Abstract
C/EBPβ is known to be important for monocytic differentiation and macrophage function. Here, we found that expression of all three C/EBPβ isoforms induced in HL60 cells by 1,25-dihydroxyvitamin D3 (1,25D) was upregulated in a sustained manner that correlates with the appearance of monocytic phenotype and with the G1 phase cell cycle arrest. In 1,25D-resistant HL60-40AF cells, isoforms β-1 and β-3 were expressed at levels comparable to 1,25D-sensitive HL60-G cells, but isoform β-2 was difficult to detect. Treatment of sensitive HL60 cells with 1,25D resulted in predominantly nuclear localization of C/EBP isoforms β-2 and β-3, while a large proportion of C/EBPβ-1 remained in the cytoplasm. Attenuation of the MEK-ERK MAPK pathway by the inhibitor PD98059 markedly reduced the expression, 1,25D-induced phosphorylation and nuclear localization of C/EBPβ-2 and C/EBPβ-3. Interestingly, only the lower molecular mass isoforms of C/EBPβ phosphorylated on Thr235 were found in the nuclei, while C/EBPβ-1 was constitutively phosphorylated and was detected principally in the cytoplasmic fraction. Although the role of C/EBPβ isoforms in 1,25D-induced differentiation is complex, our results taken together strongly suggest that the phosphorylation of C/EBPβ isoforms on Thr235 takes place mainly via the MEK-ERK pathway and that C/EBPβ-2 is the principal transcription factor in this cell system.
Keywords: Vitamin D, Monocytic differentiation, C/EBP transcription factors, MEK1-ERK MAP kinase pathway, Subcellular localization, PD98059, SB203580
Introduction
Hematopoiesis is a multistage process in which all formed elements of the blood are differentiated from multipotential hematopoietic stem cells [1,2]. The pathway of differentiation is determined by signals supplied by series of growth factors and hormones in the microenvironment of the cells in the bone marrow. These signals are translated within the cells into the expression or activation of transcription factors, which direct the differentiation process [3]. The CCAAT enhancer binding proteins (C/EBPs) are a family of basic leucine zipper transcription factors which participate in the differentiation of several cell types including myeloid cells [4,5]. Out of the six members of C/EBP family of transcription factors, C/EBPα and C/EBPε are known to be critical for normal granulocytic differentiation [6-9], while C/EBPβ is particularly important for macrophage function [10-12]. It was recently shown that in leukemic cells undergoing monocytic differentiation in response to 1,25-dihydroxyvitamin D3 (1,25D) exposure, the expression of C/EBPβ transcription factor is upregulated [13]. Of note, the expression of C/EBPβ correlated with the degree of monocytic differentiation, and the expression appeared to be under the control of MAPK pathways, though the role of these signaling pathway was not clear [13,14]. Thus, it seems that C/EBPβ is important for the function of macrophages and for differentiation of myeloid leukemic cells along the monocytic lineage, but the mechanisms require further elucidation.
It is well documented that 1,25D induces differentiation and inhibits proliferation of various cancer cells including myeloid leukemia cells [15–19]. When exposed to 1,25D, these cells acquire the functional properties and express cell surface differentiation markers of monocytes (e.g.[20]). Yet, despite many previous studies of 1,25D-induced cell differentiation, the precise molecular basis of 1,25D differentiation-inducing actions is unclear. It is known that the biological activity of 1,25D is mediated mainly by the nuclear vitamin D receptor (VDR) and that the cells lacking VDR or harboring mutated VDR are unresponsive to 1,25D [21–23]. It has also been reported that 1,25D activates several intracellular signaling pathways such as the PKC pathway [24,25], calcium-dependent pathways [26– 28], the PI3/AKT-kinase pathway [29–31], and the MAP kinase pathways [32–37]. In myeloid leukemia cells, activation of one or more of these signal transduction pathways eventually leads to regulation of transcription factors such as AP-1 [38–40], and it is likely that the transcriptional activity of C/EBPβ protein is also regulated by at least some of these pathways [13,14].
Activation of C/EBPβ function as a transcription factor is a complicated process since there are several levels of its regulation. The C/EBPβ gene is intronless, but its mRNA has three alternative AUG translation initiation sites, and three products of C/EBPβ gene can be translated [41,42]. The human C/EBPβ-1 is a full-length protein of 346 amino acids that is translated from the first in-frame methionine. CEBPβ-2, which begins at the second in-frame methionine, is truncated by 23 amino acids, and CEBPβ-3 begins at the final in-frame methionine at position 199, and thus is the truncated C-terminal part of the protein (Fig. 1). In rodents, the corresponding polypeptides are shorter and have been termed “full-length” LAP and LIP (e.g. [43]). Because the transactivation domains are at the N-termini of these proteins, the different protein products have been proposed to be functionally different [44]. Since C/EBPβ-3 lacks the transcription activating domain, yet it can dimerize and bind to DNA [41], it is believed to act as a transcriptional repressor. Another level of regulation of the function of C/EBP family members may be achieved through protein-protein interactions of C/EBPs with other proteins important for cell proliferation, such as Cdk2, Cdk4 and the Rb protein [13,45,46]. Yet, another level of regulation is obtained by multiple phosphorylations of C/EBPβ. Human C/EBPβ has several known phosphorylation sites, including those at Thr235, Thr266, Ser288 and Ser325 (Fig. 1), which are important for intracellular localization and transcriptional activity of the protein (e.g. [47-51]). Since the known phosphorylation sites are located in the C-terminal part of human C/EBPβ, all three isoforms of C/EBPβ contain these sites [41]. It has been shown that a serine in the leucine zipper of C/EBPβ was phosphorylated by a calcium-calmodulin-dependent protein kinase [52], the phosphorylation of Ser288 is carried out by protein kinase A [48], while phosphorylation at Thr266 is mediated in rat cells by the kinase p90RSK, which is downstream from the Raf-MEK1-ERK-1/2 signal transduction module [53]. The kinase responsible for phosphorylation at Thr235 has been shown in several systems to be part of the MAPK cascade, presumably ERK, and this phosphorylation confers major transcriptional activity on C/EBPβ [50,51,54].
Fig. 1.
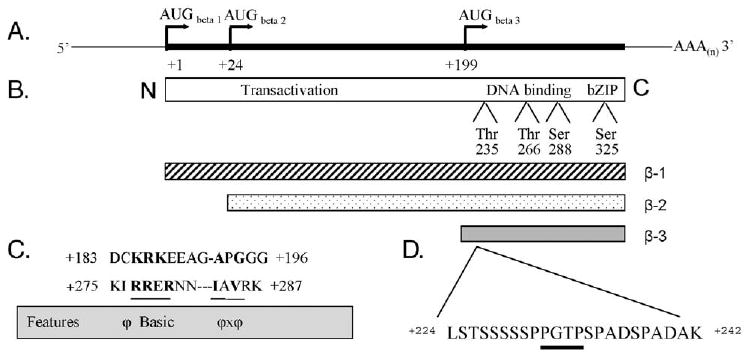
Schematic representation of the transcript of the intronless human C/EBPβ gene, protein isoforms, phosphorylation sites and hypothetical ERK/MAPK docking sites. (A) Drawing of C/EBPβ mRNA demonstrating three distinct in-frame translation initiation start sites. (B) Schematic of the full-length human C/EBPβ protein illustrating the conserved protein domains and the experimentally identified phosphorylation sites, Thr235 (ERK/MAPK), Thr266 (p90RSK), Ser288 (PKA) and Ser325 (CaM-dependent kinase IV), found in all protein isoforms. (C) Primary amino acid sequence of a human C/EBPβ fragment demonstrating hypothetical ERK/MAPK docking sites determined by in silico analysis. Bold lettering represents spatial arrangement of amino acid motifs common in known ERK/MAPK docking sites (outlined by the gray box); hyphens were inserted for alignment purposes. Region +183 to +196 is unique to the human sequence and can only be found in β-1 and β-2, while region +275 to +287 is conserved among mammalian species and can be found in all three isoforms. φ = hydrophobic amino acid, and x = any amino acid. (D) The MAPK phosphoacceptor consensus sequence (dark underline) is found in all three isoforms. The representations depicted in this figure and conclusions derived from in silico analysis were based on the review of published literature [44,81,82].
