Abstract
Although targeted mutagenesis of Streptococcus pneumoniae is readily accomplished with the aid of natural genetic transformation and chimeric donor DNA constructs assembled in vitro, the drug resistance markers often employed for selection of recombinant products can themselves be undesirable by-products of the genetic manipulation. A new cassette carrying the erythromycin-resistance marker ermAM is described that can be used as a temporary marker for selection of desired recombinants. The cassette may subsequently be removed at will by virtue of an embedded fucose-regulated Cre recombinase gene and terminal lox66 and lox71 Cre recognition sites, with retention of 34 bp from the cassette as an inert residual double-mutant lox72 site.
1. Introduction
Streptococcus pneumonia (pneumococcus), a Gram-positive (Gm+) bacterium which normally inhabits the human upper respiratory tract, is an important opportunistic pathogen that causes a variety of infections and diseases. As a naturally transformable species, it is especially accessible to genetic manipulation (Morrison, 2007) and many of its virulence factors have been identified through intensive genetic analysis (Ng et al., 2003; Porter et al., 1976). In such genetic analysis, it is sometimes desirable to construct strains with double, triple, or multiple gene disruptions. Especially in the case of multiple disruptions, it is often preferable that such disruptions do not themselves cause accumulation of heterologous proteins in the resulting mutant. While a variety of marker-less gene disruption strategies have been developed for application in pneumococcus (Iannelli et al., 2004; Sung et al., 2001; Standish et al., 2005), they involve either multiple genetic manipulation steps or tedious screening steps.
The Cre/loxP strategy for creating marker-less deletions is especially attractive, as it uses the well-characterized, naturally occurring cofactor-independent site-specific recombinase of bacteriophage P1 to delete arbitrary targets delimited by two copies of the 34-bp loxP recognition sequence (Ghosh et al., 2002; Saucer, 1987; St-Onge et al., 1996; Zuo et al., 2001). Furthermore, use of carefully chosen single-mutant loxP sites can ensure that a residual double-mutant loxP site is produced that does not participate in further Cre-mediated recombination (Albet et al., 1995; Zhang et al., 2002). Several implementations of this strategy for use in Gm+ bacteria have already been described (Banerjee et al., 2008; Lambert et all, 2007; Leibig et al., 2008; Pomerantsev et al., 2006; Yan et al., 2008). In practice, two directly repeated loxP sites are arranged to flank a selectable marker, which is substituted for the deletion target by use of targeted recombination. Then, upon expression of a Cre recombinase gene, recombination between the loxP sites excises the intervening sequence, leaving one residual recombinant loxP element in place of the deletion target. Even here, however, current bacterial implementations typically employ a four-step strategy of (I) emplacing the selectable marker and single-mutant loxP sites as a substitute for the intended deletion target, (II) introducing a heterologous cre gene into the resulting mutant, (III) allowing expression of cre and excision of the selectable marker, and (IV) removing the heterologous cre gene from the resulting deletion mutant. To simplify this process for use in pneumococcus while taking advantage of its highly efficient natural transformation system, we sought to combine steps (I) and (II) and obviate step (IV) by creating a new self-deleting lox/erm/cre/lox cassette, much as has been implemented in the plant system of Arabidopsis (Hare et al.,2002; Zhang et al., 2002). Because the excision would be irreversible, the level of expression of the cre gene in such a cassette could in principle be adjusted to provide stability high enough for steps (I) and (II), but low enough to provide easy recovery of deletions at step (III). We have chosen instead to place cre under the control of a native regulated pneumococcal promoter that has a low basal level of expression, but is readily activated. In this note, we describe construction of such a self-deleting cassette, and show that it is stable in glucose medium but is readily excised during growth in the presence of fucose.
2. Materials and Methods
2.1 Bacterial strains and media
All pneumococcal strains used in this study are described in Table 1. CP1250 and CP2000 are derivatives of the Rx strain with point mutation and in-frame deletion of capsular genes respectively. They were taken as the wild-type in our studies. CP1334 and CPM7 are derivatives of CP1250. In CPM7, the lacZ reporter plasmid pEVP3 is inserted into the ssbB gene. CP1334 has an in-frame deletion of spr1630 and spr1631 with the selective marker aphIII. All the strains were grown in complete CAT medium or its modified form with 1.5% agar. The complete CAT medium was prepared by adding 5g of tryptone (Difco Laboratories), 10g of enzymatic casein hydrolysate (ICN Nutritional Biochemicals), 1g of yeast extract (Difco), and 5g of NaCl into 1 liter H2O. After sterilizing for 40 min at 121 C, it was brought to 0.2% glucose and 1/60 (0.0167) M K2HPO4 before use.
