Abstract
In this review we summarize recent studies that demonstrate the importance of epigenetic mechanisms for maintaining genome integrity, specifically with respect to repeated DNAs within heterochromatin. Potential problems that arise during replication, recombination, and repair of repeated sequences are counteracted by post-translational histone modifications and associated proteins, including the cohesins. These factors appear to ensure repeat stability by multiple mechanisms: suppressing homologous recombination, controlling the three-dimensional organization of damaged repeats to reduce the probability of aberrant recombination, and promoting the use of less problematic repair pathways. The presence of such systems may facilitate repeat and chromosome evolution, and their failure can lead to genome instability, chromosome rearrangements, and the onset of pathogenesis.
Introduction
Eukaryotic genomes are partitioned into heterochromatin and euchromatin, which are cytologically, genomically, and functionally distinct. `Classical' heterochromatin was originally defined by differential staining, indicating constitutive condensation throughout the cell cycle. Complete or nearly complete genome sequences have elucidated distinguishing features of these regions. In most eukaryotes heterochromatin is concentrated in pericentromeric and telomeric regions, is enriched for repetitive sequences, including highly repeated tandem `satellite' sequences and transposable elements (see Glossary for terms and abbreviations used in this review), and has a relatively low gene density [1]. Surprisingly, heterochromatin comprises 20–30% of many eukaryotic genomes, including flies and humans [2,3], and can reach 90% of some genomes, generally found among the plants.
Defining heterochromatin has become much more difficult in recent years. Chromatin composition and function, specifically post-translational histone modifications, associated proteins, and epigenetic gene silencing, are currently used as `the' defining characteristics of heterochromatin. For example, in most eukaryotes, heterochromatic regions are enriched for hypoacetylated histones, di- and tri-methylated histone H3 lysine 9 (H3K9me2 and H3K9me3), and heterochromatin protein 1 (HP1). However, euchromatic regions also contain `heterochromatic' modifications and proteins. Although often described `negatively' as transcriptionally inert `junk' sequences [4], heterochromatin is essential for normal chromosome organization [5,6], centromere function [7], and telomere protection [8]. Furthermore, heterochromatin contains the highly active ribosomal RNA genes and many protein-encoding genes [3,9•], and is to some extent formed and regulated by transcripts and small RNAs via RNA interference (RNAi) pathways. Thus, defining heterochromatin based on chromatin composition or functional properties such as gene silencing is problematic. For simplicity, we will use the cytological/genomic definition of `heterochromatin' to specifically refer to the large, contiguous, repetitive DNA domains associated with centromeric and telomeric regions.
The high concentrations of repeated sequences in heterochromatin present serious challenges to genome stability and can impact both cellular and organismal viability and chromosome evolution. Tandemly repeated sequences are particularly problematic with respect to the fidelity of DNA replication, repair, and recombination (Figure 1). Replication across repeated sequences can result in sequence expansion, duplications, and replication fork stalling with resulting double-stranded breaks (DSBs). Unequal exchange between homologous repeats alters length of the repetitious region, and homologous recombination can produce dicentric or acentric chromosomes, which results in aneuploidy [10]. Simple repeat expansions cause hereditary disorders in humans, including fragile X syndrome, myotonic dystrophy, Huntington's disease, various spinocerebellar ataxias, and others [11].
Figure 1.
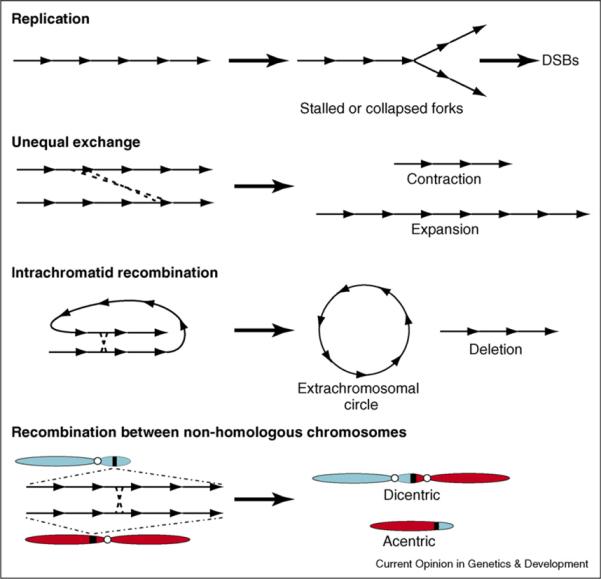
Replication, repair, or recombination of tandemly repeated DNAs can result in genome instability. Aberrant replication of repeated DNAs can produce stalled and collapsed forks that can result in DSBs. Unequal exchange between tandemly repeated DNA sequences results in contraction and expansion of tandem arrays. Intrachromatid recombination between homologous repeats produces extrachromosomal circular DNAs, as well as deletions of repeated DNAs. Recombination between homologous repeats in different chromosomes produces dicentric and acentric chromosomes, resulting in chromosome fragmentation and aneuploidy during mitosis or meiosis.
