Abstract
Human immunodeficiency virus (HIV) gene therapy offers a promising alternative approach to current antiretroviral treatments to inhibit HIV-1 infection. Various stages of the HIV life cycle including pre-entry, preintegration, and postintegration can be targeted by gene therapy to block viral infection and replication. By combining multiple highly potent anti-HIV transgenes in a single gene therapy vector, HIV-1 resistance can be achieved in transduced cells while prohibiting the generation of escape mutants. Here, we describe a combination lentiviral vector that encodes three highly effective anti-HIV genes functioning at separate stages of the viral life cycle including a CCR5 short hairpin RNA (shRNA) (pre-entry), a human/rhesus macaque chimeric TRIM5α (postentry/preintegration), and a transactivation response element (TAR) decoy (postintegration). The major focus on designing this anti-HIV vector was to block productive infection of HIV-1 and to inhibit any formation of provirus that would maintain the viral reservoir. Upon viral challenge, potent preintegration inhibition of HIV-1 infection was achieved in combination vector–transduced cells in both cultured and primary CD34+ hematopoietic progenitor cell (HPC)–derived macrophages. The generation of escape mutants was also blocked as evaluated by long-term culture of challenged cells. The ability of this combination anti-HIV lentiviral vector to prevent HIV-1 infection, in vitro, warrants further evaluation of its in vivo efficacy.
Introduction
Human immunodeficiency virus (HIV) infection continues to spread worldwide in both developed and underdeveloped countries with no effective vaccine available. Current antiretroviral drug therapies have been successful in suppressing viral infection as long as the patient is compliant with the prescribed regimen. However, with prolonged use, these treatments can become toxic and drug-resistant viral escape mutants arise.1,2,3,4,5 New and innovative therapies need to be developed that overcome the limitations of current small drug antiretrovirals. Gene therapy offers a promising alternative or supplement to current treatments due to advantages that include the possibility of a one-time treatment, controlled or constitutive anti-HIV gene expression, and long-term viral inhibition particularly if hematopoietic progenitor cells (HPCs) are targeted. Many anti-HIV genes have been evaluated for their efficacy in inhibiting viral infection including antisense RNAs, ribozymes, RNA decoys, small interfering RNAs, intrabodies, transdominant proteins, and restriction factors.6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23 These molecules have been targeted to both viral genes and proteins, and also cellular genes critical for viral infection and replication. Several groups, including ours, have been involved with human clinical trials using a select number of these anti-HIV genes transferred into HPCs by retroviral and lentiviral vectors.13,15,16,17,23 However, improvements in stem cell transduction efficiency and in the effectiveness of the vector to interfere with different stages of the HIV life cycle are still needed.
Pre-entry and preintegration inhibition of infection is an ideal method to elicit resistance to HIV-1 infection. Therapies aimed at blocking HIV-1 integration will combat the generation of provirus and further establishment of a viral reservoir, which is the main reason for the failure to cure an HIV-infected individual. A number of preintegration anti-HIV transgenes have been previously tested as individual constructs and have displayed strong resistance to HIV-1 infection.5,8,9,10,14,18,22 By combining multiple anti-HIV genes in a single vector, a novel anti-HIV vector could be produced that not only provides strong inhibition of HIV-1 infection but would also provide a block to the generation of escape mutants by creating a difficult environment for HIV to mutate around the multiple anti-HIV molecules.
In the initial stages of HIV-1 infection, attachment and fusion to target cells occur via interaction of the viral envelope gp120 and gp41 glycoproteins with the cellular major receptor CD4 and a minor co-receptor, two of these being CCR5 and CXCR4 that are members of the chemokine receptor family. CCR5 is utilized by R5-tropic strains of HIV-1 primarily during the initial stages of infection, followed by a switch in tropism to X4-tropic HIV-1 that is mainly detected during late-stage infection.24 In a small percentage of the human population, a mutant allele of the CCR5 gene, containing a 32-base-pair (bp) deletion, renders the protein defective, and therefore, the receptor is absent from the cell surface. Homozygous and heterozygous individuals harboring this allele have been reported to be resistant to HIV-1 infection and remain physiologically normal due to receptor redundancy in the chemokine system.25,26,27 Recently, long-term control of viral replication was observed in an HIV-1-infected individual who received a stem cell transplant for acute myeloid leukemia.28 The transplanted allogeneic stem cells were from an individual who was homozygous for the CCR5 Δ32-bp deletion. The results provided from this study demonstrate the importance in developing anti-HIV molecules that block the use of CCR5 during HIV-1 infection. Based on this natural phenotype of CCR5 null, CCR5 knockdown for HIV gene therapy offers a promising approach to inhibit viral infection at the level of viral entry.
The mechanism of RNA interference using small interfering RNAs is a highly potent method to silence gene expression and offers an ideal approach to knock down expression of CCR5 (ref. 29). Numerous reports have evaluated the efficacy of CCR5 gene knockdown using small interfering RNAs and have demonstrated protection from HIV-1 infection.8,9,14,18 However, the silencing of CCR5 gene expression, alone, will not be sufficient to completely inhibit HIV infection and also would not inhibit infection from X4-tropic or dual-tropic viral strains. Other anti-HIV strategies added to the CCR5 knockdown would therefore greatly enhance viral protection.
Another naturally occurring molecule, TRIM5α, has been shown to inhibit HIV-1 infection at the postentry/preintegration stage by disrupting the uncoating of the viral capsid upon entering the cytoplasm.30 Certain isoforms of TRIM5α found in Old World monkeys are capable of strongly restricting HIV-1 infection. Humans also naturally express a distinct isoform of TRIM5α, but it does not afford protection from HIV-1 infection. A recently developed human/rhesus macaque chimeric TRIM5α isoform, incorporating a small number of key HIV-restrictive amino acids, was demonstrated to inhibit HIV-1 infection in a hematopoietic stem cell gene therapy setting.22,31 If used in a clinical application, the design of this chimeric TRIM5α molecule, which consists of mainly human amino-acid sequences, will help to avoid immune rejection that would occur with the use of wild-type rhesus macaque TRIM5α. A third molecule, a transactivation response element (TAR) decoy, has been previously described to inhibit transactivation of proviral transcription.7 By mimicking the structure of the viral TAR, the TAR decoy is able to bind the viral Tat protein and sequester it away from its normal action of aiding efficient proviral HIV transcription.
In the present study, we describe the construction and preclinical evaluation of a triple combination anti-HIV lentiviral vector that focuses on the preintegration block of HIV-1 infection to minimize the formation of integrated provirus and the generation of escape mutants. The three highly potent anti-HIV transgenes, a chimeric TRIM5α molecule, a CCR5 short hairpin RNA (shRNA) capable of almost complete knockdown of CCR5 expression, and a TAR decoy, combined in a single vector, displayed complete protection from productive viral infection and integration of multiple strains of HIV-1 upon transduction into HIV target cells. Our results establish the future application of this vector for use in a clinical setting.
Results
Vector production, expression of transgenes, vector stability, and transduction efficiency
A third-generation lentiviral vector, CCLc-MNDU3-x-PGK-EGFP, was utilized to construct the triple combination of anti-HIV vector (Figure 1a). The three genes inserted into this vector include a human/rhesus macaque TRIM5α isoform, a CCR5 shRNA, and a TAR decoy (Figure 1b). By combining these three genes, a potent preintegration block to HIV-1 infection could be established due to the pre-entry block by the knockdown of CCR5 expression and the postentry/preintegration block by the chimeric TRIM5α. If infectious virus is able to circumvent the potent inhibition established by the first two anti-HIV molecules, the TAR decoy will prevent upregulation of HIV-1 transcription.