Table 1.
Strains Used in This Study
| Strain | Description | Sourcea |
|---|---|---|
| CP1250 | hex malM511 str-1 bgl-1; Rx derivative, Hex− Mal− SmR Bga− | Pestova et al.,1996 |
| CP1334 | CP1250, but Δ(spr1630spr1631)::kan; SmR KanR | Peterson et al.,2004 |
| CP1939 | CPM7, but ΔssbB::erm-PfcsK::(pEVP3):: ssbB+; SsbB+ SmR EmR CmR | This study |
| CP2000 | CP1250, but Δcps; Hex− Mal− Cps− SmR Bga− | A. Piotrowski |
| CP2052 | CP2000 but ΔssbB::erm-PfcsK::(pEVP3):: ssbB+; Cps− SmR EmR CmR | CP2000 x CP1939 |
| CP2055 | CP1334, but Δ(spr1630spr1631)::kan::Cheshire; SmR EmR | This study |
| CP2062 | CP2000, but Δ(spr1630spr1631)::Kan::Cheshire; SmR EmR | CP2000 x CP2055 |
| CPM7 | CP1250, but ssbB−::(pEVP3)::ssbB+; SsbB+ SmR CmR | Lee et al.,1999 |
Transformation crosses for strain construction indicated as: recipient x donor.
2.2 Transformation of pneumococcal cells and selection of tranformants
Culture grown in CAT medium to OD0.03 were exposed to DNA (100ng/ml) for 45 min after the treatment with CSP (250μg/ml), CaCl2 (0.05%), and bovine serum album (0.04%). Transformants were selected on complete CAT solidified with 1.5% agar containing appropriate drugs at the following concentrations: kanamycin, 200 μg/ml; erythromycin, 0.05 μg/ml. The plating method is as described by Lee et al., 1999.
2.3 Construction of new strains
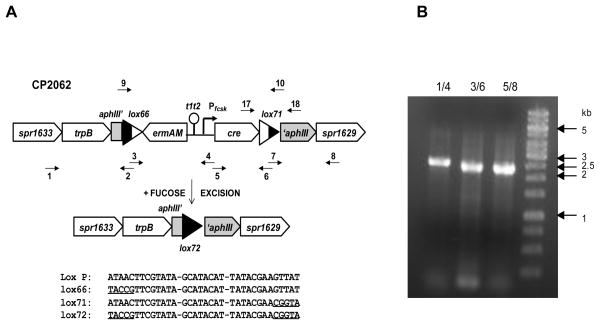
To construct strain CP2055, a loxP/ermAM/PfcsK/cre/loxP cassette was constructed. Four fragments were prepared by PCR using primer pairs 1/2 and 7/8, primer pair 3/4, primer pair 5/6, with genomic DNA of CP1334, the ermAM-t1t2-PfcsK cassette and plasmid pCrePA as templates respectively, as shown in Figure 1. Following the amplification, each fragment was purified and digested with BsaI at 37 C for 2 hours. After digestion, all fragments were repurified, mixed together, treated with T4 DNA ligase (Fermentas, Glen Burnie) and used to transform CP1334. One transformant selected with Em was retained as CP2055. Finally, CP2000 was transformed with genomic DNA of CP2055 to yield CP2062.
Figure 1.
Construction of Cheshire cassette. (A) Construction of lox/erm/cre/lox cassette as an insertion into a kanamycin resistance gene of S. pneumoniae. t1t2, E coli rRNA terminator. lox66 and lox71, left-end and right-end single-mutant loxP sites. Kan’, ‘Kan, aphIII truncations. Small arrows show positions of indicated primers. Pentagons, ORFs. Triangles, loxP sites, filled to indicate mutant 13-bp repeats within loxP. Elements not drawn to scale. Wild type, single-mutant, and double-mutant loxP sites are shown in the bottom. (B) Proof of the structure of the cassette in the EmR clone CP2062. PCR products obtained with primers pairs 1/4, 3/6, and 5/8 are shown. Expected sizes of the products were 2.6, 2.4, and 2.3 kb, respectively. In marker lane, bands of sizes 1, 2, 3, and 5 kb are indicated by arrows.
