Abstract
The absence of biological markers allowing for the assessment of the evolution and prognosis of glioblastoma (GBM) is a major impediment to the clinical management of GBM patients. The observed variability in patients' treatment responses and in outcomes implies biological heterogeneity and the existence of unidentified patient categories. Here, we define for the first time three GBM patient categories with distinct and clinically predictive three-dimensional nuclear-telomeric architecture defined by telomere number, size, and frequency of telomeric aggregates. GBM patient samples were examined by three-dimensional fluorescent in situ hybridization of telomeres using two independent three-dimensional telomere-measurement tools (TeloView program [P1] and SpotScan system [P2]). These measurements identified three patients categories (categories 1–3), displaying significant differences in telomere numbers/nucleus (P1 = .0275; P2 ≤ .0001), telomere length (P1 and P2 = .0275), and number of telomeric aggregates (P1 = .0464; P2 ≤ .0001). These categories corresponded to patients with long-term, intermediate, and short-term survival, respectively (P = .0393). The time to progression analyses showed significant differences between the three categories (P = .0167). There was a correlation between time to progression, median survival, and nuclear telomere architecture. Our study suggests a link between patient outcome and three-dimensional nuclear-telomere organization and highlights the potential clinical power of telomere signatures as a new prognostic, predictive, and potentially pharmacodynamic biomarker in GBM. Furthermore, novel automated three-dimensional high-throughput scanning as developed here permits to obtain data from 300 nuclei in 20 minutes. This method is applicable to any cell type and scanning application.
Introduction
Glioblastoma (GBM) is the most common and aggressive primary malignant astrocytic brain tumor in adults [1]. Despite decades of experimentation with a view to improving outcomes for patients with this disease, GBM remains fatal, with a median survival (MS) of approximately 12 to 15 months [2]. GBM is associated with one of the worst 5-year survival rates among all human cancers [3]. Interestingly, however, patients with one type of GBM, dubbed “GBM with long-term survival,” survive for more than 36 months, a fact that highlights the biologic heterogeneity of this disease [3].
Recent molecular studies have enhanced our understanding of the molecular pathogenesis of GBM. GBMs may develop as primary or type 1 GBM, with epidermal growth factor receptor amplification, or evolve from a low-grade diffuse astrocytoma after TP53 mutation, eventually leading to secondary or type 2 GBM [1,4,5]. However, 30% of GBM show molecular alterations that correspond to neither the type 1 nor the type 2 pathway [6–8], pointing to the limitations of this approach to molecular stratification. More so, this molecular stratification does not impact the choice of therapy or patient's outcome.
So far, the most useful prognostic tools in this disease remain clinical indices such as age, the Karnofsky Performance Status (KPS) score and the histopathologic grade, as was reported in the recursive partitioning analysis performed by the RTOG (Radiation Therapy Oncology Group) [9], as well as the Ki-67 proliferative index. Recently, attempts to categorize GBM according to the response to chemotherapy were proposed. Inactivity of the O6-methylguanine-DNA methyltransferase seems to increase the responsiveness of GBMs to alkylating agents. However, the O6-methylguanine-DNA methyltransferase promoter is methylated in only 45%of GBM[10,11]. Various molecules have been proposed as potential biomarkers, but none has met with clinical acceptance yet [12].
The three-dimensional organization of the genome and the nucleus is an essential component of tumorigenesis as stipulated by Theodore Boveri (1862–1915) a century ago [13,14]. Telomeres, the nucleoprotein complexes located at the end of eukaryotic chromosomes, have essential roles in preserving chromosomal integrity. Intact telomeres prevent end-to-end fusions, degradation of the chromosome ends, and contribute to the adequate chromosome positioning within the nucleus [15]. The telomeres consist of a tandem repeated DNA sequence (TTAGGG in vertebrates) that varies in length from 5 to 15 kb in humans. Telomere dysfunction is known to promote chromosomal instability (CIN) and carcinogenesis [16]. In most human somatic cells, telomeres act as a mitotic clock that limits indefinite cell division [17]. Telomeres are organized in a very typical way within the three-dimensional space of the nucleus, and telomeres of normal cells do not overlap [18] and are localized in microterritories [19]. In contrast, telomeres of tumor cell nuclei show an altered three-dimensional nuclear organization and form telomeric aggregates (TAs) that can be observed in the interphase nucleus [18]. Alterations in the nuclear telomere architecture and telomeric dysfunction are associated with CIN (structural and numerical CIN) [20,21], which is a hallmark of GBM [6,22]. In the context of this study, we examined whether the telomeric nuclear architecture in GBM could serve as a marker of genetic instability and a biological marker, predicting patient outcome.
In the present article, we introduce the three-dimensional telomeric signatures of GBM nuclei and propose the use of these signatures as a novel all-encompassing biomarker suitable for characterizing and stratifying GBM and allowing us to predict patient survival with increased accuracy. In addition, the novel high-throughput automated three-dimensional telomeric scanning approach that we developed and applied in a blinded study to the GBM patients permits a quick scan and analysis of hundreds of nuclei for this and any related clinical or basic research application.
