Abstract
Transcriptional targeting using a tissue-specific cellular promoter is proving to be a powerful means for restricting transgene expression in targeted tissues. In the context of cancer suicide gene therapy, this approach may lead to cytotoxic effects in both cancer and nontarget normal cells. Considering microRNA (miRNA) function in post-transcriptional regulation of gene expression, we have developed a viral vector platform combining cellular promoter–based transcriptional targeting with miRNA regulation for a glioma suicide gene therapy in the mouse brain. The therapy employed, in a single baculoviral vector, a glial fibrillary acidic protein (GFAP) gene promoter and the repeated target sequences of three miRNAs that are enriched in astrocytes but downregulated in glioblastoma cells to control the expression of the herpes simplex virus thymidine kinase (HSVtk) gene. This resulted in significantly improved in vivo selectivity over the use of a control vector without miRNA regulation, enabling effective elimination of human glioma xenografts while producing negligible toxic effects on normal astrocytes. Thus, incorporating miRNA regulation into a transcriptional targeting vector adds an extra layer of security to prevent off-target transgene expression and should be useful for the development of gene delivery vectors with high targeting specificity for cancer therapy.
Introduction
Restricted gene expression through tissue-specific cellular promoter–based transcriptional targeting is an attractive approach to study cell type–specific functions of transgenes and, in gene therapy, to reduce side effects while increasing therapeutic efficacy. This strategy is especially appealing to cancer suicide gene therapies, which employ either toxic genes or genes encoding enzymes that turn prodrugs into toxic compounds. Previous efforts to use tissue-specific promoters or tumor-selective promoters in suicide gene therapy have met with mixed success, due to the lack of the tightness of such promoters in restricting therapeutic gene expression solely to tumor cells.1,2,3 These findings are consistent with the notion that defining tissue specificity is not straightforward when the human genome is pervasively transcribed.4,5 Moreover, cellular promoters may exhibit altered induction patterns due to the altered expression of endogenous regulatory elements under different physiological and pathological conditions, thus losing their tissue or cell-type specificity.1,2,3,6 Hence, additional control strategies may be necessary to attain desired tumor selectivity for cancer suicide gene therapies.
MicroRNAs (miRNAs) have gained widespread attention for their critical functions in post-transcriptional regulation of gene expression. After base pairing to mRNAs, miRNAs mediate post-transcriptional gene repression through induction of deadenylation, decay of target mRNAs and/or repression of protein production by inhibition of translation initiation, block of elongation and/or degradation of nascent peptides.7 To date, ~540 miRNA genes have been indentified in the human genome (http://microrna.sanger.ac.uk). These miRNAs regulate 10–30% of all protein-coding genes in the human genome.8,9 A single miRNA might regulate 100–200 genes and each gene can be targeted by multiple miRNAs, suggesting a complex regulatory network between miRNAs and their mRNA targets.8,9 Recently, elegant studies by Naldini's lab demonstrated that endogenous miRNAs could be broadly exploited to regulate transgene expression in different cell lineages.10,11 The authors incorporated target sequences of endogenous miRNAs into the 3′UTR region of a transgene in a gene transfer vector and reported that when the vector was used to transduce cells with high levels of the tested endogenous miRNAs, transgene expression was effectively suppressed due to the binding of the miRNAs to the introduced miRNA target sequences. This approach opens up great opportunities to control cell type–specific expression of a therapeutic gene from gene transfer vectors, for example, tumor-selective expression of a suicide gene. However, the multiplicity and cooperativity of miRNA regulation pose big challenges for selecting endogenous miRNAs for stringent control of transgene expression.
The full potential of cancer suicide gene therapy will be realized through development of approaches that permit the precise control of suicide gene expression. We reason that the use of a tissue-specific promoter could be a means to direct transgene expression preferentially to cells in a given lineage. Furthermore, the use of miRNA regulation, adjunct to the use of the tissue-specific promoter, would provide the second layer of control to differentiate transgene expression between tumor and normal cells of the same lineage. To test the hypothesis, we constructed a baculoviral vector that harbors a glial fibrillary acidic protein (GFAP) gene promoter for glioma suicide gene therapy in the brain. The goal was to drive suicide gene expression in cells of glial origin, while sparing functionally important neurons and other types of cells in the brain and other organs, in case of local or systemic leakage of intratumorally injected vectors. The expression cassette of the vector further contained the target sequences of three miRNAs that are enriched in astrocytes but downregulated in glioma cells, largely in order to avoid side effects of the vectors on nearby normal astrocytes, without compromising suicide gene expression in tumor cells. We show that this dual control using transcriptional targeting and miRNA regulation resulted in significant reduction of nontarget transgene expression in the brain.
Results
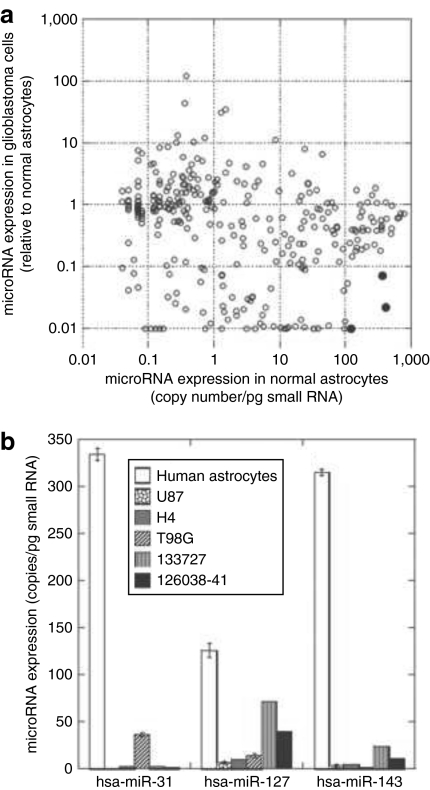
To enable rational design of vectors that are subject to miRNA regulation, we evaluated global expression changes of miRNAs in five human glioma cell lines (U87, U118, U138, H4, and T98G) versus normal human astrocytes using miRNA microarray chips. This allows miRNA profiling at a cellular level, rather than in a tissue with mixed types of cells. We identified 51 upregulated (≥2) and 129 downregulated (≤0.5) miRNAs (Supplementary Table S1). We then determined the copy number of these miRNAs in normal astrocytes based on the average signal intensity of five microarray assays and the absolute copy number of hsa-miR-31 quantified using real-time PCR. Using significant downregulation in glioma cells and high copy number in normal astrocytes as the criteria, as well as taking into consideration the published miRNA expression levels in primary glioma samples and in the normal brain,12,13,14 we selected three miRNAs, hsa-miR-31, hsa-miR-127, and hsa-miR-143, the target sequence of which can possibly be used to shut down transgene expression in normal astrocytes (Figure 1a). Quantitative real-time PCR analysis confirmed high copy numbers for these three miRNAs in normal astrocytes, ranging from around 100 copies for miR-127 to 300 copies for miR-31 and miR-143, whereas in glioma cell lines and primary human glioblastoma tissue samples, the three miRNAs were downregulated (Figure 1b).