To construct strain CP1939, a fragment was amplified with primer pair 13/14 from the ermAM-t1t2-PfcsK cassette and ligated with upstream and downstream sequences amplified by primer pairs 11/12 and 15/16 from genomic DNA of CPM7 respectively. The ligation product was transformed into CPM7 to make CP1339 by selection with Em. Eventually, CP2000 was transformed with genomic DNA of CP1339 to create CP2052. Primers used in these constructions are listed in Table 2.
Table 2.
Primers used in cassette construction:
| Primer | Location | Sequence, 5′ to 3′a | Lab name |
|---|---|---|---|
| 1 | SP1813 | gcgcatcggctattatcggtaAGCCTTGTCCGAAGAAC | DAM840 |
| 2 | kan | gcgcggtctcagtatgctatacgaacggtaAGCCATTTATTATTTCCTTCCT | DAM841 |
| 3 | ermAM | cgcgggtctcaatacattatacgaagttatTTAGCTCCTTGGAAGCTGTC | DAM842 |
| 4 | Pfcsk | gcgcatcgggtctcaAGTCTTTTCTTCTCTCTTCGTCCTTGATT | DAM843 |
| 5 | cre | gcgcatgcggtctcAGACTATGTCCAATTTACTGACCGTACAC | DAM844 |
| 6 | cre | gcgcggtctcatgctatacgaagttaTCTAATCGCCATGTTCCAGCAG | DAM845 |
| 7 | kan | gcgcggtctcaagcatacattatacgaacggtaTCGAAAAATACCGCTGCGTAAA | DAM846 |
| 8 | SP1809 | gcgctaccgataacgcaTAGCTTCTTGTGCTCTCGTCTT | DAM847 |
| 9 | kan | ggggacgcgtTGGCTTACCGTTCGTATAG | DAM868 |
| 10 | kan | ggggccatggTCGATACCGTTCGTATAATGT | DAM869 |
| 11 | oxr | CAGCGAAACACTGGACTG | DAM811 |
| 12 | ssbB | gcgcggatccAATTCGAGCTCCCATCAAAC | DAM812 |
| 13 | ermAM | gcgcggatccTTAGCTCCTTGGAAGCTGTC | DAM813 |
| 14 | Pfcsk | gcgcgaattcTTTTCTTCTCTCTTCGTCCTTGA | DAM814 |
| 15 | lacZ | gcgcgaattcTGTGGAAGTTACTGACGTAAG | DAM815 |
| 16 | cat | GTGCAGGAGCTCGTTATC | DAM816 |
| 17 | cre | CACCAGCCAGCTATCAACTC | DAM898 |
| 18 | kan | CGCAGAAGGCAATGTCATAC | DAM899 |
5′ extension bases in lower case.
2.4 PCR conditions and DNA sequencing
All the DNA fragments in this study were amplified with the PCR Supermix High Fidelity (Invitrogen, Carlsbad). The PCR conditions include: 3 min at 94 C, followed by 30 cycles of 30 seconds at 94 C, 30 seconds for annealing at around 55 C (depending on primer Tm), and 1 to 3 min (1min per kb) for extension at 72 C, and 10 min at 72 C for the final extension.
Three fragments amplified with primer pairs 1/4, 3/6, and 5/8 from CP2062 were sequenced to confirm the accuracy of the ligation junctions. Dye-terminator sequencing was carried out at ICBR, University of Florida at Gainesville.
2.5 β-galactosidase (β-Gal) activity assay
For measurement of β-galactosidase activity, a 0.4-ml sample of liquid culture was chilled on ice. After adding 100 μl 5XZ buffer (300 mM Na2HPO4, 200 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 50 mM β-mercaptoethanol, 0.5% TritonX-100) and incubation at 37 C for 10 min to lyse the cells, 150 μl of this lysate was added to 50 μl of o-nitrophenyl-β-D-galactopyranoside solution (4 mg/ml o-nitrophenyl-β-D-galactopyranoside, 60 mM Na2HPO4, 40 mM NaH2PO4) in a 96-well microplate. The plate was incubated at 37 C, and absorbance at 420 nm was read every 10 min for 90 min in Spectra Max M2 of Molecular Devices. The initial slope of the absorbance curve was used to calculate LacZ activity, reported in Miller units.
