Abstract
Translocation of nuclear-encoded preproteins across the inner envelope of chloroplasts is catalyzed by the Tic translocon, consisting of Tic110, Tic40, Tic62, Tic55, Tic32, Tic20, and Tic22. Tic62 was proposed to act as a redox sensor of the complex because of its redox-dependent shuttling between envelope and stroma and its specific interaction with the photosynthetic protein ferredoxin-NADP(H) oxidoreductase (FNR). However, the nature of this close relationship so far remained enigmatic. A putative additional localization of Tic62 at the thylakoids mandated further studies examining how this feature might be involved in the respective redox sensing pathway and the interaction with its partner protein. Therefore, both the association with FNR and the physiological role of the third, thylakoid-bound pool of Tic62 were investigated in detail. Coexpression analysis indicates that Tic62 has similar expression patterns as genes involved in photosynthetic functions and protein turnover. At the thylakoids, Tic62 and FNR form high molecular weight complexes that are not involved in photosynthetic electron transfer but are dynamically regulated by light signals and the stromal pH. Structural analyses reveal that Tic62 binds to FNR in a novel binding mode for flavoproteins, with a major contribution from hydrophobic interactions. Moreover, in absence of Tic62, membrane binding and stability of FNR are drastically reduced. We conclude that Tic62 represents a major FNR interaction partner not only at the envelope and in the stroma, but also at the thylakoids of Arabidopsis thaliana and perhaps all flowering plants. Association with Tic62 stabilizes FNR and is involved in its dynamic and light-dependent membrane tethering.
INTRODUCTION
Tic62 was discovered as a subunit of the Tic complex (translocon at the inner envelope of chloroplasts), which mediates the import of nuclear-encoded precursor proteins containing a chloroplast transit peptide in concert with the Toc complex (translocon at the outer envelope of chloroplasts) across the double membrane of the organelle. The Tic complex consists of so far seven unambiguously identified proteins with specialized properties. Tic110 is the most abundant component and functions as the channel forming subunit. Three potentially redox-active subunits form the so-called redox-regulon of the Tic complex: Tic55, Tic32, and Tic62. Tic55 is a Rieske-type protein showing homologies to the CAO/PAO-like oxygenases, with a [2Fe-2S] cluster and an additional mononuclear Fe binding site. The other two subunits of the regulon, Tic62 and Tic32, belong to the (extended) family of short-chain dehydrogenases. Both were demonstrated to be functional in vitro and to associate with the Tic complex in a redox-dependent manner (Küchler et al., 2002; Hörmann et al., 2004; Chigri et al., 2006; Stengel et al., 2008).
Tic62 is encoded by a single-copy gene in Arabidopsis thaliana (At3g18890) and has been characterized as a redox sensor of the Tic complex based on its inherent dehydrogenase activity, its ability to shuttle between the stroma and inner envelope dependent on the metabolic NADP+/NADPH ratio, and its specific and likewise redox-dependent interaction with ferredoxin-NADP+-oxidoreductase (FNR), a key photosynthetic enzyme (Küchler et al., 2002; Stengel et al., 2008). Tic62 consists of two very different modules of about equal size. The N terminus (Nt) is evolutionary well conserved in all oxyphototrophic organisms down to green sulfur bacteria (Balsera et al., 2007). It contains the dehydrogenase domain as well as a predicted hydrophobic patch, which may mediate the reversible attachment of the protein to the membrane. The Tic62 C terminus (Ct), on the other hand, is unique in its composition and found only in flowering plants. It contains a variable number of Pro/Ser-rich repeats (dependent on the species), which specifically mediate the interaction with FNR.