Previously, studies of C/EBPβ usually focused on the sum of β-1 and β-2 isoforms, while the levels of the rapidly migrating C/EBPβ-3 protein were seldom examined. In this study, we investigated the expression of the individual isoforms of C/EBPβ, and the intracellular localization and phosphorylation of these isoforms in leukemic cells differentiating along the monocytic pathway induced by 1,25D. We show that the 1,25D-induced activating phosphorylation on Thr235 of C/EBPβ isoforms β-2 and β-3 is regulated by the MEK-ERK MAPK pathway, that the phosphorylated isoforms β-2 and β-3, but not β-1, are predominantly localized in cell nuclei, and that the expression and the dependence on the MEK-ERK MAPK pathway of isoform β-2 correlate most closely with 1,25D-induced differentiation of HL60 cells.
Materials and methods
Cell cultures
HL60 cells, derived from a patient with promyeloblastic leukemia [55], were obtained for these studies from the European Collection of Cell Cultures. HL60-G [56], and HL60-40AF cells [57], derived from HL60 cells, are differentiation-sensitive and -resistant subclones of HL60 cells, respectively. These cells were propagated as a suspension culture in RPMI 1640 medium supplemented with 10% fetal calf serum (FCS, Sigma, and St. Louis, MO), 100 units/ml penicillin and 100 μg/ml streptomycin (Sigma, St. Louis, MO). The cells were kept at standard cell culture conditions, i.e. humidified atmosphere of 95% air and 5% CO2 at 37°C. The cell number and viability were determined by hemocytometer counts, and trypan blue (0.4%) exclusion and routine microbiology testing for mycoplasma were conducted. For all experiments, the cells were suspended in fresh medium containing 1,25D or the equivalent volume of ethanol as a vehicle control. Each experimental condition was repeated at least twice.
Chemicals and antibodies
1,25D was a kind gift from Dr. Peter Weber (Hoffmann-La Roche S.A., Pharma Preclinical Research, Basel, Switzerland). PD98059 and SB203580 were from Calbiochem (San Diego, CA). Protein phosphatase from λ phage was obtained from Sigma (St. Louis, MO). Chemiluminescence Blotting Substrate was from Roche Diagnostics (Mannheim, Germany). MY4-RD1 and MO1-FITC recognizing CD14 and CD11b respectively and isotype controls were from Coulter (Miami, FL). Antibodies against β-actin were obtained from Sigma (St. Louis, MO). Rabbit anti-C/EBPα, anti-C/EBPβ, anti-C/EBPε, and anti-Crk-L were from Santa Cruz Biotechnology Inc. (Santa Cruz, CA). Anti-phospho-C/EBPβ (Thr235) antibodies were from Cell Signaling Technology (Beverly, MA). Goat anti-rabbit IgG and anti-mouse IgG conjugated to peroxidase were from Jackson ImmunoResearch (West Grove, PA).
Determination of markers of differentiation and cell cycle progression
Aliquots of 1 × 106 cells were washed twice with 1× phosphate-buffered saline (PBS) then incubated with 1 μl MY4-RD1 and 1 μl MO1-FITC on ice for 45 min to analyze the expression of surface cell markers CD14 and CD11b, respectively. After the incubation, the cells were washed three times with ice-cold 1× PBS and fixed in 1.5% paraformaldehyde. The cells were then suspended in 0.5 ml 1× PBS and analyzed using FACS Calibur flow cytometer (Becton Dickinson, San Jose, CA). The acquisition parameters were set for an isotype control. Data analysis was performed with use of Cell Quest software (Becton Dickinson, San Jose, CA).
To determine cell cycle distributions, aliquots of 1 × 106 cells were washed twice and fixed in 75% ethanol at −20°C overnight, washed twice with PBS and incubated with 100 U/ml of RNase A at 37°C for 1 h. The cell pellet was washed twice and resuspended in a propidium iodide solution (10 μg/ml). The cells were analyzed using an Epics Profile II instrument (Coulter), and the cell cycle distribution was determined by Multicycle Software Program (Phoenix Flow System, San Diego, CA).
Preparation of total cell lysates
In order to obtain total cell lysates, the cells were lysed in SDS sample buffer as described before [30]. Then, samples were sonicated for 10 s twice on ice. The lysed samples were boiled and stored at −20°C for subsequent electrophoretic analyses.
λ-protein phosphatase treatment of lysates
Twenty micrograms of protein samples from the cell lysates prepared as described above was treated with 0.3 μl of λ-protein phosphatase in phosphatase buffer supplied by the manufacturer, with protease inhibitor cocktail added to each sample. All samples, including controls, were kept at 30°C for 30 min then mixed with 3× SDS sample buffer and boiled for 10 min. The samples were stored at −20°C for subsequent electrophoretic analyses to demonstrate the loss of protein phospho groups.
Cell fractionation
Cell fractionation was performed as previously described [58]. The cells were washed 3 times with PBS and lysed for 20 min on ice in lysis buffer (20 mM Tris, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100; pH 7.5) containing protease inhibitor cocktail. The lysates were separated by centrifugation for 5 min at 14,000 rpm at 4°C. Supernatants were designated the membrane/cytosolic fraction, and the nuclei remaining in pellets were sonicated for 10 s in a new portion of lysis buffer. After sonication, nuclei were centrifuged again, and the final supernatants were designated the nuclear fraction. As a control for the fractionation procedure, Western blotting for histone H3 was performed, and histone was detected in the nuclear fraction only.
Western blotting
Western blotting was performed using either whole cell extracts or cell extracts fractionated into membrane/cytosolic and nuclear fractions. Twenty five micrograms (75 μg for detection of C/EBPα and C/EBPε) of protein samples was separated on 12% SDS-PAGE gel and transferred to PVDF membranes. The membranes were blocked then blotted with primary antibodies for 1 h in room temperature, washed three times with TBS and then blotted with horseradish-peroxidase (HRP)-conjugated secondary antibody for 1 h in room temperature. The protein bands were visualized with a chemiluminescence assay system. Then, the membranes were stripped, blocked and probed with subsequent antibodies. The absorbance of each band was quantitated using an Image Quant software (Molecular Dynamics, Sunnyvale, CA).
Results
The kinetics of upregulation of C/EBP family members associated with 1,25D-induced differentiation of HL60 cells
It has been reported previously that C/EBPα and C/EBPε are principally involved in differentiation of granulocytes [6,9,59], while C/EBPβ expression is increased in several differentiation systems [10], including 1,25D-induced differentiation of HL60 cells [13], and is important for proper function of macrophages [11]. However, the expression levels of C/EBPβ isoforms have not been studied in myeloid cells. We therefore determined the kinetics of the upregulation of the protein levels of the isoforms of these transcription factors in differentiating HL60 cells (Fig. 2). In these cells, C/EBPα is upregulated transiently in the early phase of 1,25D-induced differentiation, while the upregulation of C/EBPβ is more marked and persists until late phase of differentiation. Moreover, it seems that, of the three transcription factors studied here, C/EBPβ is the most abundantly expressed because it was necessary to use three times more of cell lysates to detect C/EBPα and C/EBPε (75 μg/lane) than to detect C/EBPβ (25 μg/lane). C/EBPε-1 was also found to be upregulated with the kinetics similar to C/EBPβ, but its increase was only moderate and more transient. The results are illustrated in Fig. 2A, and the quantitation of this series of experiments is presented in Fig. 2B. The isoforms 1, 2 and 3 of C/EBPβ were identified by their molecular mass according to Eaton et al. [44]. For comparison, Fig. 2C shows that the expression of C/EBPβ-2 and C/EBPβ-3 follows a time course similar to the appearance of monocytic differentiation markers on the surface of HL60 cells treated with 1,25D. While it is likely that there is a degree of functional redundancy between the various members of C/EBP family of transcription factors, these data suggest that C/EBPβ isoforms are key players in 1,25D-induced differentiation, consistent with previous reports that C/EBPβ is required for monocyte/macrophage lineage of differentiation [60,61]. Therefore, we focused our additional studies on C/EBPβ isoforms.
Fig. 2.
