Abstract
We report an aptamer-nanoparticle strip biosensor (ANSB) for the rapid, specific, sensitive and low-cost detection of circulating cancer cells. Known for their high specificity and affinity, aptamers were first selected from live cells by the cell-SELEX (systematic evolution of ligands by exponential enrichment) process. When next combined with the unique optical properties of gold nanoparticles (Au-NPs), ANSBs were prepared on a lateral flow device. Ramos cells were used as a model target cell to demonstrate proof of principle. Under optimal conditions, the ANSB was capable of detecting a minimum of 4000 Ramos cells without instrumentation (visual judgment) and 800 Ramos cells with a portable strip reader within 15 minutes. Importantly, ANSB has successfully detected Ramos cells in human blood, thus providing a rapid, sensitive and low-cost quantitative tool for the detection of circulating cancer cells. ANSB therefore shows great promise for in-field and point-of-care cancer diagnosis and therapy.
Introduction
The non-invasive early detection of cancer and monitoring of its progress are high on the agenda of oncologists.1 Studies suggest that the circulating cancer cells found in patients are associated with short survival.2 Cancer of blood including Leukemia, Lymphoma and Myeloma is one of the most common types of cancers. Each year in the U.S., about 140,000 people are diagnosed with some type of blood cancer. About 53,000 die from it. It has been hypothesized that very sensitive monitoring of blood cancer cells could provide an easier and more effective way to monitor progression of the disease.3–7 From this perspective, identification and detection of circulating cancer cells is fundamental to early diagnosis and therapy and a means of monitoring the relevant biological processes of cancers. Various techniques, including polymerase chain reaction (PCR)-based methods, cytometric methods and cell-enrichment methods, have been developed, and some are commercially available.4 Although they have a high detection rate, many of these methods are expensive and time-consuming. They also require advanced instrumentation and enrichment of the target cells in the sample or expression of protein biomarkers or antibodies in the cells. There is, therefore, a need for an inexpensive, quick and simple tool with high sensitivity and specificity for detecting cancer cells in blood.
The most recent efforts in cancer cell detection have focused on biosensors with good sensitivity and selectivity, as well as rapid and easy operation. Various biosensors using different transducers have been reported as effective in detecting and identifying cancer cells, including those based on quartz crystal measurement (QCM)8, electrochemical measurement, 9–11 fluorescence measurement, 12, 13 single nanotube field effect transistor array 14, 15 or microfluidic devices.16, 17 Most of these reported biosensors are based on the specific interaction between antibody and protein receptor on the cell surface. Although many of them have been applied at the laboratory research level, they have not been applied in the field or for point-of-care detection because of the relatively long assay time or multiple washing and separation steps.
To solve many of these challenges, we have turned to the use of aptamers which are single-stranded oligonucletides selected by a process called SELEX (systematic evolution of ligands by exponential enrichment) from a DNA or RNA pool by repetitive binding of the target molecules.18, 19 Compared with molecular probes currently available for biomarker recognition, aptamers possess high specificity, low molecular weight, easy and reproducible production, versatility in application, and easy discovery and manipulation.20 Aptamers have had many important applications in bioanalysis, biomedicine, and biotechnology. 21–23 Most aptamers reported so far have been selected using simple targets, such as a purified protein. Recently, however, aptamer selection against complex targets, such as red blood cell membranes and endothelial cells, was also demonstrated.24–27 Using cell-SELEX, our group has created aptamers that bind to lymphocytic and myeloid leukemia, small cell and non-small cell lung cancer, and liver cancer cells.28–30 One benefit of using aptamers derived from cell-SELEX is that these aptamers can be created in the absence of any explicit molecular signature that would otherwise distinguish between cancer cells and healthy cells. More recently, the aptamer-modified microfluidic device has been developed for the enrichment of cancer cells to achieve a rapid assay without pretreatment of cells.31 Thus it shows great promise to use these aptamers as probes for the development of biosensors for cancer cell detection.
In this article, we report the development and working principle of an aptamer-nanoparticle strip biosensor (ANSB) for the rapid, sensitive, and low-cost detection of cancer cells in blood. Combining the high selectivity and affinity of aptamers with the unique optical properties of gold nanoparticles (Au-NPs), ANSB is prepared on a lateral flow device. A pair of aptamers capable of specifically binding Ramos cells is used to prepare the ANSB. A thiolated aptamer (Thiol-TD05) is immobilized on the Au-NPs, and a biotinylated aptamer (Biotin-TE02) is immobilized on the test zone of ANSB. Ramos cells interact with aptamer probes of the Au-NP-Aptamer conjugates to form the Au-NP-Aptamer-cell complex and continue to migrate along the strip. A large number of Au-NPs then accumulate on the test zone and produce a characteristic red band, which can be used for either qualitative, i.e., visual evaluation, or quantitative detection of cells by a portable strip reader. A DNA probe (complementary with the Thiol-TD05) is immobilized on the control zone to capture the excess of Au-NP-Aptamers, resulting in a second red band. The feasibility of this biosensor is evaluated by detecting Ramos cells spiked in human blood.