Why does heterochromatin persist throughout eukaryotic evolution, despite problems inherent to accurate inheritance of repeated DNA sequences? The presence of essential functions in heterochromatin provides one explanation. It is also likely that heterochromatic replication, repair, and recombination are regulated to ensure the stability of repeated DNAs. Recent studies have shown that replication of repetitious DNA and associated chromatin is tightly regulated and coordinated and that faithful DNA replication and repair rely heavily on chromatin properties [12]. Heterochromatin has been characterized as recombinationally silent, which alleviates problems that could arise from reciprocal exchange during meiosis. However, repeated DNAs display very high frequencies of sequence changes during evolution that become homogenized across genomes. These observations suggest the presence of mechanisms that balance interactions and exchange of information between heterochromatic sequences with the need to avoid negative consequences to genome stability. In this review we will summarize recent insights into conserved, chromatin-based epigenetic mechanisms that maintain heterochromatin integrity by regulating recombination and repair, and discuss the possible roles of these mechanisms in chromosome and genome evolution.
Epigenetic regulation of the stability and nuclear organization of tandem repeats
Recent investigations into epigenetic regulation of repeat integrity have focused on ribosomal DNA (rDNA). One of the best characterized DNA elements, rDNA is tandemly repeated and embedded within heterochromatin in most eukaryotes. This evolutionarily conserved positioning of rDNA within heterochromatin probably regulates important features of nucleolus formation, and also appears to prevent recombination and protect rDNA repeat integrity.
Repeated DNAs, including rDNA, can recombine to form extrachromosomal circular DNAs (eccDNAs) [13,14]. Genetic studies in S. cerevisiae identified mutations in the Sir2 histone deacetylase as a strong suppressor of ecc rDNA formation and found that ecc rDNA formation required the Rad50 and Rad52 recombination proteins [15]. This provided the first demonstration of an important role for chromatin in regulating recombination at repeated DNAs [16]; a conserved role for Sir2 in this process has recently been shown in S. pombe [17].
Recent studies in S. cerevisiae showed that other chromatin-associated proteins actively suppress rDNA recombination [18•,19•,20••,21]. In S. cerevisiae, the cohesins, proteins that ensure sister chromatid associations, are enriched in the spacers between rDNA transcription units. Cohesin mutants, or displacement of cohesins by rRNA transcription, result in elevated frequencies of ecc rDNA circles and increased recombination [18•]. Thus, cohesins are required to suppress recombination between rDNA repeats; presumably the maintenance of sister chromatid associations reduces inter- and intra-chromatid contact between rDNAs.
The Smc5–Smc6 complex, structurally related to condensins and cohesins, is required for normal DNA repair and associates with rDNA repeats and telomeres. Temperature sensitive Smc5 and Smc6 mutants display unusual, aberrant mitoses with impaired segregation of repetitive DNAs [19•]. Interestingly, these mitotic defects were suppressed by mutations in the RAD9 DNA damage checkpoint protein and by mutations in RAD52, which is required for repair of DSBs. These results suggest that the Smc5–Smc6 complex ensures proper chromosome segregation by preventing the formation of aberrant sister chromatid junctions at repeated DNAs. More recently, it has been shown that DSB sites in rDNA, visualized as RAD52 foci, are normally excluded from the nucleolus [22••]. Loss of Smc5, Smc6, the Mre11 nuclease, or SUMOylation of RAD52 results in RAD52 foci formation within the nucleolus, which is highly correlated with increased rDNA recombination and ecc rDNA circles [22••]. These results suggest that nucleolar positioning of damaged rDNA, in addition to or in combination with chromatin components, is involved in repressing recombination at repeats.
Studies in D. melanogaster and S. pombe have shown that the H3K9 methylation and RNAi pathways also regulate repeated DNA stability [20••,21]. In Drosophila, H3K9me2 levels in chromatin associated with repeated DNAs are greatly reduced in animal mutant for the Su(var)3–9 histone methyltransferase (HMTase) or the dcr-2 (dicer-2) RNAi component. Diploid and polytene nuclei from mutants displayed multiple nucleoli, dispersed rDNA and satellite DNAs, and a substantial increase in ecc-repeated DNAs [20••]. The `disorganized nucleolus' phenotype in Drosophila reflects mutations in Ligase 4 [20••] and Rad51 (Peng and Karpen, unpublished data), suggesting that repeated DNA stability involves suppression of non-homologous end joining (NHEJ) and homologous recombination (HR) pathways. Thus, as observed in S. cerevisiae, chromatin composition impacts the stability of repeated, heterochromatic sequences in Drosophila, as well as the 3D organization of chromosomal elements and nuclear organelles [20••,22••]. However, while Su(var)3–9 and H3K9 methylation were shown to be required for cohesin recruitment at repeated DNAs in Drosophila, cohesin was not essential for repressing eccDNA formation [20••]. Nevertheless, these studies demonstrated that exchange between tandem repeats is regulated epigenetically in evolutionary distant species.