Using FAD as a cofactor, FNR catalyzes the (reversible) electron transfer between ferredoxin (Fd) and NADP(H). This reaction is best known as the last step of the photosynthetic electron transport chain, producing the reducing equivalents for the reductive metabolism. In contrast with the situation in photosynthetic organisms, the reaction is driven toward Fd or flavodoxin reduction in nonphotosynthetic bacteria and eukaryotes. In Arabidopsis, this fact is reflected by a set of specialized FNR isoforms in leaves (LFNR1 and LFNR2) and roots (RFNR1 and RFNR2), allowing an efficient electron flux of the NADP(H)-FNR-Fd cascade to the respective metabolism. Besides its role in the linear electron transfer (LET), FNR has also been implicated in cyclic electron transfer (CET) processes (Guedeney et al., 1996; Quiles and Cuello, 1998; Quiles et al., 2000; Breyton et al., 2006). At least two CET routes exist, which recycle electrons from the LET around photosystem I (PSI), thereby further reducing the plastoquinone pool and leading to an enhanced proton gradient across the thylakoid membrane. This results in the production of ATP without accumulation of NADPH (for review, see Rumeau et al., 2007). FNR was supposed to interact with several thylakoidal proteins, such as the PsaE subunit of PSI, a still uncharacterized 10-kD protein called connectein, the NAD(P)H dehydrogenase, the Cytb6f complex, and a subunit initially described as part of the oxygen evolving complex of photosystem II (PSII) (Vallejos et al., 1984; Shin et al., 1985; Matthijs et al., 1986; Chan et al., 1987; Soncini and Vallejos, 1989; Andersen et al., 1992; Guedeney et al., 1996; Quiles and Cuello, 1998; Okutani et al., 2005; Zhang et al., 2001). These reports could explain the observed anchoring of the hydrophilic FNR to the thylakoid membrane but are nevertheless still disputed and many questions remain.
By generating reduction equivalents, FNR also represents a link between light-driven photosynthesis and general metabolism (e.g., carbon fixation, nitrogen metabolism, and fatty acid and chlorophyll biosynthesis). Moreover, the stromal NADP+/NADPH ratio has important regulatory and signaling functions in the chloroplast, as exemplified by the dynamic composition of the Tic complex in response to redox changes (Chigri et al., 2006; Stengel et al., 2008). Thus, FNR is a crucial enzyme at the intersection of various redox-active pathways. Its highly specific interaction with Tic62 was therefore discussed as an important feature for the redox regulation of preprotein import into the organelle, as it provides a link between photosynthetic electron transport and protein translocation. However, research on Tic62 so far focused on the characterization of the protein and its interactions with the Tic complex at the chloroplast inner envelope as well as its redox-dependent shuttling into the stromal compartment (Küchler et al., 2002; Stengel et al., 2008). The origin of the redox signal triggering the relocalization of Tic62, on the other hand, is still not entirely clear. Moreover, the nature and function of the close relationship with FNR remains enigmatic. Interestingly, Peltier et al. (2004) found the Arabidopsis homolog of Tic62 to be associated with thylakoids by proteomic analysis, indicating a triple localization of Tic62 in the chloroplast, similar to FNR. A putative function of Tic62 in the thylakoids mandated further studies examining how this localization might be involved in the respective redox sensing pathway.
To answer these open questions we undertook several new experimental approaches, making use of the model plant Arabidopsis in addition to pea (Pisum sativum). We could verify the thylakoid localization of Tic62, and our data demonstrate that Tic62 and FNR form several thylakoid-bound complexes of high molecular weight. Analysis of tic62 knockout plants revealed that the membrane attachment of FNR is severely disturbed in the mutants. The high molecular weight Tic62/FNR complexes form in neither tic62 mutants nor in lfnr1 and lfnr2 lines, indicating a hetero-oligomeric association of the three proteins. In addition, the presence of Tic62 is beneficial for the stability of FNR. It can be concluded that Tic62 represents a major FNR interaction partner not only at the envelope and in the stroma, but also at the thylakoid membranes of vascular plants, thereby acting in light-dependent regulation of the allocation of FNR between stroma and chloroplast membranes.
RESULTS
Tic62 Is Coregulated with Photosynthetic Genes and Tissue
Different databases were examined in order to identify Arabidopsis genes with a similar expression pattern as Tic62. The aim of this analysis was to develop testable hypotheses about possible regulatory pathways Tic62 might be involved in. Coexpression analyses were performed using the A. thaliana Co-Response database (AthCoR@CSB.DB) and hierarchical clustering analysis (Eisen et al., 1998) of the Arabidopsis AtGenExpress developmental series microarray data set (Schmid et al., 2005). In the intersection of both analyses, 142 genes were identified that displayed a significant coexpression behavior in both cases (ρ ≥ 0.9). The overwhelming majority of these code for proteins either predicted or shown to be localized in the chloroplast (see Supplemental Table 1 online). Interestingly, when these coexpressed genes were functionally classified according to MapMan bins (see Supplemental Figure 1A online), >50% of the annotated genes fell in only two classified groups of about similar size: photosynthesis and protein. Genes implicated in photosynthetic functions accounted for the largest group (∼30% of the annotated genes), including most enzymes of the Calvin-Benson cycle (e.g., GAPDH or fructose-1,6-bisphosphatase [FBPase]), several members of PSII (e.g., PsbQ and two PsbP-like proteins), as well as five constituents of the NDH complex (NDH-N, NDH-L, Ndf6, Ndf1, and CRR3). It is known that the genes encoding photosynthetic proteins represent a well-coordinated group (Biehl et al., 2005), to which Tic62 is apparently closely connected.