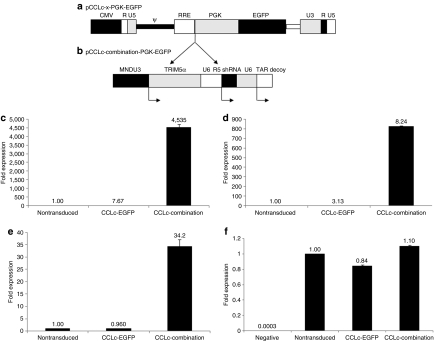
Figure 1.
Combination anti-HIV lentiviral vector and detection of transgene expression. (a) A third-generation lentiviral vector, pCCLc-x-PGK-EGFP, which contains an EGFP reporter gene was used to generate the combination anti-HIV construct. (b) The three transgenes, a chimeric human/rhesus macaque TRIM5α driven by the MNDU3 promoter, a CCR5 shRNA driven by the human polymerase-III small RNA U6 promoter, and a TAR decoy driven by the U6 promoter were inserted upstream of the EGFP reporter gene to derive pCCLc-combination-PGK-EGFP. Ghost-R5-X4-R3 cells were left nontransduced or were transduced with the EGFP alone or combination lentiviral vectors. Total cellular RNA was extracted and analyzed by quantitative real-time PCR for expression of (c) TRIM5α, (d) the CCR5 shRNA, and (e) the TAR decoy using gene-specific primers. (f) U6 snRNA was used as an internal control. Experiments were performed in triplicate. EGFP, enhanced green fluorescent protein; HIV, human immunodeficiency virus; shRNA, short hairpin RNA; TAR, transactivation response element.
With the incorporation of three anti-HIV genes into a single gene therapy vector that is based on HIV, detrimental effects on the quantity of vector production and titer due to the increased size of the vector payload and also the anti-HIV effects of the vector transgenes could be possible. Therefore, combination vector titers were compared to control enhanced green fluorescent protein (EGFP)–alone titers on HEK-293T cells. Combination vector titers, upon 100-fold concentration, were found to be, on average, ~5 × 109 transducing units/ml compared to EGFP-alone vector titers that generated ~3.5 × 109 transducing units/ml (data not shown). The vector titer results excluded any negative effects on vector titer using our combination vector construct and packaging system. Each of the transgenes is transcribed from their respective promoters for high levels of expression.
Due to the complexity of this combination vector containing three anti-HIV expression cassettes and a fourth EGFP reporter gene, expression levels of the downstream transcripts, including the two polymerase (pol)-III transcription units, may be affected. Expression of all three anti-HIV genes was confirmed by quantitative real-time PCR (QRT-PCR). The expression levels of the chimeric TRIM5α (Figure 1c), CCR5 shRNA (Figure 1d), and TAR decoy (Figure 1e) were 4,535-, 824-, and 34-fold higher, respectively, compared to control cells that do not express any of the anti-HIV genes. The levels of the internal control U6 snRNA were consistent among all cell types (Figure 1f).
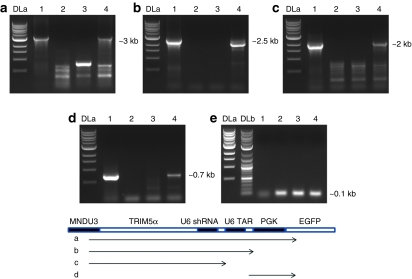
The stability of the vector in transduced cells is also a concern due to its complexity. To evaluate any vector deletions or rearrangements that may have occurred, genomic PCR was performed with vector transgene specific primers: Figure 2a shows MNDU3 forward and EGFP reverse [~3 kilobases (kb)], Figure 2b shows MNDU3 forward and TAR decoy reverse (~2.5 kb), Figure 2c shows MNDU3 forward and CCR5 shRNA reverse (~2 kb), Figure 2d shows TAR decoy forward and EGFP reverse (~0.7 kb), and Figure 2e shows albumin forward and reverse (~0.1 kb). As displayed in Figure 2, no deletions or rearrangements were detected, as PCR bands from genomic DNA from combination-transduced Ghost-R5-X4-R3 cells (lane 4) corresponded to PCR bands amplified from the combination transfer vector plasmid (lane 1) used in vector preparations. This was in contrast to nontransduced (lane 2) and EGFP-alone-transduced cells (lane 3) that did not amplify the correct PCR products and only displayed background bands. A schematic of the theoretical PCR products is displayed below the panels.
Figure 2.
Stability of the combination vector in transduced cells. Ghost-R5-X4-R3 cells were left nontransduced or were transduced with the EGFP alone or combination lentiviral vectors. Total genomic DNA was extracted and analyzed by PCR with primers specific for the respective vector transgenes. (a) MNDU3 (forward) and EGFP (reverse), (b) MNDU3 (forward) and TAR decoy (reverse), (c) MNDU3 (forward) and CCR5 shRNA (reverse), (d) TAR decoy (forward) and EGFP (reverse), and (e) albumin (forward and reverse). One kilobase DNA ladder (DLa), log DNA ladder (DLb), combination transfer vector plasmid (lane 1), nontransduced cells (lane 2), EGFP-alone vector transduced (lane 3), and combination vector transduced (lane 4). A schematic of the PCR products is below the panels. EGFP, enhanced green fluorescent protein; kb, kilobase; TAR, transactivation response element.
Due to the insertion of the three anti-HIV genes, transduction efficiencies obtained with the combination vector could also be negatively affected. In order to determine whether there was such an effect, both cultured Ghost-R5-X4-R3 cells and primary CD34+ HPCs were transduced with the control EGFP-alone vector and the anti-HIV gene combination vector. Transduction efficiencies were not affected by use of the combination vector, regardless of the multiplicity of infection (MOI) chosen, as observed by EGFP expression during fluorescence-activated cell sorting (FACS) analysis. Transduction efficiencies in cultured Ghost-R5-X4-R3 cells and primary CD34+ HPCs at an MOI of 10 were >95% and >50%, respectively, for each vector transduction (data not shown).
CCR5 downregulation in combination vector–transduced cells
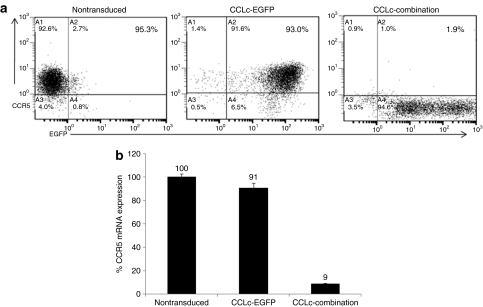
To determine whether successful transduction of cells with the combination anti-HIV lentiviral vector conferred knockdown of CCR5 expression, transduced cells were analyzed by FACS for CCR5 surface expression. As seen in Figure 3a, the combination vector–transduced Ghost-R5-X4-R3 cells displayed dramatic knockdown of CCR5 expression (>92%) compared to nontransduced and EGFP-alone-transduced cells. To further quantitate the levels of CCR5 gene silencing in combination vector–transduced cells, QRT-PCR was performed. Cells expressing the CCR5 shRNA displayed high levels of knockdown of CCR5 expression (>91%) as compared to nontransduced and EGFP-alone-transduced cells (Figure 3b). These data confirm a potent knockdown of CCR5 gene expression in cells transduced with the combination lentiviral vector.
Figure 3.
Downregulation of CCR5 surface expression and mRNA levels in transduced cells. Ghost-R5-X4-R3 cells were transduced with the EGFP alone and combination lentiviral vectors. (a) Seventy-two hours post-transduction, the cells were analyzed for CCR5 expression by fluorescence-activated cell sorting (FACS). (b) Cells were also analyzed by quantitative real-time PCR (QRT-PCR) for intracellular CCR5 mRNA levels. The FACS data are a representative of quadruple experiments. The QRT-PCR experiment was performed in triplicate. EGFP, enhanced green fluorescent protein.