Figure 1.
Selection of microRNAs (miRNAs) for regulation of transgene expression. (a) Identification of miRNAs with high copy numbers in normal human astrocytes and significantly downregulated in human glioma cells. We estimated the copy number of different miRNAs in normal human astrocytes based on the average signal intensity of astrocyte samples from five microarray assays and the absolute copy number of hsa-miR-31 quantified using real-time PCR. Three hundred and eight miRNAs that are expressed both in normal astrocytes and five human glioma cell lines (U87, U118, U138, H4, and T98G) are included in the figure. The dots of the three selected miRNAs, hsa-miRNA-31, hsa-miRNA-127, and hsa-miRNA-143, are decorated in black in the lower right corner. (b) miRNA expression analysis by real-time quantitative PCR. The expression of the three selected miRNAs was quantified in normal human astrocytes, three human glioma cell lines (U87, H4, and T98G), and two primary human glioblastoma tissue samples (133727 and 126038-41). miRNA copy numbers were calculated based on a standard curve generated using a synthetic hsa-miR-31 RNA oligonucleotide.
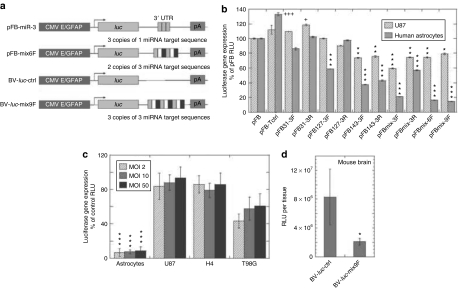
To employ both cell type–specific promoter and miRNA regulation to limit transgene expression in glioma cells, we constructed dual control vectors. We used an engineered GFAP promoter that is able to restrict transgene expression to glial cell lineage15 in the vectors and then introduced miRNA target sequences with perfect complementarity to hsa-miR-31, hsa-miR-127, and/or hsa-miR-143 into the 3′-untranslated region of a transgene (Figure 2a). Three copies of each miRNA target sequence separated by a four nucleotide linker were used as consecutive repeats (Figure 2a, Table 1) and inserted into each vector. We also generated expression cassettes with six and nine copies of the mixed three different types of target sequences (Figure 2a). Because only one restriction enzyme site (HindIII) was used for insertion, we were able to generate and select the constructs with the target sequences in either the same (forward) or opposite (reverse) direction as the endogenous miRNAs.
Figure 2.
Combinatorial effects of a GFAP promoter and miRNA regulation on luciferase transgene expression. (a) Schematic representation of the expression cassettes containing an enhanced astrocyte-specific GFAP promoter and miRNA target sequences. CMV E/GFAP: a hybrid promoter constructed by appending a 380-base pair fragment of the enhancer of human cytomegalovirus immediate-early gene 5′ to the human GFAP promoter. luc, luciferase gene. miRNA target sequences as detailed in Table 1 were inserted into the 3′UTR region. pA, polyA signal. pFastBac plasmid (pFB) vectors with the GFAP promoter and different miRNA target sequences were constructed and tested first. The pFastBac plasmids without miRNA target sequence insertion and with a mix9F sequence were used as shuttle vectors to generate baculoviral vector BV-luc-ctrl and BV-luc-mix9F, respectively. (b) Screening miRNA target sequences that downregulate transgene expression in normal human astrocytes, but not in human U87 glioma cells. The pFB vectors with different miRNA target sequences were used for transfection of U87 cells and normal human astrocytes. Luciferase gene expression was analyzed 1 day after transfection. The results are shown as the percentage of control pFB without miRNA target sequence insertion. *,+P < 0.05, **P < 0.01, ***,+++P < 0.001 versus pFB of the same cell type by analysis of variance (ANOVA). (c) In vitro luciferase transgene expression mediated by baculoviral vectors. Cells were transduced with increased MOI from 2 to 50, and luciferase gene expression was analyzed 1 day after transduction. The results from BV-luc-mix9F are shown as the percentage of BV-luc-ctrl. ***P < 0.001 versus glioma cells at the same MOI by ANOVA. (d) Luciferase transgene expression in the brain mediated by baculoviral vectors. Two days after BV-luc-ctrl or BV-luc-mix9F injection into the striatum of the mouse brain, brain tissues were collected for luciferase activity assays. The results are expressed in relative light units (RLUs) per brain. Columns, means (n = 4); bars, SD. *P < 0.05 by Student's t-test and Tukey test. CMV E, the enhancer of human cytomegalovirus immediate early gene; GFAP, glial fibrillary acidic protein; luc, the firefly luciferase gene; miRNAs, microRNAs; MOI, multiplicity of infection.
Table 1.
miRNA oligomers used in this study
After including the above regulatory elements into pFastBac expression vectors with a firefly luciferase reporter gene (pFBs), we tested the ability of different miRNA target sequences to repress luciferase gene expression after transfection in normal human astrocytes and U87 human glioma cells (Figure 2b). In the tumor cells, levels of inhibition were usually <20%, except a 40% inhibition by a construct with mixed three target sequences and forward orientation. In normal astrocytes, the levels of inhibition were low when the constructs containing three tandem copies of one type of target sequences were employed. Much greater levels of inhibition were achieved by using the constructs bearing a combination of the three miRNA-binding sites, especially when the number of binding sites increased to six or nine copies. Thus, pFBmix6F and pFBmix9F, which have two and three copies of the target sequences for each of the three selected miRNA, respectively, displayed 80% inhibition of transgene expression in normal human astrocytes (Figure 2b). To rule out the possibility of less favorable transcription caused by introduction of a long repeat sequence into the 3′-UTR, we constructed a control vector pFB-Tctrl with a mismatched miRNA targeting sequence having the same length as the mix9F in the 3′UTR of luciferase reporter gene. This sequence was designed based on the lack of significant similarity to any known miRNA (Table 1). The insertion of this mismatched miRNA targeting sequence did not downregulate transgene expression. Instead, we observed a 40% increase in luciferase expression in normal astrocytes when compared with the control vector without any targeting sequence, although the difference in U87 cells was not significant (Figure 2b). This finding demonstrates that insertion of the repeated miRNA target sequences in our vectors does not reduce transcriptional activity. Taken together, our findings suggest that the inhibition of transgene expression in normal astrocytes by pFBmix6F and pFBmix9F is possibly due to the binding of the artificially introduced reverse complements in our expression cassettes to corresponding endogenous miRNAs.