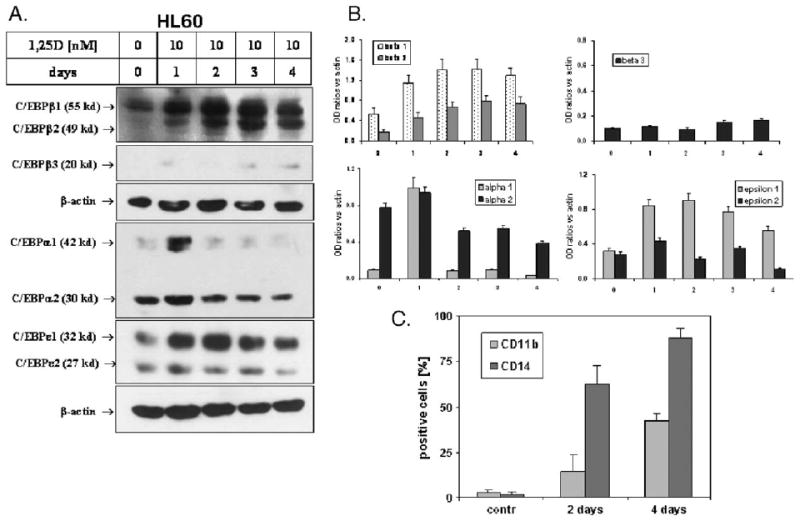
Kinetics of 1,25D-induced expression of C/EBP transcription factors in HL60 cells. (A) Western blots. HL60 cells were treated with 10 nM 1,25D for the times shown. The cell lysates were then prepared, and 25 μg of proteins for C/EBPβ, and 75 μg for C/EBPα and C/EBPε, was separated in SDS-PAGE and blotted onto the membranes. The membranes were then probed with antibodies against C/EBPα, β and ε. β-actin was used as a loading and transfer control. (B) Quantitation of these experiments. OD ratio of each band versus actin was determined. Mean values ± SD, n = 3. (C) An outline of the kinetics of the expression of monocytic cell differentiation surface markers in 1,25D-treated HL60 cells. HL60 cells were treated with 10 nM 1,25D for the indicated times, and the CD11b and CD14 cell surface markers were determined by flow cytometry. Mean values ± SE are indicated, n = 3.
Expression of isoforms of C/EBPβ is regulated by 1,25D in a concentration-dependent manner in HL60-G cells, but not in 1,25D-resistant HL60-40AF cells
The 1,25D-resistant subline of HL60 cells, designated 40AF, can be cultured continuously in the presence of 1,25D at concentrations as high as 40 nM, and differentiation markers do not appear when these cells are exposed for 2 days to even 100 nM 1,25D (Fig. 3A). Although C/EBPβ isoforms β-1 and β-3 can be detected in these cells, they are not upregulated by exposure to 1,25D (Figs. 3B and C). The isoform β-2 is difficult to detect in these differentiation-resistant cells, further showing its link to monocytic differentiation. In rapidly differentiating HL60-G cells, upregulation of C/EBPβ-2 and C/EBPβ-3 expression correlates with the degree of differentiation and with the cell cycle block in G1 phase induced by 1,25D (Figs. 3A, B and C). Although upregulation of CEBPβ-1 by 1,25D also takes place in these cells, it is less marked and a significant increase was noted only at the highest concentration of 1,25D and was accompanied by a band with reduced mobility, suggesting a post-translational modification of the β-1 protein. While the nature of this band is at present unknown, Eaton and Sealy [62] have reported that β-1, but not β-2, is conjugated to SUMO-2 and SUMO-3 [62] (Fig. 3C). A marked increase in C/EBPβ-3 was also noted, but this isoform was also present in HL60-40AF cells which did not differentiate in the presence of 1,25D. Thus, these experiments show that the expression of markers of differentiation of HL60 cells correlates best with C/EBPβ-2 expression and indicate the importance of translational control in 1,25D-induced differentiation.
Fig. 3.
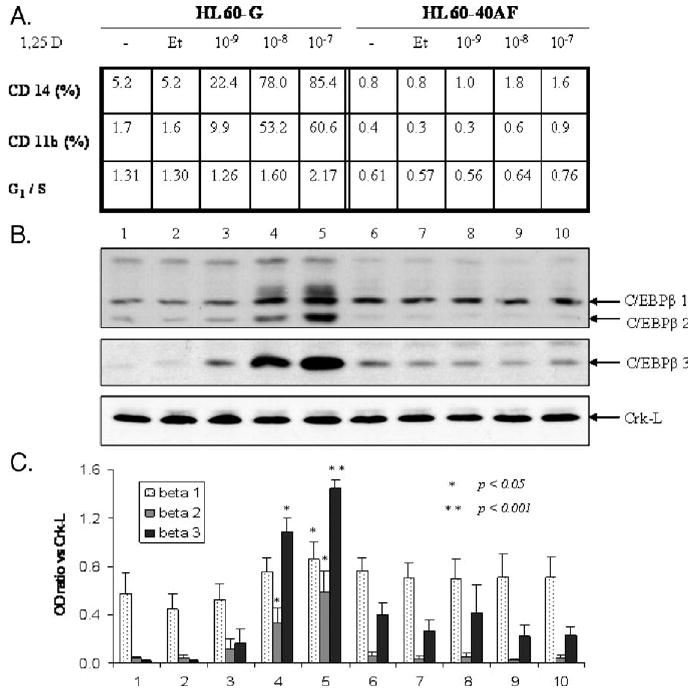
Upregulation of C/EBPβ isoform expression by 1,25D is concentration-dependent and associated with monocytic differentiation and G1 cell cycle arrest. (A) Summary of 1,25D-induced cellular differentiation and cell cycle arrest in HL60-G cells and their absence in 1,25D-resistant subclone HL60-40AF cells following 48 h of treatment with 1,25D. G1/S is the ratio of the percentage of cells in the G1 phase divided by the percentage of cells in the S phase. Note that the ratio increases in HL60-G cells as the cells differentiate, shown by the expression of monocytic surface markers CD11b and CD14. In the 1,25D-resistant HL60-40AF cells, neither the differentiation markers nor the G1 block is apparent. (B) Western blot protein analysis of C/EBPβ isoforms 1–3 in HL60-G and 40AF cells. (C) In cells sensitive to 1,25D, quantitation of the ratio of C/EBPβ signal to loading control (Crk-L) indicates a significant increase in isoforms 2 and 3 expression induced by 10 nM 1,25D (P < 0.05). Elevation of C/EBPβ-1 level requires a higher concentration of 1,25D (100 nM) (P< 0.05). In 1,25D-sensitive cells treated with the higher concentrations of 1,25D, a band appears which migrates more slowly than isoform β-1 (lanes 4 and 5) and may be a SUMO-ylated protein [62]. In HL60-40AF cells, which do not differentiate, the C/EBPβ isoform 2 is not detectable and isoform 1 and 3 levels are not upregulated by 1,25D. Means ± SE are shown, n= 3.
Intracellular localization of C/EBPβ-2 and C/EBPβ-3 is robustly regulated by 1,25D in HL60 cells
C/EBPβ activity is also regulated by its intracellular localization since as a transcription factor it must enter the cell nucleus in order to be transcriptionally active [5]. Therefore, we determined the subcellular distribution of the various C/EBPβ isoforms in HL60 cells before and after an exposure to 1,25D. As shown in Fig. 4, in 1,25D-treated HL60 cells, the majority of C/EBPβ isoforms of lower molecular mass are present in the cell nucleus, while the redistribution of C/EBPβ-1 is less marked. This further suggests that an increase in nuclear presence of C/EBPβ-2 and C/EBPβ-3 is closely linked to monocytic differentiation of HL60 cells.
Fig. 4.
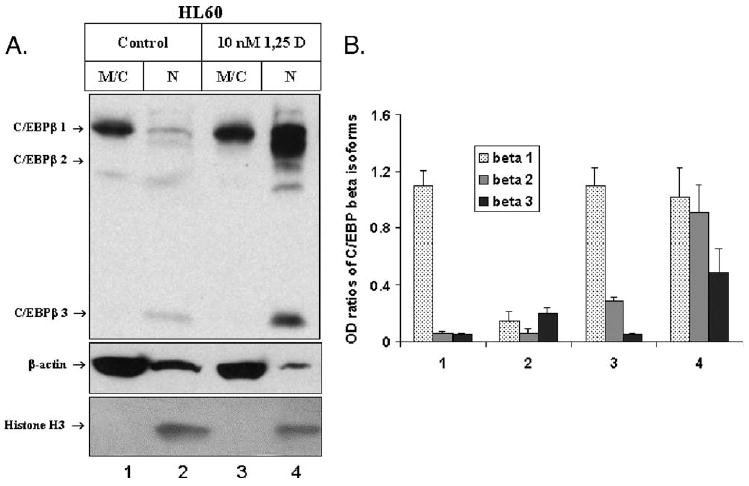
Subcellular localization of C/EBPβ isoforms in HL60 cells treated with 10 nM 1,25D. (A) Western blot. HL60 cells were treated for 3 days with 10 nM 1,25D. The cells were then fractionated into membrane/cytosol (M/C) and nuclei (N). After separation by electrophoresis and transfer, the immunoblots were probed with antibodies against C/EBPβ, histone H3 and β-actin as fractionation/loading controls. In addition to the three C/EBPβ isoforms, unidentified bands, possibly cleavage products of C/EBPβ, are present, particularly evident in the nuclear fractions. (B) Quantitation of the experiment. Mean values of normalized OD ± SE are indicated, n= 3.
