Abstract
Live imaging of the actin cytoskeleton is crucial for the study of many fundamental biological processes, but current approaches to visualize actin have several limitations. Here we describe Lifeact, a 17-amino-acid peptide, which stained filamentous actin (F-actin) structures in eukaryotic cells and tissues. Lifeact did not interfere with actin dynamics in vitro and in vivo and in its chemically modified peptide form allowed visualization of actin dynamics in nontransfectable cells.
Reliable visualization of the actin cytoskeleton is essential for various fields of biomedical research. Imaging of actin dynamics has been mostly achieved by injection of fluorescently labeled actin (technically demanding) or small amounts of fluorescently labeled phalloidin, an F-actin-binding and stabilizing compound1,2. A widely used alternative is the expression of actin-GFP fusion proteins. However, all described actin fusions are functionally impaired and rely on nontagged actin3 to buffer the defects. Recently, fusions of GFP to actin-binding domains have been used, notably from moesin in Drosophila melanogaster4, LimE in Dictyostelium discoideum5, ABP120 in D. discoideum and mammalian cells6,7, and utrophin in Xenopus laevis8. These probes consist of large domains, compete with their endogenous counterparts and are restricted to cells that can be transfected.
Abp140-GFP is the only probe that has been successfully used to label actin cables, in budding yeast9,10. Using total internal reflection (TIRF) microscopy to monitor localization of Abp140 domains fused to GFP, we found that the first 17 aa of Abp140 were sufficient to mediate actin localization comparable to the full-length protein (Fig. 1a,b). This short peptide is conserved among close relatives of Saccharomyces cerevisiae (Fig. 1c) but is absent from other organisms. This and its small size make it an attractive actin marker for higher eukaryotes. We therefore introduced the peptide, named ‘Lifeact’, as a C-terminal GFP fusion (Lifeact-GFP) into mammalian cell lines to test its suitability as an in vivo marker. We also synthesized Lifeact with an N-terminal FITC fluorophore (F-Lifeact) to test its biochemical properties.
Figure 1.
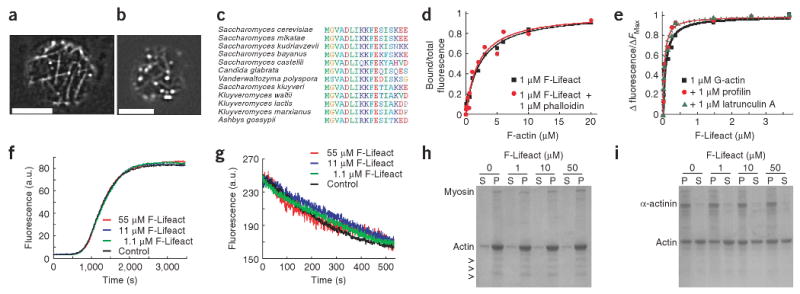
Identification and biochemical characterization of Lifeact. (a) TIRF microscopy image of Abp140-GFP in an unpolarized yeast cell. (b) Epifluorescence image of Lifeact-GFP in a yeast cell. Scale bars, 5 μm. (c) Alignment of the actin-binding sequence in fungi. (d) Bound/total fluorescence of F-Lifeact co-sedimented with rabbit muscle F-actin. (e) Fluorescence of pyrene-labeled G-actin in the presence of F-Lifeact. Fluorescence was normalized to maximum values. (f) Polymerization of 20% pyrene-labeled actin in the absence (control) or presence of indicated concentrations of F-Lifeact. (g) Depolymerization of 100% pyrene-labeled F-actin after dilution below 200 nM with F-Lifeact (control without Lifeact). (h,i) SDS PAGE of pellet (P) and supernatant (S) fractions of F-actin sedimented with myosin (h) and α-actinin (i) in the absence and presence of F-Lifeact. Arrowheads, myosin light chains.
