Abstract
Vascular endothelial growth factor (VEGF) is a homodimeric pro-angiogenic protein that induces endothelial cell migration and proliferation primarily through interactions with its major receptors, VEGFR-1 and VEGFR-2. Inhibitors of one or both of these VEGF-receptor interactions could be beneficial as therapeutics for diseases caused by dysfunctional angiogenesis (e.g., cancer). Others have reported small peptides that bind to the VEGF dimer at surface regions that are recognized by the receptors. Here we report the development of a fluorescence polarization assay based on the binding to VEGF of a derivative of one of these peptides that has been labeled with BODIPY-tetramethylrhodamine (TMR). This 384-well format assay is tolerant to DMSO (up to 4% v/v) and has a Z’ factor of 0.76, making it useful for identifying molecules that associate with the receptor-binding surface of the VEGF dimer.
Keywords: High-throughput screening, Fluorescence polarization assay, VEGF
Introduction
Vascular endothelial growth factor (VEGF) is an important regulator of angiogenesis.[1–4] VEGF promotes the migration and proliferation of endothelial cells and the formation of new blood vessels from preexisting capillaries.[5, 6] Biological responses to VEGF expression result from the binding of VEGF to two major membrane-embedded receptors, VEGFR-1 (Flt-1) and VEGFR-2 (KDR), and the subsequent intracellular signaling induced by receptor activation.[3] VEGFR-1 and -2 are found on the surface of most vascular endothelial cells.[2] The majority of proangiogenic activity of VEGF is mediated through VEGFR-2, although interaction between VEGF and VEGFR-1 may also play important roles in angiogenesis.[2, 3]
VEGF-A is a homodimeric glycoprotein that exists in several different isoforms, generated via alternative splicing of the VEGF-A mRNA. [2, 7] The four major isoforms of VEGF are VEGF121, VEGF165, VEGF189, and VEGF206; VEGF165 is predominant in vivo.[2] VEGF is active as a dimer in which the two anti-parallel monomers are linked by two disulfide bonds.[8, 9] Signaling requires the VEGF homodimer to interact with two VEGF receptor molecules.[10] Each receptor consists of an extracellular region composed of seven immunoglobulin-like domains, a transmembrane region, and an intracellular kinase domain. VEGF-induced dimerization of the receptors leads to auto-phosphorylation of the intracellular portion, which initiates cytoplasmic signaling cascades that ultimately result in increased vascular permeability, endothelial cell migration and proliferation, and angiogenesis.[2, 10] For both VEGF receptors, extracellular domains 2 and 3 provide most of the contact surface for binding to the VEGF dimer.[11–16] The importance of domain 2 of VEGFR-1 for binding VEGF is further supported by NMR and X-ray crystallographic data.[17, 18]
A balance between pro- and anti-angiogenic signaling is necessary to maintain cellular homeostasis during embryonic development and throughout life.[2, 3, 19, 20] Overexpression of VEGF causes a shift in the angiogenic equilibrium, leading to excessive vasculature formation.[10, 21] VEGF overexpression has been linked to a number of human diseases including cancer, rheumatoid arthritis, psoriasis and proliferating retinopathy.[22] Currently, there are three FDA approved drugs for treatment of diseases related to overexpression of VEGF. Each of these drugs blocks signaling by binding to VEGF and thereby preventing interaction with cell-surface receptors.[23–25] Bevacizumab (Avastin), for example, is an engineered antibody that binds to VEGF and prevents interaction with VEGFR-1 and -2;[23] Avastin is approved for anticancer chemotherapy. Ranibizumab (Lucentis) is a smaller antibody fragment with a similar mode of action that has been approved for treatment of macular degeneration.[25] Pegaptanib (Macugen), a modified oligonucleotide, binds to the heparin-binding domain of VEGF and is approved for treatment of macular degeneration.[24] In addition, a number of small molecules that inhibit the kinase activity of VEGFR-1 and/or VEGFR-2 are in clinical trials.[26] Despite the success of the therapeutic antibodies Avastin and Lucentis, inhibitors of the VEGF-VEGFR interaction with lower molecular weight (e.g., peptides or small molecules) might offer advantages in terms of production, stability and/or administration.[27] It is very challenging to identify small molecules that bind tightly and specifically to a given protein surface due to the relatively flat surface of the interaction;[28, 29] typically, thousands of inhibitor candidates must be screened.
In principle, steric inhibition of VEGF-VEGFR interaction could be accomplished via a molecule that binds to the recognition surface on VEGF or a molecule that binds to the recognition surface on a receptor. All steric inhibitors in the clinic or in clinical trials function by binding to VEGF rather than a receptor.[26] An agent that binds to VEGF can be specific for one type of biological response, while an agent that binds to a VEGF receptor may block interaction of this receptor with other natural ligands (other members of the VEGF super-family), thereby affecting processes other than angiogenesis.[30]
Several assay formats have been used to evaluate candidate molecules for the ability to bind to VEGF or to inhibit VEGF-VEGFR interaction. The most popular formats, including enzyme-linked immunosorbent assays (ELISA) and surface plasmon resonance (SPR), involve measuring the ability of candidate ligands to bind to surface-immobilized VEGF.[31, 32] Another popular assay format involves measuring competition with radioiodinated VEGF for binding to membrane-bound VEGF receptors on the surface of endothelial cells.[33] However, overexpression of one protein in a cell-based assay can affect other proteins on the cell and lead to complications in the analysis of inhibitor candidate binding targets.[34] A biochemical assay with purified proteins allows for a more conclusive identification of the target of active compounds. Fluorescence-based assays in which all components (proteins, inhibitors, probes) are in solution can be advantageous in terms of throughput relative to assay formats involving an immobilized component.[35, 36] Furthermore, the proteins are more likely to adopt conformations similar to their native state in solution than when immobilized on a surface.
We have developed a homogeneous competition fluorescence polarization (FP) assay that can be used to identify molecules that bind to the VEGF dimer in the region that is recognized by cell surface receptors. Development of this assay required a relatively small, fluorescent ligand (“tracer”) for VEGF that could be displaced by candidate inhibitors; a short peptide (v114) identified at Genentech[32] served as the basis for our tracer in these studies. We have demonstrated that the assay can rapidly and reliably distinguish compounds of different affinity for VEGF. The assay is robust and may be used in a high-throughput screening format.
Materials and Methods
General
Fmoc-L-α-amino acids, HBTU, and NovaSyn TGR resin were purchased from NovaBiochem (San Diego, CA). 6-Carboxyfluorescein was purchased from Anaspec (San Jose, CA). BODIPYTMR-SE was purchased from Invitrogen (Carlsbad, CA). (2-[2-(Fmoc-amino)-ethoxy]-ethoxy)-acetic acid (Fmoc-AEEAc-OH) was purchased from Bachem (Torrance, CA). Piperidine, HOBt, trifluoroacetic acid (TFA), HPLC-grade acetonitrile (MeCN), dimethylformamide (DMF), dichloromethane (DCM), and all other chemical reagents were purchased from Sigma-Aldrich (Milwaukee, WI).