In 1,25D-induced differentiation of HL60 cells the expression of C/EBPβ isoforms depends on ERK and p38 MAP kinase pathways
It has been shown previously that monocytic differentiation of HL60 cells can be influenced by the activity of MAPK signal transduction pathways [34,63]. It is also known from the literature that C/EBPβ can be regulated by several kinases including Ras-dependent MAP kinases such as ERK (e.g. [50,51,54]) and p90RSK, which may be a downstream target of ERK-1/2 kinases [53]. We now demonstrate that the expression of the C/EBPβ isoforms is influenced by the MAPK inhibitors that modulate 1,25D-induced HL60 cell differentiation (Fig. 5). In this system, the MEK1-ERK inhibitor PD98059 [64] markedly reduces the effect of 1,25D on the expression of all three isoforms of C/EBPβ, while the p38 MAPK inhibitor SB203580 [65] slightly increases the 1,25D-induced expression of C/EBPβ-2 (Figs. 5A and B). These effects on C/EBPβ-2 are similar to the effects of inhibitors on the CD11b marker of monocytic differentiation (Fig. 5C). The effect on CD14 is not apparent in this experiment as 1,25D alone produced maximal expression of this marker.
Fig. 5.
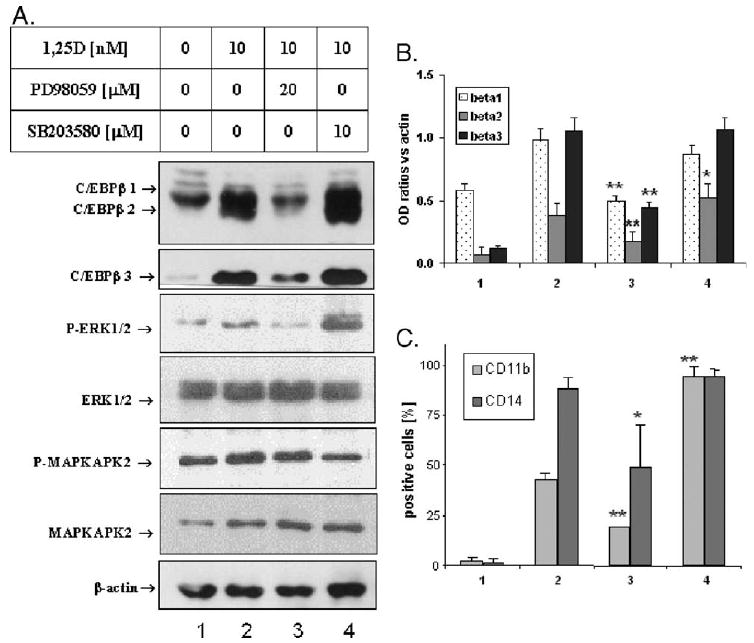
Expression of C/EBPβ isoforms and HL60 cells differentiation are regulated by MAPK pathways. (A) Western blots. HL60 cells were treated for 4 days with 10 nM 1,25D ± 20 μM PD98059 or 10 μM SB203580. The cell lysates were prepared, and equal amounts of proteins were separated in SDS-PAGE and blotted onto the membrane which was probed with the indicated antibodies. β-actin was used as a loading and transfer control. (B) Quantitation of these experiments. The OD ratio of each band was determined. (C) The CD11b and CD14 cell surface markers were determined by flow cytometry. Mean values ± SE are indicated, n= 3. PD98059 significantly decreased 1,25D-induced expression of CD11b (P = 0.006) and CD14 (P = 0.04), while SB203580 significantly increased 1,25D-induced expression of CD11b (P = 0.001), but not CD14 (P > 0.05). *P < 0.05; **P < 0.01.
In these experiments, we monitored the effects of the inhibitors by determining the phosphorylation status of the principal downstream protein targets of the kinases inhibited by the pharmacological agents, ERK 1/2 as a target of the MEK inhibitor PD98059 and MAPKAPK2, a target of p38 MAPK inhibitor SB203580. As expected, the 1,25D-induced phosphorylation was reduced by the appropriate inhibitor. The previously reported [66] increase in ERK 1/2 phosphorylation was also noted in SB203580-treated cells (Fig. 5A).
Nuclear localization of C/EBPβ isoforms is MEK-ERK- and p38-MAPK-pathway-dependent
When we examined how PD98059 and SB203580 influence nuclear localization of C/EBPβ isoforms, we found that PD98059 strongly inhibited the apparent nuclear translocation of all isoforms of C/EBPβ in 1,25D-treated cells, while SB203580 increased the nuclear presence of C/EBPβ-3 (Fig. 6A).
Fig. 6.
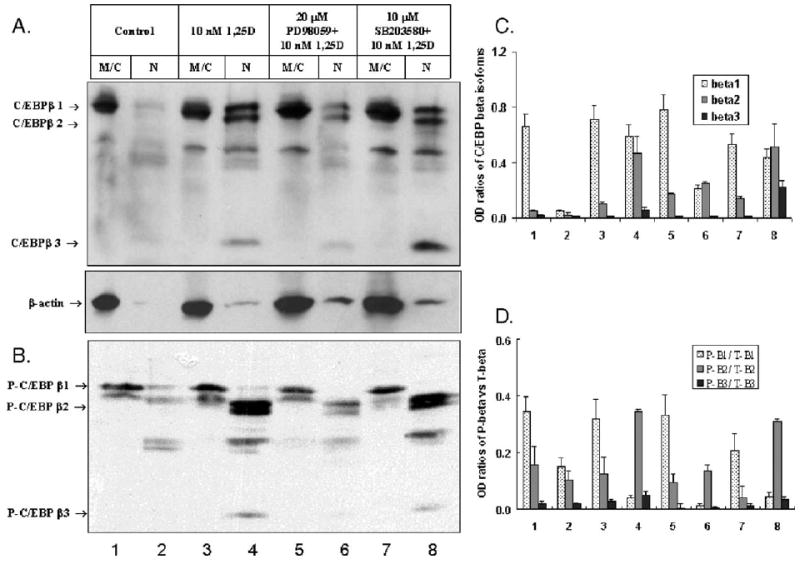
Subcellular localization of C/EBPβ isoforms in HL60 cells treated with 10 nM 1,25D is PD98059- and SB203580-dependent. (A) Western blot for total proteins. HL60 cells were treated for 3 days with 10 nM 1,25D ± 20 μM PD98059 or 10 μM SB203580. After incubation, the cells were separated into membrane/cytosol (M/C) and nuclear (N) fractions. The immunoblots were probed against C/EBPβ then stripped and reprobed against β-actin as a loading and transfer control of the cytoplasmic fractions. The band migrating at approximately 35 kDa may be a cleavage product of one of the larger isoforms. (B) Western blot probed with antibody against phospho-C/EBPβ (Thr235). Phosphorylated forms of C/EBPβ-2 and β-3 and the putative cleavage product, but not of the β-1 isoform, are localized in the nuclei of 1,25D-treated HL60 cells. (C) Quantitation of the experiments shown in panel A, mean values of OD of each band ± SE are indicated, n = 3. (D) Quantitation of the experiments shown in panel B, mean values of OD of each band ± SE are indicated, n = 3.
This figure also displays an approximately 35 kDa band which is frequently seen on Western blots from different cells types, e.g. mammary epithelial cells [44] and Wilms' tumor [67]. Since this band disappears following transfection of C/EBPβ siRNA into Wilms' tumor cells [67], and there are no AUG start sites in this region of the C/EBPβ mRNA, this band is likely to represent a cleavage product of a larger isoform of C/EBPβ. Indeed, Welm et al. reported in a murine system the generation of additional isoforms of C/EBPβ by specific cleavage of the full-length C/EBPβ [68]. Our data therefore suggest that in 1,25D-treated HL60 cells nuclear localization of C/EBPβ isoforms and of their putative cleavage product is facilitated by the activity of the ERK MAPK pathway, while the p38 MAPK pathway may inhibit the entry of C/EBPβ-3 into the nucleus.