To determine the affinity of F-Lifeact to F-actin we measured the dissociation constant (Kd) by co-sedimentation in the absence (2.2 ± 0.3 μM; Fig. 1d) and presence of 100 nM phalloidin (2.0 ± 0.4 μM). We then monitored F-Lifeact binding to globular actin (G-actin) by fluorescence enhancement of pyrene-labeled actin (Supplementary Methods online) and found a 30-fold higher Kd of 70 ± 25 nM (Fig. 1e). We confirmed the affinities to G-actin and F-actin using anisotropy (280 ± 100 nM, G-actin; 2.3 ± 0.9 μM, F-actin) and fluorescence enhancement (40 ± 10 nM, G-actin; 1.3 ± 0.5 μM, F-actin) of the FITC moiety on F-Lifeact (Supplementary Fig. 1a,b online). Addition of the G-actin–sequestering factors profilin or latrunculin A did not perturb F-Lifeact binding to G-actin (Kd = 40 ± 10 nM for both; Fig. 1e) indicating non-overlapping binding sites on actin. Next we tested the influence of F-Lifeact on polymerization and depolymerization of pyrene-labeled actin11. Nucleation and elongation phases of actin polymerization, as well as deploymerization rates, were not affected by F-Lifeact concentrations up to 55 μM (Fig. 1f,g). To test for competition of F-Lifeact with proteins binding to the sides of actin filaments, we co-pelleted myosin II and α-actinin with F-actin at different Lifeact concentrations (1.1, 11 and 55 μM) and found that both interactions remained unaffected (Fig. 1h,i). Using circular dichroism and nuclear magnetic resonance spectroscopies we found that Lifeact forms a nascent helix in water that can be stabilized by alcohol addition to a typical α helix at residues 2–10 (Supplementary Fig. 1c,d). These structural features are reminiscent of the G-actin–binding peptide thymosin β412.
To test its suitability in vivo, we introduced Lifeact-GFP into immortalized mouse embryonic fibroblasts, MDCK cells, primary rat hippocampal neurons and primary mouse dendritic cells (Fig. 2). In all these cell types we obtained clear labeling of F-actin (Fig. 2a–d) that matched previously reported patterns. In fibroblasts, Lifeact-GFP signal was comparable to staining obtained with actin-GFP or GFP-utrophin (Fig. 2e), and in double-transfected MDCK cells Lifeact-GFP completely colocalized with actin-mRFPruby13 (Fig. 2f). Consistent with its low affinity to F-actin in vitro, Lifeact-GFP recovered rapidly in photobleaching experiments (Fig. 2g), whereas actin-GFP showed the expected slower mobility (Fig. 2h). We noted no signs of cytotoxicity, abnormal morphology or of growth retardation in transiently or stably transfected cell lines (data not shown).
Figure 2.
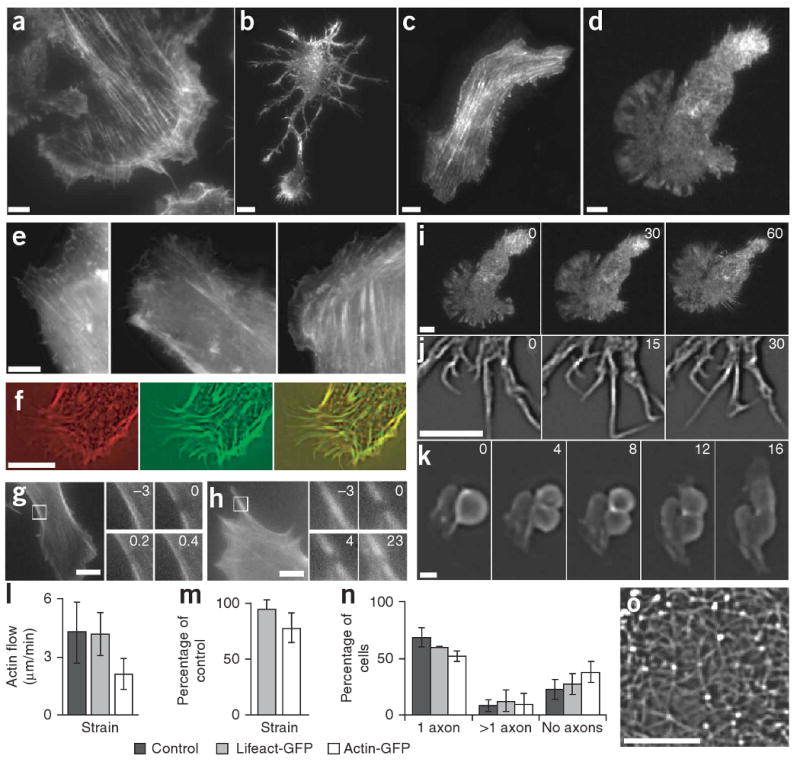
Characterization of Lifeact-GFP in vivo. (a–j) TIRF microscopy images of Lifeact-GFP transiently expressed in mouse embryonic fibroblasts (a), primary rat hippocampal neurons (b and time series of filopodial dynamics in j), MDCK cells (c) and mouse dendritic cells (d and a time series of chemotaxis in i). Epifluorescence images of actin-GFP (left), Lifeact-GFP (middle) and GFP-utrophin (right) in fibroblasts (e). MDCK cells transfected with actin-RFP and Lifeact-GFP, and imaged