Experimental Section
Apparatus
Airjet AJQ 3000 dispenser, Biojet BJQ 3000 dispenser, Clamshell Laminator and the Guillotine cutting module CM 4000 were from Biodot LTD (Irvine, CA). Portable strip reader DT1030 was purchased from Shanghai Goldbio Tech. Co., LTD (Shanghai, China).
Reagents
Streptavidin from Streptomyces avidinii, HAuCl4, sucrose, hydroxylamine, Tween-20, dithiothreitol (DTT), Triton X-100, trisodium citrate, bovine serum albumin (BSA), sodium chloride-sodium citrate (SSC) Buffer 20× concentrate (pH 7.0), and phosphate buffer saline (PBS, pH 7.4, 0.01 M) were purchased from Sigma-Aldrich. Glass fibers (GFCP000800), cellulose fiber sample pads (CFSP001700), laminated cards (HF000MC100) and nitrocellulose membranes (HFB18004 and HFB 24004) were purchased from Millipore (Billerica, MA). Human whole blood was purchased from the U. S. Food and Drug Administration.
The following aptamers were selected to prepare ANSB: TDO5 aptamer, CACCGGGAGGATAGTTCGGTGGCTGTTCAGGGTCTCCTCCCGGTG; and TE02 aptamer, TAGGCAGTGGTTTGACGTCCGCATGTTGGGAATAGCCACGCCT. Both the thiolated and biotinylated versions of the aptamer sequences were synthesized in our lab. A DNA probe (on the control zone of ANSB) was purchased from Integrated DNA Technologies, Inc. (Coralville, IA) and had the following sequence: 5'-Biotin-ATT GTA CAA AAT ACG TTT TG – 3'.
CCRF-CEM cells (CCL-119 T-cell, human acute lymphoblastic leukemia) and Ramos cells (CRL-1596, B-cell, human Burkitt's lymphoma) were obtained from ATCC (American Type Culture Association). The cells were cultured in RPMI medium supplemented with 10% fetal bovine serum (FBS) and 100 IU/mL penicillin-Streptomycin. After the cells had been dispersed in RPMI cell media buffer, they were centrifuged at 920 rpm for 5 min. They were then redispersed in cell media three times and finally redispersed again in a 1 mL cell media buffer. The cell density was determined using a hemocytometer, and this was performed prior to any experiments. During all experiments, the cells were kept in an ice bath at 4 °C.
All chemicals used in this study were analytical reagent grade. All other solutions were prepared with ultrapure (>18 MΩ) water from a Millipore Milli-Q water purification system (Billerica, MA).
Preparation of gold nanoparticles (Au-NP) and Au-NP-Aptamer conjugates
Au-NPs with average diameter 15 nm ± 3.5 nm were prepared according to the reported method with slight modifications.29 A thiolated aptamer (Thiol-TD05) was used for conjugation with Au-NPs. Before the conjugation, the thiolated olignocleotide was activated by the following procedure: 98 μL of Thiol-TD05 (1.0 OD) was mixed with 2 μL of triethylamine and 7.7 mg of DTT to react for 30 min at room temperature (RT); then the excess DTT was removed by extraction four times with 400 μL of ethylacetate solution. Conjugation reactions were carried out by adding the activated DNA probe to 1 mL of the 5-fold concentrated Au-NP solution. After standing at 4 °C for 24 h, the solution was subjected to “aging” by the addition of NaCl up to a concentration of 150 mM, and a certain quantity of 1 % sodium dodecyl sulfate (SDS) was added to reach a final concentration of 0.01 %. The solution was allowed to stand for another 24 h at 4 °C, and the excess of reagents was removed by centrifugation for 12 minutes at 12000 rpm. The supernatant was discarded, and the red pellet was redispersed in 1 mL of eluent buffer containing 20 mM Na3PO4, 5% BSA, 2.5% Tween-20 and 10% sucrose.