HIV-1 challenge of combination vector–transduced cells
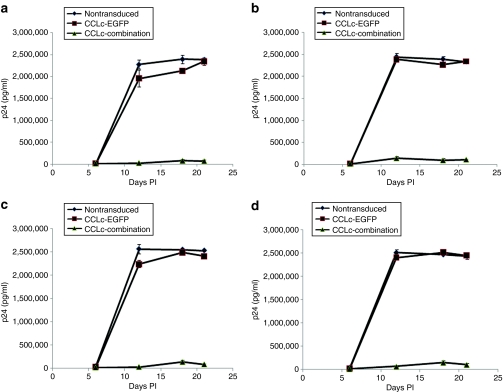
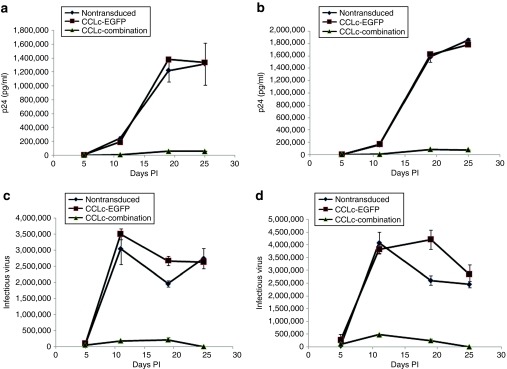
Cultured Ghost-R5-X4-R3 cells and primary macrophages are susceptible to infection from R5 and X4-tropic strains of HIV-1. However, upon transduction with the combination anti-HIV lentiviral vector, transduced cells should be resistant to HIV-1 infection. To determine whether transduction and expression of the three anti-HIV genes conferred viral resistance, cells were challenged with various strains of HIV-1. Combination vector–transduced Ghost-R5-X4-R3 cells displayed strong HIV-1 resistance and inhibition of viral replication (>1.5 log) after challenge with both R5 (BaL-1) (Figure 4a,b) and X4-tropic (NL4-3) (Figure 4c,d) strains of HIV-1 at multiple MOIs (0.01 and 0.05) as compared to nontransduced and EGFP-alone-vector-transduced cells as measured by p24 antigen enzyme-linked immunosorbent assay. CD34+ HPC–derived macrophages transduced with the combination vector also displayed strong inhibition of R5-tropic BaL-1 HIV-1 infection (>1.5 log) at multiple MOIs compared to control nontransduced and EGFP-alone-transduced cells. Inhibition was observed by p24 antigen enzyme-linked immunosorbent assay (Figure 5a,b) and by quantitating total infectious virus using a Ghost cell assay (Figure 5c,d).
Figure 4.
HIV-1 challenge of combination vector–transduced cultured cells. Ghost-R5-X4-R3 cells were transduced with the EGFP alone (closed squares) or the combination (closed triangles) lentiviral vector. Nontransduced (closed diamonds) and vector-transduced cells were subsequently challenged with the R5-tropic BaL-1 strain of HIV-1 at a multiplicity of infection (MOI) of (a) 0.01 and (b) 0.05. Cells were also challenged with the X4-tropic NL4-3 strain of HIV-1 at an MOI of (c) 0.01 and (d) 0.05. On various days postinfection, cell culture supernatants were analyzed for HIV-1 p24 antigen by enzyme-linked immunosorbent assay. The challenge experiments were performed in triplicate. EGFP, enhanced green fluorescent protein; HIV-1, human immunodeficiency virus type 1; PI, postinfection.
Figure 5.
HIV-1 challenge of combination vector–transduced CD34+ HPC–derived macrophages. CD34+ hematopoietic progenitor cells (HPCs) were transduced with the EGFP alone (closed squares) or the combination (closed triangles) lentiviral vector and cultured in a macrophage differentiation medium. Upon development of mature macrophages, nontransduced (closed diamonds) and vector-transduced cells were challenged with the R5-tropic BaL-1 strain of HIV-1. Cell culture supernatants were sampled and analyzed for HIV-1 p24 antigen by enzyme-linked immunosorbent assay (a) [multiplicity of infection (MOI) 0.01] and (b) (MOI 0.05). Challenge supernatants were also analyzed for infectious virus by a Ghost cell assay (c) (MOI 0.01) and (d) (MOI 0.05). The challenge experiments were performed in triplicate. EGFP, enhanced green fluorescent protein; HIV-1, human immunodeficiency virus type 1; PI, postinfection.
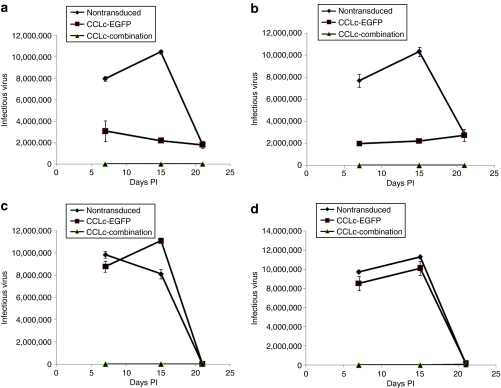
To evaluate the possible generation of escape mutants arising in long-term HIV-1-challenged combination vector–transduced cells, a secondary challenge experiment was performed. Naive cells, nontransduced, EGFP-alone, and combination vector–transduced cells were challenged using the respective viral supernatant from the last time point (day 25) of the initial challenge experiment. Cell culture supernatants were monitored for 3 weeks to identify any virus replication resulting from the generation of escape mutants. As seen in Figure 6, no viral replication was detected in challenged cells transduced with the combination lentiviral vector: BaL-1 MOI 0.01 (Figure 6a), BaL-1 MOI 0.05 (Figure 6b), NL4-3 MOI 0.01 (Figure 6c), and NL4-3 MOI 0.05 (Figure 6d). In contrast, control nontransduced and EGFP-alone-vector-transduced cells were successfully infected initially that resulted in detection of infectious virus. The levels of infectious virus then started to decrease after day 15 postinfection due to killing of infected cells within the cultures. The results from the secondary challenge experiments confirmed that no viral escape mutants could be detected in culture supernatants from combination vector–transduced cells challenged long term with HIV-1.
Figure 6.
Generation of escape mutants after long-term viral challenge. Naive nontransduced (closed diamonds), EGFP alone (closed squares), and combination (closed triangles) vector–transduced ghost-R5-X4-R3 cells were challenged with the day 25 culture supernatants from their respective initial viral challenges. (a) BaL-1 multiplicity of infection (MOI) 0.01, (b) BaL-1 MOI 0.05, (c) NL4-3 MOI 0.01, and (d) NL4-3 MOI 0.05. Cell culture supernatants were analyzed for infectious virus by a Ghost cell assay. Experiments were performed in triplicate. EGFP, enhanced green fluorescent protein; PI, postinfection.