by widefield microscopy (RFP, left; GFP, middle; overlay, right; f). Fluorescence recovery after photobleaching in fibroblasts transfected with actin-GFP (g) and Lifeact-GFP (h), where numbers in insets (magnification of the boxed areas) indicate time relative to bleaching in seconds. (k) Time series (in minutes) of an MDCK cell undergoing cytokinesis, showing Lifeact-GFP staining in the contractile ring. (l) Velocity of lamellipodial retrograde actin flow in untransfected (control) or transiently transfected (Lifeact-GFP or actin-GFP) fibroblasts measured from kymograph traces. (m) Chemotactic speed of transiently transfected dendritic cells relative to untransfected cells. (n) Quantification of neuronal polarization 3 d after transient transfection. Data are averages ± s.d. from at least three experiments. (o) Cortical actin network of a hippocampal neuron transfected with Lifeact-GFP. Scale bars, 5 μm except 1 μm in j.
To test whether Lifeact-GFP selectively associates with subsets of actin structures, we used TIRF microscopy to image dynamic cortical F-actin. In fibroblasts and dendritic cells, the flow of dynamic lamellipodial actin was prominent at the cell periphery (Fig. 2a,d,i and Supplementary Movies 1 and 2 online), but stable stress fibers formed in the cell body of tightly adherent fibroblasts (Fig. 2a). Neurons were rich in dynamic filopodia (Fig. 2b) that frequently underwent kinking (Fig. 2j and Supplementary Movie 3 online), and the surface of neuronal cell bodies was covered with an isotropic network of actin filaments (Fig. 2b,o and Supplementary Movie 4 online). MDCK cells showed stress fibers and circumferential actin belts at the cell periphery (Fig. 2c). During cytokinesis, Lifeact-GFP highlighted the typical contractile rings of MDCK cells (Fig. 2k and Supplementary Movie 5 online).
To test whether Lifeact-GFP compromises cytoskeletal functions, we measured three sensitive readouts for actin dynamics: retrograde flow within the leading lamella of fibroblasts, migration of dendritic cells along a gradient of chemokine and neuronal polarization. In all cases we compared Lifeact-GFP with actin-GFP expressed from the same vector. The speed of retrograde flow in lamellipodia of Lifeact-GFP–transfected fibroblasts was indistinguishable from that in un-transfected cells (4 μm/min), but it was reduced to about half in actin-GFP expressing cells (ANOVA: F2,134 = 53.39, P < 0.0001; post-hoc Dunnett's test: P > 0.05 for Lifeact-GFP, P < 0.05 for actin-GFP; Fig. 2l). Lifeact-GFP also had no effect on the speed of chemotactic dendritic cell migration in a collagen matrix (paired t-test, t2 = −1.05, P = 0.40, n = 3 experiments, 837 tracked cells), but actin-GFP expressing cells migrated more slowly (t3 = −3.65, P = 0.04, n = 4 experiments, 689 cells; Fig. 2m). Finally, neuronal polarization was not significantly affected by the expression of Lifeact-GFP, whereas comparable expression of actin-GFP clearly reduced polarization efficiency (ANOVA: F2,8 = 6.205, P < 0.0346; post-hoc Dunnett's test: P > 0.05 for Lifeact-GFP, P < 0.05 for actin-GFP; Fig. 2n).
