genome at a new location.15 We believe this is the method by which three of the intronic-host gene pairs we discuss arose: The Mcts2/H13 domain on mouse chromosome 2, the U2af1-rs1/Commd1 (also known as Murr1) domain on chromosome 11, and the Nap1l5/Herc3 domain on chromosome 6. The Inpp5f_v2/Inpp5f imprinted locus also likely originated this way, however at this locus, the host gene Inppf5, unlike the other examples, is not imprinted.16 The provenance of Nnat/Blcap,17,18 also located on mouse chromosome 2, is less easy to determine. Although Nnat has also been postulated to be the product of a retrotransposition event, unlike in the other domains, no homologous gene elsewhere in the mouse genome can be found from which Nnat might have originated19 so its provenance is still debatable. In addition, Nnat is multi-exonic whereas retrotransposed genes are generally expected to be intronless. See Table 1.
Table 1.
Information about the intronic genes, in the order mentioned in the text
| Intronic gene | Origin | Host gene | Orientation compared to host gene |
|---|---|---|---|
| Mcts2 | Retrogene | H13 | sense |
| U2af1-rs1 | Retrogene | Commd1 | antisense |
| Nap1l5 | Retrogene | Herc3 | antisense |
| Inpp5f_v2 | Retrogene | Inpp5f | sense |
| Nnat | Unclear | Blcap | antisense |
A consequence of the arrangement of genes into intronic-host pairs is that of marked transcript diversity. In this context, transcript diversity means the production of more than two transcripts from a intronic-host cluster. On a genome-wide level, transcript diversity is a common phenomenon in mammals, as the ~25,000 genes in the mammalian genome are predicted to be generating around 200,000 gene products using a number of mechanisms. These include alternative splicing, the use of alternative promoters and alternative poly(A). More than half of human and a third of mouse genes have multiple alternative poly(A) sites and the high degree to which they are conserved indicates their functional significance.20 The use of alternative promoters and alternative poly(A) sites are methods of generating transcript diversity that may be controlled by epigenetic means at intronic-host loci.
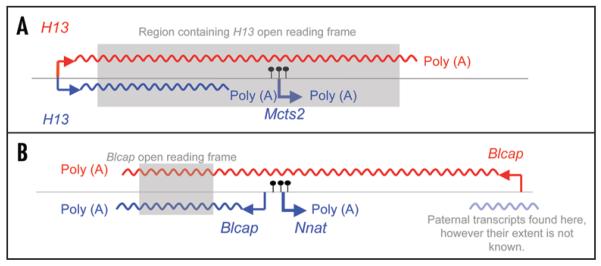
At the Mcts2/H13 locus, we showed how the choice of poly(A) site can be controlled by epigenetic mechanisms, albeit indirectly.21 This results in allele-specific transcript diversity as one set of polyadenylation sites is used on one allele of the H13 host gene, and a second set on the other allele, resulting in two sets of nascent transcripts of different lengths, and different protein-coding abilities, being transcribed from the two alleles (Fig. 2A). The removal of methylation at the Mcts2 promoter DMR abolishes this alternative poly(A). All other CpG islands in the region have been examined and the Mcts2 CpG island is the only DMR.21 Therefore all the imprinted transcripts at this locus are regulated directly or indirectly, by the methylation at the Mcts2 DMR. Studies are in progress to elucidate the precise mechanism by which this epigenetic mark mediates poly(A) site choice.
Figure 2.
Schematic showing transcripts at two imprinted intronic-host gene pairs in brain. Maternal transcripts are indicted in red, paternal in blue. Maternal methylation at the intronic gene DMR represented by black (filled) lollipops. (A) The Mcts2/H13 gene pair. (B) The Nant/Blcap gene pair.
At the Inpp5f_v2/Inpp5f imprinted locus, unusually the host gene expression is not affected by the imprinting of the intronic gene, however another transcript, Inpp5f_v3 which arises from a different promoter and unique first exon within this cluster, is exclusively paternally expressed in brain. It is likely that this is as a result of Inpp5f_v2 methylation and paternal expression, so while in this case the host gene is not the victim of Inpp5f_v2 imprinting, Inpp5f_v3 is.16 The Nnat/Blcap intronic-host cluster has an interesting history. It was initially thought than Blcap was not imprinted,22 however Blcap displays tissue-specific imprinting in the brain.18 Through the use of alternative promoters on the two alleles, Blcap transcripts of different lengths are expressed from the maternal and paternal alleles (Fig. 2B). Preliminary data have also shown that there are paternal transcripts present in the region of the promoter that produces the maternal Blcap transcript. However it is not known whether these are Blcap specific transcripts or read-through from Nnat (Fig. 2B).