Peptide Synthesis
Peptides were synthesized in 4.0-mL solid-phase extraction tubes from Alltech (Deerfield, IL) on NovaSyn TGR resin, to afford C-terminal amides upon cleavage from the resin. A vacuum manifold was used to wash the resin with DMF and DCM between coupling and deprotection steps. The following is a representative coupling-deprotection cycle for a 30 µmol scale: 115.4 mg of NovaSyn TGR resin (reported loading: 0.26 mmol/g) was swelled for 20 min in DCM. After the resin was washed with DCM and DMF, 3 equiv (90 µmol) of Fmoc-L-α-amino acid of C-terminal residue and 3 equiv of HBTU dissolved in 1.5 mL DMF were added to the resin. N,N-Diisopropylethylamine (180 µL; 1.0 M solution in DMF) and HOBt (180 µL; 0.5 M solution in DMF) were added to initiate the coupling and the reaction was put on a rocker to agitate for 1 h. After the resin was washed with DCM and DMF, Fmoc deprotection was accomplished by adding ∼2 mL 20% (v/v) piperidine in DMF and agitating the reaction on a rocker for 15 min. Fluorescein labeling for peptides 1 and 2 was accomplished by coupling 6-carboxyfluorescein overnight using the same conditions described above for coupling α-amino acids. BODIPYTMR labeling for peptide 3 was achieved after cleavage and purification (see below). All peptides were cleaved from resin using 1.5 mL cleavage solution (94% TFA, 2.5% H2O, 2.5% 1,2-ethanedithiol, 1% triisopropylsilane). After the TFA was evaporated under a stream of nitrogen, the crude peptide was dissolved in 0.2 mL TFA and precipitated in cold ether. The ether was decanted and the peptide was dissolved in 4 mL DMSO and, if a disulfide bond was needed in the peptide, 10 µL to 2 mL NH4OH (enough to make the solution basic). Peptides were purified using semipreparative, reverse-phase HPLC performed with a C4 or C18 column (Vydac, Anaheim, CA) and eluting with gradients of MeCN w/0.1% TFA (B solvent) in water w/0.1% TFA (A solvent). Fractions were lyophilized to yield the final peptide as dry powders, which were confirmed by MALDI-TOF mass spectrometry. As previously mentioned, the BODIPYTMR was coupled to the free N-terminal amine after purification as follows: 0.4 mg of the peptide was dissolved in 450 µL DMF and 18 µL triethylamine was added to the solution. The BODIPYTMR-succinimide ester was dissolved in 450 µL DMF and the two solutions were mixed. The peptide was repurified by HPLC after 7 h.
Expression and purification of VEGF [37]
A pET-3d vector (Novagen, Madison, WI) containing the gene for human VEGF (residues 2–165) was transformed into Escherichia coli BL21(DE3) cells. Cells were grown at 37°C in LB medium with 100 µg/mL ampicillin to an OD600 of ∼1.0. Protein expression was induced with 0.4 mM IPTG, and cells were grown for an additional 3 h. The cells were pelleted by centrifugation at 1,000 × g for 30 min at 4°C, resuspended in TBS, pelleted at 3,000 × g for 30 min at 4°C, and stored at −80°C. Cells were resuspended with 40 mL of resuspension buffer (20 mM Tris, 5 mM EDTA; pH 7.5) with 4 mg lysozyme and 0.5 mg DNase I. After incubating on ice for 1 h, 50 µL of a protease inhibitor cocktail (Calbiochem, #539134) was added and the cells were disrupted by pulse sonication. The cells were incubated for 1 h with 0.025 g sodium-deoxycholate and 570 µL of a 70% Tergitol solution and centrifuged at 16,000 × g for 30 min at 4°C. The pellet was resuspended in B-PER Bacterial Protein Extraction Reagent (Pierce, #78243) and centrifuged again. The inclusion bodies were solubilized in 30 mL of dissolving buffer (20 mM Tris, 7.5 M urea, 20 mM DTT; pH 7.5) and stirred for 1 h at 4°C to fully dissolve the pellet.
The concentration of VEGF was determined by measuring the A280 of the protein in a denaturing buffer (22 mM phosphate, 8 M guanidine-HCl; pH 6.5) with a final guanidine concentration of 5 M using a molar extinction coefficient of 12160 M−1cm−1 [(0.32 mg/mL)−1 cm−1].[38] The protein was diluted with dissolving buffer to a concentration of 0.75 mg/mL and dialyzed against refolding buffer (20 mM Tris, 0.4 M NaCl, 1 mM cysteine; pH 8.4). The protein was then dialyzed against ion exchange(IEX) buffer A (20 mM Tris; pH 7.5) and purified using a HiTrap SP FF 1 mL column (GE Healthcare, Piscataway, NJ) with a gradient of 0–100% IEX buffer B (20 mM Tris; pH 7.5, 1 M NaCl) over 20 min. The purified protein was dialyzed against PBS and stored at 4°C for immediate use or −80°C for long-term storage. The final isolated yield of VEGF obtained was ∼ 3 mg per liter of starting culture.
VEGF Activity Validation
In order to determine whether the VEGF we produced via bacterial expression was properly folded, we compared this protein to commercially available VEGF165 (Cell Sciences, cat # CRV000B) for the ability to enhance the proliferation of Human Umbilical Vein Endothelial Cells (HUVEC). HUVECs were cultured in Medium 200 (Cascade Biologics) containing 2% FBS with FGF2, EGF, heparin, and hydrocortisone in a 24 well tissue culture plate. The serum and growth factor containing medium was replaced with basal medium with or without VEGF (100 ng/mL). Cell proliferation was measured after about 33 hours using a colorimetric assay (Promega, cat # G3580). HUVECs growing in serum and growth factor supplemented medium constituted the positive control.
The VEGF prepared here was further tested for its ability to enhance EC migration compared to commercially available VEGF165 (R&D Biosciences and Peprotech) in a transwell migration assay. The transwell migration assay was performed as previously described[39] with some modifications. Briefly, HUVECs were serum starved overnight (2% serum containing medium), washed with 0.04% EDTA in PBS, trypsinized, and resuspended in serum-free M199 medium at 1×106 cells/mL. The transwell coated with fibronectin was rinsed with PBS and 0.5 mL of serum-free M199 or M199 containing VEGF (10 ng/ml) was added to the 24 well dish containing the transwell. Cells (0.1 mL of 1×106) were then added to the top of the transwell membrane and incubated for 16 h at 37°C in 5% CO2. The mean number of cells migrated through the membrane was determined by counting 10 high power fields (×100) and compared to the control (PBS, no VEGF).
Peptide Concentration Determination
All peptides to be tested for binding to VEGF were dissolved in DMSO for testing. The concentrations of peptides bearing attached fluorophores were quantified by measuring the fluorophore absorbance. Fluorescein-bearing tracers were diluted in 10 mM Tris, pH 8.0, and the concentration was determined from A494 using ɛ = 68,000 M−1cm−1.[40] The BODIPYTMR-bearing tracer was diluted in water, and the concentration was determined from A535 using ɛ = 50,000 M−1cm−1.[40] For peptides without a fluorophore, the extinction coefficients were calculated as described previously.[38] Peptides with two tryptophans and a cystine had an extinction coefficient of 11500 M−1cm−1, peptides with one tryptophan and a cystine had an extinction coefficient of 5810 M−1cm−1, and peptides with a single tryptophan had an extinction coefficient of 5690 M−1cm−1.
Direct Binding FP Assays
Wells of a black Costar 384-well polystyrene plate (Corning, Corning, NY) contained fluorescent peptide tracer (67 nM for tracers 1 and 2, 10 nM for tracer 3) and increasing concentrations (from 0.63 pM to 3 µM for tracers 1 and 2, 0.61 pM to 10 µM for tracer 3) of VEGF protein in FP Buffer (50 mM NaCl, 16.2 mM Na2HPO4, 3.8 mM KH2PO4, 0.15 mM NaN3, 0.15 mM EDTA, 0.5 mg/mL Pluronic; pH 7.4).[41] Plates were read after a 2 h incubation at room temperature using a PerkinElmer EnVision multi-label plate reader (Wellesley, MA) with polarized filters and optical modules for fluorescein (λexcitation: 480 nm; λemission: 535 nm) or BODIPYTMR (λexcitation: 531 nm; λemission: 595 nm). mP values were calculated from raw parallel and perpendicular fluorescence intensities.[42] GraphPad Prism 4.03 (San Diego, CA) was used to plot mP vs. VEGF concentration and the curve was fit to a single-site binding model to extract a binding dissociation constant (Kd value) for each tracer. [43] Experiments were performed in duplicate.
Competition Binding FP Assays
Wells of a 384 plate contained 10 nM BODIPYTMR tracer peptide 3, 40 nM VEGF protein, and 2 µL tested inhibitor dissolved in DMSO (final concentration from 63 pM to 300 µM) in a final volume of 50 µL in FP Buffer. Plates were read after a 5 h incubation at room temperature, the time necessary for complete equilibration. Experiments were performed in duplicate. The equilibrium dissociation constant (Ki) [44, 45] or IC50[46] was calculated and the error was given as the 95% confidence interval as calculated by GraphPad Prism.
