Abstract
Although carbon nanotubes (CNTs) have been shown to nonspecifically bind proteins through charge complementary, π-π stacking or hydrophobic interactions, they have not been shown to bind to a specific site on proteins. By generating surface molecular diversity, we created functionalized CNTs that recognize and bind to the catalytic site of α-chymotrypsin and inhibit its enzymatic activity competitively.
Enzyme and receptor proteins play essential roles in biological processes and protein dysfunction is also related to human diseases such as caspases, peroxisome proteins and matrix metalloproteases.1–3 Protein surface recognition4 and regulation of protein-protein interactions5 provide an important tool for therapeutics. However, specific regulation of such malfunctions through protein surface recognition is challenging. To block protein-protein interactions requires recognition of large protein surface areas. Peptides or protein-based therapeutics usually have poor cell permeability. Nanoparticles (NPs), which have an antibody-like protein contact surface area yet can enter cells easily, are promising candidates for regulating protein activities. A typical example is that the surface functionalized gold nanoparticles inhibited α-ChT’s enzymatic activity.6 The key issue is whether a NP can specifically bind to a protein and alter its function in a controlled fashion. Recently, binding between NPs and proteins through non-covalent interactions such as charge complementarities, π-π stacking, and mostly non-specific or unknown interactions have been published.6–10 Although protein recognizing molecules such as antibodies linked to NPs were shown to bind to protein targets specifically, site-specific binding between a NP and a protein, which is required for functional control of proteins, has not been reported. Previously, combinatorial phage-display libraries of random peptides were screened for their recognition and specific binding to semiconductor structures.11 Here, we demonstrated specific recognition of ChT and regulate its functions by screening a surface diversified CNT combinatorial library.
Carbon nanotubes (CNTs) are graphene tubes that can freely enter cells12 and mammalian organs13 with potential drug delivery13 and therapeutic14 applications in medicine. With their large surface areas, pristine CNTs have a strong tendency to bind cellular proteins nonspecifically, e.g. π-π interactions9, 15. We hypothesized that by creating molecular diversity on the surface of CNTs, we might create a molecular selective nano-environment for protein recognition. In the following study, we investigated interactions between a functionalized multiwalled carbon nanotube (f-MWNT) library containing 80 members and α-Chymotrypsin (ChT) and interrogated the binding strength, nanotube binding-induced protein conformational changes, and the effects of binding on fluorescence and the catalytic activity of ChT. We discovered four f-MWNTs that site-specifically bind to ChT’s catalytic site and competitively inhibit ChT’s enzymatic function.
To generate molecular recognition ability on f-MWNT, we made a surface diversified combinatorial f-MWNT library containing 80 members16 (see Figure S1 in Supporting Information) and investigated interactions between ChT and the library members. The loading of the functional molecules on f-MWNT is 0.4 mmol/g and the density of such molecules on the nanotube surface is approximately one molecule every 30 carbon atoms. To differentiate whether nanotubes bind to a specific site of ChT or random sites on the protein, we monitored the binding-induced changes in fluorescence and its enzymatic activity. We rationalized that if f-MWNTs’ binding strength and the bound protein amounts are all similar, random bindings between ChT and nanotubes would generate the same effects on both fluorescence and enzymatic activity.
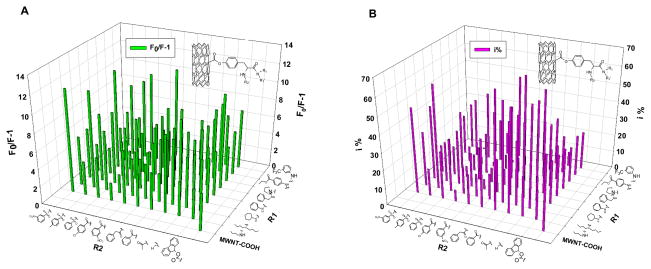
The protein intrinsic fluorescence can be quenched upon f-MWNT binding17. We first determined Fo/F, the ratio of ChT’s intrinsic fluorescence without and with f-MWNTs. This value was a measure for f-MWNTs’ allosteric effect on tryptophan fluorophores. Fluorescence of ChT was quenched to different degrees by various f-MWNTs suggesting various possibilities, such as bindings to various sites on ChT, binding in different amounts, or different binding-induced conformational changes. We also determined the enzymatic activity18 of ChT in the presence and absence of f-MWNTs. The f-MWNTs’ perturbations on enzymatic activity also varied (Figure 1). Since there is no tryptophan near the catalytic site of ChT according to its crystal structure19, specific binding of a nanotube to the catalytic site would strongly influence the catalysis, with less fluorescence quenching. On the other hand, binding at a remote site would pose varying effects on catalysis and fluorescence through allosteric interactions. Therefore, a comparison of the influence of nanotube on fluorescence and enzymatic activity would give the first indication whether a nanotube would site-specifically bind to ChT.
