Abstract
It has become clear that the products of several of the earliest identified myelin protein genes perform functions that extend beyond the myelin sheath. Interestingly, these myelin proteins, which comprise proteolipid protein, 2′,3′-cyclic nucleotide 3′-phosphodiesterase and the classic and golli MBPs (myelin basic proteins), play important roles during different stages of oligodendroglial development. These non-myelin-related functions are varied and include roles in the regulation of process outgrowth, migration, RNA transport, oligodendrocyte survival and ion channel modulation. However, despite the wide variety of cellular functions performed by the different myelin genes, the route by which they achieve these many functions seems to converge upon a common mechanism involving Ca2+ regulation, cytoskeletal rearrangements and signal transduction. In the present review, the newly emerging functions of these myelin proteins will be described, and these will then be discussed in the context of their contribution to oligodendroglial development.
Keywords: amyloid β-peptide (Aβ), calmodulin (CaM), central nervous system (CNS), extracellular matrix (ECM), oligodendrocyte (OL), proteolipid protein (PLP)
Abbreviations: Aβ, amyloid β-peptide; AMPA, α-amino-3-hydroxy-5-methylisoxazole-4-propionic acid; CaM, calmodulin; CNP, 2′,3′-cyclic nucleotide 3′-phosphodiesterase; CNS, central nervous system; ECM, extracellular matrix; ER, endoplasmic reticulum; GIP, golli-interacting protein; KO, knockout; MAG, myelin-associated glycoprotein; MARCKS, myristoylated alanine-rich C-kinase substrate; MBP, myelin basic protein; MS, multiple sclerosis; OL, oligodendrocyte; OPC, OL precursor cell; PDGF, platelet-derived growth factor; PLP, proteolipid protein; PNS, peripheral nervous system; RNAi, RNA interference; SH3 domain, Src homology 3 domain; SOCC, store-operated Ca2+ channel; VOCC, voltage-operated Ca2+ channel
INTRODUCTION
The myelin membrane provides an insulating sheath that facilitates the rapid conduction of electrical impulses along nerve fibres of the PNS (peripheral nervous system) and the CNS (central nervous system). Although mostly composed of lipids, these specialized extensions of the OL (oligodendrocyte) and Schwann cell membrane also contain a number of proteins, the so-called myelin proteins, which play a pivotal role in providing structure and integrity to the myelin sheath.
In the CNS, the major myelin proteins include myelin PLP (proteolipid protein) and the related product DM20, MBP (myelin basic protein), MAG (myelin-associated glycoprotein) and CNP (2′,3′-cyclic nucleotide 3′-phosphodiesterase). PLP and MBP are distributed within the myelin sheath and appear to be integral to the process of compaction that generates the closely apposed multilayered structure of mature myelin. MAG, on the other hand, is located in the periaxonal regions and may serve to facilitate cell–cell interactions between myelin and axonal membranes during the establishment of myelination. CNP is understood less well. Although it exhibits enzymatic activity in vitro, substrates for this action have not been identified within the myelin sheath and it has been proposed that this molecule may instead perform a structural role in the myelin membrane (Newman et al., 1995).
The products of myelin genes also appear to perform functions that extend beyond structural components of the myelin sheath. For example, as well as producing the well-known MBP and PLP products, both genes also encode a second class of protein respectively, called the golli MBP (Campagnoni et al., 1993; Pribyl et al., 1993) and, in the mouse, sr-PLP (soma-restricted PLP) proteins (Bongarzone et al., 1999) that result in alternatively spliced products with apparently different functions from the classical forms. Importantly, none of these proteins appear to be components of the myelin sheath, nor is their expression unique to myelin-forming cells, suggesting an involvement in cellular functions that are unrelated to myelination. Consistent with this idea is a growing body of work that has begun to identify specific roles for both ‘classic’ and non-classic-type myelin gene products that are unrelated to the generation of myelin. These include roles in the regulation of neurotransmitter receptors, Ca2+ channels, protein synthesis and cytoskeleton dynamics. Many of these newly described functions involve interactions with intracellular signalling pathways, placing these molecules in a prime position to influence the developmental programme of OLs. One interesting and likely possibility is that these proteins may play a role in controlling processes such as proliferation, migration and process extension in OPCs (OL precursor cells). In the present review, the newly emerging functions of four of these CNS myelin proteins: classic PLP, CNP, classic MBP and golli MBP, will be described and these findings will then be discussed with respect to the developmental processes that accompany the transition of OPCs into mature myelinating OLs.
PLP
The PLP gene produces two main isoforms, DM20 and PLP. DM20 protein expression in the CNS is highest during embryonic development, whereas PLP protein expression increases in relation to DM20 during postnatal development (LeVine et al., 1990). PLP protein differs from DM20 by the inclusion of 35 amino acids within the cytoplasmic domain of the protein. Two groups have examined the ability of DM20 to substitute the functions of the PLP isoform. Using a transgenic mouse in which the entire PLP gene was replaced by an altered gene encoding DM20, Stecca et al. (2000) found significant alterations in the structure of compacted myelin that was associated with a pronounced behavioural phenotype in aging mice. However, a second so-called DM20only mouse generated by replacing PLP exon III (responsible for generation of the PLP isoform) with a truncated exon III, lacking a critical sequence, formed essentially healthy myelin and did not exhibit a pathological phenotype, even after 48 months (Sporkel et al., 2002). Thus whether or not the DM20 and PLP isoforms perform different functions during the formation of compacted myelin remains unclear. Nonetheless, the identification of DM20 in non-myelinating cells suggests an alternative function for the protein in these cells (Campagnoni et al., 1992; Pribyl et al., 1996a, 1996b; Feng et al., 2003; Skoff et al., 2004a). Apart from its expression in mature OLs, the gene encoding PLP is also expressed in OPCs (Gudz et al., 2006), neurons (Bongarzone et al., 1999; Jacobs et al., 2003, 2004; Miller et al., 2003, 2009), embryonic neural precursors (Spassky et al., 1998; Delaunay et al., 2009) and non-neural cells (Campagnoni et al., 1992; Skoff et al., 2004a). The presence of PLP proteins in non-myelinating cells suggests an involvement in other functions unrelated to myelination. In line with this idea, PLP has been linked to the regulation of several cellular processes including ion exchange, cell migration and programmed cell death. These processes, all of which make important contributions to OPC development, will be discussed in the following sections.
Ion exchange
The earliest evidence linking PLP to transmembrane ion exchange came from studies using artificial plasma membranes. First, insertion of white-matter-derived PLP into lipid bilayers conferred these synthetic membranes with ionic currents. These conductances were voltage-dependent (Ting-Beall et al., 1979), and could be resolved at the level of single channels (Helynck et al., 1983), indicating an involvement of these proteins in the formation of ion channels in the plasma membrane. Secondly, studies using proteoliposomes containing PLP revealed that Ca2+ movements across these membranes were regulated by the interaction between PLP proteins and nucleotides such as MgATP (Diaz et al., 1990), providing further evidence of an involvement of the proteins in the exchange of ions across plasma membranes. Despite improvements in the techniques available for the examination of ion channel function in situ, few studies have attempted to examine the interactions of PLP with ion channels in intact cells (but see Gudz et al., 2006), and it is therefore unclear whether the functions that arise in synthetic preparations are representative of an endogenous role for PLP.
Transmembrane signalling, maturation and cell migration
As a membrane protein, PLP is well placed to participate in the transduction of signals between the ECM (extracellular matrix) and the interior of the cell. One potential route by which this may occur involves signalling through integrins, cell-surface proteins that function as receptors to ECM molecules. Through their communication with intracellular signalling pathways, integrins are well placed to stimulate a wide range of cellular actions, including migration, proliferation, differentiation and survival (Harburger and Calderwood, 2009). Importantly these effects are known to extend to cells in the OL family where integrins have been shown to influence survival (Frost et al., 1999), migration (Milner et al., 1996) and proliferation (Blaschuk et al., 2000). Given the multitude of cellular behaviours affected by ECM–integrin signalling, the ability to influence these interactions would confer PLP with a considerable degree of control over these key developmental processes. This possibility was advanced by the discovery that PLP forms a complex in OLs containing αvβ5 integrin and calreticulin, a Ca2+-binding protein (Gudz et al., 2002). The consequences of this complex for integrin function were revealed by studies showing enhanced levels of binding of OLs to the ECM protein fibronectin. Interestingly these actions were stimulated by the activation of mAchRs (muscarinic acetylcholine receptors), suggesting a role for neuronal signalling in controlling integrin/PLP signalling in OLs. How might these interactions between neurotransmitter signals and the PLP–αvβ5 complex regulate OL function? αvβ5 expression appears in differentiated OLs (Milner and Ffrench-Constant, 1994), and the activity of this integrin is associated with several features of maturation, including process extension and MBP protein expression (Blaschuk et al., 2000). Thus the sensitivity of PLP–αvβ5 activity to neurotransmitters such as acetylcholine may provide a means by which neuronal activity may regulate OL maturation and subsequent myelination.
Further work by Gudz et al. (2006) identified a similar complex in OPCs, this time involving an interaction between PLP and αvβ3 integrin which is involved in regulating the migration of these cells. Gudz et al. (2006) found that the ionotropic glutamate receptor agonist AMPA (α-amino-3-hydroxy-5-methylisoxazole-4-propionic acid) stimulated the formation of this complex, and that this action produced a significant enhancement in OPC migration. Importantly, this effect was absent from OPCs lacking PLP, showing that AMPA-receptor-dependent migration required the participation of PLP. Additionally, experiments measuring the ability of OPCs to bind to fibronectin indicated that binding to this ECM protein was reduced when cells were stimulated with AMPA (Gudz et al., 2006). Taken together, these results suggest that AMPA-dependent increases in migration stem in part from a reduction in ECM–integrin binding that in turn enhances the motility of these cells. These results also strengthen the hypothesis that PLP, through its actions as part of an integrin-containing complex, provides a means by which neurotransmitters may influence oligodendroglial function. Given the recent discovery of glutamatergic synaptic connections between unmyelinated callosal axons and white matter OPCs (Kukley et al., 2007; Ziskin et al., 2007), it will be intriguing to find out whether these AMPA receptor–PLP interactions also influence OPC migration in the more intact cellular environment provided by tissue slices.
OL survival
Studies of mutations in the PLP gene reveal an interesting dichotomy between effects on OL survival and myelination. While many PLP gene mutations result in the impairment of myelin sheath formation, these mutations are also associated with a substantial decrease in the survival of OLs (Knapp et al., 1986; Boison and Stoffel, 1989; Gow et al., 1998), which has been associated with an accumulation of PLP product in the ER (endoplasmic reticulum) of OLs, leading to apoptosis (Gow et al., 1998). Interestingly, the apparent effects of PLP on OL survival can occur well in advance of the generation of myelin proteins (Vermeesch et al., 1990). This functional dissociation is highlighted by the rumpshaker mouse, a PLP mutation involving a single amino acid substitution, which, on certain genetic backgrounds, produces significant deficiencies in myelination (Al-Saktawi et al., 2003), but generates healthy OLs that exhibit a normal level of survival (Schneider et al., 1992). Therefore, in some instances, mutations in the gene arise that spare functions related to survival, while impairing those that are important for myelination. One interpretation of the results obtained from PLP mutants is that these proteins perform a role in OL maturation that precedes the onset of myelin formation and which is critical to the survival of these cells. However, this interpretation is complicated by the fact that some of the effects on OL survival arising from gene mutations stem from the accumulation of misfolded PLP product in the ER (Gow et al., 1998; Dhaunchak and Nave, 2007); thus multiple factors may contribute to reduced OL survival in mice harbouring mutations in the PLP gene.