The FP signal of free BODIPYTMR tracer 3 in solution was tested for a concentration range of 0.1 to 100 nM 3 in FP Buffer in a final volume of 50 µL. DMSO tolerance was tested using the above competition protocol with peptide v107. The Z’ factor was calculated[47] for the 384-well format in the competition format using positive (40 nM VEGF, 10 nM tracer 3 in buffer, 4% DMSO) and negative (10 nM tracer 3 in buffer, 4% DMSO) controls.
Results and Discussion
VEGF Construct Selection and Validation
We considered several isoforms of VEGF-A as we were designing the FP assay. High-resolution structures of VEGF8–109 alone[48] and in complex with a soluble fragment of VEGFR-1,[17] or with the peptide v107[49] have been solved by researchers at Genentech. In addition, VEGF8–109 was used with phage-displayed libraries to identify peptide ligands v107 and v114.[32] However, VEGF165 (residues 1–165) is the most prevalent isoform in vivo. [2] This protein includes a heparin-binding domain, absent in VEGF8–109, which is necessary for mitogenic activity in cells.[2] Since VEGF165 is the most physiologically relevant protein, we chose to use this isoform in our FP assay.
We expressed VEGF165 in E. coli and assayed the purified protein for its ability to increase endothelial cell proliferation and migration, the expected biological activity of VEGF. To measure endothelial cell proliferation, human umbilical vein endothelial cells (HUVECs) were treated with VEGF and cell growth was monitored. The number of HUVECs was increased by ∼1.7-fold in the presence of VEGF165 relative to cells growing in the basal medium. This result is comparable to the ∼2.0-fold relative increase in HUVEC proliferation observed in response to commercially available VEGF165. HUVEC migration in response to VEGF treatment was monitored in a transwell migration assay. Migration of HUVECs increased ∼4-fold in the presence of our VEGF relative to the negative control; a ∼4-fold increase in HUVEC migration was also observed for commercially available VEGF. Taken together, these cell-based assays indicated that the VEGF expressed in E. coli was properly folded and biologically active. VEGF expressed in E. coli has been shown to give similar biological activity to glycosylated VEGF derived from a baculovirus expression system.[50] Our VEGF from E. coli could therefore be used to develop the FP assay.
Tracer Design and Optimization
According to previous results, peptide v114 (H-VEPNCDIHVMWEWECFERL-NH2 with the cysteines forming a disulfide) binds to VEGF8–109 with an IC50 of 0.22 µM in a competition SPR-based assay, making it one of the tightest-binding VEGF peptides in the literature.[32] Although no structural data are available for the complex between v114 and VEGF, the NMR structure of complex between VEGF8–109 and a closely related 19-mer peptide, v107 (H-GGNECDIARMWEWECFERL-NH2 with the cysteines forming a disulfide; IC50 (SPR) = 0.70 µM), shows that v107 binds to the VEGF receptor-binding surface (Table 1).[32, 49] Thus, we assumed that v114 would also target the receptor-binding surface on VEGF. This assumption is supported by the observation that peptide v114 blocks VEGF165-induced proliferation of HUVECs in culture.[32] We chose to base our FP tracer sequence on v114 instead of v107 because v114 binds more tightly to VEGF, and because v114 has been shown to be biologically active. A tracer with a low Kd was desirable in order to minimize the amount of protein and tracer needed to obtain a strong FP signal with a good dynamic range. A tighter binding tracer would also be able to measure a wider range of inhibitor potency and maintain the sensitivity to identify weakly binding inhibitors.[51]
Table 1.
19-mer Peptides that Bind VEGF (cysteines in bold form a disulfide)
| Peptide | Sequence | IC50 (µM) [32] |
|---|---|---|
| v114 | H-VEPNCDIHVMWEWECFERL-NH2 | 0.22 |
| v107 | H-GGNECDIARMWEWECFERL-NH2 | 0.70 |
Initially, we evaluated fluorescein (Flu) as the fluorophore in our FP tracer candidates. In the reported structure of the v107-VEGF complex,[49] the N-terminal segment of v107 (residues Gly-Gly-Asn-Glu) appears to be flexible when the peptide is bound to VEGF; the analogous segment in v114 (Val-Glu-Pro-Asn), to which is the fluorophore would be attached, would likely be flexible as well. Based upon this possibility, we focused on v114 tracer candidates lacking the four N-terminal residues, in order to promote local ordering of the fluorophore in the VEGF-bound conformation.
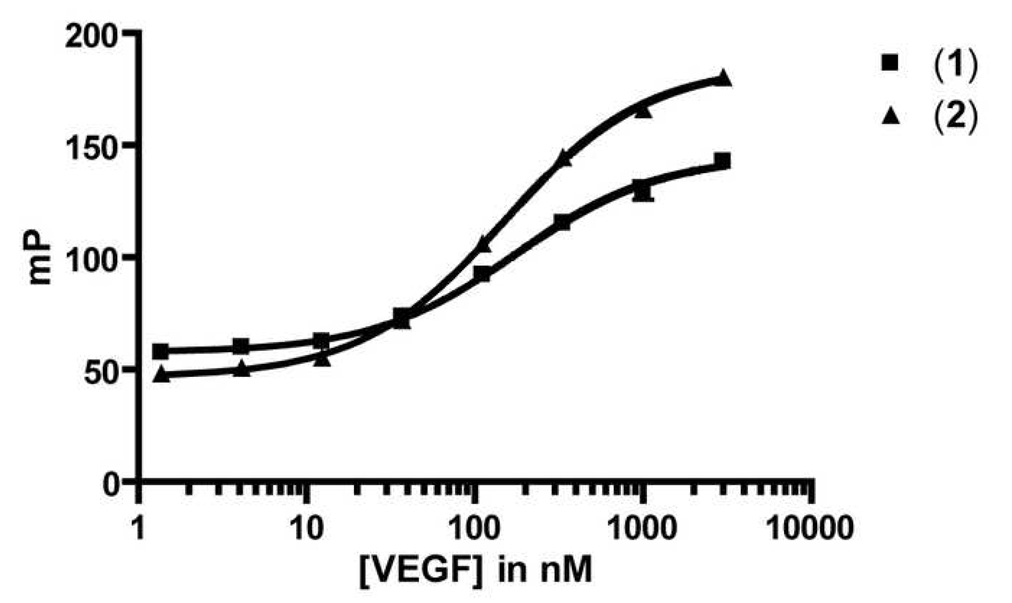
Two truncated fluorescein-bearing tracer candidates were evaluated (Table 2), one with the fluorophore directly attached to the N-terminus (1) and the other with a [2-(2-amino-ethoxy)-ethoxy]-acetic acid spacer between the fluorophore and the peptide (2). Unlike Flu-v114, these tracers displayed significant binding to VEGF in the direct titration FP assay (Figure 1). The dissociation constants (Kd) of tracers 1 and 2 were similar (170 and 140 nM, respectively). Tracer 2 was considered superior to 1 because 2 had a larger change in the fluorescence polarization signal between free and bound tracer (dynamic range = 140 ± 8 mP units for 2 vs. 88 ± 7 mP units for 1).
Table 2.
Tracer Peptide Sequences Tested (cysteines in bold form a disulfide)
| Peptide | Sequence | Kd (nM) | ΔmP |
|---|---|---|---|
| (1) | Flu-CDIHVMWEWECFERL-NH2 | 170 ± 50 | 88 ± 7 |
| (2) | Flu-X-CDIHVMWEWECFERL-NH2 | 140 ± 40 | 140 ± 8 |
| (3) | (BODIPYTMR)-X-CDIHVMWEWECFERL-NH2 | 25 ± 3 | 200 ± 7 |
X = [2-[2-amino-ethoxy]-ethoxy}-acetic acid]
Figure 1.