Materials and Methods
Patients
This study received approval by the research ethics board on human studies at the Centre Hospitalier Universitaire de Sherbrooke. Patients undergoing or having undergone surgery for GBM at the Centre Hospitalier Universitaire de Sherbrooke were enrolled in this study after the obtaining of written informed consent. The patient population included in this study was composed of 11 individuals, 8 (72.7%) men and 3 (27.3%) women ranging in age from 24 to 66 years, with a median age of 46.5 years. These patients all depicted a preoperative presumptive diagnosis of GBM that was confirmed at surgery. The fresh surgical biopsies obtained immediately after resection in the operating room were collected prospectively. All surgeries were performed on newly diagnosed, treatment-naive patients. Immediately after collection, touch preparation smear slides (TP slides) were prepared. The TP slides were performed by smearing a core biopsy onto a glass slide, which was then air-dried [23]. The TP slides were fixed using fresh Carnoy (3 vol. methanol/1 vol. glacial acetic acid), air-dried overnight in a chemical hood, and stored at -20°C until they were used for quantitative fluorescent in situ hybridization (FISH).
Histopathologic Analysis
Tumors were classified according to the World Health Organization (WHO) classification [1]. After surgical removal, tumor samples were fixed in 10% formaldehyde and embedded in paraffin. Three-micrometer sections were stained with hematoxylin and eosin and submitted to immunohistochemical reactions for the detection of glial fibrillary acidic protein (GFAP), TP53, and epidermal growth factor receptor expression. Additional sections were submitted for K-67 antibody staining to evaluate the proliferative index, which was defined as the ratio between the number of Ki-67-positive tumor cell nuclei and the total number of tumor cell nuclei counted; areas of high positivity were selected for the counts. Tumor vascularization was evaluated with the CD34 antibody. All antibodies were provided by DAKO (Carpinteria, CA). Tumors exhibiting variable GFAP-positive cells within a fibrillar matrix showing high cellularity, nuclear pleomorphism, and neoangiogenesis and displaying necrotic areas together with a high mitotic count and/or Ki-67 proliferative index were classified as GBM.
Fluorescent In Situ Hybridization Protocol for Three-dimensional Analysis
The TP slides were thawed for 1 hour at room temperature (RT). The procedure was performed as described previously [20]. Briefly, slides were incubated in 3.7% formadehyde/phosphate-buffered saline (PBS; pH 7.4) for 20 minutes and washed three times in 1x PBS for 5 minutes each. Slides were incubated in 0.5% Triton X-100 for 3 minutes followed by an incubation in 20% glycerol for 1 hour, and three-dimensionally preserved by four repeated cycles of glycerol/liquid nitrogen treatment [24] and washed twice in 1x PBS for 5 minutes each followed by a 5-minute incubation in 0.1N HCl. Before fixation in 3.7% formamide/2x SSC at pH 7.0 for 1 hour, slides were washed twice for 5 minutes in PBS. Immediately after fixation, 8 µl of PNA telomeric probe (Dako, Glostrup, Denmark) was added to the slide. For denaturation of the nuclear DNA and the probe, the slides were incubated at 80°C for 3 minutes followed by hybridization at 30°C for 2 hours using a Hybrite (Vysis; Abbott Diagnostics, Des Plains, IL). The slides were washed twice for 15 minutes in 70% formamide/10 mM Tris pH 7.4 followed by washing in 1x PBS at RT for 1 minute while shaking and in 0.1x SSC at 55°C for 5 minutes while shaking. Slides were washed in 2x SSC/0.05% Tween 20 twice for 5 minutes each at RT while shaking for 5 minutes, after which they were counterstained with 4′,6-diamino-2-phenylindole (DAPI) (0.1 µl/ml). Excess DAPI was removed with deionized distilled water before dehydration in ethanol at 70%, 90%, and 100% for 2 minutes each. The slides were then air-dried and cover slipped with Vestashield (Vector Laboratories, Burlington, Ontario, Canada) for analysis.
Image Acquisition and Three-dimensional Image Analysis Using TeloView System
Imaging data from all patient GBM tissues were obtained by two independent image acquisition methods. As a criterion standard, we performed three-dimensional image analysis on 30 interphase nuclei per slide using an AxioImager Z1 microscope (Carl Zeiss Canada Ltd, Toronto, Canada) and an AxioCam HRm charge-coupled device (Carl ZeissCanada Ltd, Toronto, Canada) [20]. A 100x oil objective lens (Carl Zeiss Canada Ltd, Toronto, Canada) was used at acquisition times of 366 milliseconds for Cy3 (telomeres) and 109 milliseconds for DAPI (nuclei). Sixty z-stacks were acquired at a sampling distance of xy: 107 nm and z: 200 nm for each slice of the stack. AxioVision 4.6 software (Carl Zeiss Canada Ltd, Toronto, Canada) and a constrained iterative algorithm [25] were used for deconvolution. Deconvolved images were converted into TIFF files and exported for three-dimensional analysis using the TeloView software program [20].
Image Acquisition and Three-dimensional Image Analysis Using the Novel SpotScan System
The slides subjected to TeloView analysis were also scanned and analyzed using a novel SpotScan system (Applied Spectral Imaging, Migdal HaEmek, Israel) and the newly developed three-dimensional TeloScan software. The system uses a fully automated Olympus BX61 microscope (Olympus, Center Valley, PA) equipped with filters for DAPI and tetramethyl rhodamine iso-thiocyanate (TRITC); the software included CaseDataManager 6.0 and ScanView (Applied Spectral Imaging, Migdal Ha-Emek, Israel). Imaging was done with a x60 magnification oil objective. Cells were detected based on DAPI nuclear stain. For each cell, images of 10 focal planes 0.7 µm apart were collected. SpotScan analyzed the three-dimensional data based on DAPI (nucleus) and TRITC (telomeres). The data collected from each scan included information on number of signals, signal intensity, and the existence of aggregates within each nucleus examined. Three hundred nuclei per 20 minutes per scanned slide were classified according to their number of telomeres and recorded. Data were saved in Excel format. This tool combined with three-dimensional quantitative FISH technique and the data analyses will categorize a GBM patient in 9 to 12 hours after getting the sample.