Further evidence connecting PLP to OL viability has emerged from cell culture studies showing opposing effects on survival when PLP protein expression is either enhanced, leading to a reduction in survival (Bongarzone et al., 2001), or reduced, leading to an increase in survival (Yang and Skoff, 1997). In the latter study, antisense molecules targeted to the initiation codon of PLP were used to decrease the level of PLP expression. In comparison with untreated cultures, cells subjected to antisense oligonucleotides showed a 90% reduction in PLP immunofluorescence, and this reduced level of PLP protein was associated with visible improvements in the number of healthy surviving OLs. The effects of PLP on survival were not restricted to cells in the OL family, or indeed neural cells. A study by Skoff et al. (2004a) shows that PLP is expressed in a number of developing non-neural cell types, and that the levels of PLP in these cells correlates with the degree of apoptosis. Taken together with the abundance of data highlighting the close relationship between OL survival and PLP mis-expression, these results from non-neural cells indicate that PLP plays a role in regulating apoptosis.
An indication of how PLP may influence cell death in OLs came from experiments in which neurons were co-cultured with a cell line expressing PLP. Levels of neuronal apoptosis were correlated with the number of PLP-expressing cells and, interestingly, this effect was associated with an acidification of the medium that was dependent on PLP expression. Since apoptosis is facilitated under low pH conditions, and expression of PLP appeared to acidify the medium, it seems possible that this influence on pH may contribute to the enhanced level of cell death associated with PLP expression. These findings were extended by experiments on CNS tissue isolated from a transgenic line overexpressing PLP in OLs (Skoff et al., 2004b). In that study, a proton flux assay revealed an enhanced release of protons from the PLP-overexpressing tissue. These results raise the possibility that decreased levels of survival in PLP-mutant OLs may be related to increased acidification of the surrounding extracellular space due to increased proton release. Further studies of PLP-overexpressing tissue have revealed several indications of mitochondrial dysfunction, including depolarization of the mitochondrial membrane potential, increased levels of cytochrome c oxidase and reductions in ATP levels (Hüttemann et al., 2009). Overall these results point towards a role for PLP in regulating OL mitochondrial function, which may in turn be responsible for the disruption in pH homoeostasis and cell survival associated with increased levels of PLP expression.
CNP
Constituting 4% of the protein content of the CNS myelin and expressed earlier than any other myelin protein, CNP remains something of an enigma: in vitro this molecule possesses enzymatic activity catalysing the cleavage of 2′,3′-cyclic nucleotides to 2′-nucleotides; however, years of research have failed to detect a substrate for this reaction within the myelin sheath. To date the function of CNP within the sheath remains unclear, although it has been posited that this enzymatic activity is now redundant and merely represents an evolutionary hangover in a molecule that has evolved to perform new roles (Gravel et al., 2009). Despite the uncertainty regarding the contribution of CNP catalytic activity to myelination, there has been significant progress in identifying functions for CNP beyond the myelin sheath, including roles in OL process outgrowth and RNA transport. Here, the focus will be on these CNP functions.
Morphological restructuring and process outgrowth
Analysis of CNP-null mice reveals that myelin forms normally in these animals (Lappe-Siefke et al., 2003). However, as these mice mature they begin to exhibit extensive neurodegeneration with a pronounced decline in axon integrity. These results point to a distinction between the function of myelin in supporting high-velocity action potential conduction and its role in providing trophic support to the axon. A later study of CNP-KO (knockout) mice revealed deficits in the formation of the paranodal loop, extensive cytoplasm-filled extensions of the OL membrane that enwrap the axon on either side of the node (Rasband et al., 2005). Importantly, these deficits preceded the onset of axon degeneration, suggesting a causative role for paranodal loop dysfunction in the decline in axonal integrity observed in these animals. This idea is supported by the fact that paranodal loops play an important role in the organization of nodal components, including voltage-gated Na+ channels (Rios et al., 2003), which are essential to axonal function. These deficits in loop formation point to a role for CNP in OL process outgrowth, a notion that is supported by the dramatic increase in membrane expansion exhibited by OLs in transgenic mice overexpressing CNP (Yin et al., 1997). The mechanism by which CNP influences process outgrowth was investigated first in cell lines transfected with CNP cDNA. Using this approach De Angelis and Braun (1994) demonstrated that expression of rat CNP in non-neural cell lines produced dramatic changes in the morphology of these cells. Exogenous CNP was found to be present at the plasma membrane, and at particularly high concentrations within processes and filopodia-like extensions. CNP was found to stimulate the extension of these filopodia, and these actions were dependent on prenylation, a reaction involving the addition of hydrophobic groups, which facilitates membrane associations. Clearly these actions on process outgrowth suggest putative interactions with the actin cytoskeleton, an idea that was confirmed in a second study by De Angelis and Braun (1996) that showed that CNP is retained within detergent-insoluble cell fractions (indicating association with the cytoskeleton) and is co-immunoprecipitated with actin. Taken collectively, the localization of CNP to the plasma membrane, the requirement for prenylation in its actions on process outgrowth and its associations with cytoskeletal elements suggest that the influence of CNP on process outgrowth involves the co-ordination of cytoskeletal dynamics with the plasma membrane.
CNP can also interact with the microtubule cytoskeleton, since it exhibits binding to soluble tubulin and can stimulate the polymerization of microtubules (Bifulco et al., 2002; Lee et al., 2005). These interactions appear to be important for CNP actions on process outgrowth. Expression of CNP in COS-7 cells leads to progressive changes in morphology, with cells passing through two clear stages. Early transformed cells adopted a flattened ‘spreading’ morphology with many short membrane protrusions, while late-transformed cells displayed an OL-like appearance with much elongated highly branched processes and a smaller cell body. Importantly, these changes in morphology were reflected by a biphasic pattern of microtubule dynamics. Early stage cells, which had begun the process of morphological remodelling, displayed a decrease in the levels of stable microtubules, whereas late-stage transformed cells that had acquired elongated OL-like processes displayed increases in the amount of stable tubulin filaments. Cell shape and rigidity are provided by cortical actin, and these F-actin cytoskeletal structures must be reorganized to permit microtubule-driven protrusions to extend the membrane. Given the previous associations identified between CNP and actin (De Angelis and Braun, 1994, 1996) it was likely that CNP-dependent changes in cell morphology would also involve dynamic changes in the F-actin cytoskeleton. In line with these expectations, phalloidin staining revealed reductions in F-actin in CNP-expressing cells during the early and late stages of transformation (Lee et al., 2005). Thus CNP-dependent changes in morphology were associated with co-ordinated changes in the microtubule and actin cytoskeleton.
To probe the requirement for CNP–actin associations in process outgrowth and to verify the relevance of these interactions in oligodendroglial cells, Lee et al. (2005) transfected OLN-93 cell lines with CNP constructs containing amino acid deletions. These constructs harboured deletions of two key C-terminal residues (Lys379 and Gly380) previously identified to be required for the action of CNP on microtubule polymerization. These experiments revealed that OLN-93 cells transfected with control CNP exhibited an increase in the number of processes bearing secondary branches and in the total number of processes, while the morphology of cells expressing double deletions remained unaltered. To link these effects in cell lines to native OL function, Lee et al. (2005) prepared primary cultures of OLs from CNP-null mice. As discussed above, these CNP-null animals exhibit deficiencies in the formation of the paranodal loop (Rasband et al., 2005), which may be related to the influence of CNP on process outgrowth. In agreement with this hypothesis, OLs cultured from these animals exhibited significantly fewer process branches compared with cells cultured from wild-type animals. Overall these studies in cell lines, primary cultures and tissues reveal important functions for CNP in the regulation of OL process outgrowth, which are likely to depend, at least in part, on the role of CNP as a regulator of cytoskeleton dynamics. Although these findings provide important insights into the function of CNP within OLs, a number of questions still remain. In particular, while CNP clearly influences morphological remodelling in mature OLs, it is not clear whether it also exerts a similar influence on precursor and immature OLs. This seems possible since CNP is expressed by OPCs (Scherer et al., 1994; Ye et al., 2003; Gobert et al., 2009), although to date no studies have examined the influence of this myelin protein on OPC function.
Roles of CNP2 in cell morphology and mitochondrial function
The CNP gene produces two isoforms, CNP1 and CNP2, both of which show robust expression in myelinating glial cells. Oligodendroglial expression of these isoforms peaks at the onset of myelination and is then sustained in the adult brain (Scherer et al., 1994). Previous work by Lee et al. (2006) revealed that the CNP2 isoform shows both cytoplasmic and mitochondrial localization. HeLa cells transfected with CNP2 constructs exhibited either predominant cytoplasmic or mitochondrial CNP2 localization. Interestingly, cells showing cytoplasmic CNP2 expression exhibited morphological remodelling similar to that previously observed in OL cell lines expressing CNP1 (Lee et al., 2005). However, cell morphology was not altered when CNP2 expression was restricted to mitochondria. Thus the function of CNP2 appears to differ depending on which cytoplasmic compartment it resides in. Currently there are no results available to explain the role of CNP2 within mitochondria. CNP2 is also expressed at low levels in many non-neural tissues (Scherer et al., 1994), where it appears to be preferentially localized to mitochondria (Lee et al., 2006). Together, the localization of CNP2 to non-neural and OL mitochondria, and the cell-compartment-specific dissociation between CNP2 functions during morphological restructuring, hint at a more general function beyond the generation of myelin.
RNA binding and translation inhibition
Several lines of evidence link CNP function to RNA metabolism. The catalytic domain of mammalian CNP contains two tetra-residue motifs, H-X-(T/S)-X, which place it within a superfamily of 2H phosphodiesterases (Kozlov et al., 2003), several members of which are associated with RNA editing. Importantly, these structural similarities appear to be reflected in shared functions since CNP is able to correct the impaired growth exhibited by yeast cells expressing mutated versions of two of these H-X-(T/S)-X residue-bearing enzymes (Schwer et al., 2008). In this study, the growth of yeast cells expressing mutated copies of tRNA ligase enzymes was rescued by expression of the rat CNP molecule. Importantly, CNP constructs harbouring mutations in the H-X-(T/S)-X motifs did not rescue yeast cell growth. This finding is significant since the same mutations render the construct catalytically dead, indicating that this activity is critical to the ability of rat CNP molecules to contribute to tRNA editing. Overall this study indicates that rat CNP is able to replace the function of tRNA ligase enzymes in yeast cells, and is suggestive of a role for CNP in RNA metabolism in animal cells. This possibility has received support from a recent study demonstrating that CNP molecules bind to RNA in vitro, and that this RNA-binding activity is also present in oligodendroglial cells lines (Gravel et al., 2009). RNA-binding activity was not compromised in a deletion mutant lacking the N-terminal region, showing that this binding was dependent on structures located within the catalytic portion of the molecule. Furthermore, the same catalytically dead mutant employed by Schwer et al. (2008) was still able to bind RNA. Thus although RNA binding (Gravel et al., 2009) and potential editing functions (Schwer et al., 2008) share a requirement for the catalytic domain, they can be distinguished by the requirement for CNP catalytic activity, suggesting that these functions arise from distinct regions within this catalytic domain.