At Nap1l5/Herc3, we found evidence for allele-specific alternative poly(A) of the host Herc3 gene (Wood A, unpublished data) and allele-specific alternative transcripts of Herc3 exit. At the U2af1-rs1/Commd1 cluster on mouse chromosome 11, allele-specific transcription of the U2af1-rs1 intronic gene through the host Commd1 promoter in an antisense orientation is thought to cause imprinting of the Commd1 host gene.23 In all intronic-host imprinted loci where imprinting of the intronic gene affects the host’s imprinting status, it seems likely that one DMR at the promoter of the intronic gene regulates the imprinted expression of the entire cluster and in three of the five cases, reciprocally imprinted transcripts of the host gene are present. This mechanism is likely to be particularly important in tissue-specific imprinting and the brain is the common tissue in which the imprinted intronic genes are expressed and the corresponding host genes affected.
What are the functional, evolutionary and practical consequences of imprinted transcript diversity at intronic-host gene clusters? While the Mcts2/H13 and Nnat/Blcap clusters both display allele-specific transcripts of different lengths with reciprocal patterns of expression, the consequences for the proteins produced may be rather different. At Mcts2/H13, the shorter nascent H13 transcript has a truncated open reading frame, so the translated protein is predicted to have a different structure, and possibly function (Fig. 2A). The maternal and paternal alleles are transcribing different ‘genes’, if a gene is regarded as encoding a particular protein product. The functionality of the truncated H13 protein should be investigated, so that conclusions about the potential evolutionary benefits, or indeed costs, of regulating imprinting in this way at this locus can be made.
In the case of Blcap, the alternative transcripts only differ in terms of the 5′ untranslated region of the mRNA, and so the structure of the Blcap protein is unchanged (Fig. 2B). In this case, there does not appear to be much functional significance to the alternative transcripts. However, there may be evolutionary consequences for transcript diversity even at Nnat/Blcap. The transcript diversity at this locus might be considered ‘noise’; a by-product of locating a gene inside the intron of another. However, this ‘biological noise’ could also be thought of as ‘raw material’, which may differ between individuals, upon which natural selection might be acting to produce biologically advantageous patterns of gene expression. The existence of large amounts of transcript diversity at small imprinted domains may contribute to the evolution of the more extensive and more complex imprinted loci that are the subject of investigation today.
Let us not overlook the practical consequences of reciprocally imprinted transcripts. The discovery of novel imprinted genes is an ongoing and popular activity for researchers in the field. If attention is not paid to the possible existence of reciprocally imprinted transcripts of a gene of interest, then both transcripts could be assayed together, resulting in the appearance of biallelic gene expression. In terms of epigenetic gene regulation, it is important to remember that while epigenetic mechanisms are intimately associated with transcript initiation, the co-transcriptional processes associated with termination are also likely to be subject to epigenetic influences. The next obvious step is to determine how widespread these mechanisms are and to what extent they influence the mammalian transcriptome.
Acknowledgements
We thank Andrew Wood, Roland G. Roberts and Reiner Schulz for their critical reading of this manuscript. We thank the Wellcome Trust for supporting this work and R.M. is supported by a Phillip Harris Studentship.
References
- 1.Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nature Reviews Genetics. 2001;2:21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- 2.Beechey CV, Cattanach BM, Blake A, Peters J. Mouse imprinting map. MRC Genetics Unit; Oxfordshire, UK: 2004. http://www.mgu.har.mrc.ac.uk/research/imprinting/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alleman M, Doctor J. Genomic imprinting in plants: observations and evolutionary implications. Plant Molecular Biology. 2000;43:147–61. doi: 10.1023/a:1006419025155. [DOI] [PubMed] [Google Scholar]