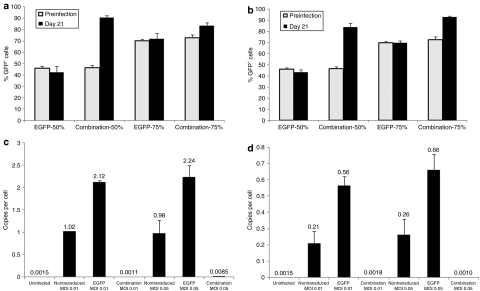
It is expected that in a human clinical HIV gene therapy application, not all of an individual's HIV-susceptible cells will be transduced with anti-HIV gene therapy vectors and will be capable of resisting viral infection and replication. Therefore, to evaluate the selective survival advantage of anti-HIV gene–transduced cells, a mixed population of nontransduced and transduced cells, either EGFP alone or combination vector transduced, were challenged with HIV-1, both BaL-1 and NL4-3. As seen in Figure 7, a selective survival advantage was conferred and maintained in cultures containing the combination vector–transduced cells demonstrated by an increase of the total percent EGFP+ cells by the end of the viral challenge. In cultures containing a 1:1 and 2:1 ratio of combination vector–transduced cells to nontransduced cells, the preinfection EGFP percent being, ~46 and ~73%, respectively, increased to >84% EGFP+ cells by day 21 postinfection for both BaL-1 (Figure 7a) and NL4-3 (Figure 7b). This increase was statistically significant (BaL-1 50%, P value = 0.001; BaL-1 75%, P value = 0.05) (NL4-3 50%, P value = 0.001; NL4-3 75%, P value = 0.001). These results are in contrast to cultures containing EGFP-alone-vector-transduced cells where the EGFP percent positive cell population remained relatively constant on day 21 postinfection (~42 and ~72% for ratios of 1:1 and 2:1, respectively) compared to the preinfection EGFP percent of 46 and 70% (BaL-1 50%, P value = 0.23; BaL-1 75%, P value = 0.69) (NL4-3 50%, P value = 0.62; NL4-3 75%, P value = 0.95). These results establish that the anti-HIV combination vector–transduced cells were able to survive and increase in number during the course of the HIV-1 infection.
Figure 7.
Selective survival advantage and detection of HIV-1 proviral integration of challenged cells. Ghost-R5-X4-R3 cells were transduced with the EGFP alone or the combination lentiviral vector. Cells were mixed at a ratio of 1:1 and 2:1 (transduced:nontransduced) and were challenged with the (a) R5-tropic BaL-1 or (b) X4-tropic strain of HIV-1 at a multiplicity of infection (MOI) of 0.01. Cells were analyzed by fluorescence-activated cell sorting for EGFP expression preinfection and on day 21 postinfection. Detection of HIV-1 integration: ghost-R5-X4-R3 nontransduced, EGFP alone, and combination vector–transduced cells were challenged with the (c) R5-tropic BaL-1 and (d) X4-tropic NL4-3 strain of HIV-1 at MOIs of 0.01 and 0.05. On day 25 postinfection, genomic DNA from virus challenged cells was analyzed by quantitative real-time PCR for integrated HIV-1 provirus using primers specific for the pol gene. Experiments were performed in triplicate. EGFP, enhanced green fluorescent protein; HIV-1, human immunodeficiency virus type 1.
Inhibition of HIV-1 proviral formation
Viral reservoirs that are maintained in an HIV-infected individual continue to be a major barrier for HIV eradication and curing patients with an established infection. To better eradicate the virus from infected individuals, the combination anti-HIV vector was designed to inhibit HIV-1 prior to integration, thus avoiding any provirus formation. To determine whether viral inhibition was occurring at the stages of pre-entry (due to the CCR5 shRNA) and postentry/preintegration (due to TRIM5α), cells challenged with both BaL-1 and NL4-3 were further analyzed by QRT-PCR for genomic HIV-1 provirus. As seen in Figure 7, a highly potent block of HIV-1 provirus formation could be demonstrated when genomic DNA from challenged cells (day 25 postinfection) was analyzed by QRT-PCR with primers specific for the HIV-1 pol gene. Combination vector–transduced cells contained undetectable levels, similar to background levels in uninfected cells, of HIV-1 provirus as compared to control nontransduced and EGFP-alone–vector-transduced infected cells at MOIs of 0.01 and 0.05 for BaL-1 (Figure 7c) and NL4-3 (Figure 7d) strains of HIV-1. These data confirm that the combination vector, indeed, conferred a strong block to HIV-1 integration, therefore preventing HIV proviral DNA formation.
Phenotypic analysis of transduced CD34+ HPC–derived macrophages
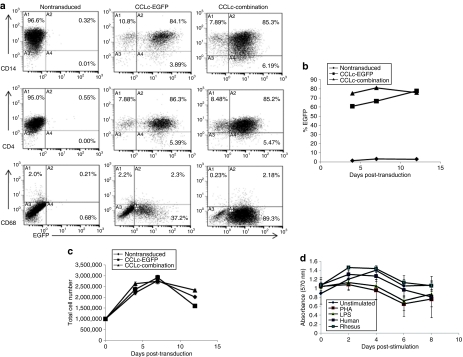
The introduction of foreign genes into cells has the potential to cause detrimental effects on their development. To evaluate whether lentiviral vector transduction of target cells and expression of the combination vector anti-HIV transgenes affected the differentiation of CD34+ HPCs into mature macrophages, cells were analyzed by FACS for macrophage-specific cell surface markers. Combination vector–transduced macrophages displayed normal levels of the surface markers CD14 (>93%), CD4 (>93%), and CD68 (an activation marker) (~2%) as compared to nontransduced and EGFP-alone-transduced cells (Figure 8a). Phenotypic analysis of these cells confirmed that normal differentiation had occurred from transduced CD34+ HPCs to mature macrophages and that transduced cells were indistinguishable from nontransduced cells.
Figure 8.
Toxicity of combination vector transduction. (a) CD34+ hematopoietic progenitor cells were transduced with the EGFP alone or combination lentiviral vector, and differentiated into mature macrophages. Cells were stained with the macrophage cell surface markers CD14, CD4, and CD68, and analyzed by fluorescence-activated cell sorting (FACS). (b) Peripheral blood mononuclear cells (PBMCs) were transduced with the EGFP alone or combination vectors. On various post-transduction, the transduced cells were analyzed for EGFP expression by FACS or (c) total cell counts. (d) Monocytes and dendritic cells from PBMCs were left unmanipulated or incubated with PHA, LPS, an 11-amino-acid human peptide, or a 13-aa rhesus macaque peptide. Lymphocytes were added back and analyzed for immune cell activation. All experiments were performed in triplicate. EGFP, enhanced green fluorescent protein; LPS, lipopolysaccharide; PHA, phytohemagglutinin.
Toxicity studies of vector-transduced cells
A previous report demonstrated that CCR5 shRNAs expressed by the U6 pol-III promoter were toxic to cells as compared to expression from the less robust H1 pol-III promoter.32 To evaluate the toxicity of the combination anti-HIV vector that contains a U6-expressed CCR5 shRNA, phytohemagglutinin-stimulated peripheral blood mononuclear cells (PBMCs) were either left nontransduced or transduced with the EGFP-alone or combination vectors. Cells were monitored on days 0, 4, 7, and 12 for both EGFP expression and total cell counts. Constant EGFP expression was observed for both the EGFP-alone and combination vector–transduced PBMCs throughout the experiment (Figure 8b). Also, total cell counts were comparable between nontransduced, EGFP-alone-transduced, and combination vector–transduced PBMCs (Figure 8c).
The expression of the chimeric TRIM5α isoform has the possibility of invoking an immune response due to the 13-aa patch from the rhesus macaque isoform that was inserted into the human isoform. To further detect immune activation in response to presentation of the rhesus macaque 13-aa patch, an immune cell activation assay was performed. As displayed in Figure 8d, no activation of immune cells had occurred in the samples that were pulsed with the rhesus macaque 13-aa peptide. The metabolic activity of the immune cells on days 6 and 8 poststimulation was not significantly different from those observed in unmanipulated cells (day 6 P value = 0.951 and day 8 P value = 0.993) and the cell samples pulsed with the human 11-aa peptide (day 6 P value = 0.938 and day 8 P value = 0.419). These results confirm that the rhesus macaque 13-aa patch located in the human/rhesus macaque chimeric TRIM5α isoform does not invoke immune cell activation when tested in a monocyte/dendritic cell culture system.