Figure 1.
The quenching of ChT’s fluorescence and inhibition of its enzymatic activity by f-MWNTs. The fluorescence quenching expressed as F0/(F-1) and enzyme inhibition (i %) are presented in a library format arranged as two dimensions of diversities of the surface molecules. Steady fluorescence spectra experimental condition: ChT concentration was 2 μM in 25 mM Tris-HCl buffer solution (pH~7.6), and final concentration of f-MWNTs in ChT solutions was 10 μg/mL. Enzyme inhibition experimental condition: final f-MWNTs’ concentration was 40 μg/mL and the ChT concentration was 3.2 μM (i% = (1 − νi/ν0) × 100%, where νi, ν0 represent the enzyme reaction velocity initial and after adding f-MWNT respectively).
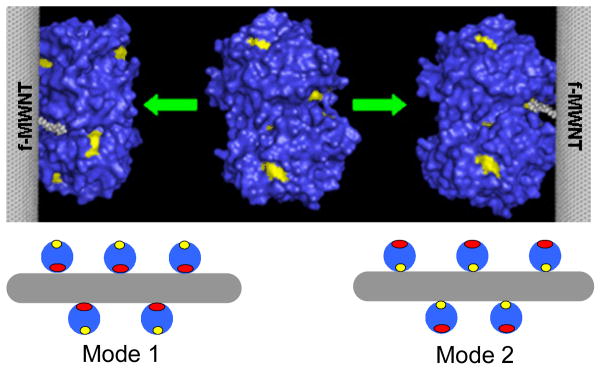
Analysis of the screening data (Figure 1 and Figure S2 in Supporting Information) identified four f-MWNTs (group 1, #30, #34, #35, #36) that showed strong enzymatic inhibition with much less fluorescence quenching (see SI, Figure S1 for f-MWNT numbering). Other f-MWNTs (group 2, #16, #20, #45, #50) influence enzyme activity as much as they affected fluorescence. This finding was not coincidence because all the four f-MWNTs of group 1 contain a surface molecules having a common side chain – dibutylamino group. On the basis of their different influence on activity and fluorescence and their structural similarity, we speculated that these f-MWNTs might site-specifically bind to or near the catalytic site and inhibited ChT competitively (Mode 2 in Figure 4).
Figure 4.
Models showing different ways f-MWNTs bind to ChT. ChT structure was taken from Protein Data Bank19. Mode 1: competitive inhibition; mode 2: non-competitive inhibition. Red: catalytic site; yellow: tryptophan fluorophores.
To test our hypothesis, especially the Mode 1 in Figure 4, we need to exclude the possibility that f-MWNTs have different binding strength to ChT and that the binding of some f-MWNTs caused changes in ChT’s secondary structures. On the other hand, if f-MWNTs have a comparable binding strength to ChT and exert similar effects on the conformation of the bound ChT, then their significantly different effects on ChT enzymatic activity and fluorescence (p<0.0005 by Student’s t-test) from these two groups of f-MWNTs must be due to the site-specific binding to ChT. Furthermore, a competitive inhibition effect will be observed if the specific binding site happens to be the catalytic site. Therefore, we first quantitatively determined the amount of ChT proteins bound to eight selected f-MWNTs (four from each of group 1 and 2) and the ChT conformational changes upon f-MWNTs’ binding.