The second biggest group (bin protein) comprised factors functioning in protein folding (e.g., four immunophilins and a cyclophilin; Romano et al., 2005), protein degradation (e.g., DEG5 and DEG8), and protein transport (e.g., SecA and Tic20-V, a distant homolog of Tic20-I). The remaining coregulated genes were found to be distributed between 18 different functional groups, many of which contribute to chloroplast or thylakoid biogenesis and maintenance.
To complement the in silico expression analysis of Tic62 with data on spatial expression, we transformed Arabidopsis plants with a transgene composed of the native TIC62 promoter region driving the β-glucuronidase (GUS) reporter gene (ProTIC62-GUS; see Supplemental Figures 1B to 1D online). GUS activity was initially detected around day 3 of seedling development in the cotyledons, which was at about the same time when the seedlings started greening (see Supplemental Figure 1B online). Signal intensity increased over the next days but was limited to the cotyledons and true leaves and was not visible in the roots and in the vascular leaf veins. Closer examination revealed that the GUS signal generally started to develop at the leaf tips (see Supplemental Figure 1C online). Fully grown plants showed a GUS expression pattern that was almost exclusively restricted to green plant tissues (see Supplemental Figure 1D online). These observations are in line with the in silico analyses, showing that Tic62 expression correlates with the development of photosynthetic tissue.
Tic62 Is Not Only Present at the Envelope and in the Stroma but Also at the Thylakoids
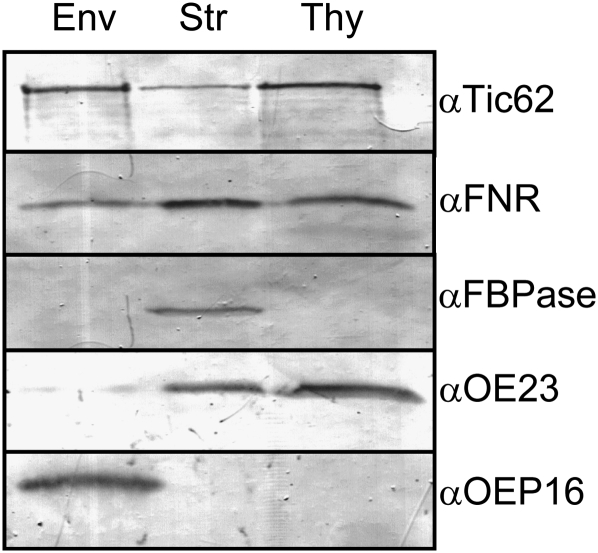
In order to verify the thylakoid localization of Tic62 observed by Peltier et al. (2004), we prepared subfractions from Arabidopsis chloroplasts and analyzed those by immunoblotting (Figure 1). Indeed, Tic62 displayed a triple localization in the envelope, stroma, and thylakoids, which was very similar to the chloroplastic distribution of FNR. However, FNR was predominantly present in the stroma, while less protein could be detected in the envelope and thylakoid membranes. By contrast, Tic62 was found to be mostly membrane associated.
Figure 1.
Localization of Tic62 and FNR in Arabidopsis Chloroplasts.
Tic62 and FNR show a triple localization in chloroplasts. Immunoblot analysis of Arabidopsis chloroplast subfractions envelope (Env), stroma (Str), and thylakoids (Thy) using antibodies generated against Tic62, FNR, FBPase (stroma marker), OE23 (thylakoid marker; soluble lumenal protein and thus also detected in minor amounts in the stroma), and OEP16 (envelope marker).
In addition to fractionation, transient transformation of Arabidopsis mesophyll protoplasts was employed to analyze the localization of Tic62 (see Supplemental Figure 2 online). For this purpose, the localization of partial or full-length Tic62 constructs, both leaf isoforms of FNR as well as Tic110 were monitored. The Tic62-green fluorescent protein (GFP) signal was visible exclusively within the chloroplasts, clearly overlapping with the red autofluorescence emitted from the thylakoid membranes. The strong signal intensity made it impossible to distinguish any additional signals from the envelopes that closely encompass the thylakoids, as can be seen with the exclusively envelope-localized construct Tic110-GFP. No signal was detected in the stromal compartment, possibly due to highly oxidized conditions present in the protoplasts. These might have been caused by the rather long incubation in the dark, leading to preferential membrane attachment of the redox-sensitive Tic62 (Stengel et al., 2008).
