Abstract
Zebrafish mutants have traditionally been obtained using random mutagenesis or retroviral insertions, methods that cannot be targeted to a specific gene and require laborious gene mapping and sequencing. Recently, we and others have shown that customized zinc finger nucleases (ZFNs) can introduce targeted frame-shift mutations with high efficiency, thereby enabling directed creation of zebrafish gene mutations. Here we describe a detailed protocol for constructing ZFN expression vectors, for generating and introducing ZFN-encoding RNAs into zebrafish embryos, and for identifying ZFN-generated mutations in targeted genomic sites. All of our vectors and methods are compatible with previously described Zinc Finger Consortium reagents for constructing engineered zinc finger arrays. Using these methods, zebrafish founders carrying targeted mutations can be identified within four months.
INTRODUCTION
Zebrafish have emerged in recent years as an important and popular vertebrate model organism for studying gene function and as an alternative model system for understanding human diseases. However, one drawback that limits the utility of this model organism is the lack of technology to efficiently generate targeted mutations in genes of interest. Engineered zinc finger nucleases (ZFNs) are broadly useful and powerful reagents for genome manipulation1–6. These artificial proteins consist of a zinc finger array fused to a non-specific nuclease domain7,8. ZFNs have been used to direct double-strand DNA breaks (DSBs) to specific genomic loci and repair of these breaks by normal cellular repair machinery pathways often leads to the introduction of insertions and deletions (indels) at the site of the breaks9–18. It has been shown that engineered ZFNs can induce targeted knockout mutations in cultured human cells and in a number of model organisms including zebrafish9–27.
Engineering high-quality zinc finger arrays is crucial to the generation of ZFNs that bind to their target sequences with high affinities and specificities9,17,28,29. Engineered ZFNs bind as dimers to specific DNA sequences7,8,30,31 and therefore two customized zinc finger arrays must be engineered for each potential target site (Fig. 1). We and others have shown that the zinc finger arrays made by the selection-based Oligomerized Pool ENgineering (OPEN) can induce indel mutations at target genomic loci in zebrafish (as well as in plants and cultured human cells) with high efficiency9,17,22,26. Alternatively, a process known as “modular assembly”32–35 can also be used to assemble arrays from archives of individual, pre-selected zinc finger domains11,19–21,36 although the efficiency of this process has been shown to be low9,37. Reagents for engineering zinc finger arrays by OPEN and modular assembly are publicly available from the plasmid distribution service Addgene (http://www.addgene.org/zfc) and detailed protocols for practicing OPEN38 and modular assembly39,40 have been previously described. In addition, an online software program named ZiFiT (http://www.zincfingers.org/software-tools.htm) can be used to identify potential ZFN target sites in any gene or sequence of interest41.
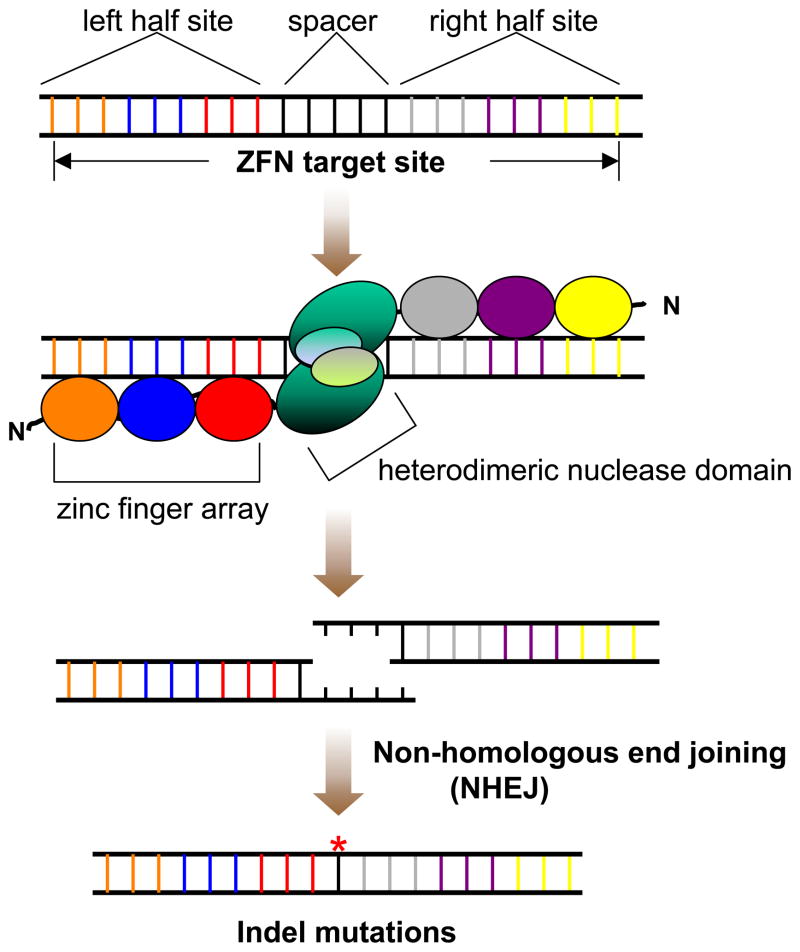
Figure 1.
A diagram of ZFN-induced indel mutations. A full ZFN target site consists of two “half-sites” separated by a 5–6 bp spacer. Each half-site contains a 9 bp sequence that can be recognized by a 3-finger zinc finger array. A ZFN consists of a zinc finger array fused to a nuclease domain. A heterodimeric pair of ZFNs binds to the left and right half sites and induces a double strand DNA break (DSB) in the spacer. Cells utilize non-homologous end joining (NHEJ) machinery to repair the DSB, an error-prone process that can lead to random insertions or deletions (indels) at the target site.
The potential “off-target” cleavage activities of engineered ZFNs have been investigated in two studies published by Doyon et al and Meng et al16,18. In the first study, the authors were not able to detect any “off-target” mutations at other genomic loci highly similar to the ZFN target site16. In the second study, the authors determined that engineered ZFNs are 770-fold more likely to introduce mutations at the ZFN target site than the other similar genomic loci in the embryos that developed normally after injection of ZFN-encoding RNAs18. However, the off-target cleavages of engineered ZFNs often cause detrimental developmental defects in the embryos injected with ZFN-encoding RNAs16–18. It is conceivable that each pair of customized ZFNs will have different degrees of specificity (and toxicity) depending on the selected zinc finger arrays. However, any potential collateral mutations in the founder can be removed by crossing the founder or out-crossing the heterozygous fish to wild-type fish.
There are several limitations to introducing targeted mutations using this method. First, a potential ZFN target site may not exist in the desired target regions of some of the zebrafish genes. Second, although all of the OPEN-selected zinc finger arrays should show satisfactory binding affinities to their target sequences in the bacterial two-hybrid (B2H) assays, some of the engineered ZFNs that use these arrays may fail to induce mutations in the target sites in vivo (see the ANTICIPATED RESULTS). In addition, ZFN-induced mutations are random insertions or deletions. Thus, the exact mutations at the target sites cannot be predetermined using this method.
Here we provide a validated protocol for using engineered ZFNs to generate targeted mutations in endogenous zebrafish genes. This protocol is designed to seamlessly integrate with other protocols for engineering zinc finger arrays by OPEN selection or modular assembly previously published by the Zinc Finger Consortium38,40. To constitute a pair of functional ZFNs, DNA fragments encoding customized zinc finger arrays are directly cloned into ZFN expression vectors designed to express engineered heterodimeric FokI nuclease domains13,42 (Fig. 1). To date, we have used this protocol to successfully identify ZFN-induced indel mutations in 12 endogenous zebrafish genes (17 and unpublished results). Starting with engineered zinc finger arrays in hand, we can obtain homozygous zebrafish mutants in just six to seven months time. Our experience demonstrates that using customized ZFNs to introduce targeted mutations in endogenous zebrafish genes is rapid and robust.