Discussion
Gene therapy for HIV offers an innovative and promising alternative to current antiretroviral drug treatments by providing long-term constitutive protection from HIV-1 infection. Single anti-HIV vectors have been previously utilized in numerous experiments and in several human clinical trials.6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23 This approach, however, is not sufficient, as the generation of escape mutants is possible due to the high mutation rate of HIV, as similarly seen when current small molecule antiretroviral drugs are used in monotherapy. Therefore, as in combination anti-HIV drug therapy, multiple anti-HIV genes inserted into a single vector can potentially inhibit HIV-1 infection long term while prohibiting the generation of escape mutants.
The production and persistence of an HIV reservoir in infected individuals is one of the main reasons for the lack of a cure, in spite of many years of research into anti-HIV small molecule drugs. When developing new and innovative therapies for HIV, the consistent high-level expression of anti-HIV activity at the stages of pre-entry and preintegration is important in blocking the productive infection of naive cells that, if infected, would allow the reservoir to persist. As an improved next step in HIV gene therapy, we have constructed a combination lentiviral vector encoding three highly potent anti-HIV genes that mainly focus on blocking infection at the pre-entry and preintegration stages of the viral life cycle (Figure 1b). A CCR5 shRNA capable of near complete knockdown of CCR5 expression was combined with a chimeric isoform of the postentry/preintegration species–specific HIV-1 restriction factor, TRIM5α. The third molecule, a TAR decoy, was added as an extra line of defense to disrupt the replication of any virus that still manages to integrate. Due to the complexity of the combination vector containing three anti-HIV expression cassettes and a fourth EGFP reporter gene, the packaging and expression of the various vector components could be compromised. There are no poly-A signals at the end of the TRIM5α or EGFP pol-II expression cassettes, and, therefore, transcription and packaging of the full-length vector RNA should not be disrupted. Also, the two U6 pol-III expression cassette downstream from the TRIM5α gene have separate promoters and do not rely on transcription machinery from other parts of the vector. Expression of all three anti-HIV genes was confirmed by QRT-PCR; however, varied levels of expression were detected. Transcription of TRIM5α was highest followed by the CCR5 shRNA and the TAR decoy. This may be due to the position of the respective transgenes in relation to the 5′ end of the vector. With four separate and active promoters in the vector, a variegated expression profile was observed with the most 5′ anti-HIV gene (TRIM5α) displaying the highest level of expression followed by the second (CCR5 shRNA) and third (TAR decoy) anti-HIV genes (Figure 1). This observation may be due to the multiple expression cassettes present. Based on these results, it was appropriate to use a more robust pol-III promoter in the U6 compared to the H1 for adequate expression of the CCR5 shRNA and the TAR decoy to potently knock down CCR5 expression and inhibit HIV-1 infection. Based on careful vector design using highly active promoters, we are able to generate consistent high-titered combination vectors including successful expression of all three anti-HIV genes and the EGFP reporter gene at levels capable of inhibiting HIV-1 infection. Due to the complexity of the combination vector, the stability of the vector was also a concern for deletions or rearrangements. The vector remained intact in transduced cells as confirmed by genomic PCR with vector transgene–specific primers (Figure 2).
This combination anti-HIV lentiviral vector was evaluated in both HIV-1-susceptible cultured cells and in primary CD34+ HPC–derived macrophages, in vitro. Combination vector–transduced cells displayed stable and substantial knockdown of CCR5 expression as compared to control cells due to the expression of the CCR5 shRNA (Figure 3). These cells were engineered to acquire a phenotype that mimics the natural Δ32-bp deletion allele of CCR5 enabling a small percentage of the human population to resist HIV-1 infection.25,26,27 The silencing of CCR5 expression as a single therapy, however, would not offer complete protection from HIV including infection from X4- and dual-tropic strains. With the addition of the other anti-HIV molecules, TRIM5α and the TAR decoy, a broader range of viral inhibition could be achieved. Our results demonstrate that viral protection from R5-, X4-, and dual-tropic strains, at increasing MOI, could be achieved in combination vector–transduced cells in both cultured cells and primary CD34+ HPC–derived macrophages (Figures 4 and 5). Combining multiple stages of inhibition in the viral life cycle also prohibited the generation of escape mutants. After long-term repeated challenge of combination vector–transduced cells, no rise in HIV replication could be detected confirming a lack of HIV-1 replication and a lack of mutant escape virus formation (Figure 6). The CCR5 shRNA (pre-entry) and the chimeric TRIM5α (postentry/preintegration) molecules combined exhibited efficient inhibition of HIV-1 integration and proviral formation (Figure 7b). These stages of the HIV life cycle are critical for a continued infection and, if blocked, are key points to target for eradicating HIV from the body.
HIV gene therapy offers a potential alternative or augmentation of traditional antiretroviral treatments that can become toxic over prolonged use and allow for the generation of escape mutants if not taken under strict compliance.1,2,3,4,5 The introduction of foreign transgenes due to the integration of the lentiviral vector, however, could cause unwanted side effects in transduced cells. As a first step in evaluating the safety of this combination vector, in vitro, CD34+ HPC–derived macrophages were analyzed for normal development and differentiation. Combination vector–transduced macrophages were phenotypically normal displaying similar levels of macrophage-specific cell surface markers as control cells, thus, confirming normal development (Figure 8a). A previous report demonstrated that CCR5 shRNAs expressed from a U6 promoter were toxic compared to those expressed from the less robust H1 promoter.32 We observed that in combination vector–transduced cells, no toxicity had occurred as measured by a constant expression of EGFP (Figure 8b) and total cell counts (Figure 8c) as compared to control cells. These results differ from the previous report for a couple of reasons. First, this particular CCR5 shRNA has also been previously tested and did not show any toxic side effects in cells transfected with large amounts of chemically synthesized shRNAs and also in cells transduced with a lentiviral vector containing the U6 promoter–driven CCR5 shRNA in contrast to other more toxic CCR5 shRNAs in that study.14 However, in the context of the combination vector, decreased levels of the CCR5 shRNA from the U6 promoter due to the complexity of the vector with numerous expression cassettes may also be contributing to the nontoxic effect. Anti-HIV vector–transduced cells were also found to be nontoxic and non-immunostimulating as evaluated by transduced PBMCs and the MTT cell activation assay. Cells expressing the human/rhesus macaque chimeric TRIM5α isoform have the potential to be recognized as foreign due to the possible presentation of the 13-aa patch inserted into the TRIM5α gene. The transduced PBMCs expressing the chimeric TRIM5α would display peptides on major histocompatibility complex I from the processed protein and would be eliminated due to immune cells recognizing the peptides as foreign antigen. A lack of combination vector–transduced cell death was observed in Figure 8 as displayed by a constant expression of EGFP and total cell counts. To further analyze whether immune activation would occur with peptides corresponding to the entire rhesus macaque 13-aa sequence, monocytes and dendritic cells were pulsed with the peptide and cell activation was analyzed. No activation of human immune cells had occurred demonstrating that the chimeric TRIM5α is tolerated and established the usefulness of this potent anti-HIV molecule in the combination vector (Figure 8d).
As HIV continues to infect millions of people worldwide, it is imperative to develop novel anti-viral strategies that aim to block productive HIV infection and help eliminate the viral reservoir, enhancing therapeutic options. Our highly effective anti-HIV combination lentiviral vector that strongly inhibits HIV infection in a preintegration manner is a much-needed tool for this purpose. Further preclinical, in vivo, studies designed to produce the necessary safety, toxicity, and efficacy data to initiate a human clinical trial using this vector are currently ongoing.