Interestingly, the Tic62-GFP signal was not distributed evenly throughout the entire thylakoids. Instead, areas with the strongest autofluorescence, which likely represent the grana stacks, were mostly free of GFP signal, indicating that the protein associates with the stroma lamellae. The N-terminal half of Tic62 fused to GFP (Tic62-Nt) produced a signal very similar to that of the full-length construct, albeit with a reduced signal intensity. Expression of Tic62-Ct, on the other hand, N-terminally fused to the native transit peptide and C-terminally to GFP, resulted in a signal reminiscent of soluble stromal proteins. These results confirm that Tic62 is indeed able to bind to the thylakoid membrane system and demonstrate that the Nt of Tic62 contains all the necessary information for the internal targeting of the protein within the chloroplast.
Expression of both leaf-type FNR isoforms (LFNR1 and LFNR2) as C-terminal red fluorescent protein fusions resulted in a very similar signal pattern for both constructs (see Supplemental Figure 2 online). The signals were associated with the thylakoid membranes, but formed a nonregular, spotted pattern and had the tendency to accumulate at the thylakoid-to-stroma border.
Taken together, our data verify the presence of a Tic62 pool at the thylakoid membranes in proximity to both FNR isoforms and support the idea of Tic62 having functions beyond its role as a translocon component, potentially affecting the fate of photosynthesis-related proteins.
Tic62 Is Part of High Molecular Weight Complexes in the Thylakoids Associated with FNR
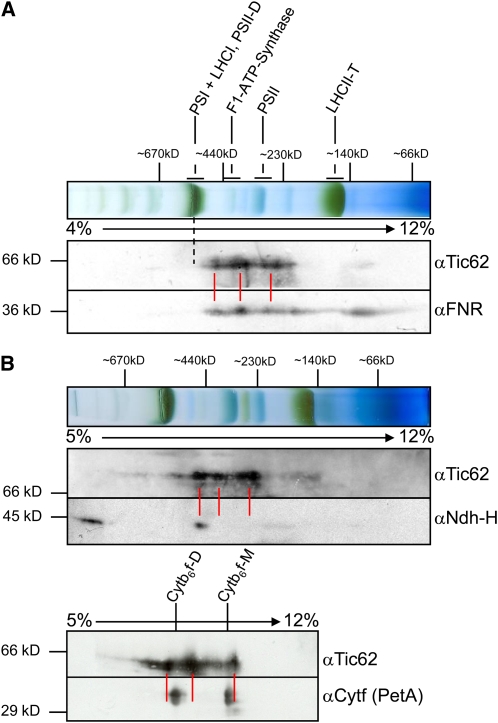
Having established that a pool of Tic62 is located at the thylakoid membranes, the question arose whether it might interact with other proteins or protein complexes present in the same compartment. For this purpose, mildly solubilized chloroplast membranes were used for two-dimensional blue native (2D BN)/SDS-PAGE analyses. As the signal intensity for Tic62 with an antibody raised against the Ct of Tic62 from pea was quite weak when used in second dimension blots from Arabidopsis, pea thylakoids were used in addition to Arabidopsis samples. The visible migration pattern of the thylakoid complexes was almost identical in samples from both organisms (cf. Figures 2A and 2B). The same was true for the Tic62 signal, which was mainly detected in three to four complexes ranging from roughly 250 to 500 kD. When using antibodies against FNR, it was found that the migration behavior of both proteins matched very well in the entire high molecular weight (HMW) range (Figure 2A, indicated by red lines).
Figure 2.
Thylakoidal Tic62 Comigrates Almost Exclusively with FNR.
(A) Comigration of Tic62 and FNR was observed in 2D-BN/SDS-PAGE of pea thylakoids solubilized with 1% n-dodecyl β-d-maltoside. The first dimension (10 μg chlorophyll) and immunoblots of the second dimension with Tic62 and FNR antibodies are shown with the positions of the major thylakoidal complexes indicated. Red lines represent the main signals detected for Tic62 and the dashed line the location of PSI, which displays a slower mobility than Tic62 and FNR.