Data Presentation and Statistical Analysis
Three types of histogram were produced: 1) line graphs showing the distribution in the intensity of the registered telomeric fluorescent signals, 2) histograms of the distribution of the number of aggregates per cell, and 3) histograms of the distribution of the number of registered signals per cell. The percentage of cells having TAs, the mean number of signals, and the mean number of aggregates per cell were calculated. The histogram data from different samples were combined into a single chart for comparison.
Overall Survival and Time to Progression
Kaplan-Meier curves were estimated for survival and time to progression (TTP). Survival curves were compared with the log-rank tests. Distribution of telomere intensities was compared by χ2 analysis, whereas summary of cell parameters was analyzed with nested factorial analysis of variance. Significance level was set at α = 0.05.
Results
Clinical Surrogates and Pathological Diagnosis
Data were collected from a total of 11 nonconsecutive patients operated on between 2004 and 2009. Patients were selected for this analysis based on the disparity in their clinical evolution, so as to create a heterogeneous clinical group. The conditions of all of these 11 patients were initially diagnosed as GBM tumor (Table 1) according to the WHO criteria [1]. These patients presented with variable clinical findings. Most of the patients presented with focal neurological symptoms related to the tumors.
Table 1.
Clinical Data of the Population Cohort.
| Patient | Category* | Age (years) | Sex | First Symptoms | Treatment | TTP (days) | MS (days) | Proliferative Index (%) | KPS Index |
| P1 | 3 | 25 | M | Headache | Craniotomy, STUPP, Temodal | 26 | 163 | 15 | 70 |
| N + Vo | |||||||||
| Seizure | |||||||||
| P2 | 2 | 26 | M | Seizure | Craniotomy, STUPP | 301 | Alive | 10 | 90 |
| P3 | 2 | 51 | M | Seizure | Craniotomy, STUPP, Temodal | 386 | 551 | 50 | 90 |
| P4 | 2 | 55 | M | Headache | Craniotomy, STUPP, Temodal, intra-arterial chemotherapy, CCNU | 149 | 308 | ND | 40 |
| Seizure | |||||||||
| P5 | 3 | 58 | M | Left hemiparesis | Craniotomy, STUPP, Temodal, intra-arterial chemotherapy | 91 | 490 | 20 | 80 |
| Seizure | |||||||||
| P6 | 1 | 56 | M | Dysphasia | Craniotomy, STUPP, Temodal | 595 | Alive | 30 | 80 |
| Headache | |||||||||
| P7 | 1 | 24 | F | Headache | Craniotomy, intra-arterial chemotherapy, Temodal | 587 | 751 | 30 | 80 |
| Right hemiparesis | |||||||||
| P8 | 3 | 46 | M | Dizziness | Craniotomy, STUPP, CCNU | 100 | 880 | 15 | 70 |
| Headache | |||||||||
| N + Vo | |||||||||
| P9 | 2 | 56 | M | Headache | Craniotomy, STUPP, Temodal, intra-arterial chemotherapy, CCNU | 227 | Alive | 12 | 50 |
| Left hemiparesis | |||||||||
| P10 | 2 | 66 | F | Dysphasia | Craniotomy, STUPP, Temodal, intra-arterial chemotherapy | 190 | 430 | 20 | 90 |
| Seizures | |||||||||
| P11 | 2 | 49 | F | Memory disturbances | Craniotomy, STUPP, Temodal | 409 | Alive | 30 | 50 |
| Headache | |||||||||
| Syncope |
CCNU indicates lomustine; CT, computed tomography; MRI, magnetic resonance imaging; N + Vo, nausea and vomiting; ND, not determined; STUPP, temozolomide and radiotherapy.
Three-dimensional telomere-defined categories.
Number of Telomeres, Nuclear Distribution of Telomeres, and TAs
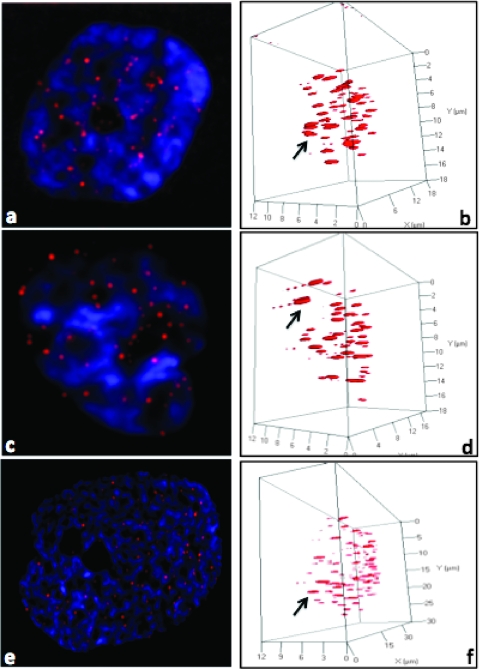
We first analyzed the total number of telomeres in each sample with two methods (TeloView and TeloScan) and thereafter observed three categories of patients (Figures 1, 2, and 3). Figure 1 illustrates telomeric nuclear signals of each patient category in two-dimensional and three-dimensional images as determined using imaging and three-dimensional reconstruction after constrained iterative deconvolution (Materials and Methods) [20,25]. Figure 2 sums up the data of 30 nuclei per category as identified in our blinded study after the use of the quantitative three-dimensional telomere analysis program TeloView [26]. Figure 3 shows screenshots of image galleries and patient histograms that were obtained after three-dimensional automated scanning of nuclear telomeric signals (see Materials and Methods).