Direct Binding of Two Fluorescein Tracers Based on v114
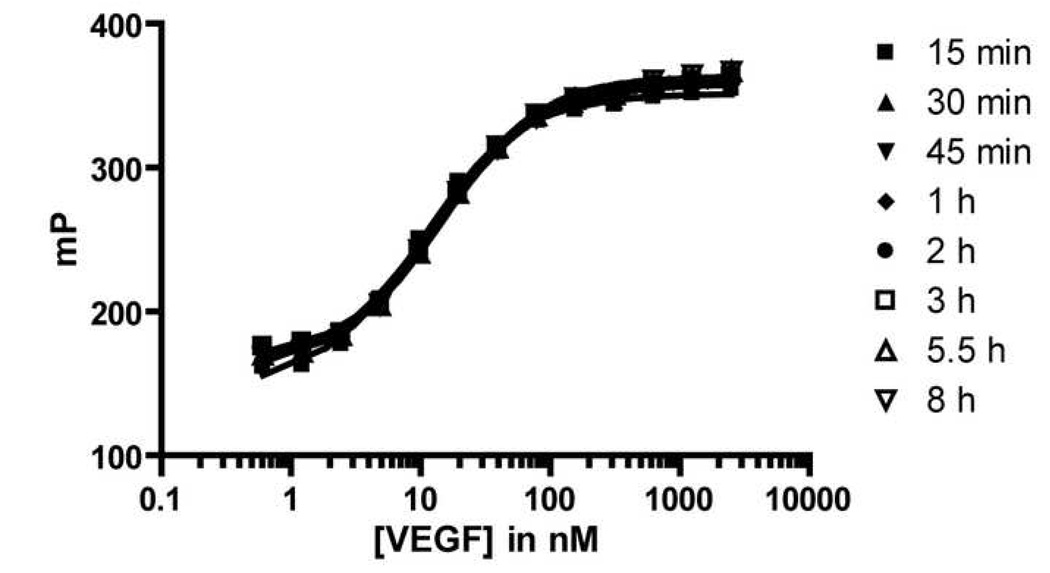
Common assay interferences, such as those arising from compound aggregation or fluorescent impurities, tend to be diminished in assays that employ red-shifted fluorophores (e.g., BODIPYTMR) as opposed to assays that employ green fluorophores (e.g., fluorescein).[52] We therefore replaced the fluorescein unit on peptide 2 with BODIPYTMR to generate peptide 3. Interestingly, we observed an increase in both the binding affinity for VEGF (Kd = 25 nM) and the dynamic range of the FP signal (∼200 mP) for tracer 3 relative to 2 (Figure 2). The assay was stable for at least 8 hours (i.e., the FP signal was constant for at least 8 hours after the reagents were mixed), which is important for testing multiple sample plates in a high-throughput format. Subsequent experiments were carried out with the optimized tracer 3.
Figure 2.
Direct Binding of BODIPYTMR Tracer 3 over Time
Competition FP Assay Assessment
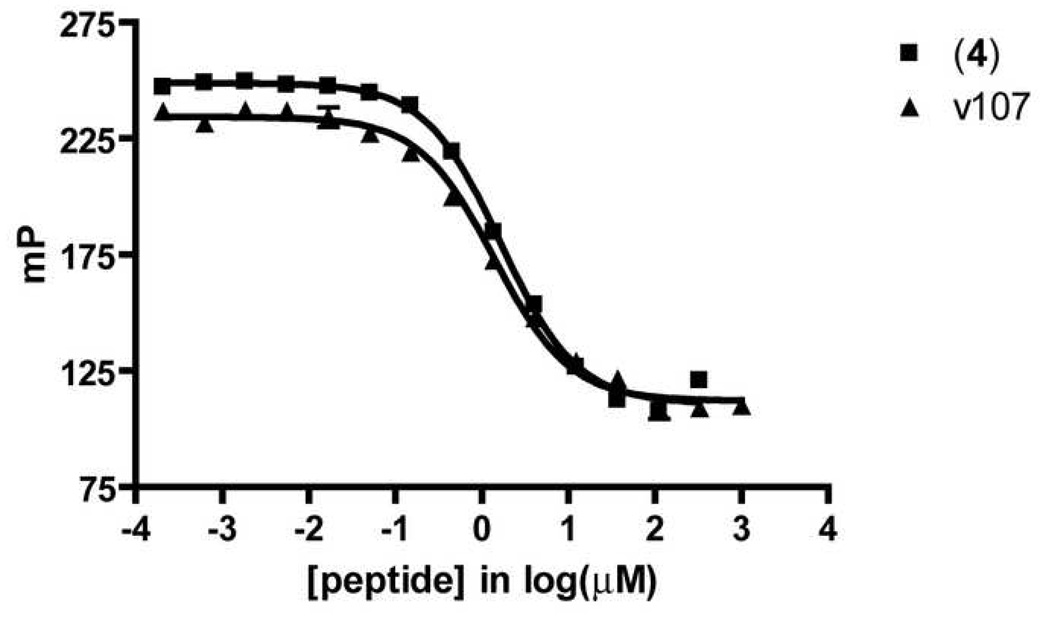
Based upon the Kd value and dynamic range determined in direct binding FP experiments described above, we established a competition FP assay in which inhibitors can be screened for their ability to displace tracer 3 from VEGF. Affinity of competitors for VEGF was quantified as an equilibrium dissociation constant (Ki value), which was calculated from the inhibition curve. [44, 45] The unlabeled truncated v114 analogue 4 was able to compete with tracer 3 for binding to VEGF; a Ki value of 0.61 ± 0.08 µM was calculated for binding of 4 to VEGF from the FP data (Table 3, Figure 3). Unlabeled v107 displayed a Ki value nearly identical to that of 4 when assessed with the competition FP assay. Peptides v107 and 4 have thirteen amino acids in common. These common residues include the ten most C-terminal residues, which contain most of the residues important for binding with VEGF (as identified through alanine scanning). [49] Thus, the fact that peptide 4 displays an affinity for VEGF similar to that of v107 in our assay suggests that the first four residues of v107 do not confer additional binding energy. These results are consistent with structural data showing that the N-terminal portion of v107 does not make close interactions with the VEGF surface.[49] The competition we observe between the tracer and v107 suggests that the tracer binds to the region of VEGF that is important for binding to VEGF receptors. This conclusion is based on published structural data, which show that v107 binds in the region of VEGF that contacts the receptors, and on the report that biotinylated v107 competes with a fragment of VEGFR-1 for binding to VEGF8–109.[49]
Table 3.
Unlabeled Peptides for Competition Experiments (cysteines in bold form a disulfide)
| Peptide | Sequence | Ki (µM) |
|---|---|---|
| (4) | H-X-CDIHVMWEWECFERL-NH2 | 0.61 ± 0.08 |
| v107 | H-GGNECDIARMWEWECFERL-NH2 | 0.62 ± 0.10 |
X = [2-[2-amino-ethoxy]-ethoxy}-acetic acid]
Figure 3.
Competitive Displacement of v107 and Unlabeled Tracer Peptides
Assay Quality Assessment
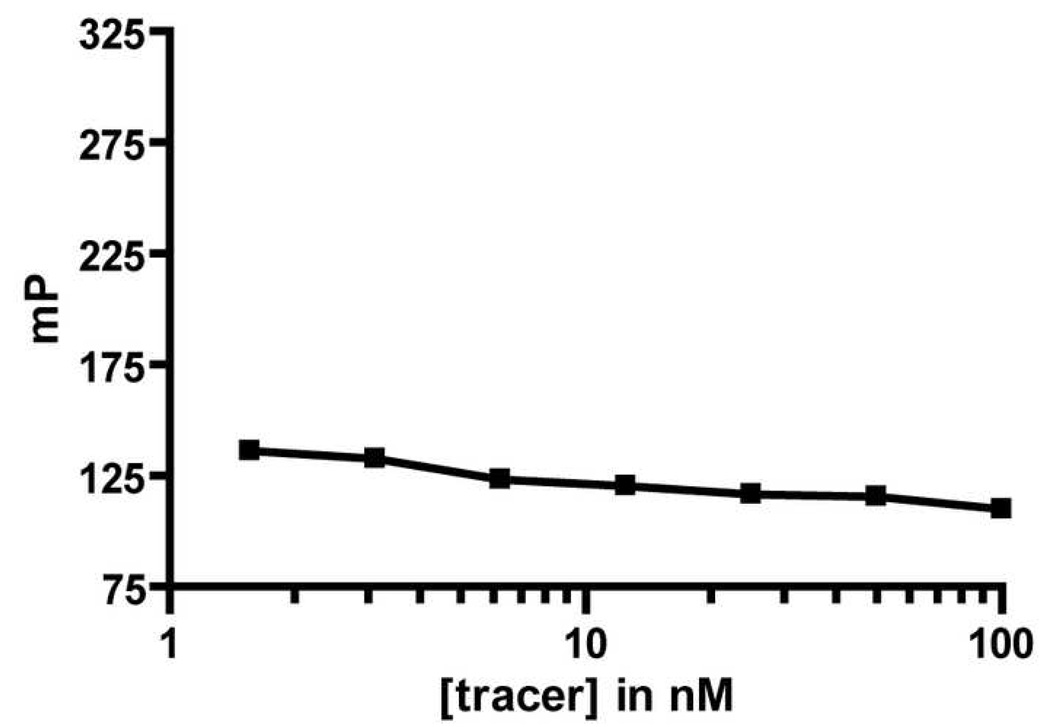
The assay based on the BODIPYTMR tracer (3) was characterized in order to determine its suitability for use in a high-throughput format.[45] First, we examined the FP signal of the tracer in the absence of VEGF as a function of tracer concentration. The mP value did not change between 6 and 25 nM 3 (Figure 4); our standard assay format involves 10 nM tracer, and the FP stability in this concentration range indicates that small fluctuations in tracer concentration from assay to assay would not affect the limiting mP value for fully displaced tracer. This stability in the mP value over a large free tracer concentration would be important if a higher tracer concentration were needed for the assay. A change in tracer concentration could be beneficial if the inhibitor candidates were expected to interfere with the assay signal, either through aggregation or intrinsic fluorescence.