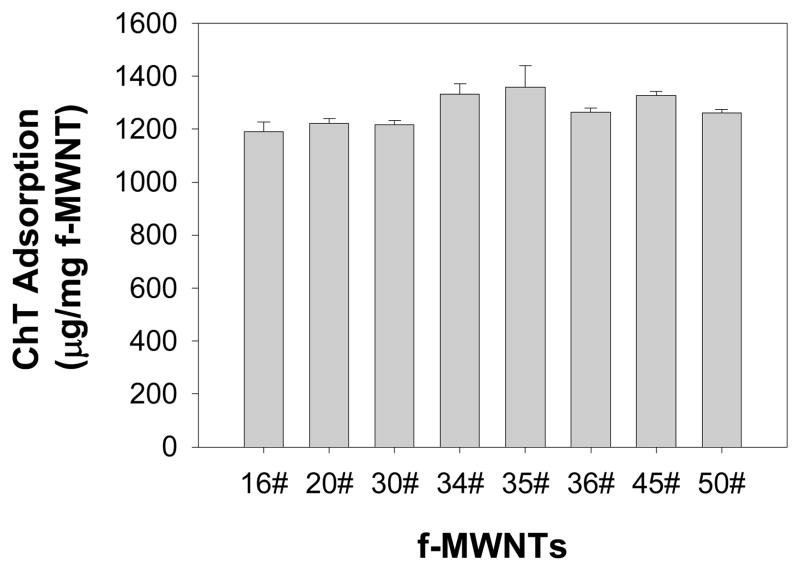
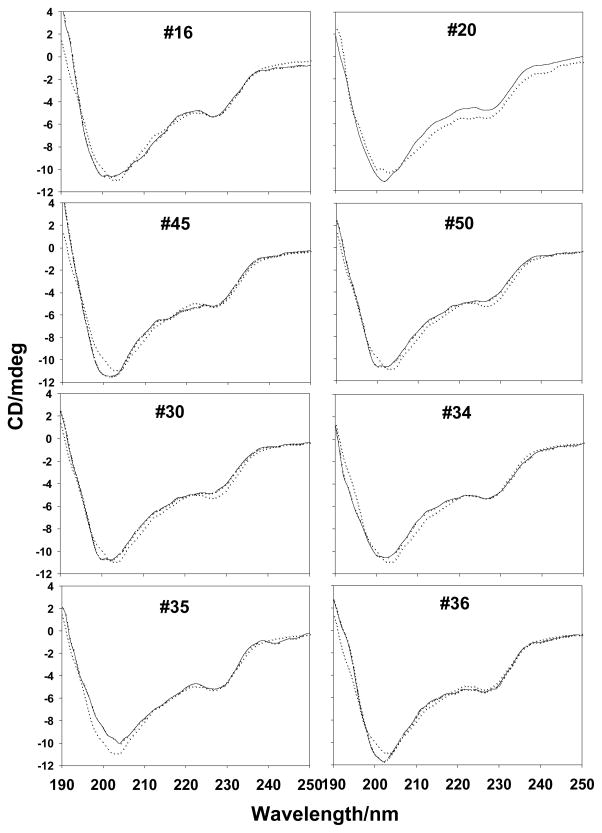
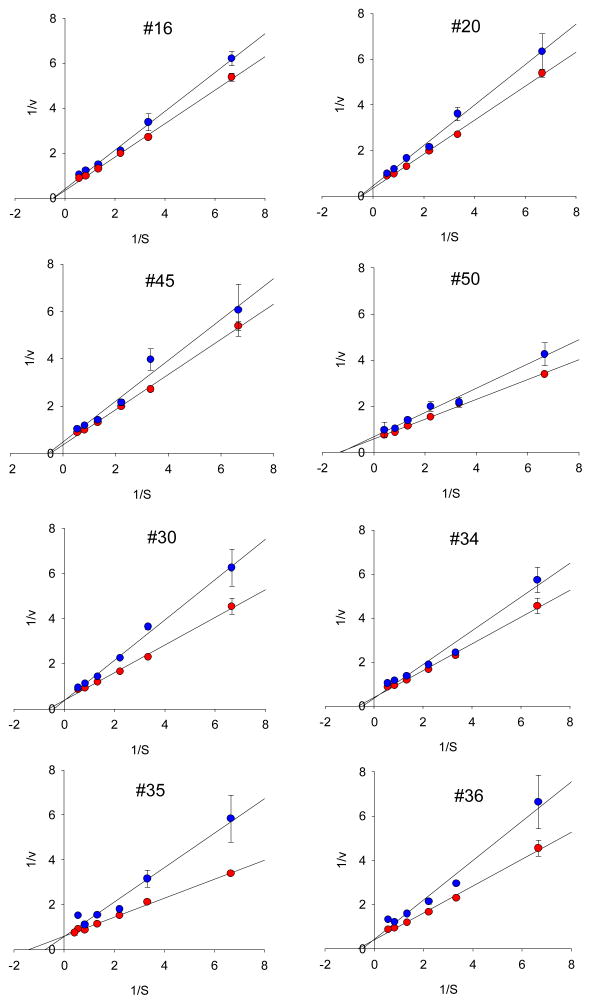
Functionalized MWNTs bound to proteins including ChT irreversibly by forming a static complex17. By determining the amount of proteins bound to f-MWNTs, we found that the binding strength between f-MWNTs and ChT are not significantly different. Therefore, nonspecific f-MWNTs bindings would yield very similar effects on fluorescence and catalytic activity of ChT. Eight f-MWNTs bound to comparable amounts of ChT proteins (186±10 μg/mg f-MWNT, see Fig. 2), suggesting that there is no significant difference in protein binding strength among these nanotubes. Circular dichroism (CD) measurements (Fig. 3) showed that although fluorescence quenching experiments under the identical conditions strongly indicated f-MWNTs/ChT binding, there is no appreciable change in the secondary structure of ChT upon f-MWNT’s binding. These results support the conclusion that the diverse influences of f-MWNTs on ChT’s fluorescence and catalytic activity result from the binding of f-MWNTs to specific sites of ChT. These results also indicate the possibility that f-MWNTs #30, #34, #35 and #36 might bind to the enzyme’s catalytic site to initiate strong inhibition of ChT’s enzymatic activity while induce less fluorescence quenching. Analysis of Michaelis-Menten enzyme kinetics and the derived Lineweaver-Burk plots further confirmed these possibilities. The Lineweaver–Burk plots (Figure 5) indicate that f-MWNTs #30, #34, #35, and #36 show a typical feature of competitive inhibition whereas #16, #20, #45 and #50 show the feature of non-competitive inhibition. The discovery that a common building block in the surface molecules in group 2 helped explain the structure-activity relationship of the f-MWNT/ChT recognition. An interesting finding was that free dibutylamine molecules did not inhibit ChT’s enzymatic activity. Considering the large f-MWNT/protein contact areas, this functional group, in combination with the nanotube and adjacent ligands, was likely crucial for targeting the catalytic site of ChT. Previously, gold nanoparticles functionalized with amino acids were reported to inhibit ChT activity through electrostatic and hydrophobic interactions,6 while site-specific binding was not reported. In our study, we demonstrated that competitive and non-competitive inhibitions of ChT enzymatic activity were mainly through site-specific interaction, and such controllable regulation of enzyme activity may provide powerful tools for diagnostic and therapeutic applications.