Chemically synthesized F-Lifeact could allow convenient labeling of fixed samples or nontransfectable cells. We stained paraformaldehyde-fixed MDCK cells with F-Lifeact and Cy3-conjugated phalloidin, and observed extensive overlap of the two markers (Fig. 3a). We observed similar co-localization of these two labels in paraformaldehyde-fixed tissue sections of mouse skeletal muscle (Fig. 3b). We obtained efficient staining with as little as 5 nM F-Lifeact, and signal intensity was not visibly reduced after extensive washing (10 times for 10 min after staining with 1 μM peptide). However, F-Lifeact rapidly exchanged on F-actin (Fig. 3c); the observed robust labeling was probably achieved because the peptide cannot exit from fixed cells. Thus the F-Lifeact peptide can be used as a cheap and nontoxic alternative to phalloidin.
Figure 3.
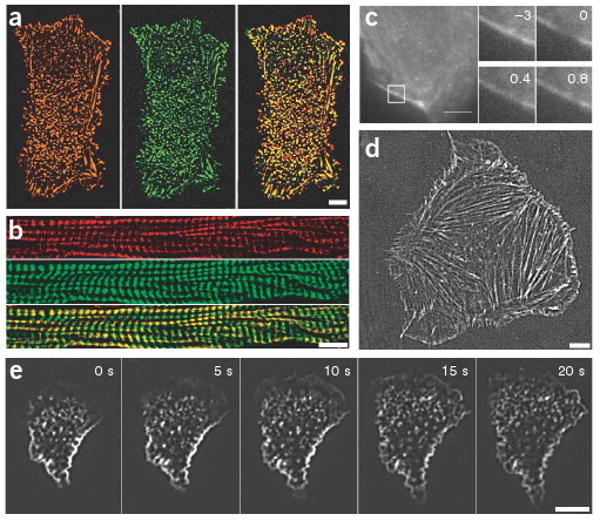
F-Lifeact staining in fixed and living samples. (a,b) MDCK cells (a) and cryo-sections of mouse skeletal muscle (b) fixed with paraformaldehyde and stained with F-Lifeact (green) and phalloidin-Cy3 (red, overlay in yellow). (c) Fluorescence recovery after photobleaching of a paraformaldehyde-fixed F-Lifeact–stained mouse embryonic fibroblast. Numbers in insets (magnification of the boxed areas) indicate time relative to bleaching in seconds. (d,e) Mouse embryonic fibroblasts (d) and human primary neutrophil granulocytes (e) were scrape-loaded with F-Lifeact, replated and cell spreading was visualized with TIRF microscopy. Scale bars, 5 μm.
We next used F-Lifeact to label live fibroblasts and human neutrophils using ‘scrape-loading’14. Fibroblasts loaded with F-Lifeact revealed the typical distribution of F-actin in stress fibers and lamellipodia (Fig. 3d). The fluorescence signal was maintained over 4–6 h before degradation of the peptide and/or fading of the fluorochrome. In scrape-loaded human neutrophils plated on immune complex–coated coverslips, we observed the formation of extensive undirected lamellar protrusions and of stable actin patches (Fig. 3e and Supplementary Movie 6 online). These structures likely result from integrin-dependent cytoskeletal reorganization in response to immune complex deposits that have been previously studied only in fixed cells, using fluorescent phalloidin15.