Epigenetic regulation of transposable element stability
In addition to tandem repetitive sequences, heterochromatin is also highly enriched for transposable elements. The H3K9 methylation and RNAi pathways have been shown to suppress transcription, transposition, and hypermutability of mobile elements, in yeast to humans (reviewed by Slotkin and Martienssen [23•]). Studies in S. pombe, plants and C. elegans demonstrated that the RNAi pathway degrades mRNAs produced by transposable elements, thereby limiting their transposition. In addition, the RNAi pathway induces transcriptional silencing of transposable elements by recruiting chromatin modifiers to transposable element loci. Mutations in components of epigenetic silencing, that is Su(var)3–9, the DNMT1 DNA methyltransferase, or the RNAi pathways, lead to increased transposable element transcription and mobility [23•].
The RNAi pathway regulated by Piwi/Aubergine directly impacts genome stability and the development of germlines in D. melanogaster, mammals, and C. elegans. Best characterized in D. melanogaster, Piwi/Aubergine regulation of repeat associated small interfering RNAs (rasiRNAs) mediates silencing of retrotransposons and the repeated Stellate locus [24•] and promotes normal embryonic axis specification and germline development [25]. Surprisingly, ATR/Chk2 mutations that disrupt DNA damage signaling suppress the axis defects but do not restore transposon silencing. The frequencies of DNA damage foci increased specifically in the germline of rasiRNA pathway mutants. These increases in the number of damage foci are independent of the Spo11 endonuclease, which is responsible for normal meiotic DSBs and recombination [25]. Thus, the defects in axis specification are a secondary effect of activating the ATR/Chk2 kinase pathway, and the primary function of the rasiRNA pathway in the Drosophila germline is to suppress transposition and its resultant DNA damage.
Mammalian piRNAs and Drosophila rasiRNAs appear to function similarly in suppressing transposable elements in the germline. The Piwi-related Argonauts in mouse (Miwi and Mili) bind piRNAs derived primarily from single-stranded RNAs [26–28]. Mutations in Miwi and Mili disrupt germline development, leading to defective spermatogenesis and increased apoptosis, phenotypes resembling those observed in Drosophila rasiRNA mutants [29]. The specific functions of piRNAs in mammals, their roles with respect to DNA damage at transposable elements, and the reason for their specificity to spermatogenesis are currently unknown.
Evolutionary plasticity of heterochromatin
The regulation of DNA recombination and repair by chromatin are likely to influence heterochromatin sequence plasticity over evolutionary time. Comparative sequence analysis in Arabidopsis and Drosophila suggests dramatic structural reorganization of genes whose euchromatic and heterochromatic locations change during evolution [30,31,32•]. For example, in Drosophila heterochromatic genes contain many more transposable elements in their introns and flanking regions, in addition to increased A-T content in the coding sequences, when compared with orthologous genes present in euchromatin in other fly species [32•]. The heterochromatic transposable elements are frequently deleted and rearranged. Thus, regulation of transposable element mobility, damage and repair, and exchange may affect the structure of genic and non-genic regions in heterochromatin.
Remarkably, tandem repeats expand and contract within and among species during evolution. Repeats also undergo homogenization, in which variant sequences spread across the genome [33,34]. Thus, information is readily exchanged among similar heterochromatic repeats on homologous and non-homologous chromosomes, at least over evolutionary timescales, despite the suppression of reciprocal recombination [35,36]. These observations suggest that repeat length changes and homogenization can occur without reciprocal exchange, for example via gene conversion and/or unequal crossing over, which would not result in rearrangements and aneuploidy.
Models for epigenetic regulation of heterochromatin stability and plasticity
How heterochromatin inhibits repeated DNA recombination, whether spontaneous or damage-induced, remains a mystery. One obvious model is that heterochromatin composition or structure physically prohibits access of recombination machinery to repeated DNAs (Figure 2, right panel). However, this hypothesis does not explain how DNA damage within heterochromatin is repaired, or how repeated sequences homogenize during evolution. Alternatives include the possibility that heterochromatin affects the frequency of DNA damage, or the nature or efficiency of repair.
Figure 2.