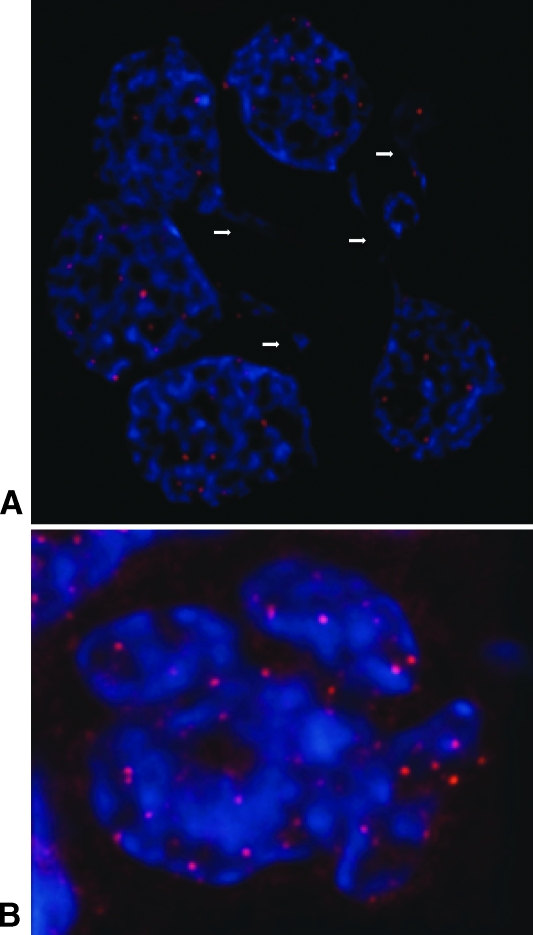
Figure 1.
Two-dimensional (A, C, E) and three-dimensional images (B, D, F) of human GBM nuclei (blue) and their telomeres (red) imaged and visualized with AxioVision 4.6. Nuclear telomere distribution patterns define TAs indicated with an arrowin the three categories of patient: (A, B) from P7 in category 1, (C, D) from P9 in category 2, and (E, F) from P1 in category 3. The arrows point to the telomere aggregates.
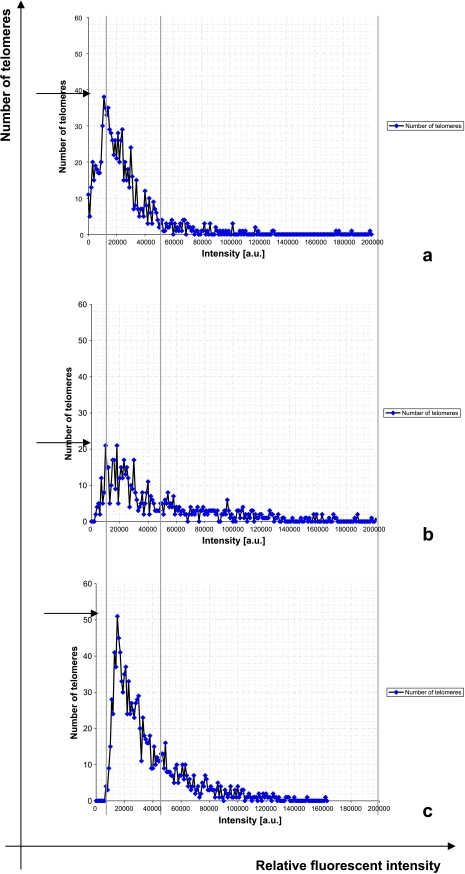
Figure 2.
Number of telomeres versus relative intensities of fluorescent signal analyzed by TeloView. Lines and arrows define the three categories of cells. (A) Category 1, P7. (B) Category 2, P9. (C) Category 3, P1. P = .0275.
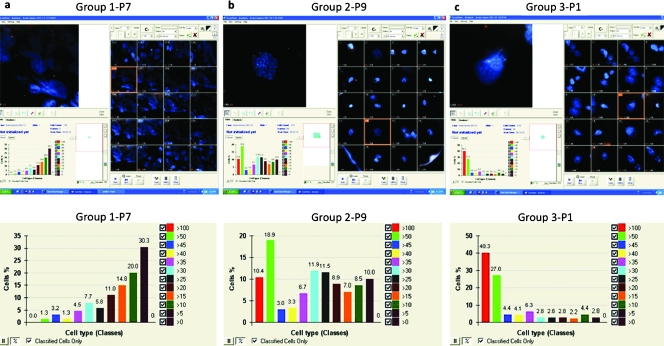
Figure 3.
Screenshots of human GBM nuclei (blue) and telomeres (red) with ScanView 6.0. Upper panel: Screenshot of a selected field of the gallery of 20 scanned cells. Bottompanel: The corresponding histogram showing the distribution of classified defined cells based on the number of telomeres/cell. (A) Category 1, P7. (B) Category 2, P9. (C) Category 3, P1. For patient details, see Table 1.
Patients were grouped into three categories that can be summarized by differences in telomere signal intensities and numbers as revealed by TeloView analysis (Figure 2) and TeloScan (Figure 3). TeloView indicated the presence of three different telomeric profiles that are shown in Figure 2, A, B, and C, respectively. These profiles became apparent after the quantitative analysis of 30 nuclei of touch preparations prepared at surgery from treatment-naive patients. In addition, these profiles indicate some diversity within the tumor cell nucleus population. For example, Figure 2, A and B, indicates that there are three sub-populations: nuclei with low relative fluorescent intensities representing cells with short telomeres, nuclei with intermediate relative fluorescent intensity indicating intermediate length of telomeres, and nuclei with telomeric signals of high intensity, that is, large telomeres. Figure 2C illustrates that this latter patient group lacks cells with very short telomeres, that is, signals with low fluorescent intensity.