Figure 4.
Fluorescence Polarization Signal for Free Tracer at Different Concentrations
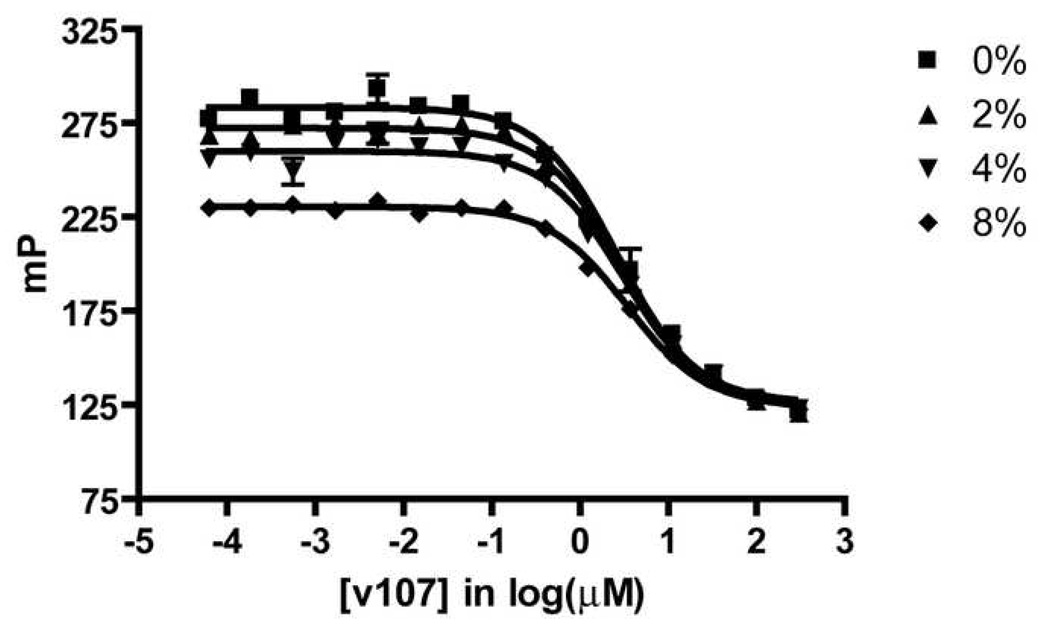
The robustness of the competition FP assay was evaluated by measuring the effect of DMSO concentration on the assay window (mP difference between bound and free tracer) and by calculating the Z’ factor, a measure assay precision. DMSO is a common solvent for preparing stock solutions of inhibitor candidates; therefore, it is important that the presence of small amounts of DMSO in the assay solution not perturb assay results. Our data show that DMSO concentrations up to 4% (v/v) have very little effect on the calculated Ki values (Figure 5). Significant effects on these values are observed with 8% (v/v) DMSO, although it is possible that useful data could be obtained under these conditions if required. The tolerance of at least 4% (v/v) DMSO makes this assay useful for screening of libraries of compounds that are dissolved in DMSO.
Figure 5.
Assay Stability with Different Concentrations of DMSO (v/v)
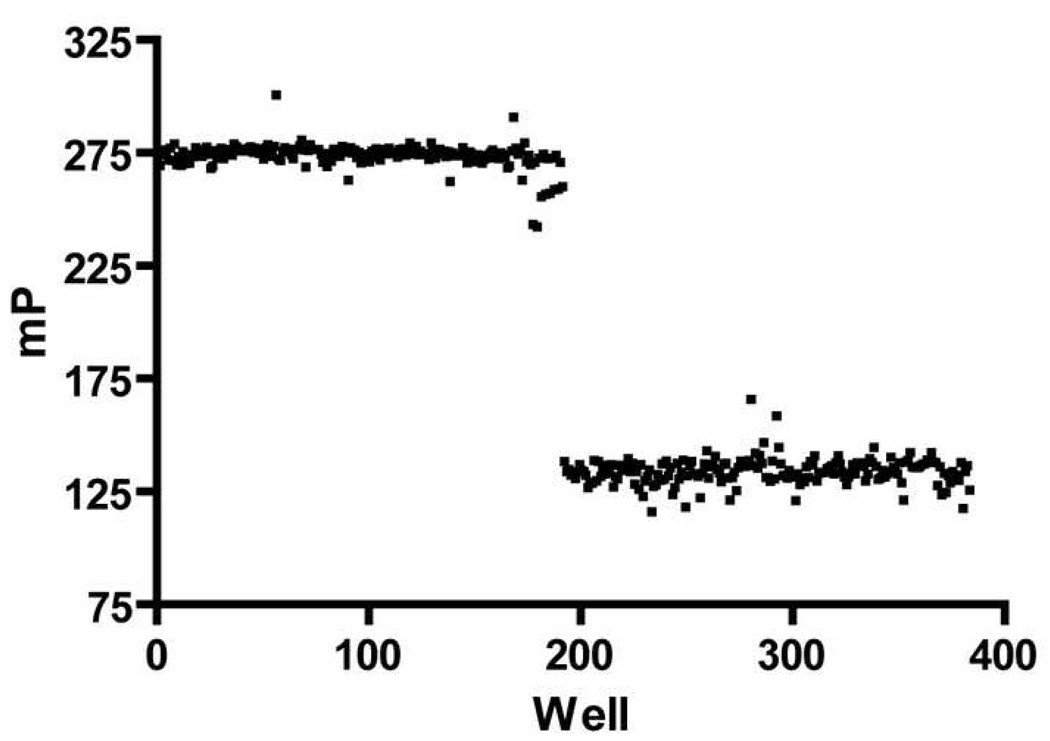
The Z’ factor provides an indication of the suitability of an assay for high-throughput screening.[45, 47] The Z’ factor is a measure of the reproducibility in the difference in signal between a free and bound tracer controls across a large number of assay wells. An assay with ideal reproducibility displays a Z’ of 1. Practically, the Z’ factor for a good high-throughput assay is 0.5 to 1.[53] The Z’ factor calculated for our FP assay is 0.76 (Figure 6), which indicates that this assay can be adapted to a high-throughput screening format.
Figure 6.