3. Results
3.1 Construction of Cheshire cassette
A loxP/ermAM/PfcsK/cre/loxP cassette, which we propose to name Cheshire, was constructed by using PCR amplification, restriction nuclease digestions, and ligation (Fig. 1). One fragment was amplified from the ermAM-t1t2-PfcsK cassette, in which the ermAM erythromycin (Em) resistance determinant from plasmid pAMβ-1 and the fcsK promoter (PfcsK) from the fucose operon of S. pneumoniae are linked to a strong rRNA transcriptional terminator t1t2 to minimize read-through transcription toward PfcsK and cre (Chan et al., 2003). The cre gene was amplified from the vector pCrePA (Bannerjee et al., 2008). Mutant loxP sites (Albert et al., 1995) were assembled at the ends of the cassette by designing half-loxP sites into the amplicons containing ermAM-t1t2-PfcsK and cre, while the other half of each loxP site was provided in the flanking fragments used to target recombination into the genome by natural transformation. PCR was used to confirm the structure of the construct and the structure of each new junction in the resulting cassette was verified by sequencing. Finally, primer pair 9/10 was designed to allow applications of the cassette using terminal MluI and NcoI sites. The sequence of the cassette as amplified using primer pair 9/10 is deposited in Genbank as Accession bankit1147842.
3.2 Regulation of promoter PfcsK in fucose
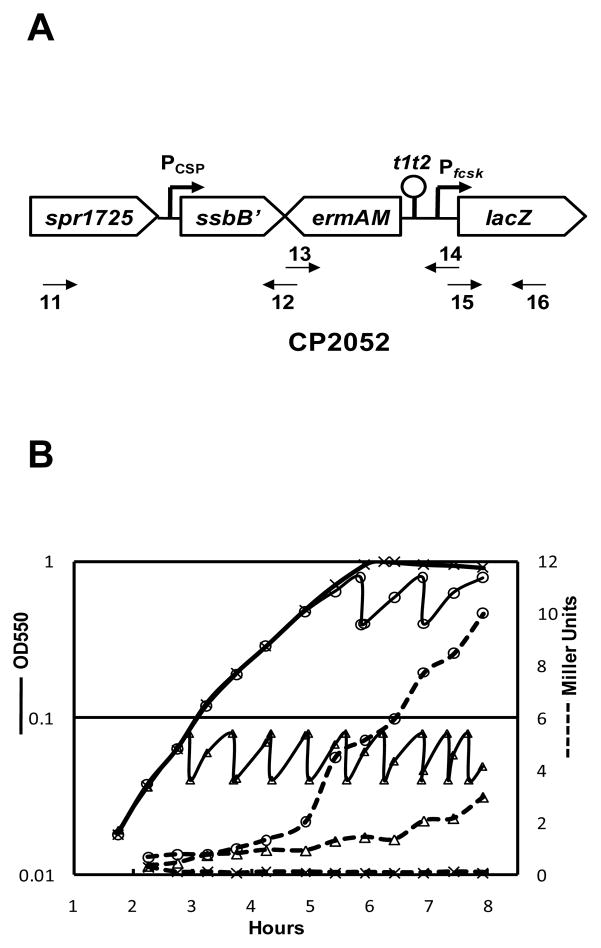
Because fucose-induced expression from PfcsK is reported to increase strongly during approach to stationary phase in S. pneumoniae (Chan et al., 2003), we sought a treatment regimen in which a high level of expression from PfcsK in the new cassette could be maintained for an arbitrary period. To characterize the transcriptional response of the fucose promoter (PfcsK), we used a PfcsK::lacZ fusion constructed at the ssbB locus in CP2052 (Fig. 2). Two portions of the reporter strain were treated with fucose, one at early log phase and one at late log phase, and maintained at respective densities by periodic dilution with fresh fucose medium. Compared to the culture without fucose induction, LacZ activity increased 5–10 fold in the culture maintained at OD 0.04–0.08, but 70–140-fold in the culture held at OD 0.4–0.8 (Fig. 2). This is consistent with the report by Chan, and suggests that the rate of excision could be highest in cultures grown with fucose to high cell densities.
Figure 2.
Effect of fucose on PfcsK-driven expression. (A) Construction of an ectopic PfcsK::lacZ transcriptional fusion in the S. pneumoniae genome. Pcsp, CSP-inducible combox promoter of ssbB. (B) Effect of fucose on expression of PfcsK::lacZ in CP2052. ———, OD550; ––––, β-gal activity; ×, no fucose; ○, △, 0.5% fucose.