This RNA-binding facility could have consequences for protein synthesis. Indeed, translation, as revealed by the activity of a luciferase reporter and through measurements of radiolabelled methionine, was significantly reduced by CNP (Gravel et al., 2009). Two additional lines of evidence suggest that the inhibitory influence of CNP on protein synthesis is likely to depend on its ability to interact with RNA. First, as was the case for the CNP–RNA-binding interactions, protein synthesis was also disrupted when the catalytically inactive mutant was included in the translation assay, and secondly, the closely related CNP homologue RICH (regeneration-induced CNP homologue), which does not exhibit RNA-binding activity, was not found to influence protein synthesis (Gravel et al., 2009). This inhibitory influence on protein synthesis could have implications for lineage progression as very recent work has identified the CNP gene as a negative regulator of OPC maturation (Gobert et al., 2009). In this study, RNAi (RNA interference) was used to selectively knock down CNP expression in the oli-neu OPC cell line. Inhibition of CNP translation increased the expression of PLP and MBP transcripts, and MBP protein, indicating increased maturation in this OPC-like cell line. One possibility therefore is that during OPC development CNP negatively regulates the translation of mRNAs that are required for OL differentiation such as PLP and MBP.
CNP may also be involved in mediating interactions between cytoskeletal elements and the translational machinery since it was found to facilitate associations between RNA and tubulin (Gravel et al., 2009). These results suggest an overlap between CNP–RNA binding and the cytoskeleton interactions described above (Lee et al., 2005), leading to the suggestion that CNP might have a role in the transport of RNA molecules along microtubules (Gravel et al., 2009). This idea has particular relevance to OLs and neurons since microtubules are known to play an important role in the binding and localization of RNA in these cells (Jansen, 1999). Future work is needed to validate this RNA transport hypothesis and to identify specific RNA molecules that can be bound by CNP and whose transport and translation may then be regulated.
THE MBP FAMILY
The MBP family comprises a variety of developmentally regulated members arising from different transcription start sites, differential splicing and post-translational modifications. The MBP gene is very large (>100 kb) and contains three transcription start sites, generating mRNAs that encode two families of proteins: classic MBPs and golli MBPs (Campagnoni et al., 1993; Pribyl et al., 1993). The classic MBPs are the products of transcription start sites 2 and 3 and are well known as major protein constituents of the myelin membrane. As such they are among the most abundant proteins in the CNS. They are also expressed almost exclusively in OLs. The classic isoforms of MBP include the 14- and 18.5-kDa isoforms, which predominate in adult human myelin and facilitate compaction of the mature myelin sheath, while the exon II-containing 17- and 21.5-kDa MBPs (MBPexII) are distributed diffusely through the cytoplasm and often accumulate in the nucleus of OLs (Allinquant et al., 1991; Hardy et al., 1996; Pedraza, 1997). In addition to membrane association, the classic MBP isoforms are able to interact with a multitude of proteins, including Ca2+-CaM (calmodulin), actin, tubulin and SH3 domain (Src homology 3 domain)-containing proteins, and thus may be signalling linkers during myelin development and remodelling (Harauz and Libich, 2009).
The golli proteins comprise the second family of proteins encoded by the MBP gene. These proteins are generated from the first transcription start site of the gene and are expressed more ubiquitously within the brain and other organs. In the mouse, three golli MBP isoforms have been identified: BG21, J37 and TP8 (Campagnoni et al., 1993). Both BG21 and J37 have segments in common with the classic 18.5 kDa isoform of MBP, but TP8 differs from this structure due to a frameshift mutation. They are expressed throughout the immune system in many cell types, including thymocytes and T-cells, and in the entire haemopoietic system, as well as in the nervous system (Feng et al., 2000; Marty et al., 2002; Feng, 2007).
All proteins in the classic/golli MBP family have intrinsically disordered structures (Harauz et al., 2004), creating a large effective surface to facilitate multiple protein associations, and are post-translationally modified to various degrees by methylation, phosphorylation and deimination, raising the possibility that MBPs might play roles as scaffolding proteins in intracellular signalling complexes.
Golli MBPs
The primary biological function of the classic MBPs appears to be that of a myelin structural component (Campagnoni and Skoff, 2001). Like the classic forms in the CNS, expression of golli proteins is generally developmentally regulated (Campagnoni et al., 1993; Landry et al., 1996). It tends to be high during embryonic development and declines with age (Landry et al., 1996, 1998). This developmental regulation does not appear to occur in the PNS, thymus or some brain regions, such as the olfactory system (Landry et al., 1997). In the nervous system, golli proteins are expressed in OLs and in specific subsets of neurons, although golli protein expression has also been reported in activated microglia, macrophages and adult OL progenitors around MS (multiple sclerosis) lesions (Filipovic et al., 2002).
Golli proteins are localized in process extensions of neural cells
Golli proteins are localized within the cytoplasm and process extensions of neural cells, and they are often first evident in these cells when they begin to migrate and extend processes (Pribyl et al., 1996c; Landry et al., 1997, 1998; Jacobs et al., 2009). Golli proteins were found in OLs within cell bodies and processes, including those processes that were contacting and surrounding axonal fibres (Pribyl et al., 1996c). Within the lateral funiculus, OLs immunopositive for golli protein can be seen along fibre tracts. In these cells, golli protein is distributed throughout the OL cell body, nucleus and the processes connecting these cells to the myelin sheath as they envelope axonal segments. Interestingly, immunohistochemistry of the embryonic spinal cord with golli antibody also reveals OLs at the earliest stages of myelin sheath production, at which point the protein is found to localize to the cell processes and terminal sheet-like structures (Pribyl et al., 1996c).
In vitro transfection studies have shown that overexpression of golli proteins, or indeed the golli domain alone, in OL cell lines can induce these cells to elaborate extensive processes and membrane sheets, adopting a morphology similar to mature OLs in culture (Reyes and Campagnoni, 2002; Paez et al., 2007). Similarly, transfection of golli into neuronal cell lines results in significant neurite extension, and in PC12 cells it induces neurite outgrowth reminiscent of that induced by NGF (nerve growth factor; Reyes and Campagnoni, 2002). These results complement the known expression of these products in vivo in a number of neuronal populations and in OL precursors during process extension and migration. Filipovic et al. (2002) have shown that golli immunoreactivity is high in adult OL progenitors surrounding MS plaques prior to remyelination attempts. All these results combine to suggest a role for golli proteins in process extension and myelin elaboration.
A role for golli in the nucleus
The golli proteins are also localized in the nucleus of many cells, and this localization is developmentally sensitive in a number of neuronal populations. For example, in cerebellar granule cells in the external granule cell layer, golli is localized in cell processes, but after their migration into the inner granule cell layer there is a shift in golli localization to the nucleus (Landry et al., 1996). It was found that the site responsible for this translocation is located within the MBP domain of the golli proteins (Reyes and Campagnoni, 2002). In other developing systems, this schedule is reversed such that golli protein expression is first seen within the nucleus or cell body and is later found only in the fibres. This later pattern of expression has been observed in neurons within the neostriatum (Landry et al., 1996) and cortical subplate (Landry et al., 1998). This behaviour is interesting because many cell signalling molecules shuttle between the nucleus and the cytoplasm.
A molecular partner for golli proteins, called GIP (golli-interacting protein), has been isolated by a yeast two-hybrid approach, which suggests that, in the nucleus, golli may be part of a transcriptional regulatory complex (Fernandes et al., 2004). GIP is a nuclear protein present in all cells in which golli has been found. It can bind to golli, and it can also bind to NLI (nuclear LIM interactor), which is known to be associated with LIM-containing nuclear complexes that are involved in regulating the transcription of some genes (Jurata et al., 1996). Thus, in the nucleus, golli is likely to be involved in the modulation of genes regulated by LIM family members.
Golli as a modulator of VOCCs (voltage-operated Ca2+ channels)
Early studies indicated that overexpression of golli proteins in cells transfected with golli–GFP (green fluorescent protein) induced immature OL cell lines (e.g. N19) to extend sheets and processes (Reyes and Campagnoni, 2002). Several studies in the literature indicate that Ca2+ is important in OL and OPC process extension (Pende et al., 1997; Stariha et al., 1997; Yoo et al., 1999). Since increased levels of both golli and intracellular Ca2+ lead to process outgrowth in OPCs, it seems likely that golli influences Ca2+ levels, leading to process extension. Pharmacological experiments demonstrate that VOCCs were involved in this golli-induced process extension. In the presence of VOCC antagonists, there was strong inhibition of process extension and membrane sheet formation induced by golli, indicating that this remodelling was mediated by VOCCs (Paez et al., 2007). High-resolution spatiotemporal analysis revealed that Ca2+ influx signals were initiated with different latencies at discrete cellular regions and were propagated along the N19 cell body and processes. High correlations between the patterns of local Ca2+ amplitudes and the patterns of golli protein distribution were consistently found. Cell regions with golli accumulations tended to co-localize with high levels of local Ca2+ signals, i.e. ‘hot spots’. These studies indicated that golli-induced process extension is associated with increases in Ca2+ influx and accumulations of golli near these regions in OPC processes (Paez et al., 2007). Additionally, a domain analysis of the golli protein using a deletion approach identified a myristoylation site at the C-terminus, which appears to be essential for these golli actions, suggesting that binding of golli to the plasma membrane is important for modulating Ca2+ homoeostasis (Paez et al., 2007).
These studies in cell lines were later confirmed using primary cultures of OPCs and acute brain slice preparations from golli-KO (Jacobs et al., 2005) and JOE (J37-golli-overexpressing) mice (P.M. Paez, D.J. Fulton, V. Spreur, V. Handley, C.W. Campagnoni and A.T. Campagnoni, unpublished work). Resting Ca2+ levels did not differ between KO and control OPCs, but the magnitudes of the Ca2+ response to several stimuli were significantly lower in KO cells. In contrast and consistent with the KO results, enhanced Ca2+ influx was found to be induced by depolarization of golli-overexpressing cells. This was the predicted result, and was opposite to the effects observed in golli-KO OPCs. In golli-overexpressing cells, increases in Ca2+ uptake were abolished in zero Ca2+ and were blocked by Cd2+, verapamil and nifedipine, confirming that this rise in [Ca2+]i (intracellular [Ca2+]) results from Ca2+ influx through VOCCs. Additionally, differences in the amplitude of Ca2+ uptake between genotypes were enhanced by Bay K 8644, an L-type Ca2+ channel agonist, which enhances channel function by prolonging single channel open times.