EXPERIMENTAL DESIGN
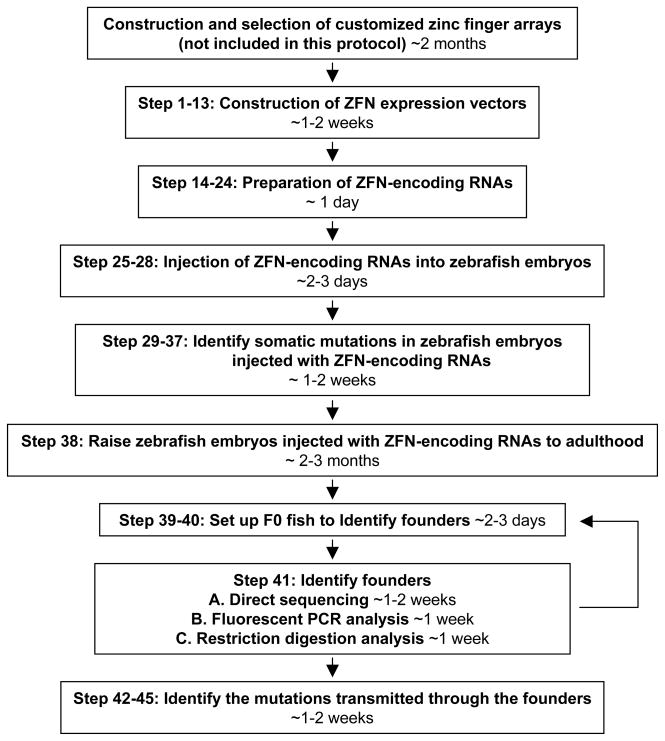
A flowchart in Figure 2 outlines the experimental procedures. First, DNA fragments encoding engineered zinc finger arrays are cloned into plasmids previously described by the Zinc Finger Consortium to create the ZFN expression vectors38,40. Next, ZFN-encoding RNAs are transcribed in vitro from these ZFN expression vectors and then introduced into early zebrafish embryos by microinjection. Subsequently, genomic DNA is isolated from a small number of the injected embryos and the somatic mutation efficiencies of the engineered ZFNs are assessed by sequencing. If ZFN-induced somatic mutations are detected with high frequency, the remaining injected embryos will be raised to adulthood and screened as potential founders. We describe below three different options for detecting ZFN-induced indel mutations in the progeny of potential founders depending on the resources available to individual labs and the sequences of targeted loci.
Figure 2.
Flowchart outlining the experimental procedures described in this protocol and expected timing for each step.
A. Direct sequencing
In this approach, PCR primers are designed to amplify approximately 500 bp of the genomic DNA sequence encompassing the expected mutation site. The resulting PCR product from each single embryo is subjected to sequencing using an internal primer residing inside the PCR fragment and at least 100–150 bp away from the ZFN target site. This approach can be used at any locus. However, the cost of this method is relatively higher than the other two options.
B. Fluorescent PCR analysis
In this method, PCR primers are designed to amplify approximately 150–350 bp of the genomic DNA sequence that flanks the expected mutation. One of the PCR primers is end-labelled with a fluorescent dye such as 6-FAM, NED, PET, or VIC. The lengths of the labeled strand of the resulting PCR products are analyzed using an ABI 3730xl DNA analyzer. DNA strands from PCR products amplified from alleles that have sustained ZFN-induced indel mutations will be longer or shorter than those amplified from wild-type alleles. This method is also universal and fast. It is our preferred method because it is sensitive enough to identify indel mutations from small numbers of pooled embryos.
C. Restriction digest analysis
In this method, the genomic DNA sequence surrounding the ZFN cleavage site is analyzed to determine whether an indel mutation will destroy the recognition site of a restriction enzyme. If such a site can be identified, PCR products encompassing the ZFN target site can be digested with the restriction enzyme and analyzed using gel electrophoresis. This method is only applicable to a subset of target sites that contain an appropriately placed restriction site. In addition, in some cases, we have found insertional mutations that retain the recognition sequences of the test restriction enzymes due to the tendency of such mutations to duplicate regions of microhomology around the cleavage site during NHEJ-mediated repair. However, this method is fast, inexpensive, and does not require any specialized equipment.
Since each pair of customized ZFNs will likely have a different degree of targeting efficiency in vivo, researchers that would like to conduct these experiments may wish to request existing zinc finger arrays that have been deposited in the Zinc Finger Database (ZiFDB; available at http://www.zincfingers.org/software-tools.htm) or published ZFNs to serve as controls16–18.
MATERIALS
REAGENTS
Zebrafish: TU (Tüebingen) or other wild-type strain can be obtained from the Zebrafish International Resource Center (http://zfin.org)
Plasmids encoding engineered zinc finger arrays (see REAGENT SETUP)
Primer OK.1677 (5′-GACGATGATGACAAATCTAGACCCG-3′)
Primer OK.1678 (5′-CTAGTCTCTAGTTACTACTGTGCAGAGG-3′)
Primer OK.567 (5′-CGCAAATGGGCGGTAGGCGTG-3′)
Expand High Fidelity PCR System (Roche, cat. no. 11732641001)
10 mM dNTPs (Invitrogen, cat. no. 18427-013)
5% non-denaturing polyacrylamide gel and/or 3% wt/vol agarose gel
QIAquick PCR Purification kit (Qiagen, cat. no. 28106)
Restriction enzymes from New Englad Biolabs (NEB): DpnI (cat. no. R0176S), XbaI (cat. no. R0145S), BamHI (cat. no. R0136S), PmeI (cat. no. R0560S)
10x BamHI buffer (see REAGENT SETUP)
MinElute PCR Purification kit (QIAGEN, cat. no. 28006)
ZFN expression vectors pMLM290 & pMLM292 (available by request from the Joung lab)
Nuclease-free water
2x Quick Ligation Reaction Buffer (NEB, cat. no. M2200S)
T4 DNA ligase (NEB, cat. no. M0202S)
Chemically competent E. coli XL-1 Blue cells (prepared as previously described38)
QIAGEN Plasmid Midi kit (cat. no. 12143)
LB/carbenicillin agar plates (see REAGENT SETUP)
LB/carbenicillin medium (see REAGENT SETUP)
LB/kanamycin agar plates (see REAGENT SETUP)
LB/kanamycin medium (see REAGENT SETUP)
mMESSAGE mMACHINE T7 Ultra kit (Ambion, cat. no. AM1345)
1x Danieau solution (see REAGENT SETUP)
0.5% phenol red solution (Sigma, cat. no. P0290)
Embryo water (see REAGENT SETUP)
Gene-specific primers (see REAGENT SETUP)
Platinum Taq DNA Polymerase High Fidelity (Invitrogen, cat. no. 11304-011)
TOPO TA kit (Invitrogen, cat. no. K4530-20)
Taq DNA polymerase (Roche, cat. no. 11146173001)
GeneScan-500 LIZ (ABI, cat. no. 4322679)
TB/kanamycin medium (see REAGENT SETUP)
2-ml 96-well pyramidal bottom block (Corning Costar, cat. no. 3960)
Solution I (see REAGENT SETUP)
Solution II (see REAGENT SETUP)
Solution III (see REAGENT SETUP)
Hyflo Super Cel Diatomaceous Earth (Aldrich 392545)
96-well 25-μm polypropylene filter block (Whatman, cat. no. 7700-1804)
96-well 0.7 μm 2-ml polypropylene GF filter block (Seahorse Bioscience, cat. no. F20060)
1-ml 96-well polypropylene block (Nalgene Nunc, cat. no. 260252)
EQUIPMENT
Thermocycler
Microcentrifuge, refrigerated and non-refrigerated
Electrophoresis apparatus for acrylamide gels and/or agarose gels
Microinjection apparatus (as previously described43)
Incubators at 37 °C & 28.5 °C
Shaker incubator at 37 °C
Sorvall RT6000D or other similar tabletop centrifuge
ABI 3730xl DNA analyzer
REAGENT SETUP
Plasmid DNA encoding zinc finger arrays engineered using Zinc Finger Consortium reagents for OPEN selection or modular assembly38,40. These plasmids encode the engineered zinc finger array between standardized XbaI and BamHI restriction sites built into the Consortium vectors. These plasmids also confer antibiotic resistance to carbenicillin. Plasmids encoding zinc finger arrays selected by OPEN must be propagated in a bacterial strain encoding the lacIQ gene to prevent toxicity due to overexpression of the zinc fingers. Plasmids encoding zinc finger arrays designed by modular assembly do not require propagation in any specialized E. coli strain.
10x BamHI Buffer 1.5 M NaCl, 100 mM Tris-HCl, 100 mM MgCl2, 10 mM DTT, pH 7.9; filter-sterilized. Store at −20 °C for 1 year.
1x Danieau solution 58 mM NaCl, 0.7 mM KCl, 0.4 mM MgSO4, 0.6 mM Ca(NO3)2, 5 mM HEPES, pH 7.6; filter-sterilized. Store at 25 °C for 1 year.
Embryo water 5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4. Store at 25°C for 1 month.
Gene-specific primers Design primers for each ZFN target site based on the genomic DNA sequence flanking it. Each primer should anneal about 150–250 bp away from the expected DSB location and extend toward it. Ideally, primers should have a melting temperature of approximately 65 °C and a length of 25 bp. Free online software such as Primer3 (http://frodo.wi.mit.edu/primer3/) and OligoPerfect™ Designer (http://tools.invitrogen.com/content.cfm?pageid=9716) can be used for designing the primers. For fluorescent PCR analysis, one of the gene-specific primers must also be labeled with a fluorescent group on the 5′ end (e.g. 6-FAM, NED, PET, or VIC).