In general, interphase nuclei display less telomeres than expected due to their organization in microterritories and/or aggregates [19,27]. Telomere numbers in our patients vary as obvious when profiles of Figure 2, A, B, and C, are compared (see arrows). Figure 2A indicates that patients within this category have a maximum number of 40 detected telomeric signals, Figure 2B indicates that that group has less telomeric signals (maximum is 20/cell), whereas Figure 2C shows that this patient group has increased numbers of telomeres (>50 per cell) when compared with the other two groups.
Automated three-dimensional scanning using TeloScan (Figure 3) performed on the identical samples examined, scanned and classified 300 cells per patient and also identified three patient categories. The first category (Figure 3A) shows telomere numbers ranging from 5 to 50, with most telomeres being in the small to intermediate telomere number groups. The second category of scanned profiles (Figure 3B) has telomere numbers that vary from 5 to 100, with most cells being in the mid size range. The third category of scanned profiles (Figure 3C) also displays 5 to 100 telomeres per nucleus; however, most of the signals are in the high number range (50–100 telomeres).
In conclusion, both assays led to the identical categorization of patients, although TeloScan (Figure 3) examine higher numbers of cells than TeloView.
After the analysis, the samples were decoded. The first category of patients was composed of two patients (P6 and P7), the second category was composed of six patients (P2, P3, P4, P9, P10, and P11), whereas the third was composed of three patients (P1, P5, and P8; Table 1). These categories displayed significant differences along three parameters: telomere numbers/nucleus (P = .0275 with TeloView data and P ≤ .0001 with TeloScan data), telomere length (P = .0275 with TeloView and TeloScan data), and number of TAs (P = .0464 with TeloView data and P ≤ .0001 with TeloScan data).
With respect to TAs, category 1 displayed the lowest level, whereas category 2 was intermediate, and category 3 presented the highest TAs level. We did not find significant differences when analyzing the average telomeric intensities (P = .3132) and the total telomeric intensities (P = .3147) between the three categories of patients.
Telomeres versus TTP and MS
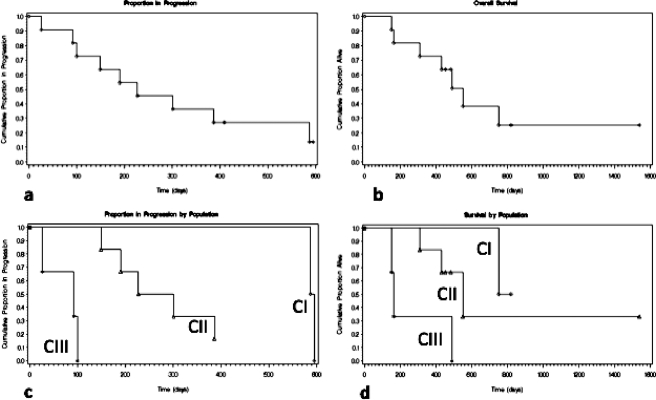
The MS for the 11 patients was 551 days (calculated from day of diagnosis to day of death or censoring), and the mean survival was 515 days (SE = ±75). As shown in Figure 4, patients in category 1 (curves CI) are characterized by longest TTP (591 days) and longest MS on Kaplan-Meier estimates (one censored; one died at 751 days). In contrast, patients in category 2 (curves CII) have an intermediate TTP (264 days) and an intermediate MS (three censored; three died at 308, 430, and 551 days, respectively; Figure 4). The patients in the third category (curves CIII) presented with the shortest TTP (92 days) and the shortest MS (one censored; two died at 149 and 163 days, respectively; Figure 4). The Kaplan-Meier survival curves display a significant difference between the three categories defined by the telomere analyses (Figure 4; P = .0393). The TTP analyses also show significant differences between the three categories of patients (P = .0167). Interestingly, survival did not correlate with the proliferation index (P = .90) or with the KPS scale (P = .86; Table 1) in this study.
Figure 4.
Kaplan-Meier curves depicting the TTP and overall survival of the three categories of population defined by telomere organization.
To our knowledge, this study describes the first molecular marker that enables such a distinction in GBM patients. Using this methodology allowed the production of Kaplan-Meier survival curves that reflected the telomere-based categorization of these patients. The three-dimensional telomeric signature-based survival curves display significant differences between the three categories defined by the telomere analyses.
Telomeres and Cell Cycle Distribution
We studied the genomic stability of the cells in each patient by determining the a/c ratio and the nuclear volume. The nuclear space occupied by telomeres is measured by two main axes, a and b, that are equal in length, and a third axis, c, that has a different length [26]. The distribution of telomeres in the three-dimensional space of the nucleus varies with cell cycle; as the specific stages of the cell cycle (G0/G1, S, and G2) phases have characteristic a/c ratios, the investigator can determine where they reside in the cell cycle [26]. The a/c ratio is a mean of defining progression through cell cycle in interphase cells. Thus, using this tool, we determined whether there was a difference in cell cycle between our patient groups. We did not find significant differences for the a/c ratio (P = .82) and for nuclear volume (P = .9244) among the three categories. The nonsignificant differences in a/c ratios are consistent with the high levels of proliferation in GBM and are consistent with the Ki-67 staining (Table 1). Moreover, we found several nuclear bridges indicative of breakage bridge fusion cycles in many of these samples (Figure 5) consistent with a high degree of genomic instability and telomere dysfunction.