(B) Tic62 does not comigrate with the high molecular weight NDH complex or the Cytb6f complex. The first dimension and immunoblots of the second dimension of 2D-BN/SDS-PAGE of Arabidopsis chloroplasts (20 μg chlorophyll) with Tic62 and Ndh-H antibodies are shown (top panel). The NDH complex was detected in two complexes, probably representing a dimeric and a monomeric form. The immunoblot of the second dimension of a 2D-BN/SDS-PAGE of pea thylakoids is depicted (bottom panel), incubated with antibodies generated against Tic62 and Cytf (PetA). The Cytb6f complex is found in a monomeric (Cytb6f-M) and a dimeric (Cytb6f-D) form. Positions of molecular weight marker bands in the first and second dimensions are indicated (in kD) as well as the acrylamide concentration gradient used for the BN-PAGEs.
Since FNR had already been found associated with components of PSI, the Cytb6f complex, and the NDH complex in various studies (e.g., Andersen et al., 1992; Guedeney et al., 1996; Okutani et al., 2005; Zhang et al., 2001), potential comigration of Tic62 and FNR with representatives from those complexes was investigated. PSI subunits generally migrate together with LHCI in a single complex at around 550 to 600 kD, which is readily visible in the first BN dimension due to its dark-green color (indicated by a dotted line in Figure 2A). As mentioned above, the largest complex containing Tic62 and FNR displayed a significantly faster movement in the first dimension and could be found at ∼500 kD. Thus, no comigration of either component with PSI could be observed in this assay.
Antibodies directed against a component of the NDH complex (Ndh-H) revealed two distinct signals: one migrating with a size of around 1000 kD and another at ∼500 kD, possibly corresponding to monomeric and dimeric NDH complexes, respectively (Figure 2B; compare Aro et al., 2005; Darie et al., 2005; Ishihara et al., 2007). The monomeric complex had a similar mobility like the largest Tic62/FNR complexes. A signal at the size of the dimeric NDH complex for Tic62 (or FNR) was, however, not detectable, arguing against a stable association. The Cytb6f complex was detected in a monomeric and a dimeric form after solubilization as well, migrating in the same molecular weight range as Tic62 (Figure 2C). At closer inspection, however, the signal peaks only partially overlapped, probably due to high signal strength rather than colocalization.
In summary, the BN-PAGE analysis suggests that Tic62 and FNR form several HMW complexes at the thylakoid membrane. Analyzing the migration behavior of thylakoid protein complexes, which have been implicated with FNR binding to the thylakoids, no specific and stable association could be verified. However, more transient or dynamic interactions with one or several of the respective complexes cannot be ruled out.
Absence of Tic62 Results in Reduction of Total FNR Amount and a Drastic Loss of FNR Binding to the Chloroplast Membranes
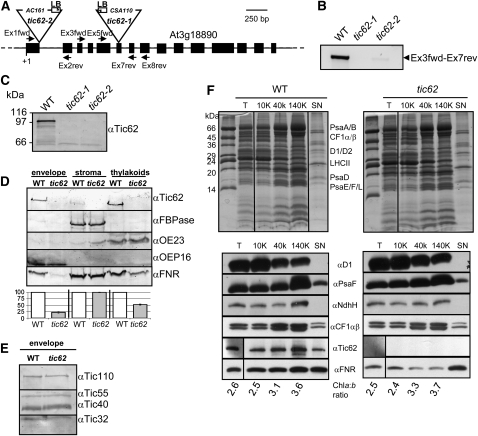
To obtain further insight into the role of Tic62, two Arabidopsis lines, tic62-1 (SAIL_124G04) and tic62-2 (GABI_439H04), containing a T-DNA insertion within the genomic sequence of the TIC62 gene (At3g18890) were analyzed (Figure 3A). Immunodecoration with Tic62 antiserum verified that no Tic62 protein was present in both lines (Figure 3C), even though RT-PCR analysis indicated a small amount (∼5%) of residual transcript in tic62-2, which nevertheless most likely represents truncated mRNA (Figure 3B). Because experiments for the following characterization of the tic62 mutant were performed with both lines and gave practically identical results, we do not present all data separately. If not stated otherwise, results obtained with tic62-1 are shown.
Figure 3.
Tic62 Knockout Lines Display a Specific Reduction of FNR in the Membrane Fractions.