Golli promotes oligodendroglial cell migration
Migration of OL progenitor cells from proliferative zones to their final location in the brain is an essential step in nervous system development. As noted above, golli proteins can modulate voltage-operated Ca2+ uptake in OPCs during process extension and retraction (Paez et al., 2007). Given the importance of process extension/retraction to movement, Paez et al. (2009a) examined the consequences of golli expression on OPC migration in vivo and in vitro using time-lapse imaging of isolated OPCs and in acute brain slice preparations from golli-KO and golli-overexpressing mice. They found that golli stimulated migration, and this enhanced motility was associated with an increase in the activity of VOCCs. Activation of VOCCs by elevated external [K+] resulted in a significant increase in the migration speed of golli-overexpressing OPCs compared with control cells, and golli-mediated modulation of OPC migration disappeared in the presence of VOCC antagonists. During migration, OPCs generated Ca2+ oscillations that were dependent on voltage-gated Ca2+ influx, and both the amplitude and frequency of these Ca2+ transients correlated positively with the rate of cell movement under a variety of pharmacological treatments. The Ca2+ transient amplitude and the rate of cell movement were significantly lower in golli-KO cells and significantly higher in golli-overexpressing cells, suggesting that the presence of golli promotes OPC migration by increasing the size of voltage-mediated Ca2+ oscillations (Paez et al., 2009a). Thus golli modulation of VOCCs accelerates cell migration by promoting Ca2+-dependent soma translocation and leading to process formation. This mechanism points to a key role for golli proteins in regulating the rate of OPC migration through spontaneous Ca2+ oscillations and for the first time provides evidence that functional voltage-gated Ca2+ channels are necessary for the migration of OPCs.
Golli enhances store-operated Ca2+ uptake in OPCs
The role of golli MBPs in regulating OPC development was examined in a recent study by Paez et al. (2009b). In OPCs lacking or overexpressing golli, differentiation induced by growth factor withdrawal was impaired. Proliferation analysis in the presence of PDGF (platelet-derived growth factor) revealed that golli enhanced the mitogen-stimulated proliferation of OPCs through the activation of SOCCs (store-operated Ca2+ channels). The PDGF treatment induced a biphasic increase in OPC intracellular Ca2+, and golli specifically increased Ca2+ influx during the second SOCC-dependent phase that followed the initial release of Ca2+ from intracellular stores. This store-operated Ca2+ uptake appeared to be essential for cell division since specific SOCC antagonists completely blocked the PDGF- and golli-dependent effects on OPC proliferation (Paez et al., 2009b). Proliferation of OPCs is clearly important for normal myelination as well as for remyelination in demyelinating diseases. The above-mentioned study indicates that golli overexpression increases the proliferation rate of OPCs as assessed by several parameters including a decrease in the cell cycle time. In contrast, golli-KO OPCs proliferated less robustly and showed a significant increase in the length of the cell cycle. Thus it is likely that golli promotes enhanced proliferation of OPCs in the presence of mitogens. In this regard, Filipovic and Zecevic (2005) reported that LPS (lipopolysaccharide)-induced inflammation promoted OPC proliferation through secretion of golli proteins from activated microglia. In summary, golli MBPs can modulate Ca2+ uptake through VOCCs and SOCCs and this regulation can directly affect early OPC functions such as process remodelling, migration and proliferation.
Golli as an adaptor protein
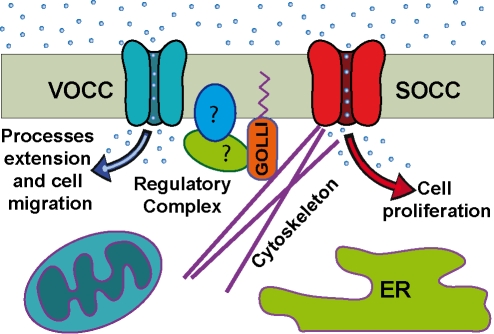
By what mechanism could golli MBP modulate the activity of Ca2+ channels? It is possible that golli serves as an adaptor protein in a complex that may directly or indirectly regulate the activity of Ca2+ channels (e.g. SOCCs and VOCCs) (Figure 1). It is possible that this complex (or a similar one) regulates cytoskeletal structures that lead to the observed morphological changes in the OPC when golli (or CNP) are overexpressed in OLs. This complex may also interact in some way with the mitochondria and ER, which have been shown by Simpson and Russell (1997) to be involved in regulating the levels of Ca2+ at Ca2+-influx microdomains in OPC processes and along the plasma-membrane surfaces of OPCs. One possible role of golli is to serve as an adaptor protein as part of a complex that might bridge the modulation of Ca2+ influx through Ca2+ channels, and the transduction of signals from events at the membrane to the cytoskeleton. Essentially all of the Ca2+-mediated events in which golli plays a role involve some change in the OPC cytoskeleton (e.g. proliferation, process extension and migration). Also, in immunoprecipitation/proteomic studies on membrane fractions, we have consistently identified cytoskeletal proteins associated with golli, particularly α- and β-tubulin (C.W. Campagnoni, unpublished work). This finding is significant since modulation of ion channels and transporters by the actin and tubulin components of the cytoskeleton is a common phenomenon (Johnson and Byerly, 1993; Galli and DeFelice, 1994; Nakamura et al., 2000).
Figure 1. Hypothetical model of a golli-associated complex that may modulate the activity of Ca2+ channels in OPCs.
Golli is shown associated with the plasma membrane through a myristoylated moiety (purple) at its N-terminus, and in association with the cytoskeleton, possibly through its MBP domain.
Golli has the appropriate physical properties to be an adaptor protein in one of these regulatory complexes. To date, no enzymatic activity has been associated with the golli proteins. However, by virtue of their high hydrophilicity and net charge, they fit the definition of intrinsically unstructured proteins. Harauz et al. (2000) have shown that golli proteins form extended structures in solution that would be ideal for binding other molecules in a signalling assembly. This notion of golli proteins serving as potential adaptor proteins in signalling complexes is supported by other studies showing that they can bind CaM in a Ca2+-dependent fashion, and that they are also able to bind phosphoinositides (Kaur et al., 2003). Additionally, the N-terminal myristoylation of golli is essential for membrane association and is critical for the ability of golli proteins to regulate Ca2+ uptake (Feng et al., 2006; Paez et al., 2007) and migration (Paez et al., 2009a) in OPCs.
The classic MBPs
The classic MBPs bind to negatively charged lipids on the cytosolic surface of the OL membrane, primarily through electrostatic interactions (Boggs and Moscarello, 1982; Boggs et al., 1982). They are present throughout compact internodal myelin and are recognized to be involved in the adhesion of the cytosolic surfaces of the multilayered myelin sheath, where they have been localized by immunoelectron microscopy and X-ray diffraction (Omlin et al., 1982; Readhead et al., 1990; Kirschner and Blaurock, 1992).
However, proteins of the classic MBP family have been found to bind to a number of molecules and targeted to many subcellular locations (Hardy et al., 1996; Pedraza, 1997), making it difficult to resolve the specific functions these myelin proteins may play in cells. For example, the roles for MBP have been suggested in the phosphoinositide signalling pathway that affect axonal cytoskeletal organization (Wood and Moscarello, 1997; Lintner and Dyer, 2000; Reyes and Campagnoni, 2002), in the nucleus (Staugaitis et al., 1996), in interactions with the cytoskeleton (Boggs et al., 2000, 2005, 2006; Boggs, 2006; Libich et al., 2003), and a regulatory function on the expression of other myelin proteins (Hardy et al., 1996; Carre et al., 2002).
There have, however, been consistent findings, suggesting a possible role for classic MBPs in signal transduction pathways and in cytoskeletal organization. Additionally, there is an intriguing comparison of both golli and classic MBPs with other proteins involved in signal transduction, such as the MARCKS (myristoylated alanine-rich C-kinase substrate) proteins (Arbuzova et al., 2002): classic and golli MBPs and MARCKS are intrinsically unstructured, are phosphorylated by both PKC (protein kinase C) and MAPK (mitogen-activated protein kinase), bind acidic lipids strongly, and also bind CaM and actin (Arbuzova et al., 2002).
Classic MBP interactions with cytoskeletal proteins
Like golli proteins, classic MBPs interact with the cytoskeleton in OLs, in cytosolic inclusions in myelin, and even in compact myelin, where classic MBP, actin and tubulin occur in the radial component, a series of tight junctions that pass through many layers of myelin (Pereyra et al., 1988; Gillespie et al., 1989; Karthigasan et al., 1994). MBP in solution binds to F-actin in a 1:1 molar ratio and induces the formation of ordered bundles of F-actin filaments (Barylko and Dobrowolski, 1984). It also binds to G-actin in solution at an MBP/actin molar ratio of 1:2 and causes its polymerization into filaments under non-polymerizing low ionic strength conditions (Dobrowolski et al., 1986; Roth et al., 1993). MBP also binds Ca2+-CaM (Grand and Perry, 1980; Chan et al., 1990) and this interaction results in dissociation of MBP from actin bundles (Barylko and Dobrowolski, 1984) and in depolymerization of actin filaments bound to MBP in solution (Dobrowolski et al., 1986). Binding to CaM has been shown to be Ca2+-dependent (Barylko and Dobrowolski, 1984, Dobrowolski et al., 1986). Although MBP can interact with actin in solution, it is not known whether it can also interact with actin when MBP is bound to the membrane surface. Along these lines, Boggs and Rangaraj (2000) have shown that MBP can still interact with actin filaments when it is bound to acidic lipid vesicles in vitro and that CaM can still regulate binding of actin filaments under these experimental conditions.
It is not yet known whether the interaction of classic MBP with actin is a non-specific electrostatic interaction or if it has a physiological role. However, the interaction of membrane-bound MBP with actin suggests that MBP might be able to bind actin to the cytosolic surface of the OL or myelin plasma membrane. Co-localization studies by immunohistochemistry indicate that MBP is closely associated with microtubules and actin microfilaments in immature OLs (Wilson and Brophy, 1989). Recently, Boggs et al. (2006) have shown that some MBPs are also co-localized with actin at the edge of OL membrane sheets where they might bind actin to the membrane and participate in membrane sheet extension.
It has been repeatedly proposed that MBP may have a role in the interactions of microtubules with membrane extensions, raising the possibility that MBP may affect microtubule functions essential for cell morphogenesis (Wilson and Brophy, 1989; Barry et al., 1996). In shiverer mice OLs, which lack MBP, actin microfilaments are not co-localized with microtubular structures as they are in the wild-type OLs (Dyer et al., 1995). MBP is attached to the membrane through lipids and apparently aligns with microtubules in sheet extensions (Lunn et al., 1997; Song et al., 1999). Additionally, classic MBP has been suggested to be involved in microtubule reorganization induced by anti-Gal C antibody (Dyer and Benjamins, 1988). Consistent with a role for classic MBP in microtubule regulation, it has been shown that MBP has microtubule-stabilizing activity in vitro (Pirollet et al., 1992), and that MBP can promote tubulin assembly into microtubules and can bundle microtubules in vitro (Hill et al., 2005). Previously, using RNAi techniques in cells derived from control and MBP-deficient shiverer mice, Galiano et al. (2006) have tested the microtubule-stabilizing activity of MBP at late stages of OL differentiation. They showed that MBP functions as a microtubule-stabilizing protein when expressed and localized in cell processes.