Figure 5.
(A) Human GBM nuclei presenting breakage bridge fusion (BBF), with their telomere distribution. Arrows point to nuclear bridges indicative of breakage bridge fusion cycles. (B) Human GBM nuclei presenting nuclear fusion. These data are observed in all three patient categories. A qualitative evaluation showed that these characteristic figures of GBM are most prominent in category 3, followed by category 2 and category 1.
Discussion
GBM is a devastating disease, with only minor treatment breakthroughs in the last decades. We possess only a limited understanding of the disease process, as exemplified by the fact that patients whose conditions were diagnosed with common histopathologic criteria display an extremely heterogeneous clinical evolution and response to treatment. Molecular characterization has failed so far in providing a stratification rationale. The only distinctive factors with some prognostic value so far have been the age at diagnosis and the KPS at presentation. Proliferative indices have been used with limited success to provide a pathological tool to refine biological stratification; however, a reliable biological and prognostic marker for GBM patients is clearly lacking.
Our previous data showed altered three-dimensional telomeric organization in plasmacytoma [20,28], cervical cancer [27,29], Burkitt lymphoma [18], and head and neck cancer [18]. More recently, we were able to gain a mechanistic understanding of the transition of mononuclear Hodgkin cells to multinuclear Reed-Sternberg cells [30]. Encouraged by these data, we investigated whether three-dimensional nuclear telomeric architecture could predict disease aggressiveness in GBM and thus provide a new basis for defining clinically relevant prognostic patient categories. To this end, three-dimensional FISH analyses of telomeres were carried out in a blind fashion on touch preparations of fresh GBM samples (Table 1). We examined three-dimensional nuclear telomeric parameters that included the total number of telomeres, their nuclear distribution, and the presence of TAs (Figure 1).
Telomere numbers and their nuclear distribution differed markedly among patients within our GBM cohort (Figures 2 and 3). Two three-dimensional quantitative telomere analysis tools allowed for this conclusion. We first used TeloView, a quantitative program that measures telomeres within the three-dimensional nuclear space in an interactive manner [26]. TeloView analysis of the sample slides was followed by the use of a novel tool we developed, three-dimensional TeloScan. This imaging program allows for automated and high-throughput three-dimensional telomere scanning and analysis and automatically displays telomere numbers, intensities, and aggregates. Both methods led to the same basic conclusions. On the basis of defined telomere parameters, we identified three patient categories (1–3) within our GBM patient cohort (Table 1; Figures 2 and 3).
Decoding of the samples allowed us to compare the clinical data with the three-dimensional telomere signature-based patient categories. The three categories corresponded to patients with long-term, intermediate, and short-term survival respectively (P = .0393; Table 1 and Figure 4). The TTP analyses showed significant differences between the three categories of patients as well (P = .0167; Table 1 and Figure 4). There was thus an expected correlation between TTP and MS. In this study, the natural history of GBM evolution was therefore predicted by nuclear telomere architecture distinguishing long-term survival GBM (category 1) from short-term survival GBM (category 3). This prognostic value of the three-dimensional nuclear telomere architecture analysis will need to be validated with a larger set of patients. However, if this observation is confirmed, this methodology could eventually provide valuable guidance in clinical treatment decisions by identifying and monitoring patient responses to existing and newly emerging therapies. The three-dimensional nuclear telomere architecture characterization in GBM could be used as an individual GBM patient biomedical profile index in tailored therapy approaches acting as an important novel predictive biomarker. This is the case in PML-RARA translocation in promyelocytic leukemia, which responds to all-trans retinoic acid, and BCR-ABL translocation in chronic myelocytic leukemia, which responds to imatinib mesylate [31]. Finally, we suggest that three-dimensional nuclear telomere architecture could measure the near-term treatment effects of a drug on the GBM tumor cells directly as a pharmacodynamic biomarker. This could be performed by an initial three-dimensional FISH on a touch preparation slide of the tumor on surgery to categorize the GBM patient. Three-dimensional FISH could then be repeated during a follow-up on touch preparation slide derived from stereotactic biopsies after a given treatment. This stepwise analysis could allow recategorizing a patient and monitoring the evolution of his tumor. The remodeling of the nuclear telomere architecture and the consequent change of category of the GBM patient could then aid to adjust therapeutic options. Because the telomere aggregates represent a clear distinction between normal and tumor cells, we hypothesize that fewer TAs in the tumor would reflect tumor response to treatment. The newly established high-throughput semiautomatic nuclear scanning method (TeloScan) allows categorizing GBM patients in 9 to 12 hours after surgery or stereotactic biopsy. Therefore, the telomere biomarker can be used with different clinical surrogates and could eventually be incorporated in clinical trials as a monitoring tool providing a new basis for more effective therapies aimed at improving the dire prognosis of GBM.
Clinical management of patients bearing GBM is greatly influenced by the KPS scale. In our study, the three-dimensional telomeric stratification signature was infinitely more reliable than the KPS, the most useful current clinical prognostic factor, in predicting MS and TTP. Cohort MS in this series failed to correlate with either KPS (P = .86) or proliferation index (P = .90), which is clear evidence of the usefulness of our methods of analysis. Some studies have pointed to the value of the proliferative index as a prognostic tool [32,33], but this remains highly controversial [34].