(A) Genomic structure of Tic62 from Arabidopsis (At3g18890). Black boxes denote exons, black lines introns, and dotted lines 5′ and 3′ untranslated regions (not to scale). The insertion sites of T-DNAs in lines SAIL_124G04 (tic62-1) and GABI_439H04 (tic62-2) are indicated by triangles. Furthermore, binding sites for Tic62 gene-specific primers and T-DNA–specific left border primers used for screening for homozygous plants are depicted.
(B) Tic62 transcript is almost completely absent in tic62 knockout lines. An RT-PCR experiment performed with the Ex3fwd-Ex7rev primers (being 3′ of the tic62-2 and flanking the tic62-1 T-DNA insertion site) of the wild type, tic62-1, and tic62-2 RNA is shown.
(C) No Tic62 protein can be detected in tic62 mutants by an immunoblot analysis of wild-type, tic62-1, and tic62-2 chloroplast extracts. Note that the Arabidopsis Tic62 protein displays an aberrant mobility and is found at ∼98 kD in SDS-PAGE gels.
(D) FNR is specifically lost from membrane fractions of tic62 chloroplasts. Five micrograms of protein were used for envelope and thylakoid fractions and 10 μg protein in stroma samples. The immunoblot shows the signals obtained with antibodies generated against Tic62, FNR, FBPase (stroma marker), OE23 (thylakoid marker, soluble lumenal protein), and OEP16 (envelope marker) of envelope, stroma, and thylakoids from wild-type and tic62 chloroplasts. Beneath the blot the quantification of FNR in the various fractions is shown, in which the wild-type amount was arbitrarily set to 100% (±sd; n = 2).
(E) Tic subunits are not affected in tic62 plants. Immunoblot of wild-type and tic62 envelopes with Tic110, Tic55, Tic40, and Tic32 antibodies.
(F) Tic62 is located at the stroma-lamellae of thylakoids. Arabidopsis thylakoids from wild-type and tic62 plants were subfractionated by differential centrifugation. A Coomassie-stained 15% urea-SDS gel (major thylakoid proteins are indicated) and an immunoblot of the obtained fractions is shown: T, untreated thylakoids; 10K, centrifugation at 10,000g, containing grana thylakoids; 40K, centrifugation at 40,000g, representing margins; 140K, centrifugation at 140,000g, enriching stroma lamellae. SN represents the final trichloroacetic acid-precipitated supernatant. For the immunoblots, 5 μg of chlorophyll and 10 μg protein of the supernatant were used per lane and probed with antibodies against Tic62, FNR, and representatives of the main thylakoidal protein complexes: PSII (D1), PSI (PsaF), NDH (Ndh-H), and the ATPase (CF1αβ). The indicated chlorophyll a:b ratio is a measure for the successful enrichment of grana (low a:b ratio) or stroma (high a:b ratio) thylakoids. The figure shows one of two independent repetitions with essentially identical results.
Knockout plants had wild-type appearance under all tested conditions including long-day (16 h light), short-day (8 h light), constant light (24 h light), low light (<30 μmol photons m−2 s−1), high light (>300 μmol photons m−2 s−1), cold stress (+10°C on soil, +4°C on plates), or addition of methylviologen. The same was found for the ultrastructure of mesophyll chloroplasts (from long-day plants) using transmission electron microscopy (see Supplemental Figure 3 online). However, after fractionation and immunoblotting of isolated chloroplasts from wild-type and mutant plants, a distinct molecular phenotype was observed (Figure 3D). In particular the membrane-bound pools of FNR were severely affected: the envelope-bound fraction of FNR was barely visible in the mutant samples (<20% of the wild type), while the thylakoid-localized pool was about halved (∼50% of the wild type). Interestingly, the soluble pool of FNR in the stroma remained unchanged. When testing for components of the Tic complex, the subunits were likewise found to be present at unchanged levels in the mutants compared with the wild type (Figure 3E). In addition to immunoblotting, 2D isoelectric focusing/SDS-PAGE of thylakoids from both the wild type and tic62 was performed to screen for further changes in the proteome of this compartment. However, only one spot with a markedly lower steady state level was detectable, which was identified by mass spectrometry as LFNR2, corroborating a very specific loss of FNR (see Supplemental Figure 4 online).