Proteinase activity of classic MBPs
It was reported that purified MBP possesses endogenous serine proteinase activity and can undergo autocatalytic cleavage (D’Souza et al., 2005). Ser151 was identified as the active serine residue involved in autocatalysis (D’Souza et al., 2005). Hoos et al. (2007, 2009) have shown that MBP binds Aβ (amyloid β-peptide) and inhibits Aβ fibril formation. The progressive accumulation of fibrillar Aβ in senile plaques and in the cerebral vasculature is the hallmark of Alzheimer's disease and related disorders. Recently, the same group has reported that Aβ peptides and assembled fibrillar Aβ are degraded by purified human brain MBP and recombinant human MBP, both in vitro and in situ (Liao et al., 2009). MBP-mediated Aβ degradation is inhibited by serine proteinase inhibitors, and cell lines expressing MBP degrade exogenous Aβ peptides in vitro (Liao et al., 2009). In addition, it has also been proposed that MBP could counteract the surface structure of Aβ fibril-mediated cytotoxicity (Yoshiike et al., 2007). In any case, future in vivo studies are warranted to further examine the potential functions of classic MBPs in the inhibition of Aβ fibril assembly, Aβ degradation and the modulation of Aβ-mediated cytotoxicity that may play a role in the pathogenesis associated with amyloid deposition.
Classic MBP may serve in signalling complexes
Classic MBP has been classified as an intrinsically disordered protein (Harauz et al., 2004). This unordered structure confers on it the flexibility to interact with an array of negatively charged surfaces and ligands, and to acquire whatever local conformation is necessary to optimize binding to several different targets. In addition to negatively charged lipids and cytoskeletal proteins, the MBP classic isoforms are able to interact with SH3-domain-containing proteins (Harauz and Libich, 2009). Using SH3 domain microarrays, Polverini et al. (2008) have found that the unmodified recombinant murine 18.5 kDa MBP isoform shows in vitro binding to the following proteins containing SH3 domains: Yes1, PSD-95 (postsynaptic density-95), cortactin, PexD, Abl, Fyn and c-Src. Recently, Homchaudhuri et al. (2009) have shown that increasing the negative surface charge of the membrane by increasing the proportion of phosphatidylinositol reduces the amount of Fyn-SH3 domain that binds to membrane-associated MBP. These results suggest that changes in membrane-negative surface charge due to protein or lipid modifications, which could occur during cell signalling, can regulate the binding of the Fyn-SH3 domain to MBP and thus could regulate the activity of Fyn at the OL plasma membrane.
The ability of classic MBPs to bind cytoskeletal and signalling proteins containing SH3 domains to a lipid bilayer suggests that, by functioning as a scaffolding protein in OLs or myelin, it could act to organize these proteins into signalling complexes. The interaction of MBP with the SH3 domains of proteins, in addition to its already well-established interactions with cytoskeletal proteins, points to a more defined role for MBP in signal transduction. Although all of these studies have been suggestive it is not yet known whether these findings underlie a novel physiological function in cells.
SUMMARY
In conclusion, considerable progress has been made in identifying alternative functions for some myelin protein genes, and it is now clear that their products influence a diverse array of cellular processes that are unrelated to their well-known functions within the myelin membrane. In particular, these proteins are able to interact with signalling elements located at multiple levels in the pathways regulating oligodendroglial function. For example, through their influence on membrane receptors and ion channels PLP and golli MBP regulate responses to external stimuli and modulate the ensuing intracellular signals, while extensive coupling to cytoskeletal structures places all of these molecules in a prime position to regulate a wide variety of cellular behaviours, including process extension, migration, protein translation, proliferation and cell survival. Given this multitude of influences, and a capacity to integrate within signalling complexes, it appears that the products of myelin protein genes contribute substantially to the physiological development and function of cells at several stages in the differentiation of the OL lineage. Finally, several of these myelin proteins show prominent expression in developing tissues located outside of the nervous system (O’Neill et al., 1997; Marty et al., 2002; Feng et al., 2003, 2006; Skoff et al., 2004a), suggesting that they play a wider role in the regulation of processes contributing to development.
REFERENCES
- Al-Saktawi K, McLaughlin M, Klugmann M, Schneider A, Barrie JA, McCulloch MC, Montague P, Kirkham D, Nave KA, Griffiths IR. Genetic background determines phenotypic severity of the PLP rumpshaker mutation. J Neurosci Res. 2003;72:12–24. doi: 10.1002/jnr.10561. [DOI] [PubMed] [Google Scholar]
- Allinquant B, Staugaitis SM, D’Urso D, Colman DR. The ectopic expression of myelin basic protein isoforms in Shiverer oligodendrocytes: implications for myelinogenesis. J Cell Biol. 1991;113:393–403. doi: 10.1083/jcb.113.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbuzova A, Schmitz AA, Vergeres G. Cross-talk unfolded: MARCKS proteins. Biochem J. 2002;362:1–12. doi: 10.1042/0264-6021:3620001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry C, Pearson C, Barbarese E. Morphological organization of oligodendrocyte processes during development in culture and in vivo. Dev Neurosci. 1996;18:233–242. doi: 10.1159/000111413. [DOI] [PubMed] [Google Scholar]
- Barylko B, Dobrowolski Z. Ca2+-calmodulin-dependent regulation of F-actin-myelin basic protein interaction. Eur J Cell Biol. 1984;35:327–335. [PubMed] [Google Scholar]
- Bifulco M, Laezza C, Stingo S, Wolff J. 2′,3′-Cyclic nucleotide 3′-phosphodiesterase: a membrane-bound, microtubule-associated protein and membrane anchor for tubulin. Proc Natl Acad Sci USA. 2002;99:1807–1812. doi: 10.1073/pnas.042678799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaschuk KL, Frost EE, Ffrench-Constant C. The regulation of proliferation and differentiation in oligodendrocyte progenitor cells by αV integrins. Development. 2000;127:1961–1969. doi: 10.1242/dev.127.9.1961. [DOI] [PubMed] [Google Scholar]
- Boggs JM, Moscarello MA. Interdigitation of fatty acid chains of dipalmitoylphosphatidylglycerol due to intercalation of myelin basic protein. Biophys J. 1982;37:57–59. doi: 10.1016/S0006-3495(82)84597-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggs JM, Stamp D, Moscarello MA. Effect of pH and fatty acid chain length on the interaction of myelin basic protein with phosphatidylglycerol. Biochemistry. 1982;21:1208–1214. doi: 10.1021/bi00535a016. [DOI] [PubMed] [Google Scholar]
- Boggs JM, Rangaraj G. Interaction of lipid-bound myelin basic protein with actin filaments and calmodulin. Biochemistry. 2000;39:7799–7806. doi: 10.1021/bi0002129. [DOI] [PubMed] [Google Scholar]
- Boggs JM, Rangaraj G, Koshy KM, Mueller JP. Adhesion of acidic lipid vesicles by 21.5 kDa (recombinant) and 18.5 kDa isoforms of myelin basic protein. Biochim Biophys Acta. 2000;1463:81–87. doi: 10.1016/s0005-2736(99)00181-9. [DOI] [PubMed] [Google Scholar]
- Boggs JM, Rangaraj G, Hill CM, Bates IR, Heng YM, Harauz G. Effect of arginine loss in myelin basic protein, as occurs in its deiminated charge isoform, on mediation of actin polymerization and actin binding to a lipid membrane in vitro. Biochemistry. 2005;44:3524–3534. doi: 10.1021/bi0473760. [DOI] [PubMed] [Google Scholar]
- Boggs JM. Myelin basic protein: a multifunctional protein. Cell Mol Life Sci. 2006;63:1945–1961. doi: 10.1007/s00018-006-6094-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggs JM, Rangaraj G, Gao W, Heng YM. Effect of phosphorylation of myelin basic protein by MAPK on its interactions with actin and actin binding to a lipid membrane in vitro. Biochemistry. 2006;45:391–401. doi: 10.1021/bi0519194. [DOI] [PubMed] [Google Scholar]
- Boison D, Stoffel W. Myelin-deficient rat: a point mutation in exon III (A----C, Thr75----Pro) of the myelin proteolipid protein causes dysmyelination and oligodendrocyte death. EMBO J. 1989;8:3295–3302. doi: 10.1002/j.1460-2075.1989.tb08490.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongarzone ER, Campagnoni CW, Kampf K, Jacobs EC, Handley VW, Schonmann V, Campagnoni AT. Identification of a new exon in the myelin proteolipid protein gene encoding novel protein isoforms that are restricted to the somata of oligodendrocytes and neurons. J Neurosci. 1999;19:8349–8357. doi: 10.1523/JNEUROSCI.19-19-08349.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongarzone ER, Jacobs E, Schonmann V, Kampf K, Campagnoni CW, Campagnoni AT. Differential sensitivity in the survival of oligodendrocyte cell lines to overexpression of myelin proteolipid protein gene products. J Neurosci Res. 2001;65:485–492. doi: 10.1002/jnr.1178. [DOI] [PubMed] [Google Scholar]
- Campagnoni CW, Garbay B, Micevych P, Pribyl T, Kampf K, Handley VW, Campagnoni AT. DM20 mRNA splice product of the myelin proteolipid protein gene is expressed in the murine heart. J Neurosci Res. 1992;33:148–155. doi: 10.1002/jnr.490330119. [DOI] [PubMed] [Google Scholar]
- Campagnoni AT, Pribyl TM, Campagnoni CW, Kampf K, Amur-Umarjee S, Landry CF, Handley VW, Newman SL, Garbay B, Kitamura K. Structure and developmental regulation of golli-MBP, a 105-kilobase gene that encompasses the myelin basic protein gene and is expressed in cells in the oligodendrocyte lineage in the brain. J Biol Chem. 1993;268:4930–4938. [PubMed] [Google Scholar]