LB/carbenecillin agar plate Add 15 g agar into 1 liter of LB. Autoclave. Add 2 ml of 50 mg/ml carbenecillin stock solution and pour into 40 10-cm Petri dishes. Plates can be stored at 4°C for up to two months.
LB/carbenecillin medium Add 0.5 ml of 50 mg/ml carbenecillin stock solution to 500 ml of autoclaved LB. Store at 4 °C for up to two months.
LB/kanamycin agar plate Add 15 g agar into 1 liter of LB. Autoclave. Add 1 ml of 30 mg/ml kanamycin stock solution and pour into 40 10-cm Petri dishes. Store at 4 °C for up to two months.
LB/kanamycin medium Add 0.5 ml of 30 mg/ml kanamycin stock solution to 500 ml of autoclaved LB. Store at 4 °C for up to two months.
TB/kanamycin medium Add 12 g Bacto tryptone, 24 g Bacto yeast extract, 4 ml glycerol and H2O to 900 ml. Autoclave. Add 100 ml of a sterile solution of 0.17 M KH2PO4 and 0.72 M K2HPO4 and 1 ml of 30 mg/ml kanamycin stock solution. Store at 4 °C for up to two months.
Solution I 50 mM Tris, pH 8.0, 10 mM EDTA, 500 μg/ml RNase A. Store at 4 °C for 1 year.
Solution II 200 mM NaCl, 1% SDS(w/v). Store at 25 °C for 1 year.
Solution III For 250 ml solution, add 58.88 g potassium acetate, 71.25 ml of glacial acetic acid, and adjust pH to 4.8 using HCl. Store at 25 °C for 1 year.
Acid-washed silica slurry made from Hyflo Super Cel Diatomaceous Earth Add 100 g Hyflo Super Cel Diatomaceous Earth to a 2-liter flask and then add sterile ddH2O to ~900 ml and then add 100 ml of HCl. Mix well and incubate overnight at room temperature. Pour out the acid solution into a beaker (Note: this solution must be neutralized to pH 5.5–10.5 using NaOH before going down the drain). Refill flask with 1 liter of H2O and let it settle for ~1 hr. Pour off the supernatant containing fines. Repeat H2O rinses thee additional times. Put a sterile stir bar into the flask and adjust the pH to 6–8 using NaOH. Allow to settle and pour off supernatant again. Resuspend in sterile ddH2O to a total of 500 ml. The silica should comprise ~40–50% of the volume. Store in a glass bottle at 25 °C for 1 year.
PROCEDURE
Construction of ZFN expression vectors Timing 1~2 weeks
- 1| Amplify DNA fragments encoding zinc finger arrays from expression plasmids by setting up the PCR reaction as tabulated below.
Component Amount Final Plasmid encoding zinc finger array 1 μl ~ 100 ng 10x Expand High Fidelity Buffer with MgCl2 5 μl 1x 10 mM dNTPs 4 μl 0.8 mM Primer OK.1677 (10 μM) 1 μl 0.2 μM Primer OK.1678 (10 μM) 1 μl 0.2 μM Expand High Fidelity Enzyme Mix (3.5 U μl−1) 0.375 μl 1.3125 U Nuclease-free water to 50 μl - 2| Run the PCR using the following conditions:
Cycle number Denature Anneal Extend 1 95 °C, 5 minutes 2–36 95 °C, 30 seconds 55 °C, 30 seconds 72 °C, 45 seconds 37 72 °C, 3 minutes 3| Run 10 μl of the PCR product on a 5% non-denaturing polyacrylamide gel or a 3% agarose gel to verify successful amplification. The amplified PCR product should be approximately 300 bp. Purify the remainder of the PCR product using a QIAquick PCR purification kit, eluting the DNA with 50 μl of a 1:10 dilution of the supplied Buffer EB in nuclease-free water.
-
4| Digest the eluted PCR product with DpnI as tabulated below at 37 °C for 1 hour to eliminate the template plasmid DNA. Steps 4–7 can be performed in parallel with steps 8–10.
Component Amount Final Eluted PCR product 15 μl unknown 10x NEBuffer 4 2.5 μl 1x DpnI (20 U/μl) 1 μl 20 U Nuclease-free water to 25 μl CRITICAL STEP DpnI is a 4-cutter that cleaves only when its substrate is methylated, such as plasmid DNA isolated from dam+ bacteria. This step prevents the contamination of colonies that contain the PCR template plasmid after transformation (Step 12).
- 5| Digest the sample from Step 4 with XbaI as tabulated below at 37 °C for 1 hour.
Component Amount Final DpnI digested DNA (from Step 4) 25 μl unknown 10x NEBuffer 2 3.5 μl 1x 10x BSA (1 mg/ml) 3.5 μl 1x XbaI (20 U/μl) 1 μl 20 U Nuclease-free water to 35 μl - 6| Digest the sample from Step 5 with BamHI as tabulated below at 37 °C for 1 hour.
Component Amount Final DpnI/XbaI digested DNA (from Step 5) 35 μl unknown 10x NEBuffer BamHI (see REAGENT SETUP) 4.5 μl 1x 10x BSA (1 mg/ml) 1 μl 1x BamHI (20 U/μl) 1 μl 20 U Nuclease-free water to 45 μl 7| Purify the triple-digested DNA using a MinElute PCR purification kit by following the manufacturer’s instructions except eluting the DNA with 10 μl of a 1:10 dilution of the Buffer EB (supplied in the kit) in nuclease-free water.
- 8| In parallel with the work of Steps 4–7, prepare the ZFN heterodimer vectors by digesting pMLM290 and pMLM292 with XbaI as tabulated below at 37 °C for 1 hour.
Component Amount Final pMLM 290 or pMLM 292 plasmid DNA 1 μg 0.04 μg/μl 10x NEBuffer 2 2.5 μl 1x 10x BSA (1 mg/ml) 2.5 μl 1x XbaI (20 U/μl) 1 μl 20 U Nuclease-free water to 25 μl - 9| Digest the sample from Step 8 with BamHI as tabulated below at 37 °C for 1 hour to create an XbaI/BamHI sticky-end vector.
Component Amount Final XbaI digested DNA (from Step 8) 25 μl 1 μg 10x NEBuffer BamHI (see REAGENT SETUP) 3.5 μl 1x 10x BSA (1 mg/ml) 1 μl 1x BamHI (20 U/μl) 1 μl 20 U Nuclease-free water to 35 μl 10| Run the entirety of the reaction from Step 9 on a 5% polyacrylamide gel. Excise the 5.7-kb band. Elute the DNA from the cut gel piece as previously described38 and resuspend the DNA pellet with 20 μl of nuclease-free water. (Alternatively, run the entirety of the reaction from Step 9 on a 1% agarose and purify the digested DNA using a QIAquick Gel Extraction kit.)
-
11| Ligate the purified PCR product from Step 7 and the purified vector backbone from Step 10 together as tabulated below, making sure to add the components in the order listed. Mix well and incubate the reaction at room temperature for 10 minutes.
Component Amount Final triple-digested PCR product (from Step 7) 9 μl unknown XbaI/BamHI-digested pMLM290 or pMLM292 (from Step 10) 1 μl unknown 2x Quick Ligation Reaction Buffer 10 μl T4 DNA ligase (400 U/μl) 1 μl CRITICAL STEP pMLM290 and pMLM292 encode matched heterodimeric ZFN frameworks. Therefore, it is important to ensure that zinc finger arrays for the two half-sites in each full ZFN target site are cloned into these vectors (i.e. make sure that each ZFN pair to be injected encodes one of each type of heterodimeric ZFN).
PAUSE POINT Ligations can be frozen at −20°C indefinitely.
12| Transform 200 μl of chemically competent XL-1 Blue cells with all of the ligation from step 11 using standard transformation conditions as previously described38, and plate 1/3 of the transformation on an LB/carbenicillin agar plate. (Alternatively, transform commercially available high-efficiency competent cells.) Incubate the plate at 37°C overnight.
-
13| Inoculate a 50-ml LB/carbenicillin culture with a single colony from the transformation plate. Grow the culture in a shaker at 37 °C overnight. Isolate the plasmid DNA using the QIAGEN Plasmid Midi kit. The sequence of the purified plasmid DNA should be verified by sequencing using primer OK.567. We re-verify that only one insert was taken up by the plasmid and that all of the sequence that lies between the XbaI and BamHI cloning sites is correct. Clones that meet these criteria are designated as ZFN expression vector DNAs.