3.3 Excision of the Cheshire cassette upon treatment with fucose
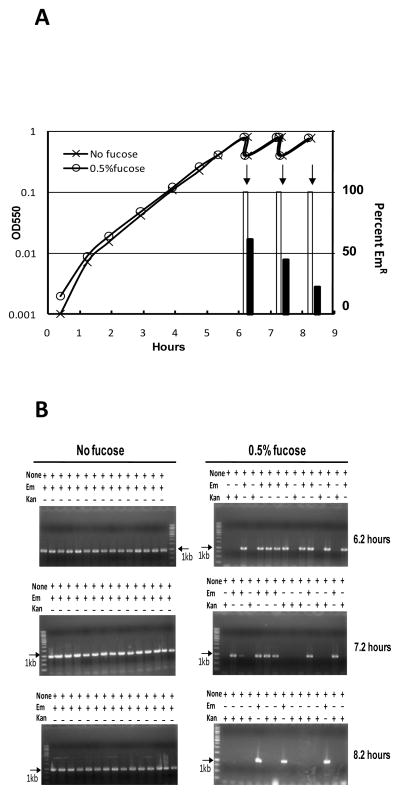
To characterize the stability and inducible excision efficiency of the new cassette, we examined cultures maintained under conditions for basal or for maximally elevated expression of Cre. Two parallel cultures of mutant CP2062 were examined: one culture was incubated without fucose, while another was incubated with fucose from OD 0.001 and then maintained by repeated dilution at late log phase between OD 0.4 and OD 0.8 (Fig. 3). As expected, the two cultures grew at the same rates up to OD 0.8. Samples were taken from both cultures at OD 0.8, as well as 1 or 2 hrs after the first dilution. Each sample was plated in non-selective CAT agar for single-colony isolation. Fifteen colonies obtained from each sample were individually used to inoculate 2 ml of CAT broth. On reaching visible turbity (OD ~ 0.02), a sample of each subculture was diluted 1:100 into three tubes containing CAT of CAT supplemented with 0.05 μg/ml Em or 200 μg/ml Kan. After 5 hrs incubation at 37C, all cultures were visibly turbid, but some of the Em cultures of clones from the fucose-treated culture failed to grow, in a proportion that was higher for longer periods of exposure to fucose (Fig. 3). PCR amplification of samples of the CAT cultures using primers to detect the presence of a spr1631-aphIII junction fragment revealed that the cassette was present only in subclones that exhibited growth in CAT + Em; all EmS clones had lost the cassette (Fig. 3). The construct was quite stable during growth without fucose, as all subclones from the culture were EmR and had maintained the Cheshire cassette during 8 hours incubation (~12 generations). In contrast, the percentage of erythromycin resistant colonies decreased rapidly during exposure to fucose. On reaching OD 0.8 in fucose medium, 40% of cfu were already free of the ermAM cassette, while after two further hours of growth with fucose at high cell density, an additional 40% of the cfu had become erythromycin sensitive and had lost the cre-aphIII junction fragment, indicating excision of the cassette from the genome. To determine whether the EmS cultures carried the predicted lox72 recombinant loxP site, two of the resulting EmS progeny were characterized by amplifying and sequencing the spr1632/aphIII region. In both cases, the results revealed that the entire Cheshire cassette was indeed precisely replaced by a single lox72 site.
Figure 3.
Fucose-driven excision of Cheshire. (A) Growth and EmR composition of cultures of CP2062. Parallel cultures were inoculated in CAT broth with or without 0.5% fucose at OD 0.001. The percentages of EmS clones were estimated from analysis of 15 isolated colonies from each sample, as shown in Panel B. Open bars, the percentage of EmR clones from the culture without fucose; filled bars, EmR from the fucose-treated culture. Results shown are repesentative of three similar experiments. (B) Clonal analysis of cultures after fucose treatment. 15 colonies obtained from each sample described in Panel A were analyzed as described in the text. Loss of the 1-kb PCR product indicates the excision of the cassette. Left images, results for the subclones from the culture grown without fucose; right images, results for those from 0.5%fucose-treated culture. +, growth in the indicated medium. −, no growth. Arrows, position of a 1-kb reference DNA band.