Free and Bound Tracer Controls for Estimating the Z’ Value
Assay Validation by Comparison with Published Results
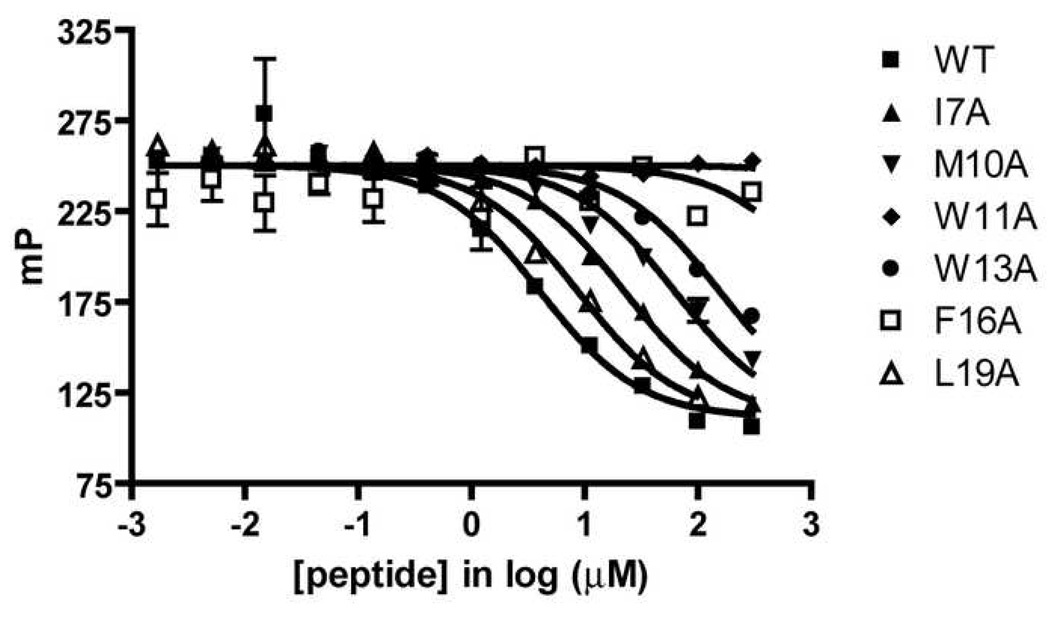
An immobilized VEGF assay analogous to an ELISA has been used previously to evaluate the effects of alanine mutations at six hydrophobic residues of v107 on affinity for VEGF. [49] A broad range of affinities for VEGF is observed among these six v107 mutants. We used this set of v107 mutants for quantitative comparison of our FP assay with the reported immobilized VEGF assay (Table 4). The comparison reveals a good correlation between the trend in IC50 values from the immobilized VEGF study and the trend from our FP assay (Figure 7, Table 4). This correlation suggests that the FP assay provides information on the binding of potential inhibitors to the receptor-binding site of VEGF equivalent to that obtained using the more cumbersome immobilized VEGF assay.
Table 4.
Alanine Mutations to Hydrophobic Residues in v107 (cysteines in bold form a disulfide)
| v107 | Sequence | Literature IC50 [49] (µM) | Experimental IC50 (µM) |
|---|---|---|---|
| WT | H-GGNECDIARMWEWECFERL-NH2 | 1 | 4.2 ± 2 |
| I7A | H-GGNECDAARMWEWECFERL-NH2 | 25 | 23 ± 6 |
| M10A | H-GGNECDIARAWEWECFERL-NH2 | 209 | 68 ± 20 |
| W11A | H-GGNECDIARMAEWECFERL-NH2 | NBa | NBb |
| W13A | H-GGNECDIARMWEAECFERL-NH2 | 256 | 178 ± 60 |
| F16A | H-GGNECDIARMWEWECAERL-NH2 | NBa | NBb |
| L19A | H-GGNECDIARMWEWECFERA-NH2 | 4 | 8.2 ± 0.8 |
IC50 > 2 mM
IC50 > 300 µM
Figure 7.
v107 Alanine Mutant Competitive Displacement
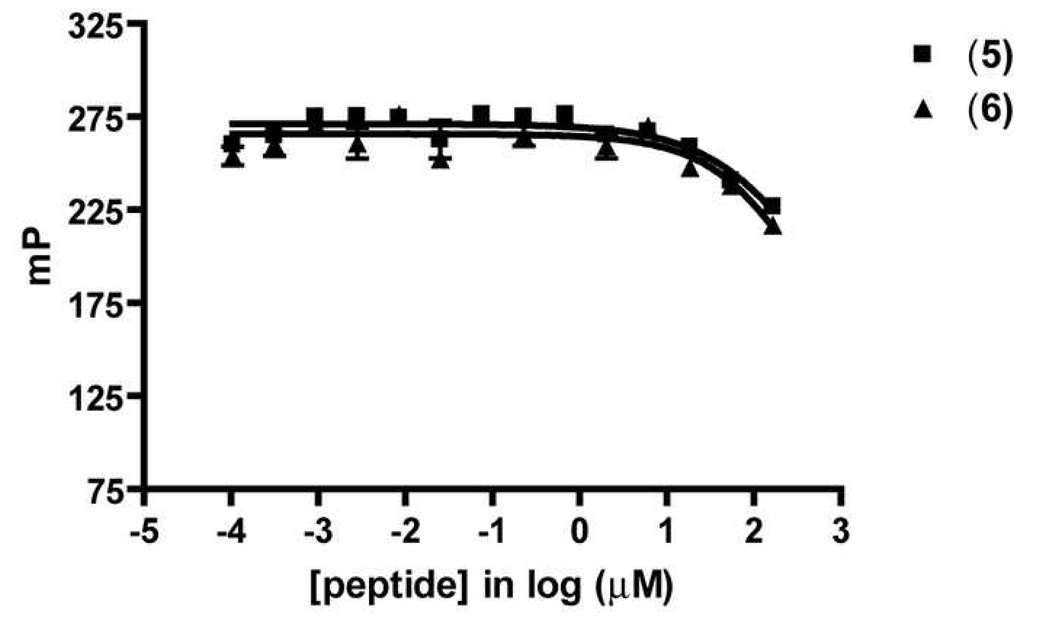
Two heptapeptides unrelated to v107, H-WHLPFKC-NH2 (5) and H-WHKPFRF-NH2 (6), have recently been reported to bind to the VEGF homodimer based on an SPR assay, and to inhibit endothelial cell proliferation.[54] Peptides 5 and 6 are proposed to block the VEGF-VEGFR interaction. We evaluated 5 and 6 in our FP assay, but neither was effective at displacing the BODIPYTMR tracer 3 from VEGF (Figure 8). Our findings suggest that peptides 5 and 6 do not bind to the surface of VEGF contacted by v114. Since related peptide v107 binds to the region of VEGF that is also recognized by VEGFR-1 and VEGFR-2,[4, 49] the inactivity of 5 and 6 in our assay raises the prospect that these peptides do not directly inhibit VEGF-VEGFR interactions. It remains possible, however, that these two peptides bind to a portion of the receptor-binding site on VEGF that does not overlap with v114-binding site on VEGF.
Figure 8.
Competitive Displacement of Heptapeptides Unrelated to v107
Conclusions
We have developed a fluorescence polarization assay that can be used to screen for molecules that bind to VEGF165 at the VEGF receptor-binding site, which is shared by small peptides v107 and v114. This is the first demonstration of a fluorescence-based assay for VEGF that is suitable for use in a high-throughput format. This assay has several advantages versus other assay modes, such as having all components free in solution and not relying on radioactive materials for quantification. Blocking the interaction between VEGF and cell surface receptors is a clinically validated therapeutic strategy for cancer and age-related macular degeneration.[24, 55] Although antibodies have been developed to inhibit this interaction and have reached the clinic, there remains a need for inhibitors with lower molecular weight. Such compounds could prove easier to produce and/or administer. The assay developed here should be useful for identifying such compounds.