CRITICAL STEP Contaminating RNase may inhibit downstream RNA synthesis. Use a QIAGEN kit that has not been contaminated with RNase (except that RNase is in the P1 solution of the kit) and clean all work surfaces with solution that inactivates RNase. Use certified RNase-free microfuge tubes to store the final DNA elution.
Preparation of ZFN-encoding RNAs Timing 1~2 days
- 14| Linearize the ZFN expression vector DNA from Step 13 with PmeI as tabulated below at 37 °C for 2 hours.
Component Amount Final ZFN plasmid DNA (from Step 13) 10 μl 10 μg 10x NEBuffer 4 10 μl 1x 100x BSA (10 mg/ml) 1 μl 1x PmeI (20 U/μl) 1 μl 20 U Nuclease-free water to 100 μl -
15| Run 2 μl of the digestion mixture on a 1% agarose gel to confirm that the digestion is complete. Purify the linearized DNA using a QIAquick PCR purification kit, eluting the DNA with 50 μl of Buffer EB. Measure the concentration of the eluted DNA with a spectrophotometer.
PAUSE POINT The eluted DNA can be stored at −20 °C indefinitely.
-
16| Transcribe ZFN-encoding RNA using a mMESSAGE mMACHINE T7 Ultra kit as tabulated below. Incubate the reaction mixture at 37 °C for 1 hour.
Component Amount Final PmeI-linearized ZFN plasmid DNA 3 μl 0.5 μg 2x NTP/ARCA 5 μl 1x 10x T7 Reaction Buffer 1 μl 1x T7 Enzyme Mix 1 μl CRITICAL STEP The reaction mixture should be constituted in RNase-free microfuge tubes.
17| Add 1 μl of TURBO DNase provided in the kit to the reaction mixture and incubate at 37 °C for 15 minutes.
- 18| Perform a poly(A) tailing reaction using the components provided in the kit as tabulated below.
Component Amount Final Transcribed RNA (from Step 17) 11 μl unknown 5x E-PAP 10 μl 1x 25 mM ATP 5 μl 2.5 mM 25 mM MnCl2 5 μl 2.5 mM Nuclease-free water to 50 μl 19| Save 2 μl of the reaction mix in a microfuge tube and store at −20 °C for subsequent analysis on an agarose gel. Add 2 μl of E-PAP to the remainder of the reaction mix and incubate at 37 °C for 30 minutes.
-
20| At the end of the reaction, remove 2 μl of the reaction mixture and run it side-by-side with the sample saved in Step 19 on a 1% agarose gel. Use the Formaldehyde Loading Dye provided in the kit. The transcribed RNA should appear as a distinct band (sometimes multiple distinct bands due to secondary structures) without smearing. The RNA with a poly(A) tail should migrate more slowly on the gel (Fig. 3).
? TROUBLESHOOTING
21| Add 25 μl of the Lithium Chloride Precipitation Solution provided in the kit to the reaction mixture. Mix well and incubate the samples at −20 °C for 30 minutes.
22| Centrifuge at 4 °C for 15 minutes at 14,000 × g. Discard the supernatant. Wash the pellet with 1 ml of 70% ethanol, and centrifuge again for 10 minutes.
23| Completely remove the supernatant. Air dry the pellet until it becomes transparent. Resuspend the pellet with 20 μl of nuclease-free water.
-
24| Incubate the resuspended RNA samples at 65 °C for 10 minutes to dissolve the RNA. Measure RNA concentrations using a spectrophotometer. The total yield of the RNA should be around 15 μg. RNA should be stored at −80 °C in 1-μl aliquots.
CRITICAL STEP Preparation of aliquots is important because RNA degrades more readily if it is subjected to multiple freeze-thaw cycles.
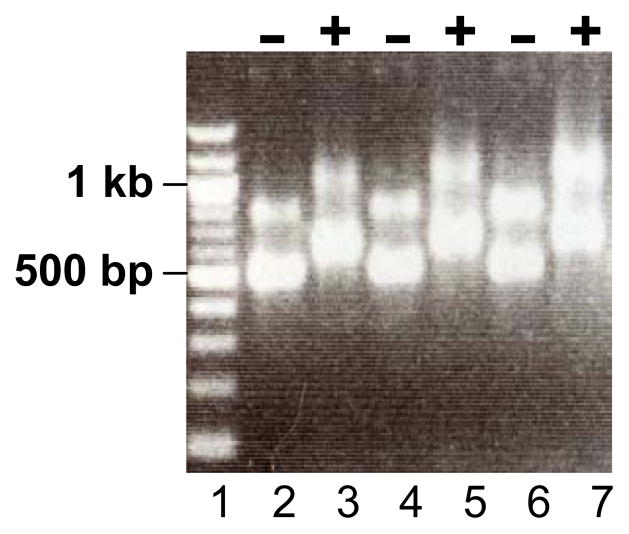
Figure 3.
A representative gel of in vitro transcribed RNAs encoding ZFNs. The first lane is DNA size standard. Lane 2–3, 4–5 & 6–7 contain 3 sets of ZFN RNAs (−, before poly(A) tailing reaction; +, after poly(A) tailing reaction).
Injection of ZFN-encoding RNAs into zebrafish embryos Timing 2~3 days
25| Set up zebrafish matings for spawning the evening before injection as described previously44.
-
26| On the day of injection, set up for the injections of zebrafish embryos as described previously43. Prepare the injection solution as tabulated below. All components should be kept on ice.
Component Amount Amount ZFN-encoding RNA (for the left half site) variable 750 ng ZFN-encoding RNA (for the right half site) variable 750 ng 1% phenol red 0.5 μl 0.08% 1x Danieau solution to 6 μl CRITICAL STEP Injection solution should be prepared fresh each time. Do not freeze the injection solutions and reuse.
-
27| Centrifuge the injection solution at 14,000 × g for 5 minutes in a refrigerated microfuge. Load the injection solution into the capillary injection needle and inject ~ 2 nl of the solution into each zebrafish embryo.
CRITICAL STEP It is important to inject the solution into the cell directly (not into the yolk) at the one-cell stage. The inclusion of phenol red in the injection solution is not essential but helps to monitor the location and the amount of solution injected. The volume of the injection can be measured using a micrometer slide45 and should not exceed 1/10 of the cell’s volume.
-
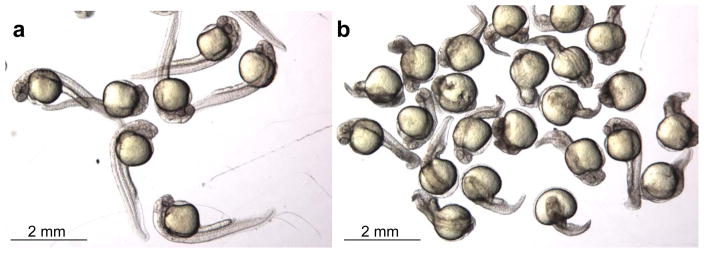
28| Save some uninjected embryos as controls. Put the injected and uninjected embryos at 28.5 °C overnight. On the next day, remove and count the dead embryos. Divide the remaining embryos into two groups — normal-looking embryos and deformed embryos that show visible development defects (Fig. 4). Count the numbers of embryos in each group. Remove deformed embryos and change the embryo water to prevent it from becoming spoiled. Place the normal-looking embryos back at 28.5 °C for another 1–2 days.
? TROUBLESHOOTING
Figure 4.
Representative results of normal-looking embryos (a) and deformed embryos (b) after injections of ZFN-encoding RNAs.
Identify somatic mutations in zebrafish embryos injected with ZFN-encoding RNAs Timing 1~2 weeks
29| 2–3 days after the injections, transfer 8 normal-looking embryos that have been injected with a pair of ZFNs into a microfuge tube. If the embryos have not come out of the chorion, remove the chorion manually. Return the remaining injected embryos back to 28.5 °C until the sequencing results are obtained.
30 | Isolate genomic DNA from the pool of embryos as described in Box 1.
- 31| Use the genomic DNA of Step 30 as template for a PCR reaction to amplify the genomic DNA encompassing the ZFN target site with gene-specific primers (see REGENT SETUP) as tabulated below.
Component Amount Final Genomic DNA (from Step 30) 2.5 μl unknown 10x High Fidelity PCR Buffer 5 μl 1x 50 mM MgSO4 2 μl 2 mM 10 mM dNTPs 1 μl 0.2 mM DMSO 2.5 μl 5% Forward primer (10 μM) 5 μl 1 μM Reverse primer (10 μM) 5 μl 1 μM Platinum Taq High Fidelity 0.2 μl Nuclease-free water to 50 μl -
32| Run the PCR using the following conditions:
Cycle number Denature Anneal Extend 1 95 °C, 5 minutes 2–25 95 °C, 30 seconds 65 °C, 30 seconds 68 °C, 45 seconds 26 68 °C, 5 minutes PAUSE POINT: PCR product can be frozen at −20°C indefinitely.