- Campagnoni AT, Skoff RP. The pathobiology of myelin mutants reveal novel biological functions of the MBP and PLP genes. Brain Pathol. 2001;11:74–91. doi: 10.1111/j.1750-3639.2001.tb00383.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carre JL, Goetz BD, O’Connor LT, Bremer Q, Duncan ID. Mutations in the rat myelin basic protein gene are associated with specific alterations in other myelin gene expression. Neurosci Lett. 2002;330:17–20. doi: 10.1016/s0304-3940(02)00709-7. [DOI] [PubMed] [Google Scholar]
- Chan KF, Robb ND, Chen WH. Myelin basic protein: interaction with calmodulin and gangliosides. J Neurosci Res. 1990;25:535–544. doi: 10.1002/jnr.490250410. [DOI] [PubMed] [Google Scholar]
- De Angelis DA, Braun PE. Isoprenylation of brain 2′,3′-cyclic nucleotide 3′-phosphodiesterase modulates cell morphology. J Neurosci Res. 1994;39:386–397. doi: 10.1002/jnr.490390405. [DOI] [PubMed] [Google Scholar]
- De Angelis DA, Braun PE. 2′,3′-Cyclic nucleotide 3′-phosphodiesterase binds to actin-based cytoskeletal elements in an isoprenylation-independent manner. J Neurochem. 1996;67:943–951. doi: 10.1046/j.1471-4159.1996.67030943.x. [DOI] [PubMed] [Google Scholar]
- Delaunay D, Heydon K, Miguez A, Schwab M, Nave KA, Thomas JL, Spassky N, Martinez S, Zalc B. Genetic tracing of subpopulation neurons in the prethalamus of mice (Mus musculus). J Comp Neurol. 2009;512:74–83. doi: 10.1002/cne.21904. [DOI] [PubMed] [Google Scholar]
- Dhaunchak AS, Nave KA. A common mechanism of PLP/DM20 misfolding causes cysteine-mediated endoplasmic reticulum retention in oligodendrocytes and Pelizaeus–Merzbacher disease. Proc Natl Acad Sci USA. 2007;104:17813–17818. doi: 10.1073/pnas.0704975104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz RS, Monreal J, Lucas M. Calcium movements mediated by proteolipid protein and nucleotides in liposomes prepared with the endogenous lipids from brain white matter. J Neurochem. 1990;55:1304–1309. doi: 10.1111/j.1471-4159.1990.tb03139.x. [DOI] [PubMed] [Google Scholar]
- Dobrowolski Z, Osinska H, Mossakowska M, Barylko B. Ca2+-calmodulin-dependent polymerization of actin by myelin basic protein. Eur J Cell Biol. 1986;42:17–26. [PubMed] [Google Scholar]
- D’Souza CA, Wood DD, She YM, Moscarello MA. Autocatalytic cleavage of myelin basic protein: an alternative to molecular mimicry. Biochemistry. 2005;44:12905–12913. doi: 10.1021/bi051152f. [DOI] [PubMed] [Google Scholar]
- Dyer CA, Benjamins JA. Antibody to galactocerebroside alters organization of oligodendroglial membrane sheets in culture. J Neurosci. 1988;8:4307–4318. doi: 10.1523/JNEUROSCI.08-11-04307.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer CA, Philibotte TM, Billings-Gagliardi S, Wolf MK. Cytoskeleton in myelin-basic-protein-deficient shiverer oligodendrocytes. Dev Neurosci. 1995;17:53–62. doi: 10.1159/000111273. [DOI] [PubMed] [Google Scholar]
- Feng JM, Givogri IM, Bongarzone ER, Campagnoni C, Jacobs E, Handley VW, Schonmann V, Campagnoni AT. Thymocytes express the golli products of the myelin basic protein gene and levels of expression are stage dependent. J Immunol. 2000;165:5443–5450. doi: 10.4049/jimmunol.165.10.5443. [DOI] [PubMed] [Google Scholar]
- Feng JM, Fernandes AO, Bongarzone ER, Campagnoni CW, Kampf K, Campagnoni AT. Expression of soma-restricted proteolipid/DM20 proteins in lymphoid cells. J Neuroimmunol. 2003;144:9–15. doi: 10.1016/j.jneuroim.2003.08.003. [DOI] [PubMed] [Google Scholar]
- Feng JM, Hu YK, Xie LH, Colwell CS, Shao XM, Sun XP, Chen B, Tang H, Campagnoni AT. Golli protein negatively regulates store depletion-induced calcium influx in T cells. Immunity. 2006;24:717–727. doi: 10.1016/j.immuni.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Feng JM. Minireview: expression and function of golli protein in immune system. Neurochem Res. 2007;32:273–278. doi: 10.1007/s11064-006-9164-1. [DOI] [PubMed] [Google Scholar]
- Fernandes AO, Campagnoni CW, Kampf K, Feng JM, Handley VW, Schonmann V, Bongarzone ER, Reyes S, Campagnoni AT. Identification of a protein that interacts with the golli-myelin basic protein and with nuclear LIM interactor in the nervous system. J Neurosci Res. 2004;75:461–471. doi: 10.1002/jnr.10882. [DOI] [PubMed] [Google Scholar]
- Filipovic R, Rakic S, Zecevic N. Expression of golli proteins in adult human brain and multiple sclerosis lesions. J Neuroimmunol. 2002;127:1–12. doi: 10.1016/s0165-5728(02)00070-x. [DOI] [PubMed] [Google Scholar]
- Filipovic R, Zecevic N. Lipopolysaccharide affects golli expression and promotes proliferation of oligodendrocyte progenitors. Glia. 2005;49:457–466. doi: 10.1002/glia.20125. [DOI] [PubMed] [Google Scholar]
- Frost EE, Buttery PC, Milner R, Ffrench-Constant C. Integrins mediate a neuronal survival signal for oligodendrocytes. Curr Biol. 1999;9:1251–1254. doi: 10.1016/s0960-9822(99)80506-5. [DOI] [PubMed] [Google Scholar]
- Galiano MR, Andrieux A, Deloulme JC, Bosc C, Schweitzer A, Job D, Hallak ME. Myelin basic protein functions as a microtubule stabilizing protein in differentiated oligodendrocytes. J Neurosci Res. 2006;84:534–541. doi: 10.1002/jnr.20960. [DOI] [PubMed] [Google Scholar]
- Galli A, DeFelice LJ. Inactivation of L-type Ca channels in embryonic chick ventricle cells: dependence on the cytoskeletal agents colchicine and taxol. Biophys J. 1994;67:2296–2304. doi: 10.1016/S0006-3495(94)80715-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie CS, Wilson R, Davidson A, Brophy PJ. Characterization of a cytoskeletal matrix associated with myelin from rat brain. Biochem J. 1989;260:689–696. doi: 10.1042/bj2600689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobert RP, Joubert L, Curchod ML, Salvat C, Foucault I, Jorand-Lebrun C, Lamarine M, Peixoto H, Vignaud C, Fremaux C, Jomotte T, Francon B, Alliod C, Bernasconi L, Abderrahim H, Perrin D, Bombrun A, Zanoguera F, Rommel C, Hooft van Huijsduijnen R. Convergent functional genomics of oligodendrocyte differentiation identifies multiple autoinhibitory signaling circuits. Mol Cell Biol. 2009;29:1538–1553. doi: 10.1128/MCB.01375-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gow A, Southwood CM, Lazzarini RA. Disrupted proteolipid protein trafficking results in oligodendrocyte apoptosis in an animal model of Pelizaeus–Merzbacher disease. J Cell Biol. 1998;140:925–934. doi: 10.1083/jcb.140.4.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grand RJ, Perry SV. The binding of calmodulin to myelin basic protein and histone H2B. Biochem J. 1980;189:227–240. doi: 10.1042/bj1890227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel M, Robert F, Kottis V, Gallouzi IE, Pelletier J, Braun PE. 2′,3′-Cyclic nucleotide 3′-phosphodiesterase: a novel RNA-binding protein that inhibits protein synthesis. J Neurosci Res. 2009;87:1069–1079. doi: 10.1002/jnr.21939. [DOI] [PubMed] [Google Scholar]
- Gudz TI, Schneider TE, Haas TA, Macklin WB. Myelin proteolipid protein forms a complex with integrins and may participate in integrin receptor signaling in oligodendrocytes. J Neurosci. 2002;22:7398–7407. doi: 10.1523/JNEUROSCI.22-17-07398.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudz TI, Komuro H, Macklin WB. Glutamate stimulates oligodendrocyte progenitor migration mediated via an αv integrin/myelin proteolipid protein complex. J Neurosci. 2006;26:2458–2466. doi: 10.1523/JNEUROSCI.4054-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harauz G, Ishiyama N, Bates IR. Analogous structural motifs in myelin basic protein and in MARCKS. Mol Cell Biochem. 2000;209:155–163. doi: 10.1023/a:1007176216360. [DOI] [PubMed] [Google Scholar]
- Harauz G, Ishiyama N, Hill CM, Bates IR, Libich DS, Fares C. Myelin basic protein-diverse conformational states of an intrinsically unstructured protein and its roles in myelin assembly and multiple sclerosis. Micron. 2004;35:503–542. doi: 10.1016/j.micron.2004.04.005. [DOI] [PubMed] [Google Scholar]
- Harauz G, Libich DS. The classic basic protein of myelin – conserved structural motifs and the dynamic molecular barcode involved in membrane adhesion and protein–protein interactions. Curr Protein Pept Sci. 2009;10:196–215. doi: 10.2174/138920309788452218. [DOI] [PubMed] [Google Scholar]
- Harburger DS, Calderwood DA. Integrin signalling at a glance. J Cell Sci. 2009;122:159–163. doi: 10.1242/jcs.018093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy RJ, Lazzarini RA, Colman DR, Friedrich VL., Jr Cytoplasmic and nuclear localization of myelin basic proteins reveals heterogeneity among oligodendrocytes. J Neurosci Res. 1996;46:246–257. doi: 10.1002/(SICI)1097-4547(19961015)46:2<246::AID-JNR13>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Helynck G, Luu B, Nussbaum JL, Picken D, Skalidis G, Trifilieff E, Van Dorsselaer A, Seta P, Sandeaux R, Gavach C, Heitz F, Simon D, Spach G. Brain proteolipids. Isolation, purification and effect on ionic permeability of membranes. Eur J Biochem. 1983;133:689–695. doi: 10.1111/j.1432-1033.1983.tb07518.x. [DOI] [PubMed] [Google Scholar]
- Hill CM, Libich DS, Harauz G. Assembly of tubulin by classic myelin basic protein isoforms and regulation by post-translational modification. Biochemistry. 2005;44:16672–16683. doi: 10.1021/bi050646+. [DOI] [PubMed] [Google Scholar]
- Homchaudhuri L, Polverini E, Gao W, Harauz G, Boggs J. Influence of membrane surface charge and post-translational modifications to myelin basic protein on its ability to tether the Fyn-SH3 domain to a membrane in vitro. Biochemistry. 2009;48:2385–2393. doi: 10.1021/bi8022587. [DOI] [PubMed] [Google Scholar]
- Hoos MD, Ahmed M, Smith SO, Van Nostrand WE. Inhibition of familial cerebral amyloid angiopathy mutant amyloid β-protein fibril assembly by myelin basic protein. J Biol Chem. 2007;282:9952–9961. doi: 10.1074/jbc.M603494200. [DOI] [PubMed] [Google Scholar]