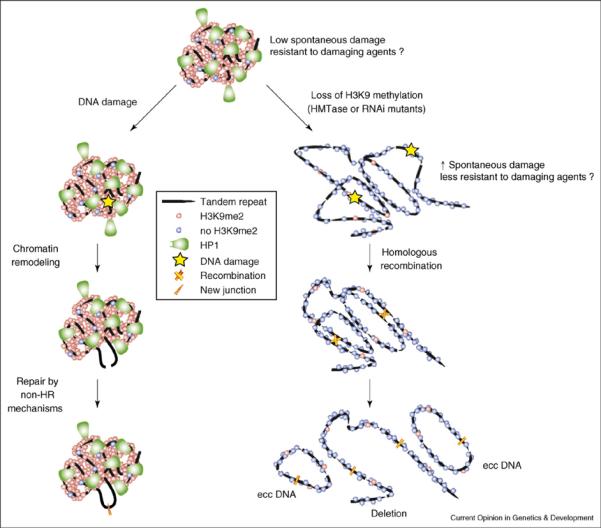
Models for epigenetic control of heterochromatic damage, repair, and exchange. Top: Heterochromatin contains tandemly repeated sequences and transposable elements (not shown). Specific histone modifications (e.g. H3K9 methylation), associated proteins (e.g. HP1), and/or a compact chromatin structure may help reduce the frequency of spontaneous DNA damage during replication, and possibly provides resistance to damaging agents. Left: Once DNA damage occurs in repeated sequences (including meiotic DSBs involved in recombination), heterochromatin structure or composition may impact the positioning or type of repair processes to reduce the probability of homologous exchange, which would lead to genome instability. Chromatin remodeling may help position damaged repeated DNA into euchromatic territories, which could reduce the probability of interactions with undamaged homologous repeats that remain heterochromatic. Alternatively, or in addition, damaged DNAs may be preferentially repaired by non-HR mechanisms, such as NHEJ, SSAR, and gene conversion. Right: Loss of heterochromatin components – due to mutations in H3K9 methyltransferases (HMTases) or RNAi pathway components – makes heterochromatic DNA sequences more prone to spontaneous and/or induced damage. In addition, homologous recombination occurs between repeats, resulting in increased extrachromosomal DNA formation, deletions, and chromosome rearrangements.
Euchromatin and heterochromatin appear to exhibit different responses to DNA damaging agents. Recent studies of ionizing radiation followed by quantitation of DNA break frequencies over time indicated that the vast majority of DNA breaks are located outside the heterochromatic `territory' in interphase cells within an hour of introducing damaging agents [37,38]. The lower frequencies of repair foci observed in heterochromatin suggest that euchromatin may be more prone to damage by ionizing radiation. Alternatively, initial damage frequencies within euchromatin and heterochromatin may be very similar, with faster repair of heterochromatic breaks. Experiments to differentiate between these two explanations are needed, such as comparing break frequencies within seconds/minutes of damage.
Another possibility is that repeated DNAs rapidly change their three-dimensional organization after DNA damage and are moved into euchromatic `territories' for repair. Live cell studies, DNA–FISH, and electron micrograph analysis of UV-irradiated cells demonstrated immediate (within seconds) chromatin expansion around individual double-stranded breaks. This process occurs in both euchromatin and heterochromatin with similar kinetics and is dependent on ATP but independent of H2AX and ATM. These results and others suggest that a rapid, energy-dependent chromatin decondensation occurs upon DNA damage, perhaps providing easier access for repair machinery and more efficient repair [39•]. While mechanisms regulating this process are still under investigation, these observations raise the possibility that rapid structural changes at heterochromatic breaks and subsequent repair may depend on chromatin composition and structure, potentially mediated by H3K9 methylation, HP1, and the RNAi pathway components.
Finally, the type of DNA damage repair may be affected by heterochromatin factors. Chromatin composition at DNA damage sites changes rapidly to facilitate recruitment of DNA repair machinery. One example is that phosphorylated H2A variants at sites of DNA damage recruit cohesins [40] and ATP-dependent chromatin remodelers. Other less well-characterized histone modifications – phosphorylation, acetylation, and methylation of histone H4 residues, H3K79 methylation, H2BK123 ubiquitination, and H2AS129 phosphorylation – are also involved in repair factor recruitment and loading [41,42]. Heterochromatin components may promote preferential associations with specific repair factors and mechanisms. For example, in S. cerevisiae γH2A does not spread into a silent mating-type locus (HML) inserted near a DSB, though it is enriched on the other side of the heterochromatic block [38]. We propose that heterochromatin components epigenetically regulate preferential utilization of non-HR mechanisms to repair DNA damage in repeated DNAs (Figure 2, left panel), for example single-strand annealing repair, non-homologous end joining, unequal exchange, or gene conversion. One appealing aspect of this model is that it can account for both maintenance of repeat stability in cells and animals, as well as repeat plasticity over evolutionary timescales.
Heterochromatin instability in human disease
Mammalian genomes are highly complex in terms of sequence composition and organization. More than 40% of the human euchromatic genome consists of repeated DNAs, and about 1% of the genome contains protein-coding genes [2]. Recombination among these repeated sequences would generate chromosome rearrangements, which are correlated with uncontrolled cell growth and tumorigenesis. Furthermore, fragile sites exist that can cause replication timing deregulation, eventually leading to gene amplification and aneuploidy [43]. How these fragile sites arise is not entirely clear, but indirect evidence suggests that one contributing factor is the high repeat content of mammalian genomes and their associated chromatin [44].