33| Purify the PCR product using a MinElute PCR purification kit into 10 μl of a 1:10 dilution of Buffer EB (provided with the kit) in nuclease-free water.
34| Subclone the PCR product into the pCR4 TOPO TA vector using a TOPO TA kit. Add 4 μl of the purified PCR product to 1 μl of the provided salt solution and 1 μl of the linearized pCR4 TOPO-TA vector, and incubate the mixture at room temperature for 30 minutes.
35| Transform a vial of chemically competent Mach-1 cells (provided in the TOPO TA kit) with the entire reaction from Step 34. Plate the entire transformation onto an LB/kanamycin plate. Incubate the plate at 37°C overnight.
36| On the next day, pick 96 colonies from the transformation plate. Inoculate each single colony into 700 μl of TB/kanamycin in a 2-ml 96-well pyramidal bottom block and incubate at 37°C with agitation overnight. Isolate 96 plasmid DNA samples as described in Box 2 or using a commercially available kit. Sequence the DNA samples using the T3 sequencing primer supplied in the TOPO TA kit.
-
37| Align the sequencing results with the genomic DNA sequence encompassing the ZFN target site from Ensembl. Identify the clones that show evidence of ZFN-induced indel mutations. Calculate the somatic mutation rates of the engineered ZFNs using the following formula. Go to Step 38 if somatic mutations are detected.
Somatic mutation rates = # of clones with indel mutations/# of clones with PCR inserts
? TROUBLESHOOTING
Box 1 . ISOLATION OF GENOMIC DNA FROM A POOL OF EMBRYOS FOR PCR.
Place 6–12 embryos (2–3 days old) in a microfuge tube. Remove any liquid in the tube.
Add 500 μl of the SDS lysis buffer (10 mM Tris, pH 8.0, 200 mM NaCl, 10 mM EDTA, 0.5% SDS, 100 μg/ml Proteinase K) and incubate at 50 °C for 2 hours to overnight.
Invert the tubes a few times during the incubation to disperse the embryos. The lysis is complete when no clumps can be seen in the tube.
Add 500 μl of 1:1 phenol/chloroform solution. Mix the solutions by inverting the tubes repeatedly for a minute. Spin the tubes in a microfuge at 14,000 × g for 5 minutes at room temperature.
Transfer the top layer of solution into a new tube and add 1 ml of 100% ethanol. Mix the solutions by inverting the tube a few times. Spin the tube in a refrigerated microfuge at 14,000 × g for 20 minutes.
Remove the supernatant. Rinse the pellet with 1 ml of 70% ethanol and spin again at 4 °C at 14,000 × g for 10 minutes.
-
Remove the supernatant and air dry the pellet. Resuspend the pellet with 40 μl of TE (10 mM Tris, pH 8, 0.5 mM EDTA).
CRITICAL STEP The dried genomic DNA pellet takes time to go into the solution. Thus, after adding TE, flick the tube to dislodge the pellet. Let the DNA dissolve at room temperature for 2 hours or at 4 °C overnight before setting up the PCR. Genomic DNA should be stored at 4 °C.
Box 2 . PLASMID DNA ISOLATION IN A 96-WELL FORMAT.
Aliquot 700 μl of Terrific Broth into each well of a 96-well 2-ml pyramidal bottom block. Inoculate each well with a single colony. Incubate the block at 37°C overnight in a Microtitertron incubator shaker at 900 rpm.
Spin down the bacteria in the block in a Sorvall RT6000D centrifuge at 900 × g (~2500 rpm) for 25 minutes. Remove the supernatant and resuspend the pellet in 150 μl Solution I (See REAGENT SETUP) using a multichannel pipet.
-
Add 150 μl Solution II (See REAGENT SETUP). Mix well with the multichannel pipet and incubate at room temperature for 5 minutes.
CRITICAL STEP: Do not let the lysis reaction proceed for longer than 5 minutes.
Neutralize the lysates with 150 μl of ice-cold Solution III (See REAGENT SETUP). Mix well.
Transfer neutralized supernatant to a 25-μm polypropylene filter block set atop of a sterile 2-ml 96-well deep well block. Centrifuge the two stacked units for 20 minutes at 4000 × g in a QIAGEN 4–15C centrifuge.
Prepare a silica filter plate by adding 250 μl of an acid-washed silica slurry made from Hyflo Super Cel Diatomaceous Earth (See REAGENT SETUP) into each well of a 96-well 0.7 μm 2-ml polypropylene GF filter block, and centrifuging at 750 × g for 3 minutes on top of a 1-ml 96-well polypropylene block. Discard flow-through.
Add 450 μl 6 M guanidine-HCl to the filtrates from Step 5. Mix thoroughly and transfer the mixture into the silica columns. Spin the silica plate over the same 1-ml polypropylene block used in Step 6 at 750 × g for 2 minutes. Discard flow-through.
Add 600 μl 80% ethanol to each well of the silica plate and spin again at 750 × g for 2 minutes. Discard flow-through and spin again for 5 minutes.
Place the silica plate on top of a sterile 96-well PCR plate. Add 60 μl of 1:10 dilution of Qiagen Buffer EB in sterile water that has been pre-warmed to 60–70 °C to each silica column. Allow to sit for 5 minutes at room temperature.
-
Spin the silica plate on top of the PCR plate at 750 × g for 3 minutes to elute DNA.
PAUSE POINT: DNA can be frozen at −20°C indefinitely.
Identify founders and the mutations transmitted through the founders Timing 2~3 months
-
38| Raise 30~60 ZFN-injected embryos (Step 29) to adulthood to screen for founders. These fish are F0 and may be used for mating at 2–3 months of age.
? TROUBLESHOOTING
39| When F0 fish reach 2–3 months of age, set up matings between F0 and wild-type zebrafish. On the next day, collect each clutch of embryos in a Petri dish. Put each F0 fish that has produced embryos into a single-housed tank. Label the F0 fish and its progeny embryos with matching numbers.
40| Let the embryos develop at 28.5 °C for 2–3 days until they naturally come out of the chorion. Continue to Step 41, 47 or 55 to identify founders.
41) Detection of ZFN-induced indel mutations can be done by direct sequencing (option A), fluorescent PCR analysis (option B) or restriction digest analysis (option C).
A. Direct sequencing Timing 1~2 weeks
i| Put one embryo into each well of a 96-well PCR plate. Analyze 12 embryos from each clutch. Isolate genomic DNA from each single embryos as described in Box 3.
- ii| Amplify the genomic DNA encompassing the ZFN target site using gene-specific primers as tabulated below.
Component Amount Final Genomic DNA 2.5 μl unknown 10x Taq DNA polymerase buffer 2 μl 1x 10 mM dNTPs 0.4 μl 0.2 mM Forward primer (10 μM) 1 μl 0.5 μM Reverse primer (10 μM) 1 μl 0.5 μM Taq DNA polymerase (5 U/μl) 0.2 μl Nuclease-free water to 20 μl - iii| Perform PCR using the following conditions:
Cycle number Denature Anneal Extend 1 94 °C, 2 minutes 2–41 94 °C, 20 seconds 65 °C, 45 seconds 72 °C, 30 seconds 42 72 °C, 5 minutes iv| Purify the PCR products using the QIAquick PCR purification kit. Elute each DNA using 50 μl of Buffer EB provided in the kit.
v| Sequence the purified PCR products using an internal primer that is at least 100 bp upstream or downstream from the expected mutation.
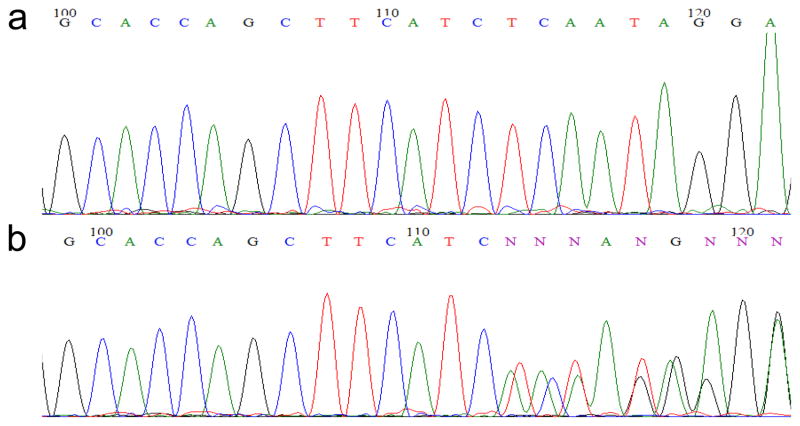
vi| Compare the sequencing results to the genomic DNA sequence encompassing the ZFN target site in Ensembl. If an embryo carries one wild-type and one mutant allele, the sequencing traces should be exactly the same as the sequence in Ensembl but become unreadable near the ZFN target site due to simultaneous amplification of both alleles (Fig. 5). Identify the F0 fish that transmit ZFN-induced mutations through the germline. If no founder is identified, repeat Step 39–41 to screen more F0 zebrafish. The genomic DNA of the mutant embryo can be used in Step 42 in order to obtain the sequence information of the exact mutation.