Phosphorylation of C/EBPβ isoforms is regulated by the MAPK pathways in 1,25D-treated HL60 cells
Transcriptional activity of C/EBPβ may be regulated not only by its expression and cellular localization but also by multiple phosphorylations. Antibodies recognizing all potential phospho-C/EBPβ sites are not available at this time, but we were able to use the antibody to phospho-Thr235 in C/EBPβ, the site reported to activate transcriptional of C/EBPβ proteins [69–71]. Surprisingly, the Thr235 phosphorylation of the full-length C/EBPβ-1 isoform was found to be constitutive, and although it was slightly increased by the exposure to 1,25D, this was not accompanied by its translocation to the nucleus (Fig. 6B). In contrast, Thr235-phosphorylated isoforms β-2 and β-3 were found only in the nuclear fractions, and their levels were reduced by the inhibition of the MEK-ERK pathway by PD98059 and, rather unexpectedly, also by SB203580, though to a lesser extent (Fig. 6B). Since isoform β-3 has no transactivating activity, these results point to C/EBPβ-2 as the functional transcription factor in this system.
In these experiments, we also pretreated the samples with λ phage protein phosphatase as a control for the specificity of the antibody for phosphate groups and found no signal for Thr235 phosphorylated proteins, confirming the specificity of the antibody (data not shown).
Discussion
This study presents several novel facets of the involvement of C/EBPβ in 1,25D-induced monocytic differentiation of myeloid leukemia cells. We found that the presence of isoforms of C/EBPβ, but not of C/EBPα or C/EBPε, correlates with the appearance of monocytic phenotype and that the low molecular mass isoforms (C/EBPβ-2 and C/EBPβ-3) increase more in rapidly differentiating HL60-G cells than in the generic HL60 cells. In contrast, there was no increase in any C/EBPβ isoform in the differentiation-resistant HL60-40AF cells. The observation that there is only modest regulation of C/EBPβ-1 by 1,25D and the predominantly cytoplasmic localization of its phosphorylated form suggest that isoforms β-2 and β-3 play a more significant role in 1,25D-induced monocytic differentiation than the full-length β-1 protein. Interestingly, the increase in the transactivation enhancing C/EBPβ-2 is accompanied by an increase in the dominant-negative C/EBPβ-3, which represses transcription because it does not possess a transactivation domain, although it can bind to proteins that activate transcription [10,41]. This implies that C/EBPβ-3 may play a part in the repression of genes the expression of which is unnecessary in differentiating cells, such as those that participate in cell proliferation. Thus, the expression of different isoforms of C/EBPβ can provide one of the mechanisms for the coupling of differentiation to cell cycle arrest. In agreement with this notion, we noted a close correlation between the onset of the G1 to S phase block and the upregulation of CEBPβ isoforms, including isoform β-3 (Fig. 3).
As is well known, transcription factors function primarily in the nucleus, and C/EBPβ isoforms are no exception [46–48]. We show here that the 1,25D-induced increases in the abundance of the C/EBPβ isoforms β-2 and β-3 are detected almost entirely in the cell nuclei. Furthermore, this localization is associated with phosphorylation of these isoforms on the transcription-activating site Thr235, present in all isoforms of C/EBPβ (see Fig. 1), but quite unexpectedly, phosphorylated C/EBPβ-1 is not found in the nuclear fraction. Nevertheless, there is clear evidence that, in 1,25D-treated cells all isoforms are, at least in part, phosphorylated by the ERK MAPK.
The lower levels of 1,25D-induced increase in C/EBPβ-3 phosphorylation on Thr235, an ERK site [50,51], as compared to phosphorylation of β-1 and β-2 C/EBP proteins, may be explained by the absence of one of the two possible ERK docking sites on this truncated protein. As illustrated in Fig. 1C, only the C-terminal potential ERK docking site is present in C/EBPβ-3, which may allow Thr235 phosphorylation by ERK2 (Fig. 6C). In this scenario, the additional docking site would increase the efficiency of Thr235 phosphorylation. From a functional aspect, the low level of transcription activating Thr235 phosphorylation is consistent with the absence of the N-terminal transactivating domain in the C/EBPβ-3 protein (Fig. 1) since such a phosphorylation would be unnecessary.
As noted above, not only the phosphorylation of C/EBPβ-2 and C/EBPβ-3 but also their protein levels are under the control of the MAPK pathways in 1,25D-treated cells, as shown by the effect of the MEK-ERK inhibitor PD98059 and the p38 MAPK inhibitor SB203580 (Figs. 5 and 6). Although PD98059 has a greater effect on the 1,25D-induced increases on the protein levels and phosphorylation of C/EBPβ isoforms than SB203580, the latter effect is significant and may be explained by the report that phosphorylation of C/EBPβ can occur by the p38 MAPK in human monocytes [72]. The mechanisms responsible for the regulation of protein levels are not clear at present, though previous studies suggest that regulation of the initiation of translation at the in-frame AUGs is a possibility (Fig. 1). For instance, it is known that a protein that binds to the GC-rich sequences (CUGBP-1) within the 5′ region of C/EBPβ mRNA induces downstream translation of LIP [42], the rodent equivalent of C/EBPβ-3 [10]. The effect of 1,25D on the C/EBPβ protein levels may therefore be explained by a requirement for the activity of the MEK-ERK pathway for the 1,25D-induced expression of CUGBP-1 or its inhibition by the p38 MAPK pathway. Alternatively, these pathways may regulate the binding of CUGBP-1 to C/EBPβ mRNA. The binding of another regulator of C/EBPβ isoform translation, such as calreticulin [43], could also provide a mechanism for 1,25D control of the expression of C/EBPβ isoform proteins.
These and our previous studies [13,14] indicate positive interactions between VDR and C/EBP transcriptional activity in myeloid leukemia cells. Such interactions are being increasingly recognized in other systems as well (e.g. [73,74,75]) and can occur by a variety of mechanisms that are likely to be cell-context-specific. In the case of myeloid cells, we have recently outlined a pathway that leads from VDR activation by 1,25D to monocytic differentiation [14]. The key elements of this pathway are transcriptional upregulation by VDR of KSR-1 expression [76], which then amplifies the signal for differentiation by increasing the efficiency of the MAPK cascade, which includes MEK and ERK. Data provided here and elsewhere (e.g. [77]) link MAPKs to C/EBPβ expression, and C/EBPs are known to contribute to the expression of several markers of myelomonocytic differentiation including CD14 [78]. In addition, the human VDR promoter has C/EBP-binding elements, and Dhawan et al. [73] have recently shown that C/EBPβ increases protein-kinase-A-mediated transcription of VDR, thus potentially closing a “feed forward” loop. However, the relative contributions of the different isoforms of C/EBPβ were not previously considered in this context, and the identification of its isoform β-2 as the principal functional transcription factor in human myeloid leukemia cells is the major novel finding of this report.
While the possibilities outlined above remain to be explored further, the elucidation of mechanisms responsible for the upregulation of C/EBPβ isoforms and their interactions with VDR during monocytic differentiation will be of great importance as it may increase the opportunities for differentiation therapy of myeloid leukemias, currently being developed at both preclinical and clinical levels (e.g. [79,80]).
Acknowledgments
We thank Dr. Robert Donnelly for comments on the manuscript. This work was supported by NIH grant R01 CA 44722-16 from the National Cancer Institute to GPS and a Research Supplement for Underrepresented Minorities to R01 CA 44722 awarded by the CMMB/NCI to EG.