- Hoos MD, Ahmed M, Smith SO, Van Nostrand WE. Myelin basic protein binds to and inhibits the fibrillar assembly of Aβ42 in vitro. Biochemistry. 2009;48:4720–4727. doi: 10.1021/bi900037s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hüttemann M, Zhang Z, Mullins C, Bessert D, Lee I, Nave KA, Appikatla S, Skoff RP. Different proteolipid protein mutants exhibit unique metabolic defects. ASN NEURO. 2009 doi: 10.1042/AN20090028. 1(3):art:e00014.doi:10.1042/AN20090028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs C, Reyes D, Campagnoni CW, Givogri IM, Kampf K, Handley V, Spreuer V, Fisher RS, Macklin W, Campagnoni AT. Targeted overexpression of a golli-myelin basic protein isoform to oligodendrocytes results in aberrant oligodendrocyte maturation and myelination. ASN NEURO. 2009 doi: 10.1042/AN20090029. 1(4):art:e00017.doi:10.1042/AN20090029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs EC, Bongarzone ER, Campagnoni CW, Kampf K, Campagnoni AT. Soma-restricted products of the myelin proteolipid gene are expressed primarily in neurons in the developing mouse nervous system. Dev Neurosci. 2003;25:96–104. doi: 10.1159/000072259. [DOI] [PubMed] [Google Scholar]
- Jacobs EC, Bongarzone ER, Campagnoni CW, Campagnoni AT. Embryonic expression of the soma-restricted products of the myelin proteolipid gene in motor neurons and muscle. Neurochem Res. 2004;29:997–1002. doi: 10.1023/b:nere.0000021244.38279.c4. [DOI] [PubMed] [Google Scholar]
- Jacobs EC, Pribyl TM, Feng JM, Kampf K, Spreur V, Campagnoni C, Colwell CS, Reyes SD, Martin M, Handley V, Hiltner TD, Readhead C, Jacobs RE, Messing A, Fisher RS, Campagnoni AT. Region-specific myelin pathology in mice lacking the golli products of the myelin basic protein gene. J Neurosci. 2005;25:7004–7013. doi: 10.1523/JNEUROSCI.0288-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen RP. RNA-cytoskeletal associations. FASEB J. 1999;13:455–466. [PubMed] [Google Scholar]
- Johnson BD, Byerly L. A cytoskeletal mechanism for Ca2+ channel metabolic dependence and inactivation by intracellular Ca2+. Neuron. 1993;10:797–804. doi: 10.1016/0896-6273(93)90196-x. [DOI] [PubMed] [Google Scholar]
- Jurata LW, Kenny DA, Gill GN. Nuclear LIM interactor, a rhombotin and LIM homeodomain interacting protein, is expressed early in neuronal development. Proc Natl Acad Sci USA. 1996;93:11693–11698. doi: 10.1073/pnas.93.21.11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karthigasan J, Kosaras B, Nguyen J, Kirschner DA. Protein and lipid composition of radial component-enriched CNS myelin. J Neurochem. 1994;62:1203–1213. doi: 10.1046/j.1471-4159.1994.62031203.x. [DOI] [PubMed] [Google Scholar]
- Kaur J, Libich DS, Campagnoni CW, Wood DD, Moscarello MA, Campagnoni AT, Harauz G. Expression and properties of the recombinant murine Golli-myelin basic protein isoform J37. J Neurosci Res. 2003;71:777–784. doi: 10.1002/jnr.10547. [DOI] [PubMed] [Google Scholar]
- Kirschner DA, Blaurock AE. Organization, phylogenetic variations, and dynamic transitions of myelin. In: Martenson RE, editor. Myelin: Biology and Chemistry. Boca Raton, FL: CRC Press; 1992. pp. 1–78. [Google Scholar]
- Knapp PE, Skoff RP, Redstone DW. Oligodendroglial cell death in jimpy mice: an explanation for the myelin deficit. J Neurosci. 1986;6:2813–2822. doi: 10.1523/JNEUROSCI.06-10-02813.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov G, Lee J, Elias D, Gravel M, Gutierrez P, Ekiel I, Braun PE, Gehring K. Structural evidence that brain cyclic nucleotide phosphodiesterase is a member of the 2H phosphodiesterase superfamily. J Biol Chem. 2003;278:46021–46028. doi: 10.1074/jbc.M305176200. [DOI] [PubMed] [Google Scholar]
- Kukley M, Capetillo-Zarate E, Dietrich D. Vesicular glutamate release from axons in white matter. Nat Neurosci. 2007;10:311–320. doi: 10.1038/nn1850. [DOI] [PubMed] [Google Scholar]
- Landry CF, Ellison JA, Pribyl TM, Campagnoni C, Kampf K, Campagnoni AT. Myelin basic protein gene expression in neurons: developmental and regional changes in protein targeting within neuronal nuclei, cell bodies, and processes. J Neurosci. 1996;16:2452–2462. doi: 10.1523/JNEUROSCI.16-08-02452.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry CF, Ellison J, Skinner E, Campagnoni AT. Golli-MBP proteins mark the earliest stages of fiber extension and terminal arboration in the mouse peripheral nervous system. J Neurosci Res. 1997;50:265–271. doi: 10.1002/(SICI)1097-4547(19971015)50:2<265::AID-JNR15>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Landry CF, Pribyl TM, Ellison JA, Givogri MI, Kampf K, Campagnoni CW, Campagnoni AT. Embryonic expression of the myelin basic protein gene: identification of a promoter region that targets transgene expression to pioneer neurons. J Neurosci. 1998;18:7315–7327. doi: 10.1523/JNEUROSCI.18-18-07315.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappe-Siefke C, Goebbels S, Gravel M, Nicksch E, Lee J, Braun PE, Griffiths IR, Nave KA. Disruption of CNP1 uncouples oligodendroglial functions in axonal support and myelination. Nat Genet. 2003;33:366–374. doi: 10.1038/ng1095. [DOI] [PubMed] [Google Scholar]
- Liao MC, Ahmed M, Smith SO, Van Nostrand WE. Degradation of amyloid beta protein by purified myelin basic protein. J Biol Chem. 2009;284:28917–28925. doi: 10.1074/jbc.M109.050856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Gravel M, Zhang R, Thibault P, Braun PE. Process outgrowth in oligodendrocytes is mediated by CNP, a novel microtubule assembly myelin protein. J Cell Biol. 2005;170:661–673. doi: 10.1083/jcb.200411047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, O’Neill RC, Park MW, Gravel M, Braun PE. Mitochondrial localization of CNP2 is regulated by phosphorylation of the N-terminal targeting signal by PKC: implications of a mitochondrial function for CNP2 in glial and non-glial cells. Mol Cell Neurosci. 2006;31:446–462. doi: 10.1016/j.mcn.2005.10.017. [DOI] [PubMed] [Google Scholar]
- LeVine SM, Wong D, Macklin WB. Developmental expression of proteolipid protein and DM20 mRNAs and proteins in the rat brain. Dev Neurosci. 1990;12:235–250. doi: 10.1159/000111853. [DOI] [PubMed] [Google Scholar]
- Libich DS, Hill CM, Haines JD, Harauz G. Myelin basic protein has multiple calmodulin-binding sites. Biochem Biophys Res Commun. 2003;308:313–319. doi: 10.1016/s0006-291x(03)01380-9. [DOI] [PubMed] [Google Scholar]
- Lintner RN, Dyer CA. Redistribution of cholesterol in oligodendrocyte membrane sheets after activation of distinct signal transduction pathways. J Neurosci Res. 2000;60:437–449. doi: 10.1002/(SICI)1097-4547(20000515)60:4<437::AID-JNR2>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Lunn KF, Baas PW, Duncan ID. Microtubule organization and stability in the oligodendrocyte. J Neurosci. 1997;17:4921–4932. doi: 10.1523/JNEUROSCI.17-13-04921.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marty MC, Alliot F, Rutin J, Fritz R, Trisler D, Pessac B. The myelin basic protein gene is expressed in differentiated blood cell lineages and in hemopoietic progenitors. Proc Natl Acad Sci USA. 2002;99:8856–8861. doi: 10.1073/pnas.122079599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MJ, Haxhiu MA, Georgiadis P, Gudz TI, Kangas CD, Macklin WB. Proteolipid protein gene mutation induces altered ventilatory response to hypoxia in the myelin-deficient rat. J Neurosci. 2003;23:2265–2273. doi: 10.1523/JNEUROSCI.23-06-02265.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MJ, Kangas CD, Macklin WB. Neuronal expression of the proteolipid protein gene in the medulla of the mouse. J Neurosci Res. 2009;87:2842–2853. doi: 10.1002/jnr.22121. [DOI] [PubMed] [Google Scholar]
- Milner R, Ffrench-Constant C. A developmental analysis of oligodendroglial integrins in primary cells: changes in α v-associated β subunits during differentiation. Development. 1994;120:3497–3506. doi: 10.1242/dev.120.12.3497. [DOI] [PubMed] [Google Scholar]
- Milner R, Edwards G, Streuli C, Ffrench-Constant C. A role in migration for the αVβ1 integrin expressed on oligodendrocyte precursors. J Neurosci. 1996;16:7240–7252. doi: 10.1523/JNEUROSCI.16-22-07240.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Sunagawa M, Kosugi T, Sperelakis N. Actin filament disruption inhibits L-type Ca2+ channel current in cultured vascular smooth muscle cells. Am J Physiol Cell Physiol. 2000;279:C480–C487. doi: 10.1152/ajpcell.2000.279.2.C480. [DOI] [PubMed] [Google Scholar]
- Newman S, Saito M, Yu RK. Myelin. In: Kettenmann H, Ransom BR, editors. Neuroglia. Oxford: Oxford University Press; 1995. pp. 535–554. [Google Scholar]
- O’Neill RC, Minuk J, Cox ME, Braun PE, Gravel M. CNP2 mRNA directs synthesis of both CNP1 and CNP2 polypeptides. J Neurosci Res. 1997;50:248–257. doi: 10.1002/(SICI)1097-4547(19971015)50:2<248::AID-JNR13>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Omlin FX, Webster HD, Palkovits CG, Cohen SR. Immunocytochemical localization of basic protein in major dense line regions of central and peripheral myelin. J Cell Biol. 1982;95:242–248. doi: 10.1083/jcb.95.1.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paez PM, Spreuer V, Handley V, Feng JM, Campagnoni C, Campagnoni AT. Increased expression of golli myelin basic proteins enhances calcium influx into oligodendroglial cells. J Neurosci. 2007;27:12690–12699. doi: 10.1523/JNEUROSCI.2381-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paez PM, Fulton DJ, Spreuer V, Handley V, Campagnoni C, Macklin WB, Colwell CS, Campagnoni AT. Golli myelin basic proteins regulate oligodendroglial progenitor cell migration through voltage-operated Ca++ influx. J Neurosci. 2009a;20:6663–6676. doi: 10.1523/JNEUROSCI.5806-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paez PM, Fulton DJ, Spreuer V, Handley V, Campagnoni C, Campagnoni AT. Regulation of store-operated and voltage operated Ca++ channels in the proliferation and death of oligodendrocyte precursor cells by Golli proteins. ASN Neuro. 2009b doi: 10.1042/AN20090003. 1:art:e00003.doi:00010.01042/AN2009003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedraza L. Nuclear transport of myelin basic protein. J Neurosci Res. 1997;50:258–264. doi: 10.1002/(SICI)1097-4547(19971015)50:2<258::AID-JNR14>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Pende M, Fisher TL, Simpson PB, Russell JT, Blenis J, Gallo V. Neurotransmitter- and growth factor-induced cAMP response element binding protein phosphorylation in glial cell progenitors: role of calcium ions, protein kinase C, and mitogen-activated protein kinase/ribosomal S6 kinase pathway. J Neurosci. 1997;17:1291–1301. doi: 10.1523/JNEUROSCI.17-04-01291.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereyra PM, Horvath E, Braun PE. Triton X-100 extractions of central nervous system myelin indicate a possible role for the minor myelin proteins in the stability in lamellae. Neurochem Res. 1988;13:583–595. doi: 10.1007/BF00973301. [DOI] [PubMed] [Google Scholar]