We have previously demonstrated that insect baculoviral vectors are efficient in transducing astrocytes and glioblastoma cells, and suppressing in vivo glioma growth by overexpression of a toxic suicide gene.16 In the current study, we used the most efficient mix9F miRNA target sequence to test whether miRNA regulation was functioning in baculoviral vectors. The baculoviral vector with a luciferase gene and the miRNA target sequence was designated as BV-luc-mix9F and the control baculoviral vector as BV-luc-ctrl (Figure 2a). When comparing the transduction efficiencies of the two vectors in glioma cell lines, we found slight to moderate reduction by incorporation of the miRNA target sequence, from 7% in U87 cells, 14% in H4 cells to 40% in T98G cells (Figure 2c). The different levels of inhibition of transgene gene expression correlated to the relative abundance of the three miRNAs in the three glioma cell lines (Figure 1b), with U87 cells that express the lowest levels of the three miRNAs being much less affected. On the contrary, in normal human astrocytes that express high levels of the three miRNAs, we observed a significant 91% reduction in gene expression level (Figure 2c). To investigate in vivo inhibition, we compared the transduction efficiencies of the two viral vectors in the brain of mice, which, owing to miRNA sequence conservation between human and mouse, express the exact homologues of the three human miRNAs tested in vitro (http://microrna.sanger.ac.uk/sequences/).17 Two days after injection of the viral vectors into the striatum of nude mice, we observed a lower level of luciferase expression from BV-luc-mix9F (Figure 2d). Although the in vivo suppressing effect of the miRNA target sequence was more moderate than that seen in vitro, the inhibition was still approximately fourfold.
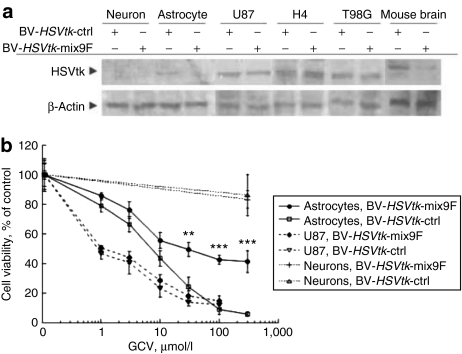
To test whether the mix9F miRNA target sequence is able to suppress the expression of a therapeutic gene, we constructed two baculoviral vectors containing the herpes simplex virus thymidine kinase (HSVtk) suicide gene under the control of the engineered GFAP promoter, one with the mix9F target sequence (BV-HSVtk-mix9F) and one without the target sequence (BV-HSVtk-ctrl). Using western blot analysis (Figure 3a), we detected HSVtk in human U87, H4, and T98G glioblastoma cells, and observed no difference in expression level after transduction of these cells using BV-HSVtk-ctrl and BV-HSVtk-mix9F. In normal human astrocytes and the mouse striatum, we observed HSVtk expression after BV-HSVtk-ctrl transduction, but not after BV-HSVtk-mix9F transduction. There was no detectable HSVtk expression in neurons derived from human embryonic stem cells after either HSVtk-ctrl or BV-HSVtk-mix9F transduction. We further investigated in vitro effects of HSVtk expression followed by ganciclovir (GCV) treatment and found a GCV dose-dependent decrease in cell viability in both normal human astrocytes and U87 cells, but not obviously in human neurons (Figure 3b). In U87 cells, there was no significant difference in cytotoxic potency between BV-HSVtk-ctrl and BV-HSVtk-mix9F, suggesting that incorporation of the mix9F miRNA target sequence does not interfere with HSVtk-related cell killing effects in glioma cells. In normal human astrocytes, the killing curves from the two baculoviral vectors were very different, with BV-HSVtk-ctrl inducing much more pronounced cell death than BV-HSVtk-mix9F at GCV concentrations above 10 µmol/l. At the highest GCV dose tested (300 µmol/l), BV-HSVtk-ctrl induced death in >90% cells, whereas BV-HSVtk-mix9F induced ~55% cell death. The IC50 values obtained by GCV treatment (concentration of GCV that inhibited cell survival by 50%) were 0.8 and 1 µmol/l in U87 glioblastoma cells, and 7 and 30 µmol/l in normal astrocytes after transduction with BV-HSVtk-ctrl and BV-HSVtk-mix9F, respectively, indicating a fourfold improvement of resistance to HSVtk/GCV-mediated cytotoxicity in normal astrocytes. The different cell killing efficiencies by BV-HSVtk-ctrl in U87 cells and normal astrocytes indicate that the inhibition of DNA synthesis by GCV is more toxic for actively proliferating tumor cells. The difference by BV-HSVtk-mix9F between U87 tumor cells and normal astrocytes is at least partially attributed to the presence of the mix9F target sequence as well as the different susceptibilities of the two types of cells to GCV. Taken together, these results demonstrate that when BV-HSVtk-mix9F is used, glioma cells and normal astrocytes display obvious different vulnerabilities to HSVtk/GCV-mediated cellular toxicity.
Figure 3.
Selective cellular effects of controlled expression of a suicide gene. (a) Western blot analysis of HSVtk expression in BV-HSVtk-ctrl- or BV-HSVtk-mix9F-transduced human neurons, astrocytes, and glioblastoma cells, and mouse brain. Human neurons derived from hES1 embryonic stem cells (neurons) were transduced with the baculoviral vectors at a multiplicity of infection (MOI) of 100. Normal human astrocytes (astrocytes) and glioblastoma cells (U87, H4, and T98G) were transduced at an MOI of 50. To prepare mouse brain samples, 107 plaque-forming units of virus particles were injected into the striatum of the mouse brain. (b) Cell viability of BV-HSVtk-ctrl- or BV-HSVtk-mix9F-transduced human neurons, astrocytes, and U87 glioblastoma cells after GCV treatment. **P < 0.01, ***P < 0.001 versus BV-HSVtk-ctrl-transduced astrocytes by analysis of variance. BV, baculoviral vector; GCV, ganciclovir; HSVtk, the herpes simplex virus thymidine kinase gene.