The short Alu repeats, consisting of 11% of the human genome, and a heterochromatin domain on human chromosome 1 (band 1q12) are well-studied examples of repeated DNAs implicated in pathogenesis. Alu repeats can recombine to cause recurrent gene mutations that result in human diseases such as breast cancer (BRCA1 deletion), glioma brain tumors (RB1 deletion), and familial hypercholesterolemia (LDL receptor deletion) [45]. Heterochromatin 1q12 contains a fragile site associated with chromosome translocations in breast, lymphoid, skin, reproductive organ, and endothelial tract cancers [46]. Comparative genome hybridization (CGH) of cancer samples suggests that satellite 2 DNA demethylation within 1q12 leads to a high incidence of chromosomal translocations [47]. In addition, heterochromatin has now been linked to human cancer progression. Recent studies show that global reductions in characteristic features of constitutive and/or facultative heterochromatin (CpG methylation, HP1, and H3K27 methylation), as well as H3K27 hypermethylation of tumor suppressor genes, are highly correlated with metastasis [48–50]. These epigenetic changes could affect cancer progression by altering gene expression, but the possibility that heterochromatin DNA stability is affected needs to be investigated.
Summary
In sum, recent studies highlight the importance of chromatin regulation of heterochromatin and repeated DNAs in maintaining the integrity of chromosomes and genomes. Future studies in this exciting, emerging field will elucidate important details of the chromatin components and reveal the mechanisms responsible for regulating recombination and repair in heterochromatin, including the impact on viability, fertility, disease, and chromosome evolution.
Acknowledgements
We thank members of the Karpen lab for useful discussions that helped us synthesize ideas presented in this review. Dr Irene Chiolo was particularly helpful in developing the hypothesis that heterochromatin may alter the nature of repair at sites of DNA damage. Our work on heterochromatin is supported by NIH grant R01HG00747.
Glossary
- ATM (ataxia telangiectasia mutated) and ATR (ATM and Rad3 related)
These phosphoinositide 3-kinases are required for activation of DNA damage checkpoints. Mutations in the human ATM gene result in defective repair, radiation sensitivity, and elevated frequencies of cancers.
- DSB
a double-strand break in the DNA double helix
- eccDNA
extrachromosomal circular DNA
- FISH
fluorescent in situ hybridization
- HR: Homologous recombination
HR is a key pathway for repair of DSBs and for meiotic recombination, which results in exchange between homologous sequences. The MRX/MRN complex processes the ends of DSBs into 3′ single-stranded DNA overhangs, which are recognized and bound by the Rad51 and Rad52 proteins to form ssDNA-Rad51 nucleoprotein filaments. This complex is required for homologous sequence searching and for initiating strand invasion, exchange, ligation, and resolution of joined molecules.
- NHEJ: Non-homologous end joining
NHEJ is an important pathway for DSB repair that results in joining of non-homologous sequences without exchange or recombination. Ku70–Ku80 heterodimers bind and hold the ends of DSBs to facilitate end-to-end ligation by the ligase 4 complex. In an alternative mechanism, DSBs bound by Ku heterodimers can be processed by the MRX/MRN exonucleases into single-stranded DNAs, which are then joined by the ligase 4 complex.
- Phosphorylated H2A variants
In response to DNA damage or cell cycle checkpoint activation, histone H2A variants are phosphorylated by phosphoinositol-3 kinases at S129 of H2A in S. cerevisiae, S139 of H2Ax in mammals, and S137 of H2Av in Drosophila. Phosphorylated H2A (designated e.g. as γH2Av) accumulates at DNA damage sites, and spreads extensively into surrounding regions.
- Post-translational histone modifications (H3K9 methylation, H3K4 methylation)
Histones and histone variants are chemically modified by enzymes post-translationally, including ubiquitylation, SUMOylation, phosphorylation, methylation, and acetylation. Modified histones are thought to affect the physical properties of the associated chromatin, which influences chromatin structure and functions such as gene expression. For example, di- and tri-methylation of the lysine 9 residue of histone H3 (H3K9me2 and me3) are associated with `silent' chromatin, whereas H3K4me2 and me3 are present in active or open chromatin at expressed genes.
- rDNA
DNA coding for ribosomal RNA
- RNAi: RNA interference; rasiRNAs: repeat-associated interfering RNAs:
In this cellular process, Dicer proteins process double-stranded RNAs into small interfering RNAs (siRNAs). rasiRNAs are produced from transcripts of repeated DNAs in heterochromatin, including TEs, and associate with the RISC complex, which then targets and cleaves transcripts to induce post-transcriptional silencing. In addition, the siRNA–RISC complex functions during heterochromatin establishment in at least some organisms by targeting H3K9 histone methyltransferases (HMTases) to heterochromatic sequences through an unknown mechanism.
- SSAR: Single-strand annealing repair
In this DNA damage repair pathway, two single-stranded homologues are generated from both ends of DSBs by exonucleases. The single-stranded sequences anneal in a Rad51-independent fashion to facilitate recombination. This process results in rapid DNA repair that inevitably deletes intervening sequences between the two homologues that recombine.