Preparation of the ANSB
The ANSB consists of four components: sample application pad, Au-NP-Aptamer conjugate pad, nitrocellulose membrane and absorbent pad (see Supporting Information, Fig. S1). All of the components were mounted on a common backing layer (typically an inert plastic, e.g., polyester) using the Clamshell Laminator (Biodot, CA, USA). The sample application pad (17 mm × 30 cm) was made from glass fiber (CFSP001700, Millipore) and saturated with a buffer (pH 8.0) containing 0.25% Triton X-100, 0.05 M Tris-HCl, 5 % Tween-20 and 0.15 M NaCl. It was then dried and stored in a desiccator at RT. The conjugate pad (8 mm× 30 cm) was prepared by dispensing a desired volume of Au-NP-Aptamer conjugates solution onto the glass fiber pad with the Airjet AJQ 3000 dispenser, leaving it to dry at RT. The pad was stored in a desiccator at 4 °C. A nitrocellulose membrane (25 mm × 30 cm) was used to immobilize the capture aptamer probes (Biotin-TE02 Aptamer probe) and control DNA probes at different zones to form the test zone and control zone, respectively. To facilitate the immobilization of biotinylated aptamers and biotinylated DNA probes on the nitrocellulose membrane, streptavidin was used to react with the biotinylated probes to form the streptavidin-biotin-aptamer and streptavdin-biotin-DNA conjugates. Briefly, 60 μL of 1 mM biotinylated aptamer (or DNA) probes and 140 μL of PBS were added to 300 μL of 1.67 mg/mL streptavidin solution, and the mixture was incubated 1 h at RT. The excess aptamer (DNA) probes were removed by centrifugation for 20 min with a centrifugal filter (cut-off 30000, Millipore) at 6000 rpm. The conjugates were washed three times with 1 mL of PBS in the same centrifugal filter. Finally, 500 μL of PBS was added into the remaining solution in the filter. The conjugates were then dispensed on the nitrocellulose membrane with the Biojet BJQ 3000 dispenser. The distance between the test zone and control zone was around 2 mm. The aptamer- and DNA probe-loaded membrane was then dried at RT for 1 h and stored at 4 °C in a dry state. Finally, the sample pad, conjugate pad, nitrocellulose membrane and absorption pad were assembled on a plastic adhesive backing (60 mm × 30 cm) using the Clamshell Laminator. Each part overlapped 2 mm to ensure migration of the solution through the strip during the assay. Strips with a 4-mm width were cut by using the Guillotine cutting module CM 4000.
Sample Assay Procedure
In a typical cell test on ANSB, 80 μL of sample solution containing a desired amount of Ramos cells in 0.01 M PBS containing 1 % BSA (PBSB) was applied to the sample application zone. After waiting for a specific time (e.g., 7 min), another 50 μL of PBSB was applied to wash the ANSB. The bands were visualized within 15 min. For quantitative measurements, the ANSB was inserted into the DT1030 strip reader. The optical intensity of the test line and control line could be recorded simultaneously by using the “GoldBio strip reader” software. In the case of detecting Ramos cells in blood, the desired amount of Ramos cells were spiked into blood, and 5 μL of blood was applied to the sample application pad. An additional 75 μl of PBSB was added to transport blood into a downstream portion of ANSB. Other procedures were the same as those described above.
Results and Discussion
Principle of ANSB measurement
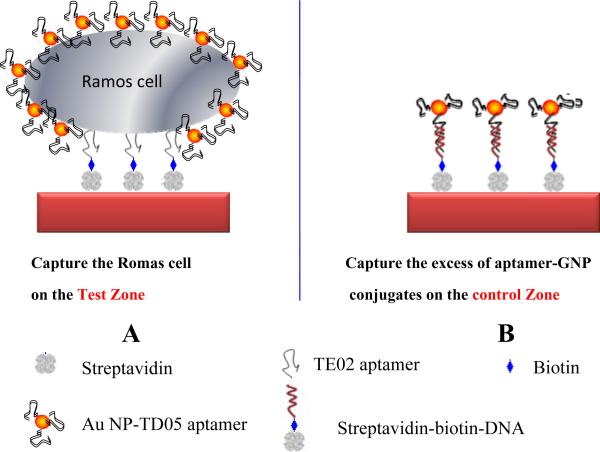
Known for their high specificity and affinity, aptamers were selected from live cells by cell-SELEX (systematic evolution of ligands by exponential enrichment). When combined with the unique optical properties of gold nanoparticles (Au-NPs), ANSB was prepared on a lateral flow device (see Supporting Information, Fig.S1). The principle of ANSB is based on the specific binding between the aptamers and cells, and the protocol is illustrated in Figure 1. Ramos cells are used as a model cell line to demonstrate the proof of principle. Briefly, a pair of aptamers capable of specifically binding Ramos cells is used to prepare the ANSB. A thiolated aptamer (Thiol-TD05) is immobilized on the Au-NPs, and a biotinylated aptamer (Biotin-TE02) is immobilized on the test zone of ANSB. In a typical assay, a sample solution containing Ramos cells is applied on the ANSB sample pad. The solution migrates by capillary action, passes the conjugate pad, and then rehydrates the Au-NP-Aptamer conjugates. Ramos cells interact with aptamer probes of the Au-NP-Aptamer conjugates to form the Au-NP-Aptamer-Cell complex and continue migrating along the strip. The Au-NP-Aptamer-Cell complexes are captured on the test zone by a second reaction between Ramos cells and the immobilized aptamers (Figure 1A). The accumulation of Au-NPs on the test zone is visualized as a characteristic red band (test line). The excess of Au-NP-Aptamer conjugates continues to migrate and pass the control zone, in which a DNA probe (complementary with the thiol-TD05) is immobilized. The excess of Au-NP-Aptamer conjugates is then captured by hybridization events between the thiolated aptamer and DNA, thus forming a second red band (control line, Figure 1B). In the absence of Ramos cells, no red band is observed in the test zone. In this case, a red control band (control line) shows that the ANSB is working properly. Qualitative analysis (visual detection) is performed by observing the color change of the test zone, and quantitative analysis is realized by reading the optical intensity of the test line with a portable strip reader.