To investigate how the loss of Tic62 in the knockout plants might affect the subthylakoidal localization of FNR in more detail, thylakoid subfractionations of both wild-type and tic62 chloroplasts were performed (adapted from Ossenbühl et al., 2002; Figure 3F). The assay resulted in thylakoid fractions enriched in grana thylakoids (10K fraction), intermediate margin regions (40K), the stroma lamellae (140K), as well as the final supernatant of the fractionation, containing those components that are attached to membrane protein complexes but readily dissociated during the procedure (e.g., as the CF1 part of the ATPase). Success of the fractionation was confirmed by Coomassie blue staining of the respective samples separated by SDS-PAGE, determination of the chlorophyll a:b ratio (increasing with enrichment of PSI/LHCI), and immunoblotting. The PSII supercomplexes are known to be preferentially located in the grana thylakoids; accordingly, components of PSII were found to be enriched in the 10K fraction (e.g., D1/D2 and LHCII).
By contrast, subunits of PSI (PsaA/B/D/F), the ATPase (CF1α/β), and the NDH complex (Ndh-H) were found to accumulate in the lower-density fractions (i.e., the stroma thylakoids), in agreement with published data (Aro et al., 2005; Dekker and Boekema, 2005). Analysis of Tic62 distribution revealed a fractionation pattern similar to the PSI and ATPase marker proteins, clearly indicating a stroma-thylakoid localization and thus supporting the data from the confocal analysis (see Supplemental Figure 2 online). Mutant tic62 thylakoids were devoid of any Tic62 signal. FNR was more evenly distributed with minor enrichment in the stroma-lamellae fraction of the wild type, equally reminiscent of the more nonregular signal in the confocal analysis. Probing the tic62 samples for FNR, the pattern differed again markedly from the wild type as the membrane-bound pool was reduced while similar amounts were found in the supernatant. Interestingly, the remaining signal was strongest in the grana fraction of tic62 and thus proportionally the least diminished, indicating that the stroma-lamellae pool of FNR was affected most by the loss of Tic62.
Taken together, the molecular phenotype of tic62 plants confirms the close link between Tic62 and FNR. The overall amount of FNR in the mutant chloroplasts is decreased, and particularly the attachment of FNR to the chloroplastic membranes is impaired.
Incorporation of FNR into HMW Complexes Is Specifically Defective in tic62 Thylakoids
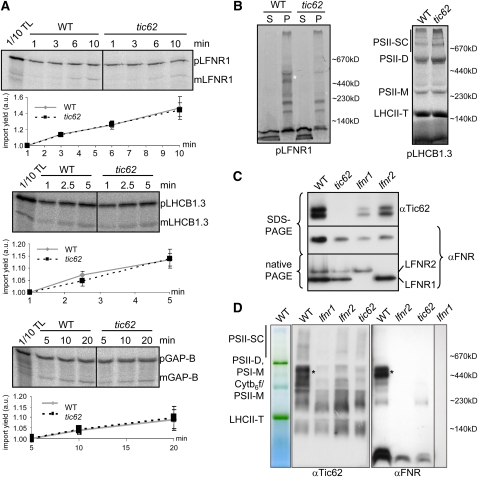
These data suggest that the binding of FNR to the internal chloroplast membrane systems is less efficient in tic62 chloroplasts. At the same time, FNR does not seem to accumulate in a soluble form in the stroma. Hence, the overall amount of FNR in the mutant chloroplasts is reduced. Assessment of the transcriptome by microarray analysis (Affymetrix GeneChip Arabidopsis ATH1) of tic62 plants harvested from the dark did not result in the detection of any significant changes when compared with the wild type. Neither of the FNR isoforms nor any other component involved in protein turnover or transport (except Tic62 itself) displayed a significant reduction in transcript level. Another possible reason for the reduction of FNR could be a defect in preprotein import, caused by loss of Tic62. To test this hypothesis, pLFNR1 was subjected to in vitro chloroplast import assays. As depicted in Figure 4A, the import efficiency of pLFNR1 in tic62 chloroplasts was equal to the wild type, as was that of two control preproteins: pLHCB1.3 (thylakoid protein) and pGAP-B (stromal protein).
Figure 4.
The Loss of Tic62 Does Not Affect FNR Import but Inhibits the Formation of FNR HMW Complexes.
(A) Import of pLFNR1 is not affected in tic62 chloroplasts. Import into isolated wild-type and tic62 Arabidopsis chloroplasts was started by the addition of translation product (pLFNR1 as well as pLHCB1.3 and pGAP-B as controls) and performed for the indicated time. Import products, including 10% of translation product (TL), were separated by SDS-PAGE, radiolabeled proteins analyzed by a phosphor imager, and the signals of the processed mature forms quantified (±sd; n = 3). p, precursor form; m, mature form; a.u., arbitrary units.