References
- 1.Danova M, Aglietta M. Cytokine receptors, growth factors and cell cycle in human bone marrow and peripheral blood hematopoietic progenitors. Haematologica. 1997;82:622–629. [PubMed] [Google Scholar]
- 2.Martyre MC, Le Bousse-Kerdiles MC. Members of the French INSERM Research Network on Myelofibrosis with Myeloid Metaplasia, Stem cell dysregulation in myelofibrosis with myeloid metaplasia: current data on growth factor and transcription factor involvement. Semin Oncol. 2005;32:373–379. doi: 10.1053/j.seminoncol.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 3.Koschmieder, Rosenbauer F, Steidl U, Owens BM, Tenen DG. Role of transcription factors C/EBPalpha and PU.1 in normal hematopoiesis and leukemia. Int J Hematol. 2005;81:368–377. doi: 10.1532/ijh97.05051. [DOI] [PubMed] [Google Scholar]
- 4.Graves BJ, Johnson PF, McKnight SL. Homologous recognition of a promoter domain common to the MSV LTR and the HSV tk gene. Cell. 1986;44:565–576. doi: 10.1016/0092-8674(86)90266-7. [DOI] [PubMed] [Google Scholar]
- 5.Johnson PF, Landschulz WH, Graves BJ, McKnight SL. Identification of a rat liver nuclear protein that binds to the enhancer core element of three animal viruses. Genes Dev. 1987;2:133–146. doi: 10.1101/gad.1.2.133. [DOI] [PubMed] [Google Scholar]
- 6.Tenen DG, Hromas R, Licht JD, Zhang DE. Transcription factors, normal myeloid development, and leukemia. Blood. 1997;90:489–519. [PubMed] [Google Scholar]
- 7.Pabst T, Mueller BU, Zhang P, Radomska HS, Narravula S, Schnittger S, Behre G, Hiddemann W, Tenen DG. Dominant-negative mutations of CEBPA, encoding CCAAT/enhancer binding protein-alpha (C/EBPalpha), in acute myeloid leukemia. Nat Genet. 2001;27:263–270. doi: 10.1038/85820. [DOI] [PubMed] [Google Scholar]
- 8.Porse BT, Bryder D, Theilgaard-Monch K, Hasemann MS, Anderson K, Damgaard I, Jacobsen SE, Nerlov C. Loss of C/EBP alpha cell cycle control increases myeloid progenitor proliferation and transforms the neutrophil granulocyte lineage. J Exp Med. 2005;202:85–96. doi: 10.1084/jem.20050067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gery S, Gombart AF, Yi WS, Koeffler C, Hofmann WK, Koeffler HP. Transcription profiling of C/EBP targets identifies Per2 as a gene implicated in myeloid leukemia. Blood. 2005;106:2827–2836. doi: 10.1182/blood-2005-01-0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lekstrom-Himes J, Xanthopoulos KG. Biological role of the CCAAT/enhancer-binding protein family of transcription factors. J Biol Chem. 1998;273:28545–28548. doi: 10.1074/jbc.273.44.28545. [DOI] [PubMed] [Google Scholar]
- 11.Poli V. The role of C/EBP isoforms in the control of inflammatory and native immunity functions. J Biol Chem. 1998;273:29279–29282. doi: 10.1074/jbc.273.45.29279. [DOI] [PubMed] [Google Scholar]
- 12.Ramji DP, Foka P. CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem J. 2002;365:561–575. doi: 10.1042/BJ20020508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ji Y, Studzinski GP. Retinoblastoma protein and CCAAT/enhancer-binding protein beta are required for 1,25-dihydroxyvitamin D3-induced monocytic differentiation of HL60 cells. Cancer Res. 2004;64:370–377. doi: 10.1158/0008-5472.can-03-3029. [DOI] [PubMed] [Google Scholar]
- 14.Studzinski GP, Wang X, Ji Y, Wang Q, Zhang Y, Kutner A, Harrison JS. The rationale for deltanoids in therapy for myeloid leukemia: role of KSR-MAPK-C/EBP pathway. J Steroid Biochem Mol Biol. 2005;97:47–55. doi: 10.1016/j.jsbmb.2005.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abe E, Miamura C, Sakagami H, Takeda M, Konno K, Yamazaki T, Yoshiki S, Suda T. Differentiation of mouse myeloid leukemia cells induced by 1-alpha,25-dihydroxyvitamin D3. Proc Natl Acad Sci U S A. 1981;78:4990–4994. doi: 10.1073/pnas.78.8.4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Studzinski GP, Bhandal AK, Brelvi ZS. A system for monocytic differentiation of leukemic cells HL 60 by a short exposure to 1,25-dihydroxycholecalciferol. Proc Soc Exp Biol Med. 1985;179:288–295. doi: 10.3181/00379727-179-42098. [DOI] [PubMed] [Google Scholar]
- 17.Suda T, Takahashi N, Martin TJ. Modulation of osteoclast differentiation. Endocr Rev. 1995;4:266–270. doi: 10.1210/edrv-13-1-66. [DOI] [PubMed] [Google Scholar]
- 18.Fogh K, Kragballe K. Recent developments in vitamin D analogs. Curr Pharm Des. 2000;6:829–972. doi: 10.2174/1381612003400128. [DOI] [PubMed] [Google Scholar]
- 19.Luong QT, Koeffler HP. Vitamin D compounds in leukemia. J Steroid Biochem Mol Biol. 2005;97:195–202. doi: 10.1016/j.jsbmb.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 20.Studzinski GP, Rathod B, Wang QM, Rao J, Zhang F. Uncoupling of cell cycle arrest from the expression of monocytic differentiation markers in HL60 cell variants. Exp Cell Res. 1997;232:376–387. doi: 10.1006/excr.1997.3484. [DOI] [PubMed] [Google Scholar]
- 21.Aslam F, McCabe L, Frenkel B, van Wijnen AJ, Stein GS, Lian JB, Stein JL. AP-1 and vitamin D receptor (VDR) signaling pathways converge at the rat osteocalcin VDR element: requirement for the internal activating protein-1 site for vitamin D-mediated trans-activation. Endocrinology. 1999;140:63–70. doi: 10.1210/endo.140.1.6429. [DOI] [PubMed] [Google Scholar]
- 22.Adachi R, Shulman AI, Yamamoto K, Shimomura I, Yamada DJS, Mangelsdorf M. Structural determinants for vitamin D receptor response to endocrine and xenobiotic signals. Mol Endocrinol. 2004;18:43–52. doi: 10.1210/me.2003-0244. [DOI] [PubMed] [Google Scholar]
- 23.Feldman D, Malloy PJ. Hereditary 1,25-dihydroxyvitamin D resistant rickets: molecular basis and implications for the role of 1,25(OH) 2D3 in normal physiology. Mol Cell Endocrinol. 1990;72:C57–C62. doi: 10.1016/0303-7207(90)90137-w. [DOI] [PubMed] [Google Scholar]
- 24.Simpson RU, O'Connell TD, Pan Q, Newhouse J, Somerman MJ. Antisense oligonucleotides targeted against protein kinase C beta and C beta II block 1,25-(OH)2D3-induced differentiation. J Biol Chem. 1998;273:19587–19591. doi: 10.1074/jbc.273.31.19587. [DOI] [PubMed] [Google Scholar]
- 25.Sitrin MD, Bissonnette M, Bolt MJ, Wali R, Khare S, Scaglione-Sewell B, Skarosi S, Brasitus TA. Rapid effects of 1,25(OH)2 vitamin D3 on signal transduction systems in colonic cells. Steroids. 1999;64:137–142. doi: 10.1016/s0039-128x(98)00102-0. [DOI] [PubMed] [Google Scholar]
- 26.Bikle DD, Oda Y, Xie Z. Calcium and 1,25(OH)2D3: interacting drivers of epidermal differentiation. J Steroid Biochem Mol Biol. 2004;90:355–360. doi: 10.1016/j.jsbmb.2004.03.020. [DOI] [PubMed] [Google Scholar]
- 27.Ellison TI, Dowd DR, MacDonald PN. Calmodulin-dependent kinase IV stimulates vitamin D receptor-mediated transcription. Mol Endocrinol. 2005;19:2309–2319. doi: 10.1210/me.2004-0382. [DOI] [PubMed] [Google Scholar]
- 28.Sergeev IN. Calcium signaling in cancer and vitamin D. J Steroid Biochem Mol Biol. 2005;97:145–151. doi: 10.1016/j.jsbmb.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 29.Hmama Z, Nandan D, Sly L, Knutson KL, Herrera-Velit P, Reiner NE. 1alpha, 25-dihydroxyvitamin D(3)-induced myeloid cell differentiation is regulated by a vitamin D receptor-phosphatidylinositol 3-kinase signaling complex. J Exp Med. 1999;190:1583–1594. doi: 10.1084/jem.190.11.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marcinkowska E, Wiedlocha A, Radzikowski C. Evidence that phosphatidylinositol 3-kinase and p70S6K protein are involved in differentiation of HL-60 cells induced by calcitriol. Anticancer Res. 1998;18:3507–3514. [PubMed] [Google Scholar]