Fig. 1.
Schematic diagram of the detection of Ramos cells on aptamer-nanoparticle strip biosensor (ANSB). (A) Capturing Au-NP-aptamer-Ramos cells on the test zone of ANSB through specific aptamer-cell interactions and (B) Capturing the excess of Au-NP-aptamer on the control zone of ANSB through aptamer-DNA hybridization reaction.
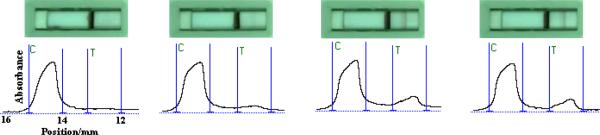
Figure 2 presents the typical photo images and corresponding optical responses of 0 Ramos cells (control, a), 8×104 CCL cells (control, b) and 8×104 Ramos cells (c) and the mixture of 8×104 CCL cells and 8×104 Ramos cells (d). Two red bands were observed in the presence of Ramos cells (Figure 2c), and only one red band (control line, top) was observed in the absence of Ramos cells (Figure 2a) and presence of CCL cells (Figure 2b). The presence of CCL cells shows slight interference on the signal of Ramos cells (Figure 2d). The intensities of the bands were recorded by the strip reader shown on the bottom of Figure 2. Well-defined peaks were observed, and the peak areas were proportional to the amount of the captured Au-NPs in the test zone (right side) and control zone (left side). The above results indicated that the ANSB would provide a rapid and simple tool for qualitative and quantitative detection of cancer cells.
Fig. 2.
Typical photo images (top) and corresponding responses (bottom) of ANSB with sample solutions containing 0 Ramos cells (a), 8×104 CCL cells (b), 8×104 Ramos cells (c) and 8×104 CCL cells and 8×104 Ramos cells. The sample solutions were prepared with 0.01 M PBS containing 1% BSA. Volume of the sample solution: 80 μL; Assay time: 15 min.
Optimization of ANSB fabrication and assay parameters
Optimal combination of the aptamer pairs for the fabrication of ANSB
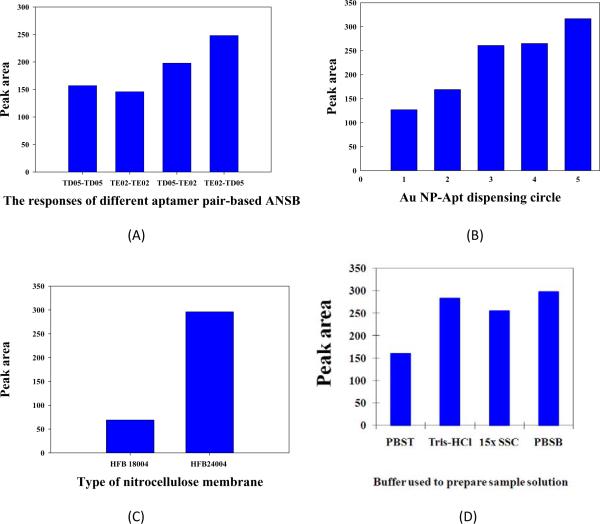
A group of aptamers with different equilibrium dissociation constants (Kd) for Ramos cells was selected by cell-SELEX, but only one pair of aptamers (TD05 and TE02) with nanomolar Kd values (high binding affinities) was used to prepare the ANSB. Considering that Ramos cells have multiple binding sites for the aptamer probes, both TD05 and TE02 could be used as both detection probe (conjugated with Au-NP) and capture probe (immobilized on the test zone). To obtain the best sensitivity of ANSB, four aptamer pairs [TD05 (capture probe)-TD05 (detection probe), TE02 (capture probe)-TE02 (detection probe), TD05 (capture probe)-TE02 (detection probe) and TE02 (capture probe)-TD05 (detection probe)] were used to prepare ANSBs. Figure 3A presents the response of 8×104 Ramos cells for different aptamer pair-based ANSBs. It can be observed that the best response was obtained by using the TE02 (capture probe)-TD05 (detection probe) pair. TE02-TE02 and TD05-TD05 pair-based ANSBs gave relatively low responses. It seems obvious that using the same aptamer as both detection probe and capture probe reduces the amount of the cells