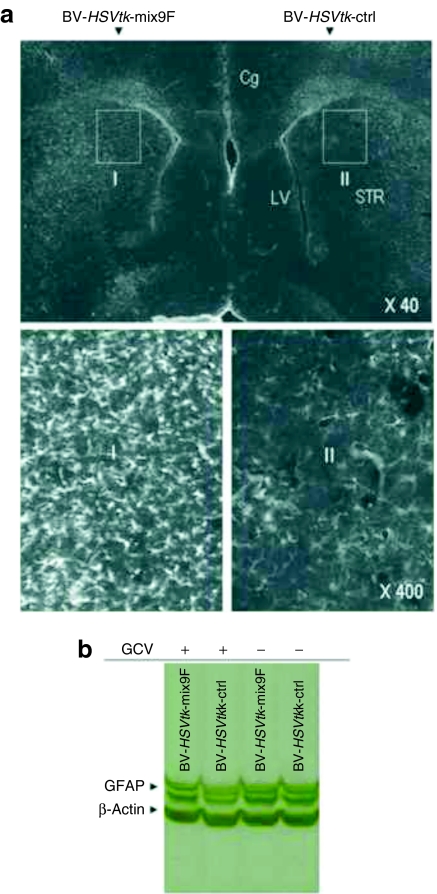
To examine whether the mix9F miRNA target sequence provides in vivo protection against HSVtk/GCV-induced cytotoxicity in the brain, BV-HSVtk-ctrl or BV-HSVtk-mix9F was injected into the striatum of nude mice followed by daily intraperitoneal (i.p.) injection of GCV for 5 days. Immunostaining of the brain sections using an antibody against the HSVtk protein demonstrated the HSVtk gene expression around the injection track, with the width of the transduced region of ~250 µm (Supplementary Figure S1). Immunostaining of the brain sections using an antibody against GFAP demonstrated a significant decrease in the number of GFAP+ cells in the striatum injected with BV-HSVtk-ctrl, but numerous cells were strongly stained with the antibodies against GFAP on the contralateral side of the same brain injected with BV-HSVtk-mix9F (Figure 4a). Similarly, western blot analysis of the striatum tissues revealed noticeable decrease in the staining intensity of GFAP bands in the animals injected with BV-HSVtk-ctrl/GCV, but no obvious change in the GFAP bands in the animals injected with BV-HSVtk-mix9F/GCV (Figure 4b). These results indicate that mix9F-mediated suppression is efficient in protecting normal astrocytes in the brain.
Figure 4.
In vivo effects of controlled expression of a suicide gene in the normal mouse brain. (a) GFAP immunostaining of brain sections from BV-HSVtk-ctrl- and BV-HSVtk-mix9F-injected mice show the protection of GFAP+ astrocytes by incorporating microRNA targeting sequences into baculoviral expression cassette. The enlarged images of region I and II are shown on the right side. (b) Western blot analysis of GFAP expression in the BV-HSVtk-ctrl- and BV-HSVtk-mix9F-injected mouse brain show incorporation of miRNA targeting sequences prevents reduction in protein content of GFAP. BV, baculoviral vector; Cg, the cingulate cortex; GCV, ganciclovir; GFAP, glial fibrillary acidic protein; HSVtk, the herpes simplex virus thymidine kinase gene; LV, the lateral ventricle; STR, the striatum.
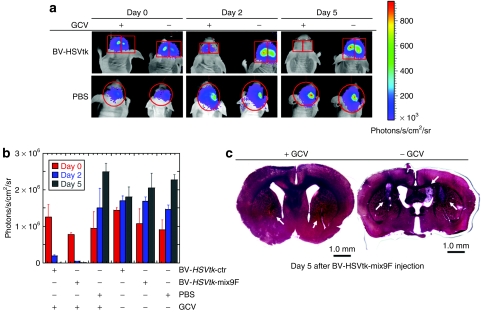
To investigate whether baculoviral vectors with miRNA targeting sequence retained in vivo antitumor effect, U87-luc cells, a modified human glioblastoma cell line that stably expresses luciferase, were inoculated into the striatum of nude mice. This was followed by a single injection of baculoviral vectors into U87 glioblastoma xenografts 7 days later and daily i.p. injection of GCV afterward for 5 days. Tumor growth in the brain of living mice was monitored using a noninvasive bioluminescent imaging method. Figure 5a shows the easily detectable luminescence activity from the inoculated U87-luc cells before virus injection and the continuous increase in luminescence intensity during the 5-day observation period in an animal injected with BV-HSVtk-ctrl and BV-HSVtk-mix9F into the tumor xenografts, but without GCV injection. In animals that received GCV injection after intratumor injection of the baculoviral vectors, there was almost no detectable luciferase activity at day 5 postinjection of the viral vectors (Figure 5a). Quantitative results from a group of four mice for each treatment are summarized in Figure 5b, showing an obvious tumor eradication by a single brain injection of either BV-HSVtk-ctrl or BV-HSVtk-mix9F followed by GCV injection for 5 days, with no significant difference in tumor suppressing effect between these two viral vectors. In two control groups that received phosphate-buffered saline (PBS) injection into the tumor xenografts in the brain, we did not observe any obvious differences in transgene expression between the one that was i.p. injected with GCV and another with PBS (Figure 5b), indicating that GCV i.p. injection alone was not enough to inhibit tumor growth. Tumor growth in the control animals that received virus injection followed by PBS i.p. injection, and tumor growth inhibition in the animals that received virus and GCV injection were confirmed by histological examination (Figure 5c).
Figure 5.
BV-HSVtk-ctrl and BV-HSVtk-mix9F displayed same therapeutic effects on glioblastomas. (a) Representative glioblastoma-bearing mice injected with BV-HSVtk-ctrl (right brain, upper panel), BV-HSVtk-mix9F (left brain, upper panel), and PBS (lower panel). A single injection was given to each of human U87 glioblastoma xenografts in the brain 7 days after tumor inoculation, followed by i.p. GCV or PBS (−GCV) injection for 5 days. Bioluminescence images are shown 0, 2, and 5 days after i.p. injection. The bioluminescence signals from the tumor cells reduced to a background level after baculovirus brain injection followed by GCV i.p. injection for 5 days, but remained after baculovirus brain injection, followed by PBS i.p. injection or PBS brain injection, followed by PBS i.p. injection for 5 days. (b) Quantitative analysis of bioluminescence signals from four animals. Note that the changes of bioluminescence signals in BV-HSVtk-mix9F-injected tumor xenografts have the same tendency as the changes in BV-HSVtk-ctrl-injected tumor xenografts. Columns, means (n = 4); bars, SD. ***P < 0.001 versus the same virus injection without GCV by analysis of variance. (c) Representative pictures of brain sections show U87 xenografts. Left: Five days after GCV injection, only small tumors (arrows) were found in the tissue sections remote from the virus injection site. Right: In the control brain without GCV injection, large and grossly visible tumor masses, with tumor dissemination in the ventricle, were found. BV, baculoviral vector; GCV, ganciclovir; HSVtk, the herpes simplex virus thymidine kinase gene; i.p., intraperitoneal injection; PBS, phosphate-buffered saline.