- Territories
DNA is highly organized in three dimensions in interphase nuclei, despite the lack of visible chromosomes. Individual chromosomes are spatially restricted to specific domains or territories. In addition, heterochromatin and euchromatin from multiple chromosomes occupy distinct territories that can be visualized by DNA FISH or antibody staining. These types of 3D organization are thought to impact nuclear functions, such as gene expression.
- TE: Transposable element
This is a term that encompasses all mobile DNAs in eukaryotic genomes, including elements that transpose through DNA or RNA intermediates. These elements can insert in protein-coding gene regions, causing mutations or misexpression of the inserted gene and/or flanking genes.
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.John B. The biology of heterochromatin. In: Verma RS, editor. Heterochromatin: Molecular Structural Aspects. Cambridge University Press; 1988. pp. 1–147. [Google Scholar]
- 2.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 3.Hoskins RA, Carlson JW, Kennedy C, Acevedo D, Evans-Holm M, Frise E, Wan KH, Park S, Mendez-Lago M, Rossi F, et al. Sequence finishing and mapping of Drosophila melanogaster heterochromatin. Science. 2007;316:1625–1628. doi: 10.1126/science.1139816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heitz E. Das heterochromatin der moose. Jahrb Wiss Bot. 1928;69:762–818. [Google Scholar]
- 5.Karpen GH, Le MH, Le H. Centric heterochromatin and the efficiency of achiasmate disjunction in Drosophila female meiosis. Science. 1996;273:118–122. doi: 10.1126/science.273.5271.118. [DOI] [PubMed] [Google Scholar]
- 6.Dernburg AF, Sedat JW, Hawley RS. Direct evidence of a role for heterochromatin in meiotic chromosome segregation. Cell. 1996;86:135–146. doi: 10.1016/s0092-8674(00)80084-7. [DOI] [PubMed] [Google Scholar]
- 7.Bernard P, Maure JF, Partridge JF, Genier S, Javerzat JP, Allshire RC. Requirement of heterochromatin for cohesion at centromeres. Science. 2001;294:2539–2542. doi: 10.1126/science.1064027. [DOI] [PubMed] [Google Scholar]
- 8.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 9•.Smith CD, Shu S, Mungall CJ, Karpen GH. The Release 5.1 annotation of Drosophila melanogaster heterochromatin. Science. 2007;316:1586–1591. doi: 10.1126/science.1139815. [DOI] [PMC free article] [PubMed] [Google Scholar]; Annotates a large amount of assembled heterochromatic sequences in Drosophila melanogaster, showing that it is composed predominantly of degenerate transposable elements (>80%) with ~250 protein coding genes. Notably, it shows that heterochromatic genes contain much longer introns on average than euchromatic genes, owing to frequent insertions of transposable elements, which also populate regulatory regions.
- 10.Pearson CE, Edamura KN, Cleary JD. Repeat instability: mechanisms of dynamic mutations. Nat Rev Genet. 2005;6:729–742. doi: 10.1038/nrg1689. [DOI] [PubMed] [Google Scholar]
- 11.Mirkin SM. DNA structures, repeat expansions and human hereditary disorders. Curr Opin Struct Biol. 2006;16:351–358. doi: 10.1016/j.sbi.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 12.Groth A, Rocha W, Verreault A, Almouzni G. Chromatin challenges during DNA replication and repair. Cell. 2007;128:721–733. doi: 10.1016/j.cell.2007.01.030. [DOI] [PubMed] [Google Scholar]
- 13.Pont G, Degroote F, Picard G. Some extrachromosomal circular DNAs from Drosophila embryos are homologous to tandemly repeated genes. J Mol Biol. 1987;195:447–451. doi: 10.1016/0022-2836(87)90665-6. [DOI] [PubMed] [Google Scholar]
- 14.Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gottlieb S, Esposito RE. A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell. 1989;56:771–776. doi: 10.1016/0092-8674(89)90681-8. [DOI] [PubMed] [Google Scholar]
- 16.Blander G, Guarente L. The Sir2 family of protein deacetylases. Annu Rev Biochem. 2004;73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- 17.Shankaranarayana GD, Motamedi MR, Moazed D, Grewal SI. Sir2 regulates histone H3 lysine 9 methylation and heterochromatin assembly in fission yeast. Curr Biol. 2003;13:1240–1246. doi: 10.1016/s0960-9822(03)00489-5. [DOI] [PubMed] [Google Scholar]
- 18•.Kobayashi T, Ganley AR. Recombination regulation by transcription-induced cohesin dissociation in rDNA repeats. Science. 2005;309:1581–1584. doi: 10.1126/science.1116102. [DOI] [PubMed] [Google Scholar]; This study shows that cohesins associated with spacers between rDNA transcription units are displaced by transcribing RNA polymerase I, leading to increased recombination and ecc rDNA formation. This paper shows how transcription and chromatin contents influence repeated DNA stability.