captured on the test zone because of limited binding sites on the surface of Ramos cells. The response difference between TD05 (capture probe)-TE02 (detection probe)-based ANSB and TE02 (capture probe)-TD05 (detection probe)-based ANSB arises from different binding affinities to Ramos cells. The Kd of TE02 (0.76±0.13 nM) is much smaller than that of TD05 (74.8±8.7 nM) and would offer high binding capability to capture Ramos cells on the test zone. Therefore, a TE02 (capture probe)-TD05 (detection probe) pair was used to prepare ANSB throughout the experiments.
Fig. 3.
(A) The responses of different aptamer pair-based ANSBs; (B) Effect of dispensing cycles of Au-NP-aptamer conjugates on the response of ANSB; (C) Effect of nitrocellulose membranes on the response of ANSB (c); Effect of the buffers used to prepare sample solutions on the response of ANSB. Amount of Ramos cells: 8×104. Volume of the sample solution: 80 μL; Assay time: 15 min.
Optimization of the amount of Au-NP-Aptamer on the conjugate pad of ANSB
The intensities of red bands (test line and control line) on the ANSB depend on the amount of captured Au-NP-Aptamer conjugates, which, in turn, corresponds to the amount of Au-NP-Aptamer on the conjugate pad. Figure 3B presents the histogram of the responses of 8×104 Ramos cells on ANSBs, which were prepared with different amounts of Au-NP-Aptamer conjugates (i.e., the dispensing of various cycles of Au-NP-Aptamer conjugates on the conjugate pads). It can be seen that the responses of ANSB increased up to 3 dispensing times on the conjugate pad, thereafter tending toward saturation. Since increase of the dispensing times causes increasingly nonspecific adsorption and assay time, three dispensing times were used to prepare the ANSB for most of the experiments.
Selection of appropriate membrane materials for the preparation of ANSB
Sandwich-type Au-NP-Aptamer-Cell-Aptamer complexes were formed on ANSB, and the reaction time, which depends on the migration time of the sample solution on the nitrocellulose membrane, plays an important role for the sensitivity of ANSB. The migration time of solution differs with different membranes. Two kinds of nitrocellulose membranes, including HFB18004 and HFB24004 (Millipore), were used to prepare ANSB. According to the manufacturer's instructions, the migration times of the buffer in HFB 18004 and HFB24004 membranes are 3 min and 4 min, respectively. Figure 3C presents the responses of 8×104 Ramos cells on the ANSBs prepared with the above nitrocellulose membranes. It can be seen that the response of ANSBs prepared with the HFB24004 is significantly higher than the response of those prepared with HFB18004. This indicates that a relatively long migration time is helpful in increasing the sensitivity of ANSB. Since the whole assay time of ANSB prepared with HFB24004 is around 15 min, the HFB24004 nitrocellulose membrane was used to prepare the ANSB.