Materials and Methods
Lentiviral vector design and production. A third-generation HIV-derived lentiviral vector containing an EGFP reporter gene was used in this study, pCCLc-MNDU3-x-PGK-EGFP (Figure 1a). A chimeric human/rhesus macaque TRIM5α gene (1.5 kb) was inserted into the lentiviral vector under the control of the MNDU3 promoter (550 bp). There is no poly-A signal at the end of the TRIM5α gene. The human pol-III U6 promoter–driven CCR5 shRNA expression cassette (340 bp) was generated, as described previously, and inserted directly downstream of the MNDU3-TRIM5α gene.33 The TAR decoy expression cassette (420 bp) was generated in a similar way to the CCR5 shRNA cassette and inserted directly downstream from the CCR5 shRNA gene. All three of these anti-HIV expression cassettes were inserted upstream of the PGK-driven EGFP reporter gene (1.3 kb) to derive pCCLc-combination-PGK-EGFP (Figure 1b). Sequencing of clones was confirmed by Laragen, Los Angeles, CA.
Lentiviral vectors were generated in HEK-293T cells. Twenty-five micrograms of the packaging construct, pΔ8.9 (packaging plasmid containing the gag and pol genes), 25 µg of pCCLc-MNDU3-x-PGK-EGFP (control empty vector), or the combination vector, pCCLc-MNDU3-TRIM5α-U6-CCR5shRNA-U6-TAR decoy-PGK-EGFP (transfer vector), and 5 µg of VSVG (envelope). DNA plasmids were transfected into cells in T225 flasks by lipofection. Vector supernatants were collected at 48 hours post-transfection and concentrated by ultrafiltration.
Transduction of cultured cells and primary CD34+ HPCs. Ghost-R5-X4-R3 cultured cells obtained from the AIDS Reference and Reagent Program were cultured in complete Dulbecco's modified Eagle's medium including 10% fetal bovine serum supplemented with hygromycin, puromycin, and G418 according to the supplier's protocol. Cells were transduced with the lentiviral vectors, either EGFP-alone or the combination vector (MOI 10), for 2 hours at 37 °C with 8 µg/ml protamine sulfate. Complete medium was then added to the cells.
CD34+ HPCs were isolated from umbilical cord blood (NDRI, Philadelphia, PA) by Ficoll-Paque (GE Healthcare, Piscataway, NJ) and purified by magnetic bead column separation (Miltenyi Biotec, Auburn, CA). CD34+ cell isolation purity (>93%) was routinely obtained. Total CD34+ cells were cultured in complete Iscove's modified Dulbecco's medium containing 10% fetal bovine serum and supplemented with 50 ng/ml stem cell factor, Flt-3 ligand, and thrombopoietin. Cells were transduced with the lentiviral vectors EGFP-alone or the combination vector (MOI 10) for 3 hours at 37 °C with 8 µg/ml protamine sulfate. Two days post-transduction, cells were sorted based on EGFP expression and cultured in semisolid methylcellulose medium with growth factors (Stem Cell Technologies, Vancouver, British Columbia, Canada) for 12 days to derive mature macrophages. After differentiation, cells were removed from the methylcellulose medium and plated in 6-well plates in complete Dulbecco's modified Eagle's medium with 10% fetal bovine serum supplemented with 10 ng/ml of granulocyte-macrophage colony-stimulating factor and macrophage colony-stimulating factor. Media were changed every 2 days for 4 days to derive mature macrophages. Both nontransduced and lentiviral vector–transduced cultured and primary CD34+ cell–derived macrophages were used for subsequent experiments.
Detection of anti-HIV gene expression. QT-PCR was utilized to detect the expression of the three anti-HIV transgenes. Total RNA was extracted from Ghost-R5-X4-R3 cells using RNA-STAT-60 (Tel-Test, Friendswood, TX). To detect the expression of the chimeric TRIM5α, first strand cDNA synthesis was performed using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA). QRT-PCR was then performed using the SYBR Green PCR Master Mix Kit (Applied Biosystems). A primer pair corresponding to the chimeric TRIM5α gene and not the native human TRIM5α gene was used: (forward) 5′-CTGGGTTGATGTGACAGTGG-3′ and (reverse) 5′-CGTGAGTGACGGAAACGTAA-3′. To detect the expression of the small RNA transcripts including the CCR5 shRNA and the TAR decoy, a QuantiMir RT kit (System Biosciences, Mountain View, CA) was utilized according to the manufacturer's protocol. Total RNA was used from the RNA-STAT-60 extraction from the Ghost-R5-X4-R3 cells. The primers for the CCR5 shRNA, TAR decoy, and control U6 small nuclear RNA are as follows: CCR5 shRNA (forward) 5′-GTAGATGTCAGTCATGCTC-3′, TAR decoy (forward) 5′-CTTGCAATGATGTCGTAATTTGCGTC-3′, and U6 snRNA (forward) 5′-CGCAAGGATGACACGCAAATTC-3′. These experiments were performed in triplicate.
Stability of combination vectors in transduced cells. To determine whether there were any deletions or rearrangements in combination vector–transduced cells, genomic PCR was performed. Total genomic DNA was extracted from nontransduced, EGFP-alone, and combination vector–transduced Ghost-R5-X4-R3 cells using the Wizard Genomic DNA Isolation System (Promega, Madison, WI). PCR was performed using high-fidelity Taq. Primers corresponding to the specific vector transgenes were used: MNDU3 (forward) 5′-CGCCCTCAGCAGTTTCTAG-3′, EGFP (reverse) 5′-CTCCTCGCCCTTGCTCACCAT-3′, TAR decoy (forward) 5′-CAATGATGTCGTAATTTGC-3′, TAR decoy (reverse) 5′-CTTG CTCAGTAAGAATTTTCGTC-3′, and CCR5 shRNA (reverse) 5′-ATGTC AGTCATGCTCGGTGTTTCG-3′ (Integrated DNA Technologies, Coralville, IA). Albumin was used as an internal control (forward) 5′-TGAAACATACGTTCCCAAAGAGTTT-3′ and (reverse) 5′-CTCTCCTTCTCAGAAAGTGTGCATAT-3′. PCR products were analyzed and visualized on an agarose gel.
Flow cytometry and QRT-PCR. To determine whether cells transduced with the combination lentiviral vector had decreased levels of CCR5 due to the expression of the CCR5 shRNA, cells were analyzed by FACS. Seventy-two hours post-transduction, transduced Ghost-R5-X4-R3 cells were stained with a phycoerythrin-conjugated antihuman CCR5 antibody (BD Biosciences, San Jose, CA). This experiment was performed in quadruplicate. CD34+ cell–derived macrophages, nontransduced, EGFP-alone-transduced, and combination vector–transduced cells were stained with antibodies to detect normal macrophage cell surface markers, including CD14-PE, CD4-PE, and CD68-PE (BD Biosciences). All FACS data were obtained on a Beckman Coulter Cytomics FC500 flow cytometer (Beckman Coulter, Fullerton, CA) and analyzed on CXP analysis software (Beckman Coulter).
To further quantitate the levels of CCR5 knockdown in combination lentiviral vector–transduced cells, QRT-PCR was performed on transduced cell RNA. Total RNA was extracted from Ghost-R5-X4-R3 cells using RNA-STAT-60. First strand cDNA synthesis was performed using the High Capacity cDNA Reverse Transcription Kit. QRT-PCR was then performed using the SYBR Green PCR Master Mix Kit with primers: 5′-ACTGCAAAAGGCTGAAGAGC-3′ and 5′-AGCATAGTGAGCCCAGA AGG-3′. GAPDH was used as an internal control. This experiment was performed in triplicate.