- 31.Zhang Y, Zhang J, Studzinski GP. AKT pathway is activated by 1,25-dihydroxyvitamin D3 and participates in its anti-apoptotic effect and cell cycle control in differentiating HL60 cells. Cell Cycle. 2006;5:447–451. doi: 10.4161/cc.5.4.2467. [DOI] [PubMed] [Google Scholar]
- 32.Beno DW, Brady LM, Bissonnette M, Davis BH. Protein kinase C and mitogen-activated protein kinase are required for 1,25-dihydroxyvitamin D3-stimulated Egr induction. J Biol Chem. 1995;270:3642–3647. doi: 10.1074/jbc.270.8.3642. [DOI] [PubMed] [Google Scholar]
- 33.Marcinkowska E, Wiedlocha A, Radzikowski C. 1,25-dihydroxyvitamin D3 induced activation and subsequent nuclear translocation of MAPK is upstream regulated by PKC in HL-60 cells. Biochem Biophys Res Commun. 1997;241:419–426. doi: 10.1006/bbrc.1997.7832. [DOI] [PubMed] [Google Scholar]
- 34.Wang X, Rao J, Studzinski GP. Inhibition of p38 MAP kinase activity up-regulates multiple MAP kinase pathways and potentiates 1,25-dihydroxyvitamin D3-induced differentiation of human leukemia HL60 cells. Exp Cell Res. 2000;258:425–437. doi: 10.1006/excr.2000.4939. [DOI] [PubMed] [Google Scholar]
- 35.Wang X, Studzinski GP. Activation of extracellular signal-regulated kinases (ERKs) defines the first phase of 1,25-dihydroxyvitamin D3-induced differentiation of HL60 cells. J Cell Biochem. 2001;80:471–482. doi: 10.1002/1097-4644(20010315)80:4<471::aid-jcb1001>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 36.Narayanan R, Sepulveda VA, Falzon M, Weigel NL. The functional consequences of cross-talk between the vitamin D receptor and ERK signaling pathways are cell-specific. J Biol Chem. 2004;279:47298–47310. doi: 10.1074/jbc.M404101200. [DOI] [PubMed] [Google Scholar]
- 37.Buitrago CG, Ronda AC, Boland AR, Boland R. MAP kinases p38 and JNK are activated by the steroid hormone 1alpha, 25 (OH)2-vitamin D3 in the C2C12 muscle cell line. J Cell Biochem. 2006;97:698–708. doi: 10.1002/jcb.20639. [DOI] [PubMed] [Google Scholar]
- 38.Chen A, Davis BH, Bissonnette M, Scaglione-Sewell B, Brasitus TA. 1,25-dihydroxyvitamin D3 stimulates activator protein-1-dependent Caco-2 cell differentiation. J Biol Chem. 1999;274:35505–35513. doi: 10.1074/jbc.274.50.35505. [DOI] [PubMed] [Google Scholar]
- 39.Johansen C, Kragballe K, Henningsen J, Westergaard M, Kristiansen K, Iversen L. 1alpha, 25-dihydroxyvitamin D3 stimulates activator protein 1 DNA-binding activity by a phosphatidylinositol 3-kinase/Ras/MEK/extracellular signal regulated kinase 1/2 and c-Jun N-terminal kinase 1-dependent increase in c-Fos, Fra1, and c-Jun expression in human keratinocytes. J Invest Dermatol. 2003;120:561–570. doi: 10.1046/j.1523-1747.2003.12095.x. [DOI] [PubMed] [Google Scholar]
- 40.Wang X, Studzinski GP. Inhibition of p38MAP kinase potentiates the JNK/SAPK pathway and AP-1 activity in monocytic but not in macrophage or granulocytic differentiation of HL60 cells. J Cell Biochem. 2001;82:68–77. doi: 10.1002/jcb.1141. [DOI] [PubMed] [Google Scholar]
- 41.Descombes P, Schibler U. A liver-enriched transcriptional activator protein, LAP, and a transcriptional inhibitory protein, LIP, are translated from the same mRNA. Cell. 1991;3:569–579. doi: 10.1016/0092-8674(91)90531-3. [DOI] [PubMed] [Google Scholar]
- 42.Timchenko NA, Welm AL, Lu X, Timchenko LT. CUG repeat binding protein (CUGBP1) interacts with the 5′ region of C/EBP beta mRNA and regulates translation of C/EBP beta isoforms. Nucleic Acids Res. 1999;27:4517–4525. doi: 10.1093/nar/27.22.4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Timchenko LT, Iakova P, Welm AL, Cai ZJ, Timchenko NA. Calreticulin interacts with C/EBPalpha and C/EBP beta mRNAs and represses translation of C/EBP proteins. Mol Cell Biol. 2002;22:7242–7257. doi: 10.1128/MCB.22.20.7242-7257.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eaton EM, Hanlon M, Bundy L, Sealy L. Characterization of C/EBP beta isoforms in normal versus neoplastic mammary epithelial cells. J Cell Physiol. 2001;189:91–105. doi: 10.1002/jcp.1139. [DOI] [PubMed] [Google Scholar]
- 45.Charles A, Tang X, Crouch E, Brody JS, Xiao ZX. Retinoblastoma protein complexes with C/EBP proteins and activates C/EBP mediated transcription. J Cell Biochem. 2001;83:414–425. doi: 10.1002/jcb.1239. [DOI] [PubMed] [Google Scholar]
- 46.Wang H, Iakova P, Wilde M, Welm A, Goode T, Roesler WJ, Timchenko NA. C/EBPalpha arrests cell proliferation through direct inhibition of Cdk2 and Cdk4. Mol Cell. 2001;4:817–828. doi: 10.1016/s1097-2765(01)00366-5. [DOI] [PubMed] [Google Scholar]
- 47.Metz R, Ziff E. cAMP stimulates the C/EBP-related transcription factor rNFIL-6 to trans-locate to the nucleus and induce c-fos transcription. Genes Dev. 1991;5:1754–1766. doi: 10.1101/gad.5.10.1754. [DOI] [PubMed] [Google Scholar]
- 48.Chinery R, Brockman JA, Dransfield DT, Coffey RJ. Antioxidant-induced nuclear translocation of CCAAT/enhancer-binding protein beta. A critical role for protein kinase A-mediated phosphorylation of Ser299. J Biol Chem. 1997;272:30356–30361. doi: 10.1074/jbc.272.48.30356. [DOI] [PubMed] [Google Scholar]
- 49.Pilipuk Piwien G, Galigniana MD, Schwartz J. Subnuclear localization of C/EBP beta is regulated by growth hormone and dependent on MAPK. J Biol Chem. 2003;278:35668–35677. doi: 10.1074/jbc.M305182200. [DOI] [PubMed] [Google Scholar]
- 50.Nakajima T, Kinoshita S, Sasagawa T, Sasaki K, Naruto M, Kishimoto T, Akira S. Phosphorylation at threonine-235 by a ras-dependent mitogen-activated protein kinase cascade is essential for transcription factor NF-IL6. Proc Natl Acad Sci U S A. 1993;90:2207–2211. doi: 10.1073/pnas.90.6.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Berg T, Didon L, Barton J, Andersson O, Nord M. Glucocorticoids increase C/EBP beta activity in the lung epithelium via phosphorylation. Biochem Biophys Res Commun. 2005;334:638–645. doi: 10.1016/j.bbrc.2005.06.146. [DOI] [PubMed] [Google Scholar]
- 52.Wegner M, Cao Z, Rosenfeld MG. Calcium-regulated phosphorylation within the leucine zipper of C/EBP beta. Science. 1992;256:370–373. doi: 10.1126/science.256.5055.370. [DOI] [PubMed] [Google Scholar]
- 53.Buck M, Poli V, Hunter T, Chojkier M. C/EBP beta phosphorylation by RSK creates a functional XEXD caspase inhibitory box critical for cell survival. Mol Cell. 2001;8:807–816. doi: 10.1016/s1097-2765(01)00374-4. [DOI] [PubMed] [Google Scholar]
- 54.Zhu S, Yoon K, Sterneck E, Johnson PF, Smart RC. CCAAT/enhancer binding protein-beta is a mediator of keratinocyte survival and skin tumorigenesis involving oncogenic Ras signaling. Proc Natl Acad Sci U S A. 2002;99:207–212. doi: 10.1073/pnas.012437299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gallagher R, Collins S, Trujillo J, McCredie K, Ahearn M, Tsai S, Metzgar R, Aulakh G, Ting R, Ruscetti F, Gallo R. Characterization of the continuous, differentiating myeloid cell line (HL-60) from a patient with acute promyelocytic leukemia. Blood. 1979;54:713–733. [PubMed] [Google Scholar]
- 56.Studzinski GP, Reddy KB, Hill HZ, Bhandal AK. Potentiation of 1-beta-d-arabinofuranosylcytosine cytotoxicity to HL-60 cells by 1,25-dihydroxyvitamin D3 correlates with reduced rate of maturation of DNA replication intermediates. Cancer Res. 1991;51:3451–3455. [PubMed] [Google Scholar]