Box 3 . ISOLATION OF GENOMIC DNA FROM SINGLE EMBRYOS FOR PCR.
Place each zebrafish embryo (2–3 days old) in a single well of a 96-well PCR plate. Remove any liquid in the well.
Add 20 μl of the PCR extraction buffer (10 mM Tris, pH 8.0, 2 mM EDTA, 0.2% Triton X-100, 100 μg/ml Proteinase K) into each well and seal the plate. Incubate the plate at 50 °C for 4 hours to overnight.
-
Heat the plate at 95 °C for 5 minutes in a thermocycler.
CRITICAL STEP This step is important to inactivate the Proteinase K in the PCR extraction buffer. Omitting this step will result in no PCR amplification in the following step.
Spin the plate at ~800–1000 × g in a tabletop centrifuge for 3 minutes at room temperature. The DNA can now be stored at 4 °C or used for PCR.
Figure 5.
Representative results of sequencing traces of a wild-type (a) and a mutant (b) embryo.
B. Fluorescent PCR analysis (our preferred method) Timing 1 week
i| Put six embryos (Step 40) into one microfuge tube. Analyze 12 embryos from each clutch. Isolate genomic DNA from a pool of embryos as described in Box 1. In the meantime, keep the remaining embryos at 28.5 °C until the analysis is finished.
-
ii| Set up PCR to generate fluorescent PCR products as described in option A. Be sure to set up a control reaction containing only genomic DNA isolated from a wild-type zebrafish embryo.
CRITICAL STEP One of the gene-specific primers should be end-labeled with either NED, 6-FAM, PET, or VIC. Minimize sample exposure to light.
iii| Aliquot 5 μl of the PCR product into each well of a 96-well PCR plate. Add 5 μl of a 1:10 dilution of the size standard GeneScan-500 LIZ in water to each well that contains sample. Seal the plate and shake down so that the samples are at the bottom of the plate.
iv| Incubate the plate at 95°C for 5 minutes and chill on ice for 5 minutes.
v| Spin the plate for 5 seconds. Run the samples on an ABI 3730xl DNA analyzer.
-
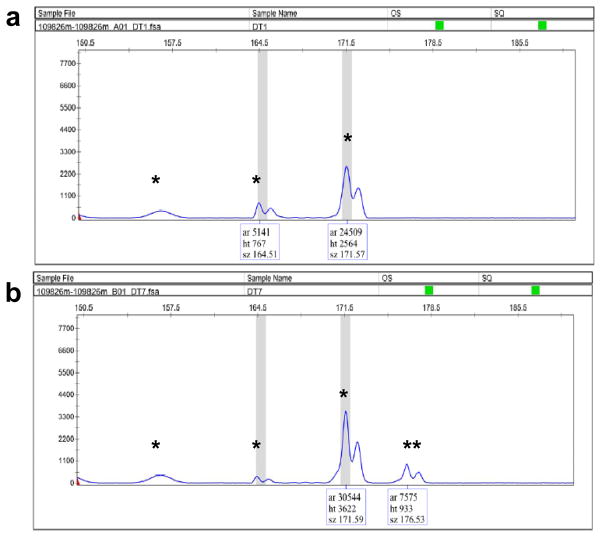
vi| Process the data using the GeneMapper software from ABI. Identify the peaks of the PCR products and determine their lengths. Compare the PCR profiles of the embryos from F0 fish to the profile of the wild-type embryo. Any additional size variants in the PCR products of the progeny of F0 fish are candidates for ZFN-induced indel mutations (Fig. 6). Go to the next step if a founder is identified. If no founder is identified, repeat Step 39–41 to screen more F0 zebrafish.
? TROUBLESHOOTING
vii| From the plate containing the progeny of the identified founder (Step i), pick additional 12 embryos and put them individually into each well of the 96-well PCR plates. Isolate genomic DNA from single embryos as described in Box 3.
viii| Set up PCR and analyze the fluorescent PCR products as described in Steps ii–vi. Identify the genomic DNA from a single embryo that contains an indel mutation. This DNA can be used in Step 42 to obtain the sequence information of the exact mutation.
Figure 6.
Representative results of fluorescent PCR analysis of a pool of wild-type embryos (a) and a pool of embryos from a founder (b). The PCR products from the wild-type embryos sometimes contain more than one peak due to PCR stutter or non-specific amplification. However, the additional peak (b, double asterisks) that is not present in the wild-type control sample indicates the presence of ZFN-induced indel mutation.
C. Restriction digest analysis Timing 1 week
i| Analyze the ZFN target site using the online software NEBcutter V2.0 (http://tools.neb.com/NEBcutter2/index.php). Identify any restriction sites that lie in or overlap the spacer between the two ZFN half sites. Ideally these sites will not occur anywhere else in the region to be amplified by PCR. Using DNA analysis software, determine the sizes of fragments that will result from restriction digest of the PCR product from both the control embryo and embryos with indel mutations that destroy the restriction site.
ii| Put one embryo (Step 40) into each well of a 96-well PCR plate. Analyze 12 embryos from each clutch. Isolate genomic DNAs from single embryos as described in Box 3.
iii| Set up PCR as described in option A.
- iv| Transfer 6 μl of each of the PCR products into separate wells of another 96-well PCR plate. Make up a restriction enzyme mix as tabulated below. Add 6 μl of the mix into each well that contains sample. Incubate the plate overnight at the optimal temperature for the restriction enzyme.
Component Amount Final 10x NEBuffer for the restriction enzyme 1.2 μl 1x Restriction enzyme (chosen from Step 55) 1 U 1 U Nuclease-free water to 6 μl -
v| Run the PCR products from Step iii alongside digested samples from Step iv on a 3% agarose gel containing 0.5 μg/ml ethidium bromide. (The percentage of the agarose gel to use depends on the sizes of the PCR product and the restricted fragments.)
CAUTION Ethidium bromide is a known mutagen and should be handled as a hazardous chemical. Wear gloves while handling.
vi| Inspect the gel using a UV light box. The PCR product from the control embryo should be digested completely into restricted fragments of anticipated sizes. Identify the samples that exhibit any additional restricted fragments. The presence of such bands provides evidence of ZFN-induced mutations.
-
vii| Identify the F0 fish that transmit ZFN-induced mutations through the germline. The genomic DNA of the mutant embryo can be used in Step 42 to obtain the sequence of the exact mutation. If no founder is identified, repeat Steps 39–41 to screen more F0 zebrafish.
? TROUBLESHOOTING
- 42| After identifying the founders and isolating genomic DNA samples from their progeny mutant embryos, set up PCR as tabulated below to amplify the DNA sequence encompassing the ZFN target site using the genomic DNA isolated from a single mutant embryo.
Component Amount Final Genomic DNA from a mutant embryo 2.5 μl unknown 10x High Fidelity PCR Buffer 5 μl 1x 50 mM MgSO4 2 μl 2 mM 10 mM dNTPs 1 μl 0.2 mM DMSO 2.5 μl 5% Forward primer (10 μM) 5 μl 1 μM Reverse primer (10 μM) 5 μl 1 μM Platinum Taq High Fidelity 0.2 μl Nuclease-free water to 50 μl - 43 | Run the PCR using the following conditions:
Cycle number Denature Anneal Extend 1 95 °C, 5 minutes 2–41 95 °C, 30 seconds 65 °C, 30 seconds 68 °C, 45 seconds 42 68 °C, 5 minutes 44| Subclone the PCR products as described in Step 33–35. Isolate and sequence the plasmid DNAs from 6 colonies.
45| Compare the sequencing results to the genomic DNA sequence in Ensembl to identify the exact mutation carried by each founder.
TIMING
Steps 1–13 Construction of ZFN expression vectors — 1~2 weeks
Steps 14–24 Preparation of ZFN-encoding RNA — 1 day
Steps 25–28 Injection of ZFN-encoding RNA into zebrafish embryos — 2~3 days
Steps 29–37 Identifying somatic mutations in zebrafish embryos injected with ZFN-encoding RNA — 1~2 weeks
Steps 38 Raise injected embryos to adulthood — 2~3 months
Steps 39–45 Identify founders and associated mutations — 2–3 weeks
TROUBLESHOOTING
Troubleshooting advice can be found in Table 1.