Discussion
Gene therapy paradigms often use selective targeting strategy to control expression of therapeutic genes. In clinical suicide gene therapy trials, direct intratumoral injection of vectors expressing a toxic suicide gene confines therapeutic gene expression to the tumor tissue. However, because natural infection spectrums of most viruses are not confined to the tumor tissue, potential leakage of intratumorally injected viral vectors into normal tissues leads to concern over collateral damage to healthy cells. Although such damage is tolerable in certain organs, it might have severe consequences in a sensitive organ like the brain.
To add further protection, tissue-specific promoters and tumor-selective promoters have been tested in cancer suicide gene therapies to minimize killing effects on nontarget normal cells. Differentiation between a tissue-specific promoter and a tumor-selective promoter relies on the relative activity of the promoter in normal and tumor tissues, but the dividing line between them is rather blurred.1 In this study, we have used an astrocyte-specific GFAP promoter, which has been extensively tested for selective expression of therapeutic genes, including the HSVtk gene, in glioma cells.16,18,19,20 GFAP+ normal glial cells have a relatively lower rate of cell division than highly proliferating tumor cells, thus being less sensitive to DNA synthesis inhibition by phosphorylated GCV. However, tumor growth will cause tissue injury, a procedure that can efficiently induce proliferation of nearby normal glial cells. Moreover, phosphorylated GCV can damage both nuclear and mitochondrial DNA;21,22 thus, HSVtk/GCV treatment is also toxic to nondividing cells or cells at low proliferation rates, for example, normal cultured astrocytes23 and glial cells in the small intestine.24 These findings emphasize the need to use other control mechanisms to protect normal glial cells, which are abundant in both the central nervous system and the peripheral nervous system, and play a crucial role in supporting the survival and physiological functions of neurons.
The tissue expression patterns of miRNAs and the roles of these small RNAs in organ development have been defined. Some miRNAs are expressed widely in the human body, whereas others display restricted expression in stage-, tissue-, or cell type–specific manners. They control almost every cellular process investigated so far, including proliferation, stem cell division, developmental timing, apoptosis, differentiation, and metabolism.25,26 Consequently, impaired miRNA expression may lead to loss of differentiation (or dedifferentiation), resulting in the occurrence of cancer.27,28 Moreover, 50% of miRNA genes map within cancer-associated genomic regions or fragile sites of chromosomes,29 providing a reasonable explanation for deregulated expression of miRNAs in various cancers.27 miRNA profiling has been found to be informative in classifying human cancers, with most of miRNAs in primary tumor tissues and cancer cell lines being downregulated compared with normal tissues.30 Consistent with the notion that miRNA expression is closely linked to differentiation, poorly differentiated tumors have lower global levels of miRNA expression and global expression of miRNAs increases with differentiation of the tumor cells.30 These expression features of miRNAs suggest the possibility of exploiting miRNA regulation to generate distinctive transgene expression patterns in normal and tumor cells.
Previous miRNA profiling studies for gliomas have consistently utilized normal brain tissue as the control sample. The earliest study by Ciafrè et al.12 compared miRNA expression in 10 glioma cell lines using normal adult brain RNA as a control for microarray profiling. The same study also compared miRNA profiles in primary gliomas to matched normal brain tissues from the same patient. Another study used primary glioma samples and human non-neoplastic brain tissues in expression profiling of 192 miRNAs by quantitative PCR.31 Yet another study did a similar comparison between primary tumor samples and normal adjacent brain using microarray technology.32 The current study identified miRNAs that are differentially expressed in glioma cell lines at a cellular level by comparing glioma cells with normal human astrocytes, the normal cellular counterpart of gliomas (Supplementary Table S1). Profiling at a cellular level, but not at a tissue level, is because of our interest in identifying differentially expressed miRNAs that can be used to regulate transgene expression in different cell types. The use of whole adult brain tissue, in which both neuronal and glial cells are predominant, precludes a direct comparison between the current study and the previous ones. Nonetheless, the expression of several miRNAs is still found to be commonly altered between the previous studies and our study. These include downregulated miRNAs: hsa-miR-128, hsa-miR-137, and hsa-miR-299 in Ciafrè et al.'s study;12 hsa-miR-31, hsa-miR-107, hsa-miR-132, hsa-miR-133a, hsa-miR-133b, hsa-miR-154*, hsa-miR-323, hsa-miR-330, hsa-miR-127, and hsa-miR-134 in Silber et al.'s study;31 and hsa-miR-181a and hsa-miR-181b in Godlewski et al.'s study.32 Among the upregulated miRNAs, hsa-miR-10b has been commonly identified by all studies including this one. Of note, even among the three miRNA profiling results previously published, there is limited overlap, suggesting that the method of profiling and sample origin can contribute to large variations in results.
MiRNAs regulation depends on the binding of the first 2–8 bases of their mature sequence (so called “miRNA seed”) to the 3′UTR of target genes, suggesting that there might be many possible target sites for one miRNA seed sequence in different 3′UTR regions. In fact, bioinformatic studies have revealed that a single miRNA might bind to as many as 200 gene targets with diverse functions.27 On the other hand, it has been demonstrated that in the 3′UTR of a single target gene there could be several sites that are complementary to different miRNAs, indicating a single gene can be subject to the regulation of multiple miRNAs. When it comes to usage of miRNA-mediated gene regulation to direct transgene expression, such multiplicity and cooperativity in miRNA regulation could make it highly challenging to select proper endogenous miRNAs suitable for targeted transgene expression. This is especially true for miRNA-regulated transgene expression in tumor tissues, where both the genetic heterogeneity of tumors and the cellular heterogeneity within a tumor tissue would contribute to global transcript profiles of a particular type of tumors. Once miRNA regulation is used adjunct to the use of a tissue-specific promoter, the miRNA selection procedure is simplified to the extent that one needs to compare only miRNA expression profiles between tumor cells and their normal counterparts in a given lineage. Thus, those miRNAs that are downregulated in abnormal tumor cells, but have a high copy number in normal cells, are selected.