To quantitate the levels of integrated HIV-1 provirus following viral challenge of transduced cells, QRT-PCR was performed on the cell's genomic DNA. Total genomic DNA was isolated from challenged cells using the Wizard Genomic DNA Isolation System. QRT-PCR was performed using the TaqMan PCR Core Reagents Kit (Applied Biosystems) using the primers 5′-CTGGCTACTATTTCTTTTGCTA-3′ and 5′-TGGCATGGGTACCAGCACA-3′ and probe 5′-TTTATCTACTTGTTCATTTCCTCCAATTCCTT-3′ (Integrated DNA Technologies). The single copy albumin gene was used as an internal control. This experiment was performed in triplicate.
HIV-1 challenge of vector-transduced cells. To determine whether the expression of the triple combination of anti-HIV genes conferred resistance to HIV-1 infection, transduced cells were challenged with R5-, X4-, and dual-tropic strains of HIV-1, BaL-1, and NL4-3, respectively. Ghost-R5-X4-R3 cells, nontransduced, EGFP-alone, and combination vector transduced were incubated with R5-tropic BaL-1 and X4-tropic NL4-3 strains of HIV-1, MOIs 0.01 and 0.05, for 2 hours at 37 °C with 8 µg/ml Polybrene. On various days postinfection, challenge supernatants were sampled for use in an HIV-1 p24 antigen enzyme-linked immunosorbent assay and infectious virus assays. Nontransduced and transduced CD34+ cell–derived macrophages were similarly challenged with BaL-1 at MOIs of 0.01 and 0.05. On various days postinfection, challenge supernatants were sampled for use in an HIV-1 p24 antigen enzyme-linked immunosorbent assay (ZeptoMetrix, Buffalo, NY) and infectious virus assays. These experiments were performed in triplicate.
Viral escape mutants can be generated after prolonged use with current antiretroviral therapies. To determine whether escape mutants were generated upon challenge of combination-transduced cells, subsequent challenges were performed with culture supernatants from the initial HIV-1 challenge experiments. Naive Ghost-R5-X4-R3 cells, nontransduced, EGFP-alone, and combination vector transduced were challenged with culture supernatants from their respective day 25 HIV-1 challenge supernatants from the initial challenge experiment. On various days postinfection, culture supernatants were sampled for infectious virus. Briefly, supernatants (75 µl) from viral challenges were incubated on naive nontransduced Ghost-R5-X4-R3 cells for 2 hours at 37 °C with 8 µg/ml Polybrene. Complete cell culture media were added. Forty-eight hours postinfection, cells were analyzed by FACS for EGFP expression to quantitate the levels of infectious virus. These experiments were performed in triplicate.
Selective survival advantage of combination vector–transduced cells. To evaluate whether combination vector–transduced cells have a selective survival advantage over nontransduced cells, a mixed culture of both cell types was challenged with HIV-1. Mixed populations of cells, with ratios of 1:1 and 2:1, transduced cells (either EGFP-alone or combination vector transduced) to nontransduced cells, were challenged with R5- and X4-tropic strains of HIV-1, BaL-1, and NL4-3, respectively, at an MOI of 0.01. Cells were analyzed by FACS for EGFP expression on day 0 (preinfection) and on day 21 postinfection to evaluate the survival advantage of the transduced cell populations. Statistics were generated using the χ2 test. These experiments were performed in triplicate.
Toxicity studies of anti-HIV genes. To determine whether the expression of the three anti-HIV genes, especially the U6-driven CCR5 shRNA, were toxic to cells, transduced PBMCs were monitored for EGFP expression and total cell counts. Freshly isolated PBMCs were stimulated for 2 days with phytohemagglutinin (1 µg/ml) and subsequently transduced with the EGFP-alone or combination vectors. Cells were monitored on days 0, 4, 7, and 12 for both EGFP expression by FACS and by total cell counts. These experiments were performed in triplicate.
The insertion of the rhesus macaque TRIM5α 13-aa patch into the C-terminal region of the human TRIM5α isoform may result in immune activation and rejection of cells expressing the chimeric TRIM5α molecule. To determine whether immune system cells were capable of responding to the presentation of the 13-aa patch, an MTT cell activation assay was performed. Fresh PBMCs were isolated by Ficoll-Paque and plated in 96-well plates in complete RPMI with 10% fetal bovine serum and supplemented with 10 ng/ml IL-2. Monocytes and dendritic cells were allowed to attach to the bottom of the wells. Lymphocytes were removed and plated in separate wells. Monocytes/dendritic cells were pulsed overnight with small peptides corresponding to either a human TRIM5α 11-aa patch (GARGTRYQTFV) or the rhesus macaque TRIM5α 13-aa patch (QAPGTLFTFPSLT) that was used to generate the chimeric TRIM5α isoform. Peptides were synthesized by GenScript (Piscataway, NJ). A separate set of monocytes/dendritic cells were pulsed with lipopolysaccharide as a positive control. A fourth set of monocytes/dendritic cells were incubated with phytohemagglutinin (1 µg/ml) to act as a positive T-cell activation control. The fifth set of monocytes/dendritic cells were left unmanipulated to serve as a negative nonactivated control. Lymphocytes were added back to the cultures the following day. On various days postaddition of the lymphocytes, the MTT cell activation assay was performed on the cells according to the manufacturer's protocol (Roche Applied Science, Indianapolis, IN). These experiments were performed in triplicate. Statistics were generated using the χ2 test.
Acknowledgments
This work was supported by the University of California–Davis Health System start-up funds from the dean's office for the Stem Cell Program and by the James B. Pendleton Charitable Trust. We acknowledge Jon Walker for his technical support and Bridget McLaughlin for help with fluorescence-activated cell sorting. The National Institutes of Health AIDS Research and Reference Reagent Program provided many reagents and cell lines used in this work. We have filed a record of invention for the combination lentiviral vector.