- Pirollet F, Margolis RL, Job D. Ca2+-calmodulin regulated effectors of microtubule stability in neuronal tissues. Biochim Biophys Acta. 1992;1160:113–119. doi: 10.1016/0167-4838(92)90044-e. [DOI] [PubMed] [Google Scholar]
- Polverini E, Rangaraj G, Libich DS, Boggs JM, Harauz G. Binding of the proline-rich segment of myelin basic protein to SH3 domains: spectroscopic, microarray, and modeling studies of ligand conformation and effects of posttranslational modifications. Biochemistry. 2008;47:267–282. doi: 10.1021/bi701336n. [DOI] [PubMed] [Google Scholar]
- Pribyl TM, Campagnoni CW, Kampf K, Kashima T, Handley VW, McMahon J, Campagnoni AT. The human myelin basic protein gene is included within a 179-kilobase transcription unit: expression in the immune and central nervous systems. Proc Natl Acad Sci USA. 1993;90:10695–10699. doi: 10.1073/pnas.90.22.10695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pribyl TM, Campagnoni C, Kampf K, Handley VW, Campagnoni AT. The major myelin protein genes are expressed in the human thymus. J Neurosci Res. 1996a;45:812–819. doi: 10.1002/(SICI)1097-4547(19960915)45:6<812::AID-JNR18>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Pribyl TM, Campagnoni CW, Kampf K, Kashima T, Handley VW, McMahon J, Campagnoni AT. Expression of the myelin proteolipid protein gene in the human fetal thymus. J Neuroimmunol. 1996b;67:125–130. doi: 10.1016/0165-5728(96)00058-6. [DOI] [PubMed] [Google Scholar]
- Pribyl TM, Campagnoni CW, Kampf K, Ellison JA, Landry CF, Kashima T, McMahon J, Campagnoni AT. Expression of the myelin basic protein gene locus in neurons and oligodendrocytes in the human fetal central nervous system. J Comp Neurol. 1996c;374:342–353. doi: 10.1002/(SICI)1096-9861(19961021)374:3<342::AID-CNE3>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Rasband MN, Tayler J, Kaga Y, Yang Y, Lappe-Siefke C, Nave KA, Bansal R. CNP is required for maintenance of axon-glia interactions at nodes of Ranvier in the CNS. Glia. 2005;50:86–90. doi: 10.1002/glia.20165. [DOI] [PubMed] [Google Scholar]
- Readhead C, Takasashi N, Shine HD, Saavedra R, Sidman R, Hood L. Role of myelin basic protein in the formation of central nervous system myelin. Ann NY Acad Sci. 1990;605:280–285. doi: 10.1111/j.1749-6632.1990.tb42401.x. [DOI] [PubMed] [Google Scholar]
- Reyes SD, Campagnoni AT. Two separate domains in the golli myelin basic proteins are responsible for nuclear targeting and process extension in transfected cells. J Neurosci Res. 2002;69:587–596. doi: 10.1002/jnr.10319. [DOI] [PubMed] [Google Scholar]
- Rios JC, Rubin M, St Martin M, Downey RT, Einheber S, Rosenbluth J, Levinson SR, Bhat M, Salzer JL. Paranodal interactions regulate expression of sodium channel subtypes and provide a diffusion barrier for the node of Ranvier. J Neurosci. 2003;23:7001–7011. doi: 10.1523/JNEUROSCI.23-18-07001.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth GA, Gonzalez MD, Monferran CG, De Santis ML, Cumar FA. Myelin basic protein domains involved in the interaction with actin. Neurochem Int. 1993;23:459–465. doi: 10.1016/0197-0186(93)90130-w. [DOI] [PubMed] [Google Scholar]
- Scherer SS, Braun PE, Grinspan J, Collarini E, Wang DY, Kamholz J. Differential regulation of the 2′,3′-cyclic nucleotide 3′-phosphodiesterase gene during oligodendrocyte development. Neuron. 1994;12:1363–1375. doi: 10.1016/0896-6273(94)90451-0. [DOI] [PubMed] [Google Scholar]
- Schneider A, Montague P, Griffiths I, Fanarraga M, Kennedy P, Brophy P, Nave KA. Uncoupling of hypomyelination and glial cell death by a mutation in the proteolipid protein gene. Nature. 1992;358:758–761. doi: 10.1038/358758a0. [DOI] [PubMed] [Google Scholar]
- Schwer B, Aronova A, Ramirez A, Braun P, Shuman S. Mammalian 2′,3′ cyclic nucleotide phosphodiesterase (CNP) can function as a tRNA splicing enzyme in vivo. RNA. 2008;14:204–210. doi: 10.1261/rna.858108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson PB, Russell JT. Role of sarcoplasmic/endoplasmic-reticulum Ca2+-ATPases in mediating Ca2+ waves and local Ca2+-release microdomains in cultured glia. Biochem J. 1997;325:239–247. doi: 10.1042/bj3250239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoff RP, Bessert DA, Cerghet M, Franklin MJ, Rout UK, Nave KA, Carlock L, Ghandour MS, Armant DR. The myelin proteolipid protein gene modulates apoptosis in neural and non-neural tissues. Cell Death Differ. 2004a;11:1247–1257. doi: 10.1038/sj.cdd.4401498. [DOI] [PubMed] [Google Scholar]
- Skoff RP, Saluja I, Bessert D, Yang X. Analyses of proteolipid protein mutants show levels of proteolipid protein regulate oligodendrocyte number and cell death in vitro and in vivo. Neurochem Res. 2004b;29:2095–2103. doi: 10.1007/s11064-004-6882-0. [DOI] [PubMed] [Google Scholar]
- Song J, O’Connor L T, Yu W, Baas PW, Duncan ID. Microtubule alterations in cultured taiep rat oligodendrocytes lead to deficits in myelin membrane formation. J Neurocytol. 1999;28:671–683. doi: 10.1023/a:1007060832459. [DOI] [PubMed] [Google Scholar]
- Spassky N, Goujet-Zalc C, Parmantier E, Olivier C, Martinez S, Ivanova A, Ikenaka K, Macklin W, Cerruti I, Zalc B, Thomas JL. Multiple restricted origin of oligodendrocytes. J Neurosci. 1998;18:8331–8343. doi: 10.1523/JNEUROSCI.18-20-08331.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporkel O, Uschkureit T, Bussow H, Stoffel W. Oligodendrocytes expressing exclusively the DM20 isoform of the proteolipid protein gene: myelination and development. Glia. 2002;37:19–30. doi: 10.1002/glia.10014. [DOI] [PubMed] [Google Scholar]
- Stariha RL, Kikuchi S, Siow YL, Pelech SL, Kim M, Kim SU. Role of extracellular signal-regulated protein kinases 1 and 2 in oligodendroglial process extension. J Neurochem. 1997;68:945–953. doi: 10.1046/j.1471-4159.1997.68030945.x. [DOI] [PubMed] [Google Scholar]
- Staugaitis SM, Colman DR, Pedraza L. Membrane adhesion and other functions for the myelin basic proteins. BioEssays. 1996;18:13–18. doi: 10.1002/bies.950180106. [DOI] [PubMed] [Google Scholar]
- Stecca B, Southwood CM, Gragerov A, Kelley KA, Friedrich VL, Jr, Gow A. The evolution of lipophilin genes from invertebrates to tetrapods: DM-20 cannot replace proteolipid protein in CNS myelin. J Neurosci. 2000;20:4002–4010. doi: 10.1523/JNEUROSCI.20-11-04002.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting-Beall HP, Lees MB, Robertson JD. Interactions of Folch–Lees proteolipid apoprotein with planar lipid bilayers. J Membr Biol. 1979;51:33–46. doi: 10.1007/BF01869342. [DOI] [PubMed] [Google Scholar]
- Vermeesch MK, Knapp PE, Skoff RP, Studzinski DM, Benjamins JA. Death of individual oligodendrocytes in jimpy brain precedes expression of proteolipid protein. Dev Neurosci. 1990;12:303–315. doi: 10.1159/000111859. [DOI] [PubMed] [Google Scholar]
- Wilson R, Brophy PJ. Role for the oligodendrocyte cytoskeleton in myelination. J Neurosci Res. 1989;22:439–448. doi: 10.1002/jnr.490220409. [DOI] [PubMed] [Google Scholar]
- Wood DD, Moscarello MA. Molecular biology of the glia: components of myelin-myelin basic protein-the implications of post-translational changes for demyelinating disease. In: Russell WC, editor. Molecular Biology of Multiple Sclerosis. Chichester: Wiley; 1997. pp. 37–54. [Google Scholar]
- Yang X, Skoff RP. Proteolipid protein regulates the survival and differentiation of oligodendrocytes. J Neurosci. 1997;17:2056–2070. doi: 10.1523/JNEUROSCI.17-06-02056.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye P, Bagnell R, D’Ercole AJ. Mouse NG2+ oligodendrocyte precursors express mRNA for proteolipid protein but not its DM-20 variant: a study of laser microdissection-captured NG2+ cells. J Neurosci. 2003;23:4401–4405. doi: 10.1523/JNEUROSCI.23-11-04401.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X, Peterson J, Gravel M, Braun PE, Trapp BD. CNP overexpression induces aberrant oligodendrocyte membranes and inhibits MBP accumulation and myelin compaction. J Neurosci Res. 1997;50:238–247. doi: 10.1002/(SICI)1097-4547(19971015)50:2<238::AID-JNR12>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Yoo AS, Krieger C, Kim SU. Process extension and intracellular Ca2+ in cultured murine oligodendrocytes. Brain Res. 1999;827:19–27. doi: 10.1016/s0006-8993(99)01282-2. [DOI] [PubMed] [Google Scholar]
- Yoshiike Y, Akagi T, Takashima A. Surface structure of amyloid-beta fibrils contributes to cytotoxicity. Biochemistry. 2007;46:9805–9812. doi: 10.1021/bi700455c. [DOI] [PubMed] [Google Scholar]
- Ziskin JL, Nishiyama A, Rubio M, Fukaya M, Bergles DE. Vesicular release of glutamate from unmyelinated axons in white matter. Nat Neurosci. 2007;10:321–330. doi: 10.1038/nn1854. [DOI] [PMC free article] [PubMed] [Google Scholar]