TABLE 1.
Troubleshooting table.
| Step | Problem | Solution |
|---|---|---|
| 20 | No RNA synthesis | Sequence the ZFN expression vector to make sure that it contains the insert |
| Poor RNA synthesis or RNA smear on the gel | Make sure the RNA transcription kit is not expired and use RNase-free reagents | |
| 28 | Too many dead or deformed embryos after RNA injection | For most ZFN pairs, we observe 30–80% of dead/deformed embryos when injected with 250 ng/μl of ZFN-encoding RNAs; if the toxicity of a ZFN pair is too high, reduce the RNA concentration to 100–150 ng/μl |
| No dead or deformed embryos after RNA injection | Make sure that the solution is injected into the cytoplasm at the 1-cell stage; if that’s the case, increase the RNA concentration 2 to 3 fold | |
| 37 | Many clones do not contain PCR inserts | Make sure that the PCR worked, and/or that the TOPO cloning kit is not expired |
| No somatic mutation is found by sequencing | If no somatic mutation is found after obtaining 96 sequences of the PCR inserts, try to inject a higher concentration of ZFN-encoding RNAs or use a different pair of ZFNs | |
| 38 | Too few zebrafish survive to adulthood | Repeat the injections at a 2-fold lower RNA concentration and raise the injected embryos |
| 52 | Too many fluorescent PCR fragments amplified from the control sample | Adjust the PCR conditions or design new pairs of gene-specific primers |
| 61 | Incomplete restriction enzyme digestion of the control sample | Some enzymes may be less efficient than the others; increase the volume of the digestion reaction and use more of the enzyme |
| No founders are identified using restriction digestion analysis | Some insertional mutations may not affect the restriction enzyme recognition sites; screen more F0 fish or try to identify founders using the other two methos |
ANTICIPATED RESULTS
Our experience has shown that introduction of targeted mutations in endogenous zebrafish genes using engineered ZFNs is rapid and robust. We have successfully targeted 12 different endogenous zebrafish genes (as judged by the introduction of somatic mutations with frequencies ranging from 1–26%) (17 and unpublished results). To date, we have also successfully identified founders for all 5 targeted loci that we have screened (5–50% founders in F0), even for one of the loci where the observed somatic mutation rate was only 6% (17 and unpublished results). The germline transmission rates of the identified founders range from 8–60% (17 and unpublished results). Although our mutagenesis frequencies are high, we note that some of the indels we have observed do not cause frameshift mutations. In this regard, one advantage of using the fluorescent PCR analysis approach to identify mutations is that the size differences of the mutant alleles may be determined and used to predict which founders are likely to carry frameshift mutations. All of the engineered zinc finger arrays that we have used to construct ZFNs have demonstrated DNA-binding activities in B2H system. We do not know why some of the engineered ZFNs do not cause mutations in vivo. It is possible that some of the genomic loci may not be accessible due to chromatin structure or modifications of the target DNA sites. If a pair of engineered ZFNs fails to introduce mutations in the target site, we recommend engineering ZFNs targeting other regions of the same gene.
Acknowledgments
We thank Dr. A.J. Giraldez for valuable suggestions and D. Cotelle for technical help on the fluorescent PCR analysis, Drs. P. Schlueter and C. Sachidanandan for helpful suggestions on the manuscript. J.E.F., M.L.M., and J.K.J. are supported by the NIH (R01GM069906, R21RR024189, and R21HL091808) and the MGH Pathology Service. R.T.P. and J.-R.J.Y. are supported by the NIH (CA118498 and GM88040) and the Ned Sahin Fund. J.-R.J.Y. is also supported by the NIH (AG031300) and the Claflin Distinguished Scholar Award.
Footnotes
CFI: The authors declare competing financial interests.
CFI statement: J.K.J. is an inventor on patent applications which describe the OPEN zinc finger engineering method. All other authors declare that they have no competing financial interests.
AUTHOR CONTRIBUTIONS: J.P. provided the protocol for plasmid DNA isolation in a 96-well format. All the other authors contributed extensively to the protocol development and preparation of the manuscript.
References
- 1.Cathomen T, Joung JK. Zinc-finger nucleases: the next generation emerges. Mol Ther. 2008;16:1200–7. doi: 10.1038/mt.2008.114. [DOI] [PubMed] [Google Scholar]
- 2.Porteus MH, Carroll D. Gene targeting using zinc finger nucleases. Nat Biotechnol. 2005;23:967–73. doi: 10.1038/nbt1125. [DOI] [PubMed] [Google Scholar]
- 3.Durai S, Mani M, Kandavelou K, Wu J, Porteus MH, Chandrasegaran S. Zinc finger nucleases: custom-designed molecular scissors for genome engineering of plant and mammalian cells. Nucleic Acids Res. 2005;33:5978–90. doi: 10.1093/nar/gki912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carroll D. Progress and prospects: zinc-finger nucleases as gene therapy agents. Gene Ther. 2008;15:1463–8. doi: 10.1038/gt.2008.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu J, Kandavelou K, Chandrasegaran S. Custom-designed zinc finger nucleases: what is next? Cell Mol Life Sci. 2007;64:2933–44. doi: 10.1007/s00018-007-7206-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Camenisch TD, Brilliant MH, Segal DJ. Critical parameters for genome editing using zinc finger nucleases. Mini Rev Med Chem. 2008;8:669–76. doi: 10.2174/138955708784567458. [DOI] [PubMed] [Google Scholar]
- 7.Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci U S A. 1996;93:1156–60. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim YG, Shi Y, Berg JM, Chandrasegaran S. Site-specific cleavage of DNA-RNA hybrids by zinc finger/FokI cleavage domain fusions. Gene. 1997;203:43–9. doi: 10.1016/s0378-1119(97)00489-7. [DOI] [PubMed] [Google Scholar]
- 9.Maeder ML, Thibodeau-Beganny S, Osiak A, Wright DA, Anthony RM, Eichtinger M, Jiang T, Foley JE, Winfrey RJ, Townsend JA, Unger-Wallace E, Sander JD, Muller-Lerch F, Fu F, Pearlberg J, Gobel C, Dassie JP, Pruett-Miller SM, Porteus MH, Sgroi DC, Iafrate AJ, Dobbs D, McCray PB, Jr, Cathomen T, Voytas DF, Joung JK. Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol Cell. 2008;31:294–301. doi: 10.1016/j.molcel.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bibikova M, Beumer K, Trautman JK, Carroll D. Enhancing gene targeting with designed zinc finger nucleases. Science. 2003;300:764. doi: 10.1126/science.1079512. [DOI] [PubMed] [Google Scholar]
- 11.Bibikova M, Golic M, Golic KG, Carroll D. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics. 2002;161:1169–75. doi: 10.1093/genetics/161.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Santiago Y, Chan E, Liu PQ, Orlando S, Zhang L, Urnov FD, Holmes MC, Guschin D, Waite A, Miller JC, Rebar EJ, Gregory PD, Klug A, Collingwood TN. Targeted gene knockout in mammalian cells by using engineered zinc-finger nucleases. Proc Natl Acad Sci U S A. 2008;105:5809–14. doi: 10.1073/pnas.0800940105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller JC, Holmes MC, Wang J, Guschin DY, Lee YL, Rupniewski I, Beausejour CM, Waite AJ, Wang NS, Kim KA, Gregory PD, Pabo CO, Rebar EJ. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol. 2007;25:778–85. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- 14.Lombardo A, Genovese P, Beausejour CM, Colleoni S, Lee YL, Kim KA, Ando D, Urnov FD, Galli C, Gregory PD, Holmes MC, Naldini L. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat Biotechnol. 2007;25:1298–306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- 15.Perez EE, Wang J, Miller JC, Jouvenot Y, Kim KA, Liu O, Wang N, Lee G, Bartsevich VV, Lee YL, Guschin DY, Rupniewski I, Waite AJ, Carpenito C, Carroll RG, Orange JS, Urnov FD, Rebar EJ, Ando D, Gregory PD, Riley JL, Holmes MC, June CH. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat Biotechnol. 