- 19•.Torres-Rosell J, Machin F, Farmer S, Jarmuz A, Eydmann T, Dalgaard JZ, Aragon L. SMC5 and SMC6 genes are required for the segregation of repetitive chromosome regions. Nat Cell Biol. 2005;7:412–419. doi: 10.1038/ncb1239. [DOI] [PubMed] [Google Scholar]; Demonstrates that the Smc5–Smc6 complex physically associates with repeated DNAs during mitosis and is required to prevent the formation of aberrant sister chromatid junctions and chromosome missegregation.
- 20••.Peng JC, Karpen GH. H3K9 methylation and RNA interference regulate nucleolar organization and repeated DNA stability. Nat Cell Biol. 2007;9:25–35. doi: 10.1038/ncb1514. [DOI] [PMC free article] [PubMed] [Google Scholar]; Shows that the H3K9 methylation and RNAi pathway regulate repeated DNA stabilization and three-dimensional organization, and that this regulation occurs via DNA repair pathways.
- 21.Cam HP, Sugiyama T, Chen ES, Chen X, FitzGerald PC, Grewal SI. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat Genet. 2005;37:809–819. doi: 10.1038/ng1602. [DOI] [PubMed] [Google Scholar]
- 22••.Torres-Rosell J, Sunjevaric I, De Piccoli G, Sacher M, Eckert-Boulet N, Reid R, Jentsch S, Rothstein R, Aragon L, Lisby M. The Smc5–Smc6 complex and SUMO modification of Rad52 regulates recombinational repair at the ribosomal gene locus. Nat Cell Biol. 2007;9:923–931. doi: 10.1038/ncb1619. [DOI] [PubMed] [Google Scholar]; Demonstrates that the Smc5–Smc6 complex and components of the homologous recombination pathway physically separate DSBs within rDNA away from the nucleolus, presumably to prevent erroneous recombination between damaged rDNA units. Failure of this process causes increased rDNA recombination and ecc rDNA formation.
- 23•.Slotkin RK, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet. 2007;8:272–285. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]; This excellent review explains in more details how epigenetically mechanisms stabilize transposable in different species.
- 24•.Theurkauf WE, Klattenhoff C, Bratu DP, McGinnis-Schultz N, Koppetsch BS, Cook HA. rasiRNAs, DNA damage, and embryonic axis specification. Cold Spring Harb Symp Quant Biol. 2006;71:171–180. doi: 10.1101/sqb.2006.71.066. [DOI] [PubMed] [Google Scholar]; Demonstrates that mutants in the rasiRNA pathway affect the response to DNA damage in the germline, which in turn affects embryonic axis specification.
- 25.Klattenhoff C, Bratu DP, McGinnis-Schultz N, Koppetsch BS, Cook HA, Theurkauf WE. Drosophila rasiRNA pathway mutations disrupt embryonic axis specification through activation of an ATR/Chk2 DNA damage response. Dev Cell. 2007;12:45–55. doi: 10.1016/j.devcel.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 26.Girard Al, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- 27.Grivna ST, Pyhtila B, Lin H. Miwi associates with translational machinery and Piwi-interacting RNAs (piRNAs) in regulating spermatogenesis. Proc Natl Acad Sci. 2006;103:13415–13420. doi: 10.1073/pnas.0605506103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aravin A, Gaidatzis D, Pfeffer SB, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. A novel class of small RNAs bind to Mili protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- 29.Kuramochi-Miyagawa S, Kimura T, Yomogida K, Kuroiwa A, Tadokoro Y, Fujita Y, Sato M, Matsuda Y, Nakano T. Two mouse Piwi-related genes: Miwi and Mili. Mech Dev. 2001;108:121–133. doi: 10.1016/s0925-4773(01)00499-3. [DOI] [PubMed] [Google Scholar]
- 30.Lippman Z, Gendrel A-V, Black M, Vaughn MW, Dedhia N, Richard McCombie W, Lavine K, Mittal V, May B, Kasschau KD, et al. Role of transposable elements in heterochromatin and epigenetic control. Nature. 2004;430:471–476. doi: 10.1038/nature02651. [DOI] [PubMed] [Google Scholar]
- 31.Carvalho AB, Clark AG. Y chromosome of D. pseudoobscura is not homologous to the ancestral Drosophila Y. Science. 2005;307:108–110. doi: 10.1126/science.1101675. [DOI] [PubMed] [Google Scholar]
- 32•.Yasuhara JC, DeCrease CH, Wakimoto BT. Evolution of heterochromatic genes of Drosophila. PNAS. 2005;102:10958–10963. doi: 10.1073/pnas.0503424102. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study compares the sequence organization of orthologous genes located in heterochromatin versus euchromatin in different Drosophila species. The heterochromatic orthologs contain transposon insertions in promoters and introns, which are not present in species where the gene is located in euchromatin.