Figure 2.
Quantification of adsorbed ChT onto MWNTs by HPLC assay. The free ChT was determined by HPLC based on a standard calibration curve. The adsorbed proteins were derived from the difference between the total proteins used in the assay and the free proteins. Error bars are the standard deviations from three measurements. The final f-MWNTs’ concentration was 150 μg/mL and the ChT concentration was 250 μg/mL. Experimental details are in section 1.5 of Supporting Information.
Figure 3.
CD spectra of ChT in the absence or presence of f-MWNTs. The solid line was for ChT and doted line for ChT in the presence of f-MWNTs. The ChT concentration was 2μM in 25 mM Tris-HCl buffer solution (pH~7.6). The final concentration of f-MWNTs was 10 μg/mL.
Figure 5.
Lineweaver–Burk plots of ChT in the presence (red circle) and absence (blue circle) of f-MWNTs. Final concentration of f-MWNTs was 10 μg/mL, and concentrations of SPNA were 0.15, 0.45, 0.75, 1.2, 1.8, 2.4mM, respectively.
In summary, enzyme kinetics and an array of spectroscopic studies provided compelling evidence that four f-MWNTs (#30, #34, #35, #36) site-specifically bind to the catalytic site of ChT and inhibit its enzymatic activity competitively. Although many studies discussed non-specific nanotube/protein interactions, we proved that surface molecular diversity can generate site-specific nanotube/enzyme recognition. This important property of f-MWNTs is now being tested in other NP systems in our laboratory. A wide range of potential applications from targeted enzyme inhibition to molecular sensing are becoming feasible.
Supplementary Material
Acknowledgments
We thank Aifeng Liu and Zhaochun Wu for technical assistance in protein adsorption quantification and Drs. Andrew Lemoff, Lei Yang and Xi Li for their valuable comments. This work was supported by National Basic Research Program of China (973 Program 2009CB930103), the American Lebanese Syrian Associated Charities (ALSAC) and St. Jude Children’s Research Hospital.