Acknowledgements
This research was supported by the NIH [GM56414 (S.H.G.), DK50107 (E.H.B.), and EY16995 (N.S.). R.C.W. was supported by the NIH and the Welch Foundation. We thank Dr. W. Seth Horne and Melissa D. Boersma for assistance with FP assay design and helpful discussions. The plate reader used for FP assays is in the UW-Madison W.M. Keck Center for Chemical Genomics. K.J.P. was supported in part by the NIH Chemistry-Biology Interface Training Program (T32 GM008505), J.D.S. was supported in part by an NSF predoctoral fellowship and S.P. was supported by a postdoctoral fellowship from the American Heart Association.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ferrara N, Kerbel RS. Angiogenesis as a Therapeutic Target. Nature. 2005;438:967–974. doi: 10.1038/nature04483. [DOI] [PubMed] [Google Scholar]
- 2.Ferrara N, Gerber HP, LeCouter J. The Biology of VEGF and its Receptors. Nat. Med. 2003;9:669–676. doi: 10.1038/nm0603-669. [DOI] [PubMed] [Google Scholar]
- 3.Ferrara N. Vascular Endothelial Growth Factor: Basic Science and Clinical Progress. Endorine Reviews. 2004;25:581–611. doi: 10.1210/er.2003-0027. [DOI] [PubMed] [Google Scholar]
- 4.Li B, Fuh G, Meng G, Xin X, Gerritsen ME, Cunningham B, de Vos AM. Receptor-selective Variants of Human Vascular Endothelial Growth Factor. J. Biol. Chem. 2000;275:29823–29828. doi: 10.1074/jbc.M002015200. [DOI] [PubMed] [Google Scholar]
- 5.D'Andrea LD, Del Gatto A, Pedone C, Benedetti E. Peptide-based Molecules in Angiogenesis. Chem. Biol. Drug Des. 2006;67:115–126. doi: 10.1111/j.1747-0285.2006.00356.x. [DOI] [PubMed] [Google Scholar]
- 6.Carmeliet P. VEGF as a Key Mediator of Angiogenesis in Cancer. Oncology. 2005;69 suppl 3:4–10. doi: 10.1159/000088478. [DOI] [PubMed] [Google Scholar]
- 7.Cross MJ, Dixelius J, Matsumoto T, Claesson-Welsh L. VEGF-Receptor Signal Transduction. TRENDS Biochem. Sci. 2003;28:488–494. doi: 10.1016/S0968-0004(03)00193-2. [DOI] [PubMed] [Google Scholar]
- 8.Muller YA, Christinger HW, Keyt BA, de Vos AM. The Crystal Structure of Vascular Endothelial Growth Factor (VEGF) Refined to 1.93Å Resolution: Multiple Copy Flexibility and Receptor Binding. Structure. 1997;5:1325–1338. doi: 10.1016/s0969-2126(97)00284-0. [DOI] [PubMed] [Google Scholar]
- 9.Muller YA, Li B, Christinger HW, Wells JA, Cunningham BC, de Vos AM. Vascular Endothelial Growth Factor: Crystal Structure and Functional Mapping of the Kinase Domain Receptor Binding Site. Proc. Natl. Acad. Sci. 1997;94:7192–7197. doi: 10.1073/pnas.94.14.7192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kowanetz M, Ferrara N. Vascular Endothelial Growth Factor Signaling Pathways: Therapeutic Perspectives. Clin. Cancer Res. 2006;12:5018–5022. doi: 10.1158/1078-0432.CCR-06-1520. [DOI] [PubMed] [Google Scholar]
- 11.Davis-Smyth T, Chen H, Park J, Presta LG, Ferrara N. The Second Immunoglobulin-like Domain of the VEGF Tyrosine Kinase Receptor Flt-1 Determines Ligand Binding and May Initiate a Signal Transduction Cascade. EMBO Journal. 1996;15:4919–4927. [PMC free article] [PubMed] [Google Scholar]
- 12.Herley MT, Yu Y, Whitney RG, Sato JD. Characterization of the VEGF Binding Site on the Flt-1 Receptor. Biochem. Biophys. Res. Commun. 1999;262:731–738. doi: 10.1006/bbrc.1999.1282. [DOI] [PubMed] [Google Scholar]
- 13.Barleon B, Totzke F, Herzog C, Blanke S, Kremmer E, Siemeister G, Marmé D, Martiny-Baron G. Mapping of the Sites for Ligand Binding and Receptor Dimerization at the Extracellular Domain of the Vascular Endothelial Growth Factor Receptor Flt-1. J. Biol. Chem. 1997;272:10382–10388. doi: 10.1074/jbc.272.16.10382. [DOI] [PubMed] [Google Scholar]
- 14.Ma L, Wang X, Zhang Z, Zhou X, Chen A, Yao L. Identification of the Ligand-Binding Domain of Human Vascular-Endothelial-Growth-Factor Receptor Flt-1. Biotechnol. Appl. Biochem. 2001;34:199–204. doi: 10.1042/ba20010043. [DOI] [PubMed] [Google Scholar]
- 15.Fuh G, Li B, Crowley C, Cunningham B, Wells JA. Requirements for Binding and Signaling of the Kinase Domain Receptor for Vascular Endothelial Growth Factor. J. Biol. Chem. 1998;273:11197. doi: 10.1074/jbc.273.18.11197. [DOI] [PubMed] [Google Scholar]
- 16.Shinkai A, Ito M, Anazawa H, Yamaguchi S, Shitara K, Shibuya M. Mapping of the Sites Involved in Ligand Association and Dissociation at the Extracellular Domain of the Kinase Insert Domain-containing Receptor for Vascular Endothelial Growth Factor. J. Biol. Chem. 1998;273:31283–31288. doi: 10.1074/jbc.273.47.31283. [DOI] [PubMed] [Google Scholar]
- 17.Wiesmann C, Fuh G, Christinger HW, Eigenbrot C, Wells JA, de Vos AM. Crystal Structure at 1.7Å Resolution of VEGF in Complex with Domain 2 of the Flt-1 Receptor. Cell. 1997;91:695–704. doi: 10.1016/s0092-8674(00)80456-0. [DOI] [PubMed] [Google Scholar]
- 18.Starovasnik MA, Christinger HW, Wiesmann C, Champe MA, de Vos AM, Skelton NJ. Solution Structure of the VEGF-binding Domain of Flt-1: Comparison of its Free and Bound States. J. Mol. Biol. 1999;293:531–544. doi: 10.1006/jmbi.1999.3134. [DOI] [PubMed] [Google Scholar]
- 19.Yancopoulos GD, Davis S, Gale NW, Rudge JS, Wiegand SJ, Holash J. Vascular-Specific Growth Factors and Blood Vessel Formation. Nature. 2000;407:242–248. doi: 10.1038/35025215. [DOI] [PubMed] [Google Scholar]
- 20.Carmeliet P. Angiogenesis in Health and Disease. Nat. Med. 2003;9:653–660. doi: 10.1038/nm0603-653. [DOI] [PubMed] [Google Scholar]
- 21.Ferrara N, Davis-Smyth T. The Biology of Vascular Endothelial Growth Factor. Endocrine Reviews. 1997;18:4–25. doi: 10.1210/edrv.18.1.0287. [DOI] [PubMed] [Google Scholar]
- 22.Carmeliet P, Jain RK. Angiogenesis in Cancer and Other Diseases. Nature. 2000;407:249–257. doi: 10.1038/35025220. [DOI] [PubMed] [Google Scholar]
- 23.Muhsin M, Graham J, Kirkpatrick P. Bevacizumab. Nat. Rev. Drug Discov. 2004;3:995–996. doi: 10.1038/nrd1601. [DOI] [PubMed] [Google Scholar]
- 24.Ng EWM, Shima DT, Calias P, Cunningham ET, Jr, Guyer DR, Adamis AP. Pegaptanib, a Targeted anti-VEGF Aptamer for Ocular Vascular Disease. Nat. Rev. Drug Discov. 2006;5:123–132. doi: 10.1038/nrd1955. [DOI] [PubMed] [Google Scholar]
- 25.Narayanan R, Kuppermann BD, Jones C, Kirkpatrick P. Ranibizumab. Nat. Rev. Drug Discov. 2006;5:815–816. doi: 10.1038/nrd2157. [DOI] [PubMed] [Google Scholar]
- 26.Andreoli CM, Miller JW. Anti-Vascular Endothelial Growth Factor Therapy for Ocular Neovascular Disease. Curr. Opin. Ophthalmol. 2007;18:502–508. doi: 10.1097/ICU.0b013e3282f0ca54. [DOI] [PubMed] [Google Scholar]
- 27.Arkin MR, Wells JA. Small-Molecule Inhibitors of Protein-Protein Interactions: Progressing Towards the Dream. Nat. Rev. Drug Discov. 2004;3:301. doi: 10.1038/nrd1343. [DOI] [PubMed] [Google Scholar]