The aphIII::lox72 gene is predicted to encode a protein with 11 amino acid residues inserted between Ala2 and Ile14 of the native AphIII enzyme. None of the 90 subcultures in CAT+Kan broth were turbid after 6 hrs incubation at 37C, nor did diluted samples form colonies in agar with 200 μg/ml Kan, indicating that the extended AphIII protein is inactive. However, by 20 hrs’ incubation at 37C, all CAT+Kan cultures of the EmS subclones had become turbid (Fig. 3). Sequencing of the aphIII gene in pure subclones of two of these cultures revealed an intragenic suppressor mutation that apparently had restored sufficient AphIII activity to confer Kan resistance. Both had a G5A substitution within the lox72 element, which causes an R4H substitution in its predicted protein product. As all EmS revertants examined were similarly revertible (Fig. 3), but none of the EmR clones were, this pattern further confirms the absence of the ermAM insert, and restoration of the aphIII ORF in the EmS revertants. We conclude that the Cheshire cassette is quite stable in the pneumococcal genome, but is readily excised during a single cycle of culture with supplemental fucose.
4. Conclusion
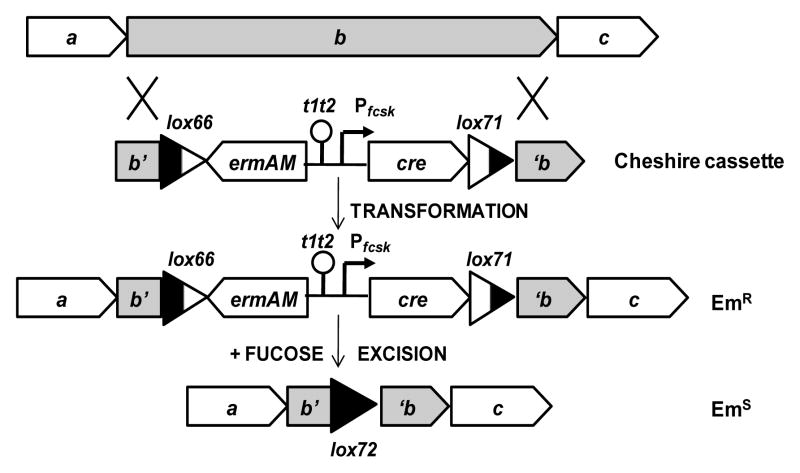
Cheshire is designed to be used as a temporary EmR marker for effecting various genetic rearrangements in pneumococcus. To delete a target sequence, for example, two sequences flanking the deletion target can be amplified from the genome and linked to the cassette to create a chimeric donor for transformation of competent cells (Fig. 4). After integration of the cassette in place of the target gene through homologous double cross-over recombination, and selection with erythromycin, the resulting EmR mutant can be grown without erythromycin but in the presence of fucose for expression of the cre gene. Cre-catalyzed excisive recombination between the two single-mutant loxP sites, lox66 and lox71, leaves a residual double-mutant loxP site known as lox72, reminiscent of the residual smile of the Cheshire cat (Dodgson, 1866). Because excision is efficient, minimal screening will readily identify EmS excision clones. As the lox72 ‘scar’ carries only a single stop codon in each strand, it permits the design of in-frame deletions and, as it is not recognized by Cre recombinase (Albet et al., 1995; Zhang et al., 2002), further loxP/Cre manipulations may be carried out in the new strain without the danger of mis-targeting of Cre.
Figure 4.
Application of the Cheshire cassette to create deletions in two steps in b, the gene targeted for deletion. a and c, the corresponding upstream and downstream genes. b’, ‘b, residual portions of gene b. Cassette elements indicated as in Fig. 1.
5. Discussion
Marker-less gene deletion methods are valuable in the construction of null mutants or other targeted rearrangements, especially in making multiple modifications in a single strain. In S. pneumoniae, several methods had been used to achieve this goal, but each is somewhat more complex than use of the present construct. Iannelli et al. and Standish et al., for example have used a method for introducing unmarked mutation into the genome of S. pneumoniae that employs SOEing PCR to construct markerless donor DNA and screening for integration of the deletion by numerous PCR assays. Although successful, this approach entails considerable lab work and depends on achieving a high level of competence. A drop-in/pop-out method for transferring arbitrary unmarked mutations into the pneumococcal genome was developed by Kloosterman et al., using a blue/white colony discrimination step to facilitate the screening for rare spontaneous pop-out excision clones, but as the mutation must first be assembled in a shuttle plasmid, it requires the added step of plasmid cloning of the mutant construct. Another method, developed by C.K. Sung, et al using the JANUS cassette, which contains the recessive streptomycin resistance gene rpsL, entails less labor, as it utilizes direct selection at both steps of transformation, but still requires two steps of transformation and a preexisting SmR mutation in the genome, and can be complicated by a significant background rate of SmR gene conversion events.