REFERENCES
- Martinez-Picado J, DePasquale MP, Kartsonis N, Hanna GJ, Wong J, Finzi D, et al. Antiretroviral resistance during successful therapy of HIV type 1 infection. Proc Natl Acad Sci USA. 2000;97:10948–10953. doi: 10.1073/pnas.97.20.10948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winters MA, Baxter JD, Mayers DL, Wentworth DN, Hoover ML, Neaton JD, et al. Frequency of antiretroviral drug resistance mutations in HIV-1 strains from patients failing triple drug regimens. The Terry Beirn Community Programs for Clinical Research on AIDS. Antivir Ther (Lond) 2000;5:57–63. [PubMed] [Google Scholar]
- Bacheler LT, Anton ED, Kudish P, Baker D, Bunville J, Krakowski K, et al. Human immunodeficiency virus type 1 mutations selected in patients failing efavirenz combination therapy. Antimicrob Agents Chemother. 2000;44:2475–2484. doi: 10.1128/aac.44.9.2475-2484.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafeuillade A, Poggi C, Hittinger G., and , Chadapaud S. Phenotypic and genotypic resistance to nucleoside reverse transcriptase inhibitors in HIV-1 clinical isolates. HIV Med. 2001;2:231–235. doi: 10.1046/j.1468-1293.2001.00078.x. [DOI] [PubMed] [Google Scholar]
- Marks K., and , Gulick RM. New antiretroviral agents for the treatment of HIV infection. Curr HIV/AIDS Rep. 2004;1:82–88. doi: 10.1007/s11904-004-0012-0. [DOI] [PubMed] [Google Scholar]
- Bai J, Gorantla S, Banda N, Cagnon L, Rossi J., and , Akkina R. Characterization of anti-CCR5 ribozyme-transduced CD34+ hematopoietic progenitor cells in vitro and in a SCID-hu mouse model in vivo. Mol Ther. 2000;1:244–254. doi: 10.1006/mthe.2000.0038. [DOI] [PubMed] [Google Scholar]
- Michienzi A, Li S, Zaia JA., and , Rossi JJ. A nucleolar TAR decoy inhibitor of HIV-1 replication. Proc Natl Acad Sci USA. 2002;99:14047–14052. doi: 10.1073/pnas.212229599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordelier P, Morse B., and , Strayer DS. Targeting CCR5 with siRNAs: using recombinant SV40-derived vectors to protect macrophages and microglia from R5-tropic HIV. Oligonucleotides. 2003;13:281–294. doi: 10.1089/154545703322616961. [DOI] [PubMed] [Google Scholar]
- An DS, Donahue RE, Kamata M, Poon B, Metzger M, Mao SH, et al. Stable reduction of CCR5 by RNAi through hematopoietic stem cell transplant in non-human primates. Proc Natl Acad Sci USA. 2007;104:13110–13115. doi: 10.1073/pnas.0705474104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding SF, Lombardi R, Nazari R., and , Joshi S. A combination anti-HIV-1 gene therapy approach using a single transcription unit that expresses antisense, decoy, and sense RNAs, and trans-dominant negative mutant Gag and Env proteins. Front Biosci. 2002;7:a15–a28. doi: 10.2741/ding. [DOI] [PubMed] [Google Scholar]
- Novina CD, Murray MF, Dykxhoorn DM, Beresford PJ, Riess J, Lee SK, et al. siRNA-directed inhibition of HIV-1 infection. Nat Med. 2002;8:681–686. doi: 10.1038/nm725. [DOI] [PubMed] [Google Scholar]
- Lee NS, Dohjima T, Bauer G, Li H, Li MJ, Ehsani A, et al. Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat Biotechnol. 2002;20:500–505. doi: 10.1038/nbt0502-500. [DOI] [PubMed] [Google Scholar]
- Anderson J, Li MJ, Palmer B, Remling L, Li S, Yam P, et al. Safety and efficacy of a lentiviral vector containing three anti-HIV genes—CCR5 ribozyme, tat-rev siRNA, and TAR decoy—in SCID-hu mouse-derived T cells. Mol Ther. 2007;15:1182–1188. doi: 10.1038/sj.mt.6300157. [DOI] [PubMed] [Google Scholar]
- Anderson J., and , Akkina R. Complete knockdown of CCR5 by lentiviral vector-expressed siRNAs and protection of transgenic macrophages against HIV-1 infection. Gene Ther. 2007;14:1287–1297. doi: 10.1038/sj.gt.3302958. [DOI] [PubMed] [Google Scholar]
- Kohn DB, Bauer G, Rice CR, Rothschild JC, Carbonaro DA, Valdez P, et al. A clinical trial of retroviral-mediated transfer of a rev-responsive element decoy gene into CD34(+) cells from the bone marrow of human immunodeficiency virus-1-infected children. Blood. 1999;94:368–371. [PubMed] [Google Scholar]
- Humeau LM, Binder GK, Lu X, Slepushkin V, Merling R, Echeagaray P, et al. Efficient lentiviral vector-mediated control of HIV-1 replication in CD4 lymphocytes from diverse HIV+ infected patients grouped according to CD4 count and viral load. Mol Ther. 2004;9:902–913. doi: 10.1016/j.ymthe.2004.03.005. [DOI] [PubMed] [Google Scholar]
- Mitsuyasu RT, Merigan TC, Carr A, Zack JA, Winters MA, Workman C, et al. Phase 2 gene therapy trial of an anti-HIV ribozyme in autologous CD34+ cells. Nat Med. 2009;15:285–292. doi: 10.1038/nm.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez MA, Gutiérrez A, Armand-Ugón M, Blanco J, Parera M, Gómez J, et al. Suppression of chemokine receptor expression by RNA interference allows for inhibition of HIV-1 replication. AIDS. 2002;16:2385–2390. doi: 10.1097/00002030-200212060-00002. [DOI] [PubMed] [Google Scholar]
- ter Brake O, Legrand N, von Eije KJ, Centlivre M, Spits H, Weijer K, et al. Evaluation of safety and efficacy of RNAi against HIV-1 in the human immune system (Rag-2(-/-)gammac(-/-)) mouse model. Gene Ther. 2009;16:148–153. doi: 10.1038/gt.2008.124. [DOI] [PubMed] [Google Scholar]
- Kumar P, Ban HS, Kim SS, Wu H, Pearson T, Greiner DL, et al. T cell-specific siRNA delivery suppresses HIV-1 infection in humanized mice. Cell. 2008;134:577–586. doi: 10.1016/j.cell.2008.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonyhadi ML, Moss K, Voytovich A, Auten J, Kalfoglou C, Plavec I, et al. RevM10-expressing T cells derived in vivo from transduced human hematopoietic stem-progenitor cells inhibit human immunodeficiency virus replication. J Virol. 1997;71:4707–4716. doi: 10.1128/jvi.71.6.4707-4716.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J., and , Akkina R. Human immunodeficiency virus type 1 restriction by human-rhesus chimeric tripartite motif 5alpha (TRIM 5alpha) in CD34(+) cell-derived macrophages in vitro and in T cells in vivo in severe combined immunodeficient (SCID-hu) mice transplanted with human fetal tissue. Hum Gene Ther. 2008;19:217–228. doi: 10.1089/hum.2007.108. [DOI] [PubMed] [Google Scholar]
- Bauer G, Selander D, Engel B, Carbonaro D, Csik S, Rawlings S, et al. Gene therapy for pediatric AIDS. Ann N Y Acad Sci. 2000;918:318–329. doi: 10.1111/j.1749-6632.2000.tb05501.x. [DOI] [PubMed] [Google Scholar]
- Berger EA, Murphy PM., and , Farber JM. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu Rev Immunol. 1999;17:657–700. doi: 10.1146/annurev.immunol.17.1.657. [DOI] [PubMed] [Google Scholar]
- Liu R, Paxton WA, Choe S, Ceradini D, Martin SR, Horuk R, et al. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- Huang Y, Paxton WA, Wolinsky SM, Neumann AU, Zhang L, He T, et al. The role of a mutant CCR5 allele in HIV-1 transmission and disease progression. Nat Med. 1996;2:1240–1243. doi: 10.1038/nm1196-1240. [DOI] [PubMed] [Google Scholar]
- Naif HM, Cunningham AL, Alali M, Li S, Nasr N, Buhler MM, et al. A human immunodeficiency virus type 1 isolate from an infected person homozygous for CCR5Delta32 exhibits dual tropism by infecting macrophages and MT2 cells via CXCR4. J Virol. 2002;76:3114–3124. doi: 10.1128/JVI.76.7.3114-3124.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hütter G, Nowak D, Mossner M, Ganepola S, Müssig A, Allers K, et al. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N Engl J Med. 2009;360:692–698. doi: 10.1056/NEJMoa0802905. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE., and , Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P., and , Sodroski J. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- Sawyer SL, Wu LI, Emerman M., and , Malik HS. Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain. Proc Natl Acad Sci USA. 2005;102:2832–2837. doi: 10.1073/pnas.0409853102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An DS, Qin FX, Auyeung VC, Mao SH, Kung SK, Baltimore D, et al. Optimization and functional effects of stable short hairpin RNA expression in primary human lymphocytes via lentiviral vectors. Mol Ther. 2006;14:494–504. doi: 10.1016/j.ymthe.2006.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanotto D, Li H., and , Rossi JJ. Functional siRNA expression from transfected PCR products. RNA. 2002;8:1454–1460. doi: 10.1017/s1355838202021362. [DOI] [PMC free article] [PubMed] [Google Scholar]