Selection of appropriate buffers for the fabrication of ANSB and cell tests
Another important factor affecting the sensitivity and reproducibility of ANSB involves the components of buffers during the fabrication of ANSB and cell tests. One of the most important issues for nanoparticle-based bioassays and biosensors is nonspecific adsorption of nanoparticles, which causes high background and low sensitivity of the tests. The use of appropriate buffers would minimize nonspecific adsorption of Au-NP-Aptamer on the membrane and increase the sensitivity and reproducibility of the ANSB. In the current study, three kinds of buffers were used in ANSB fabrication and cell tests. The sample pad of ANSB was treated for 1 hr with the buffer (pH 8.0) containing 0.25 % Triton X-100, 0.05 M Tris-HCl, 5 % Tween-20 and 0.15 M NaCl. This treatment facilitates the transport of cells into a downstream portion of the lateral flow device and reduces entrapment of cells in the sample pad. During the preparation of Au-NP-Aptamer conjugates, the Au-NP-Aptamer conjugate pellets were dispersed in the buffer containing 20 mM Na3PO4, 5 % BSA, 2.5 % Tween-20 and 10 % sucrose. The addition of BSA, Tween-20 and sucrose stabilizes the Au-NPs and facilitates the release of the Au-NP-Aptamer conjugates from the conjugate pad. The addition of these components in the buffer also reduces the nonspecific adsorption of Au-NP-Aptamer conjugates on the nitrocellulose membrane during cell tests. After applying a sample solution on ANSB, the Au-NP-Aptamer conjugates on the conjugate pad are rehydrated. The components (BSA, Tween-20 and sucrose) are dispersed in the sample solution, and, migrating along the strip, will block the nitrocellulose membrane naturally without additional blocking steps. The buffer used to prepare the sample solutions also has a strong effect on the performance of ANSB. Four kinds of buffers, including 0.01 M PBS containing 1 % BSA (PBSB), 0.01 M PBS containing 0.1% Tween-20 (PBST), 0.1 M Tris-HCl and SSC buffers, were tested. Figure 3D presents the responses of ANSB with the sample solutions prepared with the above buffers. Since the best response of ANSB was obtained with PBSB, a 0.01 M PBSB was used for the following experiments.
Analytical performance
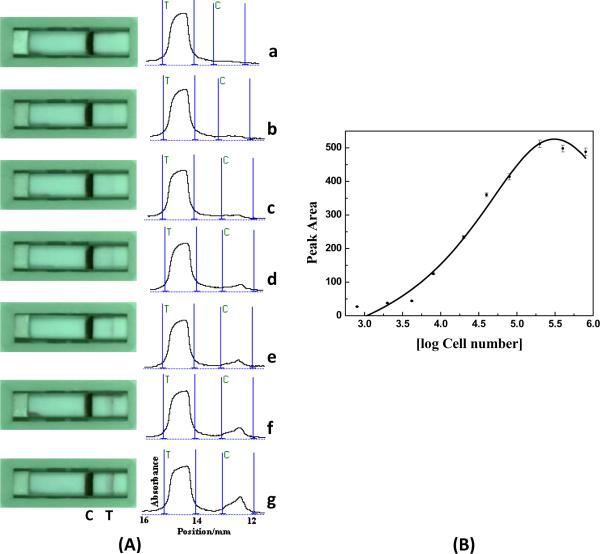
Under optimal experimental conditions, we examined the performance of ANSB with the sample solutions containing different amounts of Ramos cells. The visual judgment of the test line (color change of the test line) could be used for the qualitative detection of Ramos cells. Quantitative detections were performed by recording the intensities of the test lines with the portable strip reader. Figure 4(A) displays the typical photo images (left) and corresponding optical responses (right; the intensities of test line and control line) of ANSBs with the sample solutions containing different amounts of Ramos cells. For qualitative analysis (visual judgment), the red band in the test zone of ANSB was observed with as low as 4000 Ramos cells in the sample solution (Figure 4A, b). The red bands appearing on the control zone of ANSBs indicated that the ANSBs were working properly. Well-defined peaks were observed with a portable strip reader, and the peak areas increased with the increase of target cell amount (Figure 4A, curve a to g). The resulting calibration plot (Figure 4B) of the peak areas versus the logarithm of Ramos cell number is linear over the 4×103 to 2×105 range and is suitable for quantitative work. The S-shaped curve of the calibration plot on a log-linear scale is due to saturation at high concentration or variations in aptamer-cell affinity. The detection limit of 800 Ramos cells (based on S/N=3) was estimated in connection with the 15-min assay time. The specific response was coupled with high reproducibility. A series of measurements of 2×104 Ramos cells with 8 ANSBs yielded a reproducible signal with an RSD of 6.3 % (data not shown).
Fig. 4.
(A) The typical responses of ANSB with increasing amounts of Ramos cells. From a to g, the amounts of target cells are 0, 4×103, 8×103, 2×104, 4×104, 8×104, 2×105 and (B) the resulting calibration curve. Other conditions are the same as those shown in Figure 2.
Detection of Ramos cells in human blood
To evaluate the feasibility of ANSB for the detection of cancer cells in biological fluids, ANSB was used to detect Ramos cells in human blood. First we studied the matrix effect by adding different volumes of human blood in the sample solution. It was found there was no matrix effect on the responses of ANSB when the volume of added blood is less than 5 μL (data not shown). However, a significant decrease of the signal was observed after adding 10 μL of blood. The signal would be masked in the presence of more blood because of the nonspecific adsorption of erythrocyte cells on the membrane. Therefore, 5 μL of human blood was used in the following experiments. The resulting calibration plot (data not shown) of the peak areas versus the logarithm of the number of the Ramos cells is linear over an 8×103 to 4×105 range and is suitable for quantitative work. The assay time with ANSB is less than that of most reported methods, which require 30 min to several hours.34–36 A series of measurements of Ramos cells with 8 ANSBs yielded a reproducible signal with an RSD of 7.3% (data not shown).