2008;26:808–16. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doyon Y, McCammon JM, Miller JC, Faraji F, Ngo C, Katibah GE, Amora R, Hocking TD, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Amacher SL. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat Biotechnol. 2008;26:702–8. doi: 10.1038/nbt1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foley JE, Yeh JR, Maeder ML, Reyon D, Sander JD, Peterson RT, Joung JK. Rapid mutation of endogenous zebrafish genes using zinc finger nucleases made by Oligomerized Pool ENgineering (OPEN) PLoS ONE. 2009;4:e4348. doi: 10.1371/journal.pone.0004348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meng X, Noyes MB, Zhu LJ, Lawson ND, Wolfe SA. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat Biotechnol. 2008;26:695–701. doi: 10.1038/nbt1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beumer KJ, Trautman JK, Bozas A, Liu JL, Rutter J, Gall JG, Carroll D. Efficient gene targeting in Drosophila by direct embryo injection with zinc-finger nucleases. Proc Natl Acad Sci U S A. 2008;105:19821–6. doi: 10.1073/pnas.0810475105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beumer K, Bhattacharyya G, Bibikova M, Trautman JK, Carroll D. Efficient gene targeting in Drosophila with zinc-finger nucleases. Genetics. 2006;172:2391–403. doi: 10.1534/genetics.105.052829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morton J, Davis MW, Jorgensen EM, Carroll D. Induction and repair of zinc-finger nuclease-targeted double-strand breaks in Caenorhabditis elegans somatic cells. Proc Natl Acad Sci U S A. 2006;103:16370–5. doi: 10.1073/pnas.0605633103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Townsend JA, Wright DA, Winfrey RJ, Fu F, Maeder ML, Joung JK, Voytas DF. High-frequency modification of plant genes using engineered zinc-finger nucleases. Nature. 2009;459:442–5. doi: 10.1038/nature07845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moehle EA, Rock JM, Lee YL, Jouvenot Y, DeKelver RC, Gregory PD, Urnov FD, Holmes MC. Targeted gene addition into a specified location in the human genome using designed zinc finger nucleases. Proc Natl Acad Sci U S A. 2007;104:3055–60. doi: 10.1073/pnas.0611478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, Holmes MC. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–51. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 25.Shukla VK, Doyon Y, Miller JC, DeKelver RC, Moehle EA, Worden SE, Mitchell JC, Arnold NL, Gopalan S, Meng X, Choi VM, Rock JM, Wu YY, Katibah GE, Zhifang G, McCaskill D, Simpson MA, Blakeslee B, Greenwalt SA, Butler HJ, Hinkley SJ, Zhang L, Rebar EJ, Gregory PD, Urnov FD. Precise genome modification in the crop species Zea mays using zinc-finger nucleases. Nature. 2009;459:437–41. doi: 10.1038/nature07992. [DOI] [PubMed] [Google Scholar]
- 26.Zou J, Maeder ML, Mali P, Pruett-Miller SM, Thibodeau-Beganny S, Chou BK, Chen G, Ye Z, Park IH, Daley GQ, Porteus MH, Joung JK, Cheng L. Gene targeting of a disease-related gene in human induced pluripotent stem and embryonic stem cells. Cell Stem Cell. 2009;5:97–110. doi: 10.1016/j.stem.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cai CQ, Doyon Y, Ainley WM, Miller JC, Dekelver RC, Moehle EA, Rock JM, Lee YL, Garrison R, Schulenberg L, Blue R, Worden A, Baker L, Faraji F, Zhang L, Holmes MC, Rebar EJ, Collingwood TN, Rubin-Wilson B, Gregory PD, Urnov FD, Petolino JF. Targeted transgene integration in plant cells using designed zinc finger nucleases. Plant Mol Biol. 2009;69:699–709. doi: 10.1007/s11103-008-9449-7. [DOI] [PubMed] [Google Scholar]
- 28.Hurt JA, Thibodeau SA, Hirsh AS, Pabo CO, Joung JK. Highly specific zinc finger proteins obtained by directed domain shuffling and cell-based selection. Proc Natl Acad Sci U S A. 2003;100:12271–6. doi: 10.1073/pnas.2135381100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cornu TI, Thibodeau-Beganny S, Guhl E, Alwin S, Eichtinger M, Joung JK, Cathomen T. DNA-binding specificity is a major determinant of the activity and toxicity of zinc-finger nucleases. Mol Ther. 2008;16:352–8. doi: 10.1038/sj.mt.6300357. [DOI] [PubMed] [Google Scholar]
- 30.Smith J, Bibikova M, Whitby FG, Reddy AR, Chandrasegaran S, Carroll D. Requirements for double-strand cleavage by chimeric restriction enzymes with zinc finger DNA-recognition domains. Nucleic Acids Res. 2000;28:3361–9. doi: 10.1093/nar/28.17.3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mani M, Smith J, Kandavelou K, Berg JM, Chandrasegaran S. Binding of two zinc finger nuclease monomers to two specific sites is required for effective double-strand DNA cleavage. Biochem Biophys Res Commun. 2005;334:1191–7. doi: 10.1016/j.bbrc.2005.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beerli RR, Barbas CF., 3rd Engineering polydactyl zinc-finger transcription factors. Nat Biotechnol. 2002;20:135–41. doi: 10.1038/nbt0202-135. [DOI] [PubMed] [Google Scholar]
- 33.Segal DJ. The use of zinc finger peptides to study the role of specific factor binding sites in the chromatin environment. Methods. 2002;26:76–83. doi: 10.1016/S1046-2023(02)00009-9. [DOI] [PubMed] [Google Scholar]
- 34.Liu Q, Xia Z, Zhong X, Case CC. Validated zinc finger protein designs for all 16 GNN DNA triplet targets. J Biol Chem. 2002;277:3850–6. doi: 10.1074/jbc.M110669200. [DOI] [PubMed] [Google Scholar]
- 35.Bae KH, Kwon YD, Shin HC, Hwang MS, Ryu EH, Park KS, Yang HY, Lee DK, Lee Y, Park J, Kwon HS, Kim HW, Yeh BI, Lee HW, Sohn SH, Yoon J, Seol W, Kim JS. Human zinc fingers as building blocks in the construction of artificial transcription factors. Nat Biotechnol. 2003;21:275–80. doi: 10.1038/nbt796. [DOI] [PubMed] [Google Scholar]
- 36.Kim HJ, Lee HJ, Kim H, Cho SW, Kim JS. Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Res. 2009;19:1279–88. doi: 10.1101/gr.089417.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramirez CL, Foley JE, Wright DA, Muller-Lerch F, Rahman SH, Cornu TI, Winfrey RJ, Sander JD, Fu F, Townsend JA, Cathomen T, Voytas DF, Joung JK. Unexpected failure rates for modular assembly of engineered zinc fingers. Nat Methods. 2008;5:374–5. doi: 10.1038/nmeth0508-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maeder ML, Thibodeau-Beganny S, Sander JD, Voytas DF, Joung JK. An “open-source” protocol for making customized zinc finger arrays. Nat Protoc. 2009;4:1471–1501. doi: 10.1038/nprot.2009.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carroll D, Morton JJ, Beumer KJ, Segal DJ. Design, construction and in vitro testing of zinc finger nucleases. Nat Protoc. 2006;1:1329–41. doi: 10.1038/nprot.2006.231. [DOI] [PubMed] [Google Scholar]
- 40.Wright DA, Thibodeau-Beganny S, Sander JD, Winfrey RJ, Hirsh AS, Eichtinger M, Fu F, Porteus MH, Dobbs D, Voytas DF, Joung JK. Standardized reagents and protocols for engineering zinc finger nucleases by modular assembly. Nat Protoc. 2006;1:1637–52. doi: 10.1038/nprot.2006.259. [DOI] [PubMed] [Google Scholar]
- 41.Sander JD, Zaback P, Joung JK, Voytas DF, Dobbs D. Zinc Finger Targeter (ZiFiT): an engineered zinc finger/target site design tool. Nucleic Acids Res. 2007;35:W599–605. doi: 10.1093/nar/gkm349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Szczepek M, Brondani V, Buchel J, Serrano L, Segal DJ, Cathomen T. Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nat Biotechnol. 2007;25:786–93. doi: 10.1038/nbt1317. [DOI] [PubMed] [Google Scholar]
- 43.Rembold M, Lahiri K, Foulkes NS, Wittbrodt J. Transgenesis in fish: efficient selection of transgenic fish by co-injection with a fluorescent reporter construct. Nat Protoc. 2006;1:1133–9. doi: 10.1038/nprot.2006.165. [DOI] [PubMed] [Google Scholar]
- 44.Westerfield M. The zebrafish book: A guide for the laboratory use of zebrafish (Danio Rerio) The University of Oregon Press; Eugene: 2000. [Google Scholar]
- 45.Yuan S, Sun Z. Microinjection of mRNA and morpholino antisense oligonucleotides in zebrafish embryos. J Vis Exp. 2009 doi: 10.3791/1113. [DOI] [PMC free article] [PubMed] [Google Scholar]