The Cre/lox system has enabled efficient routes to making marker-less deletions for broad applications in a large variety of species, including plants, the mouse, yeasts and bacteria (Albert et al., 1995; St-Onge et al., 1996; Sauer, 1987; Lambert et al., 2007). In this paper, we describe a new loxP/cre/loxP cassette suitable for use in pneumococcus that simplifies the process by saving two steps commonly used in bacterial Cre/loxP strategies: introduction of a Cre plasmid by transformation and the removal of the Cre plasmid after completion of lox recombination. The new cassette itself carries the tools for its own removal: the Cre recombinase gene and a promoter that places the cre gene expression under control of the fucose-responsive regulator, FcsR. Homology searches among 1354 sequenced bacterial genomes revealed that the fucose operon is widely conserved in S. pneumoniae, but is generally absent among other streptococci (except S. suis), and that the putative fucose regulator gene fcsR (spr1974 in strain R6) has a similar distribution. Thus, it is to be expected that this cassette will be widely useful in S. pneumoniae, but rarely in other hosts. However, the simple steps of replacing the fucose promoter by other inducible promoters, with suitably low basal expression according to the specific species of interest, or inclusion of fcsR in the cassette could easily adapt this lox/ermAM/cre/lox design to other bacteria.
Acknowledgments
We thank Gregory Roberston for kindly providing the ermAM-PfcsK cassette, Andrew Piotrowski for strain CP2000, and Stephen Leppla for the pCrePA plasmid. This work was supported in part by the National Science Foundation under Grant No. MCB-0543187.
References
- Albert H, Dale EC, Lee E, Ow DW. Site-specific integration of DNA into wild-type and mutant lox sites placed in the plant genome. Plant J. 1995;7:649–659. doi: 10.1046/j.1365-313x.1995.7040649.x. [DOI] [PubMed] [Google Scholar]
- Banerjee A, Biswas I. Markerless multiple-gene-deletion system for Streptococcus mutans. Appl Environ Microbiol. 2008;74:2037–2042. doi: 10.1128/AEM.02346-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan PF, O’Dwyer KM, Palmer LM, Ambrad JD, Ingraham KA, So C, Lonetto MA, Biswas S, Rosenberg M, Holmes DJ, Zalacain M. Characterization of a novel fucose-regulated promoter (PfcsK) suiy for gene essentiality and antibacterial mode-of-action studies in Streptococcus pneumoniae. J Bacteriol. 2003;185:2051–2058. doi: 10.1128/JB.185.6.2051-2058.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodgson Charles Lutwidge. Alice’s adventures in Wonderland. Macmillan & co; London: 1866. [Google Scholar]
- Ghosh K, Van Duyne GD. Cre-loxP biochemistry. Methods. 2002;28:374–383. doi: 10.1016/s1046-2023(02)00244-x. [DOI] [PubMed] [Google Scholar]
- Hare PD, Chua NH. Excision of selectable marker genes from transgenic plants. Nat Biotechnol. 2002;20:575–580. doi: 10.1038/nbt0602-575. [DOI] [PubMed] [Google Scholar]
- Iannelli F, Pozzi G. Method for introducing specific and unmarked mutations into the chromosome of Streptococcus pneumoniae. Mol Biotechnol. 2004;26:81–86. doi: 10.1385/MB:26:1:81. [DOI] [PubMed] [Google Scholar]
- Kloosterman TG, Bijlsma JJ, Kok J, Kuipers OP. To have neighbour’s fare, extending the molecular toolbox for Streptococcus pneumoniae. Microbiology. 2006;152:351–359. doi: 10.1099/mic.0.28521-0. [DOI] [PubMed] [Google Scholar]