Footnotes
Supporting Information Available. Detailed experimental procedures, f-MWNT’s naming, protein binding data, and CD results are available free of charge via the Internet.
References
- 1.Longthorne VL, Williams GT. Embo J. 1997;16:3805–3812. doi: 10.1093/emboj/16.13.3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singh I. Mol Cell Biochem. 1997;167:1–29. doi: 10.1023/a:1006883229684. [DOI] [PubMed] [Google Scholar]
- 3.D’Armiento J. Trend Cardiov Med. 2002;12:97–101. doi: 10.1016/s1050-1738(01)00160-8. [DOI] [PubMed] [Google Scholar]
- 4.Peczuh MW, Hamilton AD. Chem Rev. 2000;100:2479–2494. doi: 10.1021/cr9900026. [DOI] [PubMed] [Google Scholar]
- 5.Veselovsky AV, YDIASIAIAPLPJ J Mol Recogn. 2002;15:405–422. doi: 10.1002/jmr.597. [DOI] [PubMed] [Google Scholar]
- 6.You CC, De M, Han G, Rotello VM. J Am Chem Soc. 2005;127:12873–12881. doi: 10.1021/ja0512881. [DOI] [PubMed] [Google Scholar]
- 7.Shang W, Nuffer JH, Dordick JS, Siegel RW. Nano Lett. 2007;7:1991–1995. doi: 10.1021/nl070777r. [DOI] [PubMed] [Google Scholar]
- 8.Cedervall T, Lynch I, Lindman S, Berggard T, Thulin E, Nilsson H, Dawson KA, Linse S. Proc Natl Acad Sci U S A. 2007;104:2050–2055. doi: 10.1073/pnas.0608582104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li X, Chen W, Zhan Q, Dai L, Sowards L, Pender M, Naik RR. J Phys Chem B. 2006;110:12621–12625. doi: 10.1021/jp061518d. [DOI] [PubMed] [Google Scholar]
- 10.Yi CQ, Fong CC, Chen WW, Qi SJ, Tzang CH, Lee ST, Yang MS. Nanotechnology. 2007;18:6. [Google Scholar]
- 11.Whaley SR, English DS, Hu EL, Barbara PF, Belcher AM. Nature. 2000;405:665–668. doi: 10.1038/35015043. [DOI] [PubMed] [Google Scholar]
- 12.Porter AE, Gass M, Muller K, Skepper JN, Midgley PA, Welland M. Nat Nanotechnol. 2007;2:713–717. doi: 10.1038/nnano.2007.347. [DOI] [PubMed] [Google Scholar]
- 13.Liu Z, Chen K, Davis C, Sherlock S, Cao QZ, Chen XY, Dai HJ. Cancer Res. 2008;68:6652–6660. doi: 10.1158/0008-5472.CAN-08-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gannon CJ, Cherukuri P, Yakobson BI, Cognet L, Kanzius JS, Kittrell C, Weisman RB, Pasquali M, Schmidt HK, Smalley RE, Curley SA. Cancer. 2007;110:2654–2665. doi: 10.1002/cncr.23155. [DOI] [PubMed] [Google Scholar]
- 15.Nepal D, Geckeler KE. Small. 2006;2:406–412. doi: 10.1002/smll.200500351. [DOI] [PubMed] [Google Scholar]
- 16.Zhou HY, Mu QX, Gao NN, Liu AF, Xing YH, Gao SL, Zhang Q, Qu GB, Chen YY, Liu G, Zhang B, Yan B. Nano Lett. 2008;8:859–865. doi: 10.1021/nl0730155. [DOI] [PubMed] [Google Scholar]
- 17.Mu QX, Liu W, Xing YH, Zhou HY, Li ZW, Zhang Y, Ji LH, Wang F, Si ZK, Zhang B, Yan B. J Phys Chem C. 2008;112:3300–3307. [Google Scholar]
- 18.Kawai Y, Matsuo T, Ohno A. J Chem Soc-Perkin Trans 2. 2000;4:887–891. [Google Scholar]
- 19.Tsukada H, Blow DM. J Mol Biol. 1985;184:703–711. doi: 10.1016/0022-2836(85)90314-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.