Conclusions
We have successfully developed an aptamer-nanoparticle strip biosensor (ANSB) for rapid, sensitive and quantitative detection of cancer cells. Under optimal conditions, the ANSB was capable of detecting a minimum of 4000 Ramos cells without instrumentation (visual detection) and 800 Ramos cells with a portable strip reader within 15 min. Compared with our previous works based on fluorescent nanoparticles28, gold nanorods29 and the microfluidic channel14 in association with aptamer probes, ANSB offers a simple, rapid and low-cost tool for both the qualitative and quantitative detection of cancer cells circulating in the bloodstream. ANSB shows great promise for field use, as well as point-of-care cancer diagnosis and therapy. The adoption of multiple aptamers, which can bind specific cancer cells, would expand the capabilities of ANSB for multiplex cancer cell detection. The sensitivity of ANSB has not met the requirement of detecting circulating tumor cells from solid tumors in general (found at very low concentrations35). Further improvements in the sensitivity of ANSB could be achieved by using more sensitive detectors (fluorescence readers) and dye-doped polystyrene nanosphere or silica nanoparticle labels.
Supplementary Material
Acknowledgments
GL acknowledges financial support from the North Dakota Experimental Program to Stimulate Competitive Research (EPSCoR) and new faculty startup funds of North Dakota State University. WT thanks NIH grants for this work.
References
- 1.Uhr JW. Nature. 2007;450:1168–1169. doi: 10.1038/4501168a. [DOI] [PubMed] [Google Scholar]
- 2.Chang Y, Di Tomaso E, McDonald DM, Jones R, Jain RK, Munn LL. Proc. Natl. Acad. Sci. U. S. A. 2000;97(26):14608–14613. doi: 10.1073/pnas.97.26.14608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vlems FA, Ruers TJ, Punt CJ, Wobbes T, van MGN. Eur J Surg Onco. 2003:289–302. doi: 10.1053/ejso.2002.1394. [DOI] [PubMed] [Google Scholar]
- 4.Paterlini-Brechot P, Benali NL. Cancer letters. 2007;253(2):180–204. doi: 10.1016/j.canlet.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 5.Pantel K, Woelfle U. Biochimica et Biophysica Acta. 2005;1756(1):53–64. doi: 10.1016/j.bbcan.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 6.Zieglachmid V, Hollmann C, Bocher O. Critical Reviews in clinical Laboratory Sciences. 2005;42(2):155–196. doi: 10.1080/10408360590913696. [DOI] [PubMed] [Google Scholar]
- 7.Mocellin S, Keilholz U, Rossi CR, Nitti D. Trends in Molecular Medicine. 2006;12(3):130–139. doi: 10.1016/j.molmed.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 8.Hayden O, Bindeus R, Dickert FL. Meas. Sci. Technol. 2003;14:1876–1881. [Google Scholar]
- 9.He F, Shen Q, Jiang H, Zhou J, Cheng J, Guo DD, Li QN, Wang XM, Fu DG, Chen BA. Talanta. 2009;77(3):1009–1014. doi: 10.1016/j.talanta.2008.07.063. [DOI] [PubMed] [Google Scholar]
- 10.Liu QJ, Yu JJ, Xiao L, Tang JCO, Zhang Y, Wang P, Yang M. Biosensors and Bioelectronics. 2009;24(5):1305–1310. doi: 10.1016/j.bios.2008.07.044. [DOI] [PubMed] [Google Scholar]
- 11.Andreescu S, Sakik OA. Methods. 2005;37(1):84–93. doi: 10.1016/j.ymeth.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 12.Tibbe AGJ, Grooth BGD, Greve J, Liberti PA, Dolan GJ, Terstappen LWMM. Nature biotechnology. 1999;17(12):1210–1213. doi: 10.1038/70761. [DOI] [PubMed] [Google Scholar]
- 13.Kang CC, Chang CC, Chang TC, Liao LJ, Lou PJ, Xie WJ, Yeung ES. Analyst. 2007;232:745–749. doi: 10.1039/b617733f. [DOI] [PubMed] [Google Scholar]