- Lambert JM, Bongers RS, Kleerebezem M. Cre-lox-based system for multiple gene deletions and selectable-marker removal in Lactobacillus plantarum. Appl Environ Microbiol. 2007;73:1126–1135. doi: 10.1128/AEM.01473-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MS, Dougherty BA, Madeo AC, Morrison DA. Construction and analysis of a library for random insertional mutagenesis in Streptococcus pneumoniae, use for recovery of mutants defective in genetictransformation and for identification of essential genes. Appl Environ Microbiol. 1999;65:1883–1890. doi: 10.1128/aem.65.5.1883-1890.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibig M, Krismer B, Kolb M, Friede A, Gotz F, Bertram R. Marker removal in staphylococci via Cre recombinase and different lox sites. Appl. Environ. Microbiol. 2008;74:1316–1323. doi: 10.1128/AEM.02424-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison DA. Genetic Transformation in Streptococcus pneumoniae: Strategies for genome modification. In: Hakenbeck R, Chhatwal S, editors. Molecular Biology of Streptococci. Horizon Scientific Press; 2007. pp. 3–24. [Google Scholar]
- Ng WL, Robertson GT, Kazmierczak KM, Zhao J, Gilmour R, Winkler ME. Constitutive expression of PcsB suppresses the requirement for the essential VicR (YycF) response regulator in Streptococcus pneumoniae R6. Mol Microbiol. 2003;50:1647–1663. doi: 10.1046/j.1365-2958.2003.03806.x. [DOI] [PubMed] [Google Scholar]
- Pestova EV, Havarstein LS, Morrison DA. Regulation of competence for genetic transformation in Streptococcus pneumoniae by an auto-induced peptide pheromone and a two-component regulatory system. Mol Microbiol. 1996;21:853–862. doi: 10.1046/j.1365-2958.1996.501417.x. [DOI] [PubMed] [Google Scholar]
- Peterson SN, Sung CK, Cline R, Desai BV, Snesrud EC, Luo P, Walling J, Li H, Mintz M, Tsegaye G, Burr PC, Do Y, Ahn S, Gilbert J, Fleischmann RD, Morrison DA. Identification of competence pheromone responsive genes in Streptococcus pneumoniae by use of DNA microarrays. Mol Microbiol. 2004;51:1051–1070. doi: 10.1046/j.1365-2958.2003.03907.x. [DOI] [PubMed] [Google Scholar]
- Piotrowski A, Luo P, Morrison DA. Competence for Genetic Transformation in Streptococcus pneumoniae, Termination of Activity of the Alternative Sigma Factor ComX Is Independent of the Proteolysis of ComX and ComW. J Bacteriol. 2009 doi: 10.1128/JB.01750-08. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerantsev AP, Sitaraman R, Galloway CR, Kivovich V, Leppla SH. Genome engineering in Bacillus anthracis using Cre recombinase. Infect Immun. 2006;74:682–693. doi: 10.1128/IAI.74.1.682-693.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer B. Functional expression of the cre-lox site-specific recombination system in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:2087–2096. doi: 10.1128/mcb.7.6.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Onge L, Furth PA, Gruss P. Temporal control of the Cre recombinase in transgenic mice by a tetracycline responsive promoter. Nucleic Acids Res. 1996;24:3875–3877. doi: 10.1093/nar/24.19.3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Standish AJ, Stroeher UH, Paton JC. The two-component signal transduction system RR06/HK06 regulates expression of cbpA in Streptococcus pneumoniae. P N A S USA. 2005;102:7701–7706. doi: 10.1073/pnas.0409377102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung CK, Li H, Claverys JP, Morrison DA. An rpsL cassette, JANUS, for gene replacement through negative selection in Streptococcus pneumoniae. Appl Environ Microbiol. 2001;67:5190–5196. doi: 10.1128/AEM.67.11.5190-5196.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Yu H-J, Hong Q, Li SP. Cre/lox System and PCR-Based Genome Engineering in Bacillus subtilis. Appl Environ Microbiol. 2008;74:5556–5562. doi: 10.1128/AEM.01156-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Lutz B. Cre recombinase-mediated inversion using lox66 and lox71: method to introduce conditional point mutations into the CREB-binding protein. Nucleic Acids Res. 2002;30:e90. doi: 10.1093/nar/gnf089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J, Niu QW, Moller SG, Chua NH. Chemical-regulated, site-specific DNA excision in transgenic plants. Nat Biotechnol. 2001;19:157–161. doi: 10.1038/84428. [DOI] [PubMed] [Google Scholar]