With only 17 amino acids, Lifeact can be synthesized quickly and cost-effectively, either as an oligonucleotide for the generation of fusion proteins or as a chemically labeled peptide. This makes Lifeact-derived probes widely accessible and opens up possibilities for the generation of actin-directed markers or drugs. Our results suggest that Lifeact can be used as a universal marker for actin imaging and as an alternative to phalloidin. However, although the low affinity of Lifeact to F-actin reduces unwanted perturbations, it also limits its use for the measurement of intra-filament dynamics that are possible with speckle microscopy or photobleaching of labeled actin subunits. The most important advantage of Lifeact is its lack of interference with cellular processes. We could not measure significant effects of Lifeact-GFP expression on sensitive processes such as neuronal polarization, lamellipodial flow or leukocyte chemotaxis. This is consistent with Lifeact's low binding affinity to F-actin in vitro, its lack of effects on actin polymerization and depolymerization, and its lack of competition with major actin-binding proteins. In addition, Lifeact has no homologous sequences in higher eukaryotes, which makes competition with endogenous proteins less likely.
Supplementary Material
Acknowledgments
We thank S. Uebel and the core facility of the Max Planck Institute of Biochemistry for peptide synthesis and labeling, and S. Cremer and S. Wedlich for helpful comments on the manuscript. We obtained latrunculin A from P. Crews (University of California Santa Cruz). This study was supported by funds from the Max Planck Society (R.W.-S.), German Research Foundation SI1323/1-1 (M.S.) and the Alexander von Humboldt Stiftung (Z.W.).
Footnotes
Note: Supplementary information is available on the Nature Methods website.
Competing Interests Statement: The authors declare competing financial interests: details accompany the full-text HTML version of the paper at http://www.nature.com/naturemethods/.
References
- 1.Schmit AC, Lambert AM. Plant Cell. 1990;2:129–138. doi: 10.1105/tpc.2.2.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Waterman-Storer CM, Desai A, Bulinski JC, Salmon ED. Curr Biol. 1998;8:1227–1230. doi: 10.1016/s0960-9822(07)00515-5. [DOI] [PubMed] [Google Scholar]
- 3.Yamada S, Pokutta S, Drees F, Weis WI, Nelson WJ. Cell. 2005;123:889–901. doi: 10.1016/j.cell.2005.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edwards KA, Demsky M, Montague RA, Weymouth N, Kiehart DP. Dev Biol. 1997;191:103–117. doi: 10.1006/dbio.1997.8707. [DOI] [PubMed] [Google Scholar]
- 5.Bretschneider T, et al. Curr Biol. 2004;14:1–10. doi: 10.1016/j.cub.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 6.Lenart P, et al. Nature. 2005;436:812–818. doi: 10.1038/nature03810. [DOI] [PubMed] [Google Scholar]
- 7.Pang KM, Lee E, Knecht DA. Curr Biol. 1998;8:405–408. doi: 10.1016/s0960-9822(98)70159-9. [DOI] [PubMed] [Google Scholar]
- 8.Burkel BM, von Dassow G, Bement WM. Cell Motil Cytoskeleton. 2007;64:822–832. doi: 10.1002/cm.20226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Asakura T, et al. Oncogene. 1998;16:121–130. doi: 10.1038/sj.onc.1201487. [DOI] [PubMed] [Google Scholar]
- 10.Yang HC, Pon LA. Proc Natl Acad Sci USA. 2002;99:751–756. doi: 10.1073/pnas.022462899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cooper JA, Walker SB, Pollard TD. J Muscle Res Cell Motil. 1983;4:253–262. doi: 10.1007/BF00712034. [DOI] [PubMed] [Google Scholar]
- 12.Czisch M, Schleicher M, Horger S, Voelter W, Holak TA. Eur J Biochem. 1993;218:335–344. doi: 10.1111/j.1432-1033.1993.tb18382.x. [DOI] [PubMed] [Google Scholar]
- 13.Fischer M, Haase I, Wiesner S, Muller-Taubenberger A. FEBS Lett. 2006;580:2495–2502. doi: 10.1016/j.febslet.2006.03.082. [DOI] [PubMed] [Google Scholar]
- 14.McNeil PL, Murphy RF, Lanni F, Taylor DL. J Cell Biol. 1984;98:1556–1564. doi: 10.1083/jcb.98.4.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang T, et al. J Exp Med. 1997;186:1853–1863. doi: 10.1084/jem.186.11.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