- 4.Morison IM, Paton CJ, Cleverley SD. The imprinted gene and parent of origin effect database. Nucleic Acids Research. 2001;29:275–6. doi: 10.1093/nar/29.1.275. http://igc.otago.ac.nz/home.html. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lian Z, Karpikov A, Lian J, Mahajan MC, Hartman S, Gerstein M, Snyder M, Weissman SM. A Genomics Analysis of RNA polymerase II modification and chromatin architecture related to 3′ end RNA polyadenylation. Genome Res. 2008 doi: 10.1101/gr.075804.107. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tian B, Pan Z, Lee JY. Widespread mRNA polyadenylation events in introns indicate dynamic interplay between polyadenylation and splicing. Genome Res. 2007;17:156–65. doi: 10.1101/gr.5532707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kass SU, Landsberger N, Wolffe AP. DNA methylation directs a time-dependent repression of transcription initiation. Curr Biol. 1997;7:157–65. doi: 10.1016/s0960-9822(97)70086-1. [DOI] [PubMed] [Google Scholar]
- 8.Kass SU, Pruss D, Wolffe AP. How does DNA methylation repress transcription? Trends Genet. 1997;13:444–9. doi: 10.1016/s0168-9525(97)01268-7. [DOI] [PubMed] [Google Scholar]
- 9.Lorincz MC, Dickerson DR, Schmitt M, Groudine M. Intragenic DNA methylation alters chromatin structure and elongation efficiency in mammalian cells. Nat Struct Mol Biol. 2004;11:1068–75. doi: 10.1038/nsmb840. [DOI] [PubMed] [Google Scholar]
- 10.Fedoriw AM, Stein P, Svoboda P, Schultz RM, Bartolomei MS. Transgenic RNAi reveals essential function for CTCF in H19 gene imprinting. Science. 2004;303:238–40. doi: 10.1126/science.1090934. [DOI] [PubMed] [Google Scholar]
- 11.Lin SP, Youngson N, Takada S, Seitz H, Reik W, Paulsen M, Cavaille J, Ferguson-Smith AC. Asymmetric regulation of imprinting on the maternal and paternal chromosomes at the Dlk1-Gtl2 imprinted cluster on mouse chromosome 12. Nat Genet. 2003;35:97–102. doi: 10.1038/ng1233. [DOI] [PubMed] [Google Scholar]
- 12.Sleutels F, Zwart R, Barlow DP. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415:810–3. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- 13.Lewis A, Mitsuya K, Umlauf D, Smith P, Dean W, Walter J, Higgins M, Feil R, Reik W. Imprinting on distal chromosome 7 in the placenta involves repressive histone methylation independent of DNA methylation. Nat Genet. 2004;36:1291–5. doi: 10.1038/ng1468. [DOI] [PubMed] [Google Scholar]
- 14.Plagge A, Gordon E, Dean W, Boiani R, Cinti S, Peters J, Kelsey G. The imprinted signaling protein XL alpha s is required for postnatal adaptation to feeding. Nat Genet. 2004;36:818–26. doi: 10.1038/ng1397. [DOI] [PubMed] [Google Scholar]
- 15.Esnault C, Maestre J, Heidmann T. Human LINE retrotransposons generate processed pseudogenes. Nat Genet. 2000;24:363–7. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
- 16.Wood AJ, Roberts RG, Monk D, Moore GE, Schulz R, Oakey RJ. A screen for retrotransposed imprinted genes reveals an association between X chromosome homology and maternal germ-line methylation. PLoS Genet. 2007;3:20. doi: 10.1371/journal.pgen.0030020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kikyo N, Williamson CM, John RM, Barton SC, Beechey CV, Ball ST, Cattanach BM, Surani MA, Peters J. Genetic and functional analysis of neuronatin in mice with maternal or paternal duplication of distal Chr 2. Dev Biol. 1997;190:66–77. doi: 10.1006/dbio.1997.8681. [DOI] [PubMed] [Google Scholar]
- 18.Schulz R, McCole RB, Woodfine K, Wood AJ, Chahal MC, Monk D, Moore GE, Oakey RJ. [Advance Access published on October 4, 2008];Hum Mol Genet. doi: 10.1093/hmg/ddn322. doi: doi:10.1093/hmg/ddn322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Evans HK, Weidman JR, Cowley DO, Jirtle RL. Comparative phylogenetic analysis of blcap/nnat reveals eutherian-specific imprinted gene. Mol Biol Evol. 2005;22:1740–8. doi: 10.1093/molbev/msi165. [DOI] [PubMed] [Google Scholar]
- 20.Zhao J, Hyman L, Moore C. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev. 1999;63:405–45. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wood AJ, Schulz R, Woodfine K, Koltowska K, Beechey CV, Peters J, Bourc’his D, Oakey RJ. Regulation of alternative polyadenylation by genomic imprinting. Genes and Development. 2008:22. doi: 10.1101/gad.473408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Evans HK, Wylie AA, Murphy SK, Jirtle RL. The neuronatin gene resides in a “micro-imprinted” domain on human chromosome 20q11.2. Genomics. 2001;77:99–104. doi: 10.1006/geno.2001.6612. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y, Joh K, Masuko S, Yatsuki H, Soejima H, Nabetani A, Beechey CV, Okinami S, Mukai T. The mouse Murr1 gene is imprinted in the adult brain, presumably due to transcriptional interference by the antisense-oriented U2af1-rs1 gene. Mol Cell Biol. 2004;24:270–9. doi: 10.1128/MCB.24.1.270-279.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]