- 28.Fry DC. Protein-Protein Interactions as Targets for Small Molecule Drug Discovery. Biopolymers (Pept Sci) 2006;84:535–552. doi: 10.1002/bip.20608. [DOI] [PubMed] [Google Scholar]
- 29.Whitty A, Kumaravel G. Between a Rock and a Hard Place? Nat. Chem. Biol. 2006;2:112–118. doi: 10.1038/nchembio0306-112. [DOI] [PubMed] [Google Scholar]
- 30.Ferrara N. VEGF as a Therapeutic Target in Cancer. Oncology. 2005;69 suppl 3:11–16. doi: 10.1159/000088479. [DOI] [PubMed] [Google Scholar]
- 31.Lee SJ, Lee HM, Ji ST, Lee SR, Mar W, Gho YS. 1,2,3,4,6-Penta-O-galloyol-beta-D-glucose Blocks Endothelial Cell Growth and Tube Formation Through Inhibition of VEGF binding to VEGF Receptor. Cancer Letters. 2004;208:89–94. doi: 10.1016/j.canlet.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 32.Fairbrother WJ, Christinger HW, Cochran AG, Fuh G, Keenan CJ, Quan C, Shriver SK, Tom JYK, Wells JA, Cunningham BC. Novel Peptides Selected to Bind Vascular Endothelial Growth Factor Target the Receptor-Binding Site. Biochemistry. 1998;37:17754–17764. doi: 10.1021/bi981931e. [DOI] [PubMed] [Google Scholar]
- 33.D'Andrea LD, Iaccarino G, Fattorusso R, Sorriento D, Carannante C, Capasso D, Trimarco B, Pedone C. Targeting Angiogenesis: Structural Characterization and Biological Properites of a de novo Engineered VEGF Mimicking Peptide. Proc. Natl. Acad. Sci. 2005;102:14215–14220. doi: 10.1073/pnas.0505047102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goncalves V, Gautier B, Garbay C, Vidal M, Inguimbert N. Development of a Chemiluminescent Screening Assay for Detection of Vascular Endothelial Growth Factor Receptor 1 Ligands. Anal. Biochem. 2007;366:108–110. doi: 10.1016/j.ab.2007.03.027. [DOI] [PubMed] [Google Scholar]
- 35.Zhang H, Nimmer P, Rosenberg SH, Ng SC, Joseph M. Development of a High-Throughput Fluorescence Polarization Assay for Bcl-xL. Anal. Biochem. 2002;307:70–75. doi: 10.1016/s0003-2697(02)00028-3. [DOI] [PubMed] [Google Scholar]
- 36.Sörme P, Kahl-Knutsson B, Huflejt M, Nilsson UJ, Leffler H. Fluorescence Polarization as an Analytical Tool to Evaluate Galectin-Ligand Interactions. Anal. Biochem. 2004;334:36–47. doi: 10.1016/j.ab.2004.06.042. [DOI] [PubMed] [Google Scholar]
- 37.Combination of Willson protocol. Christinger HW, et al. Proteins: Structure, Function, and Genetics. 1996;26:353–357. [Google Scholar]
- 38.Gill SC, von Hippel PH. Calculation of Protein Extinction Coefficients from Amino Acid Sequence Data. Anal. Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 39.Scheef EA, Huang Q, Wang S, Sorenson CM, Sheibani N. Isolation and Characterization of Corneal Endothelial Cells from Wild Type and Thrombospondin-1 Deficient Mice. Mol. Vis. 2007;13:1483–1495. [PubMed] [Google Scholar]
- 40.Amine-reactive Probes. Invitrogen. 2007 http://probes.invitrogen.com/.
- 41.Sadowsky JD, Schmitt MA, Lee HS, Umezawa N, Wang S, Tomita Y, Gellman SH. Chimeric (α/β+α)-Peptide Ligands for the BH3-Recognition Cleft of Bcl-xL: Critical Role of the Molecular Scaffold in Protein Surface Recognition. J. Am. Chem. Soc. 2005;127:11966–11968. doi: 10.1021/ja053678t. [DOI] [PubMed] [Google Scholar]
- 42.Banks P, Harvey M. Consideration for Using Fluorescence Polarization in the Screening of Protein-Coupled Receptors. J. Biomol. Screen. 2002;7:111–117. doi: 10.1177/108705710200700203. [DOI] [PubMed] [Google Scholar]
- 43.Zhang R, Mayhood T, Lipari P, Wang Y, Durkin J, Syto R, Gesell J, McNemar C, Windsor W. Fluorescence Polarization Assay and Inhibitor Design for MDM2/p53 Interaction. Anal. Biochem. 2004;331:138–146. doi: 10.1016/j.ab.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 44.Roehrl MHA, Wang JY, Wagner G. A General Framework for Development and Data Analysis of Competitive High-Throughput Screens for Small-Molecule Inhibitors of Protein-Protein Interactions by Fluorescence Polarization. Biochemistry. 2004;43:16056–16066. doi: 10.1021/bi048233g. [DOI] [PubMed] [Google Scholar]
- 45.Assay Guidance Manual Version 4.1. Eli Lilly and Company and NIH Chemical Genomics Center; 2005. Available online at: http://www.ncgc.nih.gov/guidance/manual_toc.html (last accessed 05/10/06). [PubMed] [Google Scholar]
- 46.Sigmoidal Dose Response curve fitting, GraphPad.
- 47.Zhang JH, Chung T, Oldenburg K. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J. Biomol. Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 48.Christinger HW, Muller YA, Berleau LT, Keyt BA, Cunningham BC, Ferrara N, de Vos AM. Crystallization of the Receptor Binding Domain of Vascular Endothelial Growth Factor. Proteins: Structure, Function, and Genetics. 1996;26:353–357. doi: 10.1002/(SICI)1097-0134(199611)26:3<353::AID-PROT9>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 49.Pan B, Li B, Russell SJ, Tom JYK, Cochran AG, Fairbrother WJ. Solution Structure of a Phage-derived Peptide Antagonist in Complex with Vascular Endothelial Growth Factor. J. Mol. Biol. 2002;316:769–787. doi: 10.1006/jmbi.2001.5370. [DOI] [PubMed] [Google Scholar]
- 50.Siemeister G, Schnurr B, Mohrs K, Schächtele C, Marmé D, Martiny-Baron G. Expression of Biologically Active Isoforms of the Tumor Angiogenesis Factor VEGF in Escherichia coli. Biochem. Biophys. Res. Commun. 1996;222:249–255. doi: 10.1006/bbrc.1996.0730. [DOI] [PubMed] [Google Scholar]
- 51.Huang X. Fluorescence Polarization Competition Assay: The Range of Resolvable Inhibitor Potency Is Limited by the Affinity of the Fluroescent Ligand. J. Biomol. Screen. 2003;8:34–38. doi: 10.1177/1087057102239666. [DOI] [PubMed] [Google Scholar]
- 52.Banks P, Gosselin M, Prystay L. Impact of a Red-Shifted Dye Label for High Throughput Fluorescence Polarization Assays of G Protein-Coupled Receptors. J. Biomol. Screen. 2000;5:329–334. doi: 10.1177/108705710000500504. [DOI] [PubMed] [Google Scholar]
- 53.Owicki JC. Fluorescence Polarization and Anisotropy in High Throughput Screening: Perspectives and Primer. J. Biomol. Screen. 2000;5:297–306. doi: 10.1177/108705710000500501. [DOI] [PubMed] [Google Scholar]
- 54.Erdag B, Balcioglu KB, Kumbasar A, Celikbicak O, Zeder-Lutz G, Altschuh D, Salih B, Baysal K. Novel Short Peptides Isolated from Phage Display Library Inhibit Vascular Endothelial Growth Factor Activity. Mol. Biotechnol. 1997;35:51–63. doi: 10.1385/mb:35:1:51. [DOI] [PubMed] [Google Scholar]
- 55.Ferrara N, Hillan KJ, Gerber HP, Novotny W. Discovery and Development of Bevacizumab, an Anti-VEGF Antibody for Treating Cancer. Nat. Rev. Drug Discov. 2004;3:391–400. doi: 10.1038/nrd1381. [DOI] [PubMed] [Google Scholar]