The proliferation index measured by Ki-67 staining was confirmed by the a/c ratio as measured with TeloView [26]. The a/c ratio defines cell cycle progression in interphase cells and is based on the dynamic organization of the telomeres in the three-dimensional space of the interphase nucleus during the cell cycle [26]. We did not find significant differences between the three patient categories for the a/c ratio (P = .82) or the nuclear volume (P = .92). Specific features of three-dimensional telomeric signatures, such as telomere numbers/nucleus, telomere length, and number of TAs, are, as mentioned previously, causal factors in genomic instability in cancer cells. Importantly, many of the patient samples from all three categories displayed multiple nuclear bridges indicative of breakage bridge fusion cycles, which is consistent with a high degree of genomic instability in GBM(Figure 5). The highest incidence of multiple nuclear fusions and breakage bridge fusions was evident in GBMof category 3, thus supporting the notion that this new methodology is predictive of genomic instability and that this genomic instability directly correlates with survival.
Conclusions
Malignant GBMs have a dire prognosis. We describe for the first time the three-dimensional nuclear telomeric architecture as a suitable biomarker for predicting the prognosis of GBMs and linking progression to patient survival. Telomeric architecture seems to be a more accurate parameter for monitoring GBMs than any of those in current common use for their clinical stratification. The identification of three-dimensional telomeric signatures for clinical categorization, combined with our new high-throughput method for monitoring these signatures, opens the door to future routine use of this marker as a tool for the clinical management of gliomas. This marker needs to be validated with a more extensive set of GBM patients, as well as with WHO grade 2 and grade 3 astrocytomas, to allow for a differential comparison across the whole spectrum of astrocytic malignancy. Our findings provide a new analysis tool, with results from 9 to 12 hours after surgery, and add important information to the molecular mechanisms on the evolution and progression of GBMs.
Acknowledgments
The authors thank David Eisenstat for critical review of the data, Marie Boudrias and Jana Harizanova for their excellent technical help, Mary Cheang for statistical analyses, and Rina Kampeas for the excellent editing work. The authors also thank the patients who made this study possible.
Footnotes
M.G. was supported by a short-term fellowship under the Canadian Institutes of Health Research Strategic Training Program “Innovative Technologies in Multidisciplinary Health Research Training” (S.M.). The study was partly supported by the CancerCare Manitoba Foundation to S.M. D.F. holds the National Bank of Canada Chair for the Treatment of Brain Cancer. T.K. acknowledges the financial support from the Natural Sciences and Engineering Research Council of Canada and the Manitoba Health Research Council. R.D. holds a Canada Research Chair in Genetics, Mutagenesis, and Cancer. S.M. is the Director of the Genomic Centre for Cancer Research and Diagnosis.
References
- 1.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Louis DN, Pomeroy SL, Cairncross JG. Focus on central nervous system neoplasia. Cancer Cell. 2002;1:125–128. doi: 10.1016/s1535-6108(02)00040-5. [DOI] [PubMed] [Google Scholar]
- 3.Krex D, Klink B, Hartmann C, von Deimling A, Pietsch T, Simon M, Sabel M, Steinbach JP, Heese O, Reifenberger G, et al. Long-term survival with glioblastoma multiforme. Brain. 2007;130:2596–2606. doi: 10.1093/brain/awm204. [DOI] [PubMed] [Google Scholar]
- 4.Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 5.Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359:492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 6.Martinez R, Schackert HK, Plaschke J, Baretton G, Appelt H, Schackert G. Molecular mechanisms associated with chromosomal and microsatellite instability in sporadic glioblastoma multiforme. Oncology. 2004;66:395–403. doi: 10.1159/000079488. [DOI] [PubMed] [Google Scholar]
- 7.Kleihues P, Ohgaki H. Primary and secondary glioblastomas: from concept to clinical diagnosis. Neuro Oncol. 1999;1:44–51. doi: 10.1093/neuonc/1.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holland EC. Gliomagenesis: genetic alterations and mouse models. Nat Rev Genet. 2001;2:120–129. doi: 10.1038/35052535. [DOI] [PubMed] [Google Scholar]
- 9.Scott CB, Scarantino C, Urtasun R, Movsas B, Jones CU, Simpson JR, Fischbach AJ, Curran WJ., Jr Validation and predictive power of Radiation Therapy Oncology Group (RTOG) recursive partitioning analysis classes for malignant glioma patients: a report using RTOG 90-06. Int J Radiat Oncol Biol Phys. 1998;40:51–55. doi: 10.1016/s0360-3016(97)00485-9. [DOI] [PubMed] [Google Scholar]
- 10.Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, Kros JM, Hainfellner JA, Mason W, Mariani L, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 11.Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, Ludwin SK, Allgeier A, Fisher B, Belanger K, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 12.Riemenschneider MJ, Reifenberger G. Novel insights into the pathogenesis of gliomas based on large-scale molecular profiling approaches. Curr Opin Neurol. 2009;22:619–624. doi: 10.1097/WCO.0b013e32833245b0. [DOI] [PubMed] [Google Scholar]