- 33.Elder JF, Jr, Turner BJ. Concerted evolution of repetitive DNA sequences in eukaryotes. Q Rev Biol. 1995;70:297–320. doi: 10.1086/419073. [DOI] [PubMed] [Google Scholar]
- 34.Jorgensen AL, Laursen HB, Jones C, Bak AL. Evolutionarily different alphoid repeat DNA on homologous chromosomes in human and Chimpanzee. PNAS. 1992;89:3310–3314. doi: 10.1073/pnas.89.8.3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stern C. Somatic crossing over and segregation in Drosophila melanogaster. Genetics. 1936;21:625–730. doi: 10.1093/genetics/21.6.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mehrotra S, McKim KS. Temporal analysis of meiotic DNA double strand break formation and repair in Drosophila females. PLoS Genetics. 2006 doi: 10.1371/journal.pgen.0020200. preprint. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cowell IG, Sunter NJ, Singh PB, Austin CA, Durkacz BW, Tilby MJ. gammaH2AX Foci form preferentially in euchromatin after ionising-radiation. PLoS ONE. 2007;2:e1057. doi: 10.1371/journal.pone.0001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim J-A, Kruhlak M, Dotiwala F, Nussenzweig A, Haber JE. Heterochromatin is refractory to {gamma}-H2AX modification in yeast and mammals. J Cell Biol. 2007;178:209–218. doi: 10.1083/jcb.200612031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39•.Kruhlak MJ, Celeste A, Dellaire G, Fernandez-Capetillo O, Muller WG, McNally JG, Bazett-Jones DP, Nussenzweig A. Changes in chromatin structure and mobility in living cells at sites of DNA double-strand breaks. J Cell Biol. 2006;172:823–834. doi: 10.1083/jcb.200510015. [DOI] [PMC free article] [PubMed] [Google Scholar]; Performs live cell, immunofluorescence, fluorescence in situ hybridization, and electron micrograph analyses to show that chromatin surrounding double-stranded breaks expands seconds after damage occurs; this phenomenon takes place in euchromatin and heterochromatin with similar kinetics.
- 40.Unal E, Arbel-Eden A, Sattler U, Shroff R, Lichten M, Haber JE, Koshland D. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol Cell. 2004;16:991–1002. doi: 10.1016/j.molcel.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 41.van Attikum H, Fritsch O, Hohn B, Gasser SM. Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell. 2004;119:777–788. doi: 10.1016/j.cell.2004.11.033. [DOI] [PubMed] [Google Scholar]
- 42.Morrison AJ, Highland J, Krogan NJ, Arbel-Eden A, Greenblatt JF, Haber JE, Shen X. INO80 and gamma-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell. 2004;119:767–775. doi: 10.1016/j.cell.2004.11.037. [DOI] [PubMed] [Google Scholar]
- 43.Debatisse M, Coquelle A, Toledo F, Buttin G. Gene amplification mechanisms: the role of fragile sites. Recent Results Cancer Res. 1998;154:216–226. doi: 10.1007/978-3-642-46870-4_13. [DOI] [PubMed] [Google Scholar]
- 44.Flores-Rozas H, Kolodner RD. Links between replication, recombination and genome instability in eukaryotes. Trends Biochem Sci. 2000;25:196–200. doi: 10.1016/s0968-0004(00)01568-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kolomietz E, Meyn MS, Pandita A, Squire JA. The role of Alu repeat clusters as mediators of recurrent chromosomal aberrations in tumors. Genes Chromosomes Cancer. 2002;35:97–112. doi: 10.1002/gcc.10111. [DOI] [PubMed] [Google Scholar]
- 46.Rupa DS, Hasegawa L, Eastmond DA. Detection of chromosomal breakage in the 1cen-1q12 region of interphase human lymphocytes using multicolor fluorescence in situ hybridization with tandem DNA probes. Cancer Res. 1995;55:640–645. [PubMed] [Google Scholar]
- 47.Wong N, Lam W-C, Lai PB-S, Pang E, Lau W-Y, Johnson PJ. Hypomethylation of chromosome 1 heterochromatin DNA correlates with q-Arm copy gain in human hepatocellular carcinoma. Am J Pathol. 2001;159:465–471. doi: 10.1016/S0002-9440(10)61718-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Norwood LE, Moss TJ, Margaryan NV, Cook SL, Wright L, Seftor EA, Hendrix MJ, Kirschmann DA, Wallrath LL. A requirement for dimerization of HP1Hsalpha in suppression of breast cancer invasion. J Biol Chem. 2006;281:18668–18676. doi: 10.1074/jbc.M512454200. [DOI] [PubMed] [Google Scholar]
- 49.Tan J, Yang X, Zhuang L, Jiang X, Chen W, Lee PL, Karuturi RK, Tan PB, Liu ET, Yu Q. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev. 2007;21:1050–1063. doi: 10.1101/gad.1524107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ding L, Kleer CG. Enhancer of Zeste 2 as a marker of preneoplastic progression in the breast. Cancer Res. 2006;66:9352–9355. doi: 10.1158/0008-5472.CAN-06-2384. [DOI] [PubMed] [Google Scholar]