Our findings in the current study indicate that the level of repression related to a mixed miRNA target sequence increased from approximately three- to sixfold as the number of the miRNA-binding site increased from three copies to six or nine copies. These results suggest the number of miRNA-binding sites in a target mRNA as a determinant in controlling transgene repression, most likely by increasing the probability of a single necessary repression event to take place. The high efficiency of the mix9F target sequence, in principle, closely mirrored that of miRNA regulation in metazoan cells, where repression of target mRNAs has been shown to be accomplished by the combinatorial action of different miRNA species.33 Each miR-31-binding site placed into a plasmid vector and tested in the present study contributed ~8% inhibition, miR-127 ~14%, and miR-143 ~21% in normal astrocytes. Assuming that the relative effectiveness of each site remained the same, as the number of binding sites increases, the expected total repression achieved by the mix9F target sequence will be ~43%. However, the actual level of repression achieved by using this combined miRNA target sequence was ~80% in normal astrocytes, suggesting that different miRNAs can act cooperatively to reduce transgene expression.
Although only a handful of miRNA:mRNA interaction pairs are known in animals, there are already instances in which different miRNA species have been found to regulate the same targets, for example, the miRNAs lin-4 and let-7, and their target mRNAs lin-14, lin-28, lin-41, and hbl-1.34,35,36,37 The silencing complexes that were recruited could either mutually stabilize one another or cooperatively interact to more effectively inhibit transgene expression, or both mechanisms might be operative at the same time.38 These examples, as with other biological regulatory systems, most notably cooperative interactions in gene regulation, would allow a cell to fine-tune the expression of an mRNA by regulating the degree of binding of different miRNAs to the 3′UTRs of the mRNA.34 Our present study further shows that this property of miRNA regulation can be beneficially used to enhance the suppression of transgene gene expression.
In conclusion, this study provides evidence that incorporating miRNA regulation into a transcriptional targeting vector can provide an important layer of control over transgene expression. This dual regulation system for transgene expression is closely analogous to regulation of gene expression by natural mechanisms at the transcriptional and post-transcriptional levels. Crucial steps to developing such a system include the selection of a relevant tissue-specific promoter and the determination of relative miRNA expression in tumor cells and their normal counterparts, thus selecting those miRNAs that are downregulated in tumor cells, but have a high copy number in normal cells. Looking ahead, our approach can be expanded for other tumor types, for example, using the target sequences of the selected miRNAs together with the tyrosinase promoter for melanoma or the prostate-specific antigen promoter for prostate cancer.
Materials and Methods
Cell culture and tissue samples. Human cells used in this study were U87, H4, T98G, U118, and U138 glioblastoma cell lines from American Type Culture Collection (Manassas, VA), normal human astrocytes from Clonetics primary cell systems (Lonza, Basel, Switzerland). Tumor cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum at 37 °C, 5% CO2. Normal human astrocytes were cultured in special astrocyte basal medium supplemented with the EGM SingleQuots (Lonza). U87-luc cells that stably express luciferase were established as described before.16 HES-1 human embryonic stem cells were used to generate human neurons as described previously.39 Two primary human glioblastoma tissue samples, Glioblastoma 133727 and Glioblastoma 126038-41, were obtained from Capital Biosciences (Gaithersburg, MD).
miRNA expression analysis. Small RNAs were isolated using PureLink miRNA isolation kit (Invitrogen, Carlsbad, CA). The NCode multi-species miRNA microarray version 2 (Invitrogen) was used for high-throughput analysis. The array consists of optimized probe sequences for all mature miRNAs catalogued in miRBase Sequence Database, release 9.0 (ref. 40), with 553 triplicate probes corresponding to human miRNAs. Intensity data were acquired using GenePix Pro 6.0 software (Molecular Devices, Sunnyvale, CA) and the resultant *.GPR files were imported to GeneSpring GX 7.3 (Silicon Genetics, Redwood City, CA) for further analysis. Biological triplicate slides for each combination of glioblastoma cell line versus normal human astrocytes were grouped and averaged. To obtain reliable data only for miRNA probes that are considered present, each data file under consideration was filtered on “Flag filter” to exclude absent probe spots. To identify differentially expressed miRNAs in glioblastoma cell lines, a fold-change cutoff of 2×, (intensity ratio is <0.5 or >2), was used, and further stringency filter for statistical significance (t-test P value <0.05) was applied.
Real-time PCR of miRNAs was performed based on a reported protocol.41 Total RNA was isolated and treated with TURBO DNA-free DNase (Ambion, Austin, TX). The DNAse-treated total RNA (1 µg) was polyadenylated with ATP by poly(A) polymerase using Poly(A) Tailing Kit (Ambion). The purified polyadenylated RNA was reverse transcribed with 200 U Superscript III Reverse Transcriptase (Invitrogen) and 0.5 µg poly (T) adapter [3′rapid amplification of complementary DNA ends (RACE) adapter in the FirstChoice RLM-RACE kit; Ambion]. The forward primer for real-time PCR analysis was designed based on entire known mature miRNA sequence, with additional 3 “A”s at the 3′ end to improve amplification specificity. The sequence of the primers used are as follows: hsa-miR-31 (5′-GGCAAGAUGCUGGCAUAGCUGAAA), hsa-miR-127 (5′-UCGGA UCCGUCUGAGCUUGGCUAAA), hsa-miR-143 (5′-AGAUGAAGCAC UGUAGCUCAAA), and 5S (5′-CCGCCTGGGAATACCGGGTGCTGT AGGCTTT). The reverse primer used was a 3′ adapter primer (3′ RACE outer primer in the FirstChoice RLM-RACE kit). 5S rRNA was selected as the internal reference gene for PCR quantification. To determine absolute copy number, a standard curve was generated using a synthetic miR-26a RNA oligonucleotide. Real-time PCR was performed on Rotor-Gene 6000 (Corbett Life Science, Sydney, Australia). All reactions were run in triplicate.