- 14.Shao N, Wickstrom E, Panchapakesan B. Nanotechnology. 2008;46(19):465101. doi: 10.1088/0957-4484/19/46/465101. [DOI] [PubMed] [Google Scholar]
- 15.Teker K. Materials Science and Engineering B. 2008;153(1–3):83–87. [Google Scholar]
- 16.Du Z, Colls N, Cheng KH, Vaughn MW, Gollahon L. Biosensors and Bioelectronics. 2006;21(10):1991–1995. doi: 10.1016/j.bios.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 17.Phillips JA, Xu Y, Xia Z, Fan H, Tan W. Anal. Chem. 2009;81:1033–1039. doi: 10.1021/ac802092j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ellington AD, Szostak JW. Nature. 1990;346(30):818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 19.Tuerk C, Gold L. Science. 1990;249(4968):505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 20.Jayasena SD. Clin. Chem. 1999;45(9):1628–1650. [PubMed] [Google Scholar]
- 21.Fang X, Sen A, Vicens M, Tan W. ChemBioChem. 2003;4(9):829–834. doi: 10.1002/cbic.200300615. [DOI] [PubMed] [Google Scholar]
- 22.Guo K, Wendel HP, Scheideler L, Ziemer G, Scheule AM. J. Cell. Mol. Med. 2005;9(3):731–736. doi: 10.1111/j.1582-4934.2005.tb00503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang CJ, Jockusch S, Vicens M, Turro N, Tan W. Proc. Natl. Acad. Sci. USA. 2005;102(48):17278–17283. doi: 10.1073/pnas.0508821102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morris KN, Jensen KB, Julin CM, Weil M, Gold L. Proc. Natl. Acad. Sci. USA. 1998;95(6):2902–2907. doi: 10.1073/pnas.95.6.2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blank M, Weinschenk T, Priemer M, Schluesener H. J. Biol. Chem. 2001;276:16464–16468. doi: 10.1074/jbc.M100347200. [DOI] [PubMed] [Google Scholar]
- 26.Daniels DA, Chen H, Hicke BJ, Swiderek KM, Gold L. Proc. Natl. Acad. Sci. USA. 2003;100(26):15416–15421. doi: 10.1073/pnas.2136683100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang C, Zhang M, Yang G, Zhang D, Ding H, Wang H, Fan M, Shen B, Shao NJ. Biotechnol. 2003;102(2):15–22. doi: 10.1016/s0168-1656(02)00360-7. [DOI] [PubMed] [Google Scholar]
- 28.Shangguan D, Li Y, Tang Z, Cao ZC, Chen HW, Mallikaratchy P, Sefah K, Yang CJ, Tan W. Proc. Natl. Acad. Sci. U.S.A. 2006;103(32):11838–11843. doi: 10.1073/pnas.0602615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shangguan D, Cao ZC, Li Y, Tan W. Clin. Chem. 2007;53(6):1153–1158. doi: 10.1373/clinchem.2006.083246. [DOI] [PubMed] [Google Scholar]
- 30.Shangguan D, Meng L, Cao ZC, Xiao Z, Fang X, Li Y, Cardona D, Witek RP, Liu C, Tan W. Anal. Chem. 2008;80(3):721–728. doi: 10.1021/ac701962v. [DOI] [PubMed] [Google Scholar]
- 31.Herr JK, Smith JE, Medley CD, Shangguan DH, Tan WH. Anal. Chem. 2006;78:2918–2924. doi: 10.1021/ac052015r. [DOI] [PubMed] [Google Scholar]
- 32.Huang YF, Chang HT, Tan WH. Anal. Chem. 2008;80:567–572. doi: 10.1021/ac702322j. [DOI] [PubMed] [Google Scholar]
- 33.Hosokawa M, Arakaki A, Takahashi M, Mori T, Takeyama H, Matsunaga T. Anal. Chem. 2008;81:5308–5313. doi: 10.1021/ac900535h. [DOI] [PubMed] [Google Scholar]
- 34.Smith JE, Medley CD, Tang ZW, Shangguan D, Lofton C, Tan W. Anal. Chem. 2007;79:3075–3082. doi: 10.1021/ac062151b. [DOI] [PubMed] [Google Scholar]
- 35.Medley CD, Smith JE, Tang Z, Wu Y, Bamrungsap S, Tan W. Anal. Chem. 2008;80:1067–1072. doi: 10.1021/ac702037y. [DOI] [PubMed] [Google Scholar]
- 36.Cristofanilli M, Budd T, Ellis MJ, Stopeck A, Matera J, Miller C, Reuben JM, Doyle GV, Allard WJ, Terstappen LWMM, Hayes DF. NEJM. 2004;351(8):781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.