- 57.Studzinski GP, Rathod B, Rao J, Kheir A, Wajchman HJ, Zhang F, Finan JB, Nowell PC. Transition to tetraploidy in 1,25-dihydroxyvitamin D3-resistant HL60 cells is preceded by reduced growth factor dependence and constitutive up-regulation of Sp1 and AP-1 transcription factors. Cancer Res. 1996;56:5513–5521. [PubMed] [Google Scholar]
- 58.Wiedlocha A, Nilsen T, Wesche J, Sorensen V, Malecki J, Marcinkowska E, Olsnes S. Phosphorylation-regulated nucleocytoplasmic trafficking of internalized fibroblast growth factor-1. Mol Biol Cell. 2005;16:794–810. doi: 10.1091/mbc.E04-05-0389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Morosetti R, Park DJ, Chumakov AM, Grillier I, Shiohara M, Gombart AF, Nakamaki T, Weinberg K, Koeffler HP. A novel, myeloid transcription factor, C/EBP epsilon, is upregulated during granulocytic, but not monocytic, differentiation. Blood. 1997;90:2591–2600. [PubMed] [Google Scholar]
- 60.Natsuka S, Akira S, Nishio Y, Hashimoto S, Sugita T, Isshiki H, Kishimoto T. Macrophage differentiation-specific expression of NF-IL6, a transcription factor for interleukin-6. Blood. 1992;79:460–466. [PubMed] [Google Scholar]
- 61.Tanaka T, Akira S, Yoshida K, Umemoto M, Yoneda Y, Shirafuji N, Fujiwara H, Suematsu S, Yoshida N, Kishimoto T. Targeted disruption of the NF-IL6 gene discloses its essential role in bacteria killing and tumor cytotoxicity by macrophages. Cell. 1995;80:353–361. doi: 10.1016/0092-8674(95)90418-2. [DOI] [PubMed] [Google Scholar]
- 62.Eaton EM, Sealy L. Modification of CCAAT/enhancer-binding protein-beta by the small ubiquitin-like modifier (SUMO) family members, SUMO-2 and SUMO-3. J Biol Chem. 2003;278:33416–33421. doi: 10.1074/jbc.M305680200. [DOI] [PubMed] [Google Scholar]
- 63.Marcinkowska E. Evidence that activation of MEK1,2/erk 1,2 signal transduction pathway is necessary for calcitriol-induced differentiation of HL-60 cells. Anticancer Res. 2001;2:499–504. [PubMed] [Google Scholar]
- 64.Alessi DR, Cuenda A, Cohen P, Dudley DT, Saltiel AR. PD 098059 is a specific inhibitor of the activation of mitogen-activated protein kinase kinase in vitro and in vivo. J Biol Chem. 1995;270:27489–27494. doi: 10.1074/jbc.270.46.27489. [DOI] [PubMed] [Google Scholar]
- 65.Cuenda A, Rouse J, Doza YN, Meier R, Cohen P, Gallagher TF, Young PR, Lee JC. SB 203580 is a specific inhibitor of a MAP kinase homologue which is stimulated by cellular stresses and interleukin-1. FEBS Lett. 1995;364:229–233. doi: 10.1016/0014-5793(95)00357-f. [DOI] [PubMed] [Google Scholar]
- 66.Wang X, Rao J, Studzinski GP. Inhibition of p38 MAP kinase activity up-regulates multiple MAP kinase pathways and potentiates 1,25-dihydroxyvitamin D3-induced differentiation of human leukemia HL60 cells. Exp Cell Res. 2000;258:425–437. doi: 10.1006/excr.2000.4939. [DOI] [PubMed] [Google Scholar]
- 67.Li W, Kessler P, Yeger H, Alami J, Reeve AE, Heathcott R, Skeen J, Williams BR. A gene expression signature for relapse of primary Wilms tumors. Cancer Res. 2005;65:2592–2601. doi: 10.1158/0008-5472.CAN-04-1532. [DOI] [PubMed] [Google Scholar]
- 68.Welm AL, Timchenko NA, Darlington GJ. C/EBPalpha regulates generation of C/EBPbeta isoforms through activation of specific proteolytic cleavage. Mol Cell Biol. 1999;19:1695–1704. doi: 10.1128/mcb.19.3.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nakajima T, Kinoshita S, Sasagawa T, Sasaki K, Naruto M, Kishimoto T, Akira S. Phosphorylation at threonine-235 by a ras-dependent mitogen-activated protein kinase cascade is essential for transcription factor NF-IL6. Proc Natl Acad Sci U S A. 1993;90:2207–2211. doi: 10.1073/pnas.90.6.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Piwien-Pilipuk G, MacDougald O, Schwartz J. Dual regulation of phosphorylation and dephosphorylation of C/EBPbeta modulate its transcriptional activation and DNA binding in response to growth hormone. J Biol Chem. 2002;277:44557–44565. doi: 10.1074/jbc.M206886200. [DOI] [PubMed] [Google Scholar]
- 71.Zhu S, Yoon K, Sterneck E, Johnson PF, Smart RC. CCAAT/enhancer binding protein-beta is a mediator of keratinocyte survival and skin tumorigenesis involving oncogenic Ras signaling. Proc Natl Acad Sci U S A. 2002;99:207–212. doi: 10.1073/pnas.012437299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stoffels K, Overbergh L, Giulietti A, Verlinden L, Bouillon R, Mathieu C. Immune regulation of 25-hydroxyvitamin-D3-1alpha-hydroxylase in human monocytes. J Bone Miner Res. 2006;21:37–47. doi: 10.1359/JBMR.050908. [DOI] [PubMed] [Google Scholar]
- 73.Dhawan P, Peng X, Sutton AL, MacDonald PN, Croniger CM, Trautwein C, Centrella M, McCarthy TL, Christakos S. Functional cooperation between CCAAT/enhancer-binding proteins and the vitamin D receptor in regulation of 25-hydroxyvitamin D3 24-hydroxylase. Mol Cell Biol. 2005;25:472–487. doi: 10.1128/MCB.25.1.472-487.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Centrella M, Christakos S, McCarthy TL. Skeletal hormones and the C/EBP and Runx transcription factors: interactions that integrate and redefine gene expression. Gene. 2004;342:13–24. doi: 10.1016/j.gene.2004.06.036. [DOI] [PubMed] [Google Scholar]
- 75.Song CS, Echchgadda I, Seo YK, Oh T, Kim S, Kim SA, Cho S, Shi L, Chatterjee B. An essential role of the CAAT/Enhancer Binding Protein-alpha in the vitamin D induced expression of the human steroid/bile acid-sulfotransferase (SULT2A1) Mol Endocrinol. 2005 Dec 15; doi: 10.1210/me.2005-0428. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 76.Wang X, Studzinski GP. Kinase suppressor of RAS (KSR) amplifies the differentiation signal provided by low concentrations 1,25-dihydroxyvitamin D3. J Cell Physiol. 2004;198:333–342. doi: 10.1002/jcp.10443. [DOI] [PubMed] [Google Scholar]
- 77.Kortum RL, Costanzo DL, Haferbier J, Schreiner SJ, Razidlo GL, Wu MH, Volle DJ, Mori T, Sakaue H, Chaika NV, Chaika OV, Lewis RE. The molecular scaffold kinase suppressor of Ras 1 (KSR1) regulates adipogenesis. Mol Cell Biol. 2005;25:7592–7604. doi: 10.1128/MCB.25.17.7592-7604.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pan Z, Hetherington CJ, Zhang DE. CCAAT/enhancer-binding protein activates the CD14 promoter and mediates transforming growth factor beta signaling in monocyte development. J Biol Chem. 1999;274:23242–23248. doi: 10.1074/jbc.274.33.23242. [DOI] [PubMed] [Google Scholar]
- 79.Banwell CM, O'Neil LP, Uskokovic MR, Campbel MJ. Targeting 1alpha,25-dihydroxyvitamin D3 antiproliferative insensitivity in breast cancer cells by co-treatment with histone deacetylation inhibitors. J Steroid Mol Biol. 2004;89–90:245–249. doi: 10.1016/j.jsbmb.2004.03.081. [DOI] [PubMed] [Google Scholar]
- 80.Muindi JR, Potter DM, Peng Y, Johnson CS, Trump DL. Pharmacokinetics of liquid calcitriol formulation in advanced solid tumor patients: comparison with caplet formulation. Cancer Chemother Pharmacol. 2005;56:492–496. doi: 10.1007/s00280-005-1015-2. [DOI] [PubMed] [Google Scholar]
- 81.Sharrocks AD, Yang SH, Galanis A. Docking domains and substrate specificity determination for MAP kinases. Trends Biochem Sci. 2000;25:448–453. doi: 10.1016/s0968-0004(00)01627-3. [DOI] [PubMed] [Google Scholar]
- 82.Tanoue T, Nishida E. Docking interactions in the mitogen-activated protein kinase cascades. Pharmacol Ther. 2002;93:193–202. doi: 10.1016/s0163-7258(02)00188-2. [DOI] [PubMed] [Google Scholar]