Construction of gene transfer vectors. To construct plasmid vectors, a recombinant pFastBac1 vector, pFB-CMV E/GFAP-luc previously generated by our lab and containing an engineered cell type–specific promoter,15,16 was used as a starting backbone. This construct contains the firefly luciferase (luc) gene under the control of the promoter of GFAP encoding gene, the activity of which is augmented by placing the enhancer of human cytomegalovirus immediate early gene (CMV E) upstream of the GFAP promoter.15 The HindIII site right after the luc gene in the pFB-CMV E/GFAP-luc vector was used to insert miRNA target sequences. Four types of oligonucleotides were designed as 3X-miR-31, 3X-miR-127, 3X-miR-143, and Mix-mir. The first three contains three tandem copies of a sequence (in lowercase letters in Table 1) designed to be perfectly complementary to the respective miRNA, whereas the Mix-mir construct contains one copy each of the three different miRNA complementary sequence in the order given (Table 1). Each miRNA target sequence is separated by a four nucleotide linker. The respective phosphorylated top and bottom strands of the oligonucleotides were annealed and subcloned into the Hind III site. Constructs containing varying copies of miRNA complementary sequence in both the forward and reverse orientations were selected after sequencing. A control miRNA target sequence (Tctrl, 237 base-pair long) was generated by repeating a 73 base-pair fragment of oligoneucleotides that was designed based on the lack of significant similarity to any known miRNA three times with HindIII sites between repeats. To generate expression cassettes containing the HSVtk gene, its complementary DNA was obtained through PCR amplification from a pFastBac1 vector we constructed previously42 and used to replace the luc gene in the above cassettes. Baculoviral vectors were generated using the Bac-to-Bac baculovirus expression system (Invitrogen) as described.16
In vitro transfection, transduction, transgene expression analysis, and cytotoxicity assay. Cells were seeded in 48-well plate at a density of 20,000 per well one day before transfection. Transfection was performed using Lipofectamine 2000 (Invitrogen). For baculoviral transduction, cells were incubated with baculoviruses for 1 hour, followed by removal of viruses and addition of normal growth medium. Cells were further cultured at 37 °C for 1 day before used for transgene expression analysis. Ten microliters of the cell extract was used for luciferase assay in a single-tube luminometer (Berthold Lumat LB 9507; Berthold Technologies, Bad Wildbad, Germany). The HSVtk gene expression was examined using western blot analysis. Samples of cell extracts and the supernatants of tissue homogenates were taken for protein determination using a protein assay kid from Bio-Rad Laboratories (Hercules, CA). Five micrograms of total protein was separated on 4–12% NuPAGE gradient gels (Invitrogen). The primary antibody used was a rabbit polyclonal antibody for HSVtk (kindly provided by William Summers, Yale University, New Haven, CT). β-Actin was used as a loading control and was detected using mouse monoclonal antibody against β-actin (Sigma, St Louis, MO).
For cytotoxicity assays, cells were seeded in 48-well plates at a density of 10,000 cells per well. After allowing cells to grow overnight, the medium was changed to fresh growth medium containing various concentrations of GCV (InvivoGen, San Diego, CA). After 1-day incubation at 37 °C, cells were infected with 50 multiplicity of infection of baculoviral vectors (BV-HSVtk-ctrl and BV-HSVtk-mix9F). Cell proliferation assays, with triplicate determinations for each treatment, were performed 6 days after virus infection. The relative cell growth (%) related to control cells (infected cells without GCV treatment) was calculated by (absorbance of sample − absorbance of blank)/(absorbance of control − absorbance of blank) × 100%.
Animal experiments. Adult female Balb/c nude athymic immunoincompetent nude mice (weighing 20 g, 6–8 weeks old) were used. For in vivo transgene expression analysis, 3 µl of baculovirus (107 plaque-forming units of virus particles) in PBS was injected into each side of the striatum of the mouse brain (anteroposterior: 0.0 mm, mediolateral: +2.0 mm, and dorsoventral: −3.0 mm from bregma and dura) using a 10-µl Hamilton syringe connected with a 30G needle. Brain tissue samples were collected 2 days later for analysis. For experiments investigating effects of HSV-tk/GCV treatment on the normal brain, brain injection of baculoviral vector was followed by daily i.p. injection of either GCV at a dose of 50 mg/kg or PBS for 5 days. For western blotting analysis of GFAP expression in the mouse brain, samples were prepared as described above, and 5 µg of total protein was separated on 4–12% NuPAGE gradient gels (Invitrogen). The primary antibody used was a rabbit polyclonal antibody against GFAP (Chemicon, Temecula, CA). β-Actin was used as a loading control and was detected using mouse monoclonal antibody against β-actin (Sigma). For experiments investigating therapeutic effects, mice were first inoculated with 0.5 × 106 of human glioma U87-luciferase (U87-luc) cells per side into both sides of the striatum. One week later, baculoviral vectors were injected into the tumors, followed by daily i.p. injection of GCV for 5 days. Tumor growth in the mouse brains were monitored by bioluminescent imaging of U87-luc cells after i.p. injection of D-luciferin (Promega, Madison, WI) at 100 mg/kg. Bioluminescent imaging was then performed with the IVIS imaging system coupled with cool CCD camera (Xenogen, Alameda, CA), and bioluminescent signals were measured by subtracting the background signal from the ROI signal. In the handling and care of animals, the Guidelines on the Care and Use of Animals for Scientific Purposes issued by National Advisory Committee for Laboratory Animal Research, Singapore was followed.
Statistical analysis. All data are represented as mean ± SD. The statistical significance of differences was determined by Student's t-test or the two-factor analysis of variance with replication followed by Tukey post hoc analysis. A P value of <0.05 was considered to be statistically significant.
SUPPLEMENTARY MATERIALFigure S1. Baculovirus-mediated transgene expression in the brain of nude mice.Table S1. Up- and down-regulated miRNAs in U87, U118, U138, H4 and T98G human glioma cell lines.
Supplementary Material
Baculovirus-mediated transgene expression in the brain of nude mice.
Up- and down-regulated miRNAs in U87, U118, U138, H4 and T98G human glioma cell lines.
Acknowledgments
This work was supported by Institute of Bioengineering and Nanotechnology, Biomedical Research Council, and Agency for Science, Technology and Research in Singapore.