- 13.Boveri T. Über mehrpolige Mitosen als Mittel zur Analyse des Zellkerns. Verh Phys Med Ges Wurzbg. 1902;35:67–90. (Ger). [Google Scholar]
- 14.Boveri T. Zur Frage der Entstehungmaligner Tumoren. Jena, Germany: Fischer; 1914. (Ger). [Google Scholar]
- 15.Callen E, Surralles J. Telomere dysfunction in genome instability syndromes. Mutat Res. 2004;567:85–104. doi: 10.1016/j.mrrev.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 16.DePinho RA, Polyak K. Cancer chromosomes in crisis. Nat Genet. 2004;36:932–934. doi: 10.1038/ng0904-932. [DOI] [PubMed] [Google Scholar]
- 17.Lansdorp PM. Telomeres and disease. EMBO J. 2009;28:2532–2540. doi: 10.1038/emboj.2009.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chuang TC, Moshir S, Garini Y, Chuang AY, Young IT, Vermolen B, van den Doel R, Mougey V, Perrin M, Braun M, et al. The three-dimensional organization of telomeres in the nucleus of mammalian cells. BMC Biol. 2004;2:12. doi: 10.1186/1741-7007-2-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Vos WH, Hoebe RA, Joss GH, Haffmans W, Baatout S, Van Oostveldt P, Manders EM. Controlled light exposure microscopy reveals dynamic telomere microterritories throughout the cell cycle. Cytometry A. 2009;75:428–439. doi: 10.1002/cyto.a.20699. [DOI] [PubMed] [Google Scholar]
- 20.Louis SF, Vermolen BJ, Garini Y, Young IT, Guffei A, Lichtensztejn Z, Kuttler F, Chuang TC, Moshir S, Mougey V, et al. c-Myc induces chromosomal rearrangements through telomere and chromosome remodeling in the interphase nucleus. Proc Natl Acad Sci USA. 2005;102:9613–9618. doi: 10.1073/pnas.0407512102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mai S, Garini Y. Oncogenic remodeling of the three-dimensional organization of the interphase nucleus: c-Myc induces telomeric aggregates whose formation precedes chromosomal rearrangements. Cell Cycle. 2005;4:1327–1331. doi: 10.4161/cc.4.10.2082. [DOI] [PubMed] [Google Scholar]
- 22.Saito T, Hama S, Izumi H, Yamasaki F, Kajiwara Y, Matsuura S, Morishima K, Hidaka T, Shrestha P, Sugiyama K, et al. Centrosome amplification induced by survivin suppression enhances both chromosome instability and radiosensitivity in glioma cells. Br J Cancer. 2008;98:345–355. doi: 10.1038/sj.bjc.6604160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gadji M, Fortin D, Tsanaclis AM, Drouin R. Is the 1p/19q deletion a diagnostic marker of oligodendrogliomas? Cancer Genet Cytogenet. 2009;194:12–22. doi: 10.1016/j.cancergencyto.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 24.Solovei I, Walter J, Cremer M, Habermann FA, Cremer T. FISH on three-dimensionally preserved nuclei. In: Beatty B, Mai S, Squire J, editors. FISH A Practical Approach. New York, NY: Oxford University Press; 2002. pp. 119–157. [Google Scholar]
- 25.Schaefer LH, Schuster D, Herz H. Generalized approach for accelerated maximum likelihood based image restoration applied to three-dimensional fluorescence microscopy. J Microsc. 2001;204:99–107. doi: 10.1046/j.1365-2818.2001.00949.x. [DOI] [PubMed] [Google Scholar]
- 26.Vermolen BJ, Garini Y, Mai S, Mougey V, Fest T, Chuang TC, Chuang AY, Wark L, Young IT. Characterizing the three-dimensional organization of telomeres. Cytometry A. 2005;67:144–150. doi: 10.1002/cyto.a.20159. [DOI] [PubMed] [Google Scholar]
- 27.Mai S, Garini Y. The significance of telomeric aggregates in the interphase nuclei of tumor cells. J Cell Biochem. 2006;97:904–915. doi: 10.1002/jcb.20760. [DOI] [PubMed] [Google Scholar]
- 28.Guffei A, Lichtensztejn Z, Goncalves Dos Santos Silva A, Louis SF, Caporali A, Mai S. c-Myc-dependent formation of Robertsonian translocation chromosomes in mouse cells. Neoplasia. 2007;9:578–588. doi: 10.1593/neo.07355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guijon FB, Greulich-Bode K, Paraskevas M, Baker P, Mai S. Premalignant cervical lesions are characterized by dihydrofolate reductase gene amplification and c-Myc overexpression: possible biomarkers. J Low Genit Tract Dis. 2007;11:265–272. doi: 10.1097/LGT.0b013e31803c4df0. [DOI] [PubMed] [Google Scholar]
- 30.Knecht H, Sawan B, Lichtensztejn D, Lemieux B, Wellinger RJ, Mai S. The 3D nuclear organization of telomeres marks the transition from Hodgkin to Reed-Sternberg cells. Leukemia. 2009;23:565–573. doi: 10.1038/leu.2008.314. [DOI] [PubMed] [Google Scholar]
- 31.Sawyers CL. The cancer biomarker problem. Nature. 2008;452:548–552. doi: 10.1038/nature06913. [DOI] [PubMed] [Google Scholar]
- 32.Faria MH, Goncalves BP, do Patrocinio RM, de Moraes-Filho MO, Rabenhorst SH. Expression of Ki-67, topoisomerase IIα and c-MYC in astrocytic tumors: correlation with the histopathological grade and proliferative status. Neuropathology. 2006;26:519–527. doi: 10.1111/j.1440-1789.2006.00724.x. [DOI] [PubMed] [Google Scholar]
- 33.Vaquero J, Zurita M, Coca S, Oya S. Imbalance between apostain expression and proliferative index can predict survival in primary glioblastoma. Acta Neurochir (Wien) 2002;144:151–155. doi: 10.1007/s007010200018. discussion 155–156. [DOI] [PubMed] [Google Scholar]
- 34.Lafuente JV, Alkiza K, Garibi JM, Alvarez A, Bilbao J, Figols J, Cruz-Sanchez FF. Biologic parameters that correlate with the prognosis of human gliomas. Neuropathology. 2000;20:176–183. doi: 10.1046/j.1440-1789.2000.00329.x. [DOI] [PubMed] [Google Scholar]