REFERENCES
- Harrington KJ, Linardakis E., and , Vile RG. Transcriptional control: an essential component of cancer gene therapy strategies. Adv Drug Deliv Rev. 2000;44:167–184. doi: 10.1016/s0169-409x(00)00093-4. [DOI] [PubMed] [Google Scholar]
- Robson T., and , Hirst DG. Transcriptional targeting in cancer gene therapy. J Biomed Biotechnol. 2003;2003:110–137. doi: 10.1155/S1110724303209074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saukkonen K., and , Hemminki A. Tissue-specific promoters for cancer gene therapy. Expert Opin Biol Ther. 2004;4:683–696. doi: 10.1517/14712598.4.5.683. [DOI] [PubMed] [Google Scholar]
- Yang J, Su AI., and , Li WH. Gene expression evolves faster in narrowly than in broadly expressed mammalian genes. Mol Biol Evol. 2005;22:2113–2118. doi: 10.1093/molbev/msi206. [DOI] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, Margulies EH, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoff-Khalili MA, Rivera AA, Nedeljkovic-Kurepa A, DeBenedetti A, Li XL, Odaka Y, et al. Cancer-specific targeting of a conditionally replicative adenovirus using mRNA translational control. Breast Cancer Res Treat. 2008;108:43–55. doi: 10.1007/s10549-007-9587-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN., and , Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight. Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C., and , Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB., and , Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Brown BD, Venneri MA, Zingale A, Sergi Sergi L., and , Naldini L. Endogenous microRNA regulation suppresses transgene expression in hematopoietic lineages and enables stable gene transfer. Nat Med. 2006;12:585–591. doi: 10.1038/nm1398. [DOI] [PubMed] [Google Scholar]
- Brown BD, Gentner B, Cantore A, Colleoni S, Amendola M, Zingale A, et al. Endogenous microRNA can be broadly exploited to regulate transgene expression according to tissue, lineage and differentiation state. Nat Biotechnol. 2007;25:1457–1467. doi: 10.1038/nbt1372. [DOI] [PubMed] [Google Scholar]
- Ciafrè SA, Galardi S, Mangiola A, Ferracin M, Liu CG, Sabatino G, et al. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun. 2005;334:1351–1358. doi: 10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- Saito Y, Liang G, Egger G, Friedman JM, Chuang JC, Coetzee GA, et al. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CY., and , Wang S. Astrocytic expression of transgene in the rat brain mediated by baculovirus vectors containing an astrocyte-specific promoter. Gene Ther. 2006;13:1447–1456. doi: 10.1038/sj.gt.3302771. [DOI] [PubMed] [Google Scholar]
- Wang CY, Li F, Yang Y, Guo HY, Wu CX., and , Wang S. Recombinant baculovirus containing the diphtheria toxin A gene for malignant glioma therapy. Cancer Res. 2006;66:5798–5806. doi: 10.1158/0008-5472.CAN-05-4514. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S., and , Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Bezdek T, Chang J, Kherzai AW, Willingham T, Azzara M, et al. A glial-specific, repressible, adenovirus vector for brain tumor gene therapy. Cancer Res. 1998;58:3504–3507. [PubMed] [Google Scholar]
- McKie EA, Graham DI., and , Brown SM. Selective astrocytic transgene expression in vitro and in vivo from the GFAP promoter in a HSV RL1 null mutant vector—potential glioblastoma targeting. Gene Ther. 1998;5:440–450. doi: 10.1038/sj.gt.3300621. [DOI] [PubMed] [Google Scholar]
- Horst M, Brouwer E, Verwijnen S, Rodijk M, de Jong M, Hoeben R, et al. Targeting malignant gliomas with a glial fibrillary acidic protein (GFAP)-selective oncolytic adenovirus. J Gene Med. 2007;9:1071–1079. doi: 10.1002/jgm.1110. [DOI] [PubMed] [Google Scholar]
- Herraiz M, Beraza N, Solano A, Sangro B, Montoya J, Qian C, et al. Liver failure caused by herpes simplex virus thymidine kinase plus ganciclovir therapy is associated with mitochondrial dysfunction and mitochondrial DNA depletion. Hum Gene Ther. 2003;14:463–472. doi: 10.1089/104303403321467225. [DOI] [PubMed] [Google Scholar]
- van der Eb MM, Geutskens SB, van Kuilenburg AB, van Lenthe H, van Dierendonck JH, Kuppen PJ, et al. Ganciclovir nucleotides accumulate in mitochondria of rat liver cells expressing the herpes simplex virus thymidine kinase gene. J Gene Med. 2003;5:1018–1027. doi: 10.1002/jgm.450. [DOI] [PubMed] [Google Scholar]
- Maron A, Havaux N, Le Roux A, Knoops B, Perricaudet M., and , Octave JN. Differential toxicity of ganciclovir for rat neurons and astrocytes in primary culture following adenovirus-mediated transfer of the HSVtk gene. Gene Ther. 1997;4:25–31. doi: 10.1038/sj.gt.3300353. [DOI] [PubMed] [Google Scholar]
- Bush TG, Savidge TC, Freeman TC, Cox HJ, Campbell EA, Mucke L, et al. Fulminant jejuno-ileitis following ablation of enteric glia in adult transgenic mice. Cell. 1998;93:189–201. doi: 10.1016/s0092-8674(00)81571-8. [DOI] [PubMed] [Google Scholar]
- Bushati N., and , Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- Kloosterman WP., and , Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A., and , Slack FJ. Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- Rosenfeld N, Aharonov R, Meiri E, Rosenwald S, Spector Y, Zepeniuk M, et al. MicroRNAs accurately identify cancer tissue origin. Nat Biotechnol. 2008;26:462–469. doi: 10.1038/nbt1392. [DOI] [PubMed] [Google Scholar]
- Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Silber J, Lim DA, Petritsch C, Persson AI, Maunakea AK, Yu M, et al. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med. 2008;6:14. doi: 10.1186/1741-7015-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godlewski J, Nowicki MO, Bronisz A, Williams S, Otsuki A, Nuovo G, et al. Targeting of the Bmi-1 oncogene/stem cell renewal factor by microRNA-128 inhibits glioma proliferation and self-renewal. Cancer Res. 2008;68:9125–9130. doi: 10.1158/0008-5472.CAN-08-2629. [DOI] [PubMed] [Google Scholar]
- Bartel DP., and , Chen CZ. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet. 2004;5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Abrahante JE, Daul AL, Li M, Volk ML, Tennessen JM, Miller EA, et al. The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Dev Cell. 2003;4:625–637. doi: 10.1016/s1534-5807(03)00127-8. [DOI] [PubMed] [Google Scholar]
- Lin SY, Johnson SM, Abraham M, Vella MC, Pasquinelli A, Gamberi C, et al. The C elegans hunchback homolog, hbl-1, controls temporal patterning and is a probable microRNA target. Dev Cell. 2003;4:639–650. doi: 10.1016/s1534-5807(03)00124-2. [DOI] [PubMed] [Google Scholar]
- Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E., and , Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG, Petersen CP., and , Sharp PA. siRNAs can function as miRNAs. Genes Dev. 2003;17:438–442. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng J, Du J, Zhao Y, Palanisamy N., and , Wang S. Baculoviral vector-mediated transient and stable transgene expression in human embryonic stem cells. Stem Cells. 2007;25:1055–1061. doi: 10.1634/stemcells.2006-0616. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A., and , Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R., and , Chiang VL. Facile means for quantifying microRNA expression by real-time PCR. BioTechniques. 2005;39:519–525. doi: 10.2144/000112010. [DOI] [PubMed] [Google Scholar]
- Balani P, Boulaire J, Zhao Y, Zeng J, Lin J., and , Wang S. High mobility group box2 promoter-controlled suicide gene expression enables targeted glioblastoma treatment. Mol Ther. 2009;17:1003–1011. doi: 10.1038/mt.2009.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Baculovirus-mediated transgene expression in the brain of nude mice.
Up- and down-regulated miRNAs in U87, U118, U138, H4 and T98G human glioma cell lines.