(B) LFNR1 integration into HMW complexes is defective in tic62 thylakoids. The pLFNR1 protein (and pLHCB1.3 as control) was first imported into wild-type and tic62 chloroplasts for 30 min, and the membranes were subsequently separated from the stroma compartment by disruption of the chloroplasts and centrifugation. The resulting fractions of supernatant (S) and pellet (P) were separated by a BN-PAGE gel (5 to 12% acrylamide) and analyzed on a phosphor imager. The approximate position of BN size markers is indicated, and the HMW Tic62/FNR complexes are marked by an asterisk.
(C) The amount of Tic62 is reduced in lfnr1 and lfnr2 plants. An immunoblot with αTic62 and αFNR antibodies of total protein extract from wild-type, tic62, lfnr1, and lfnr2 plants is shown. Ten micrograms of total leaf protein extract was used in SDS-PAGE and 30 μg of protein for native PAGE.
(D) The HMW Tic62/FNR complexes are similarly absent in tic62, lfnr1, and lfnr2 thylakoids. BN-PAGE (5 to 13.5%) of thylakoids isolated from wild-type, tic62, lfnr1, and lfnr2 plants. An unstained gel lane indicating the major thylakoidal complexes and immunoblots with αTic62 and αFNR antibodies is shown. The approximate position of BN size markers is indicated, and the HMW Tic62/FNR complexes are marked by an asterisk.
[See online article for color version of this figure.]
Having established that the steady state level of FNR at the membranes is reduced in tic62 mutants, it was analyzed if the initial binding to the thylakoid membranes after import was affected or whether the effect is caused at a later stage (e.g., decreased complex stability). For this purpose, pLFNR1 was imported into wild-type and tic62 chloroplasts, followed by separation of the membranes from the stroma compartment. The resulting fractions were analyzed on a BN-PAGE gel (Figure 4B). Comparison of the samples showed that the overall binding of FNR to membranes was diminished in the Tic62-depleted chloroplasts, while pLHCB1.3 was faithfully assembled into light-harvesting complexes in both wild-type and tic62 chloroplasts in the control reaction. In particular, the HMW FNR complexes were completely absent. Two complexes at lower molecular weight were detected that seemed unchanged, and the monomeric/dimeric pool of FNR was present, albeit in reduced amounts. The signal in the soluble fraction was stronger in tic62 chloroplasts than in the wild type, indicating that the excess FNR that could not be incorporated into the HMW complexes at least initially accumulates in the stroma, where it might be proteolytically degraded later on.
To verify the observed effect in a steady state situation, we solubilized chloroplasts of wild-type and both tic62 lines without prior import and separated protein complexes in a first dimension using BN-PAGE followed by a denaturing SDS-PAGE in a second dimension. Examination of the first dimensions after BN-PAGE revealed no differences in the running behavior or amount of any visible protein complexes (see Supplemental Figure 5A online; also see Figure 5A, first two lanes). However, immunodetection of FNR in the second dimension confirmed the results from the import experiments: the complexes with the highest molecular weight were not detectable in the mutant thylakoids (see Supplemental Figure 5B online; also first dimension in Figure 4D, compare lanes 6 and 8) and only smaller complexes remained present, as well as the low molecular weight signals most likely representing monomers or dimers of FNR.
Figure 5.
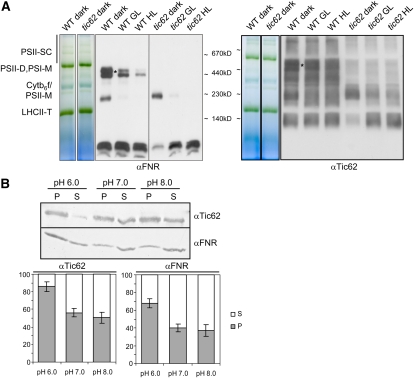
Membrane Attachment of the FNR/Tic62 Complexes Is Regulated by Light and Stromal pH.
(A) The amount of thylakoid-bound Tic62/FNR complexes is dependent on ambient growth light of the plants. Thylakoids were prepared from dark-adapted plants and compared with growth light (GL; 4 h under 100 μmol photons m−2 s−1) or high light (HL; 2 h under 1000 μmol photons m−2 s−1) treated samples. A BN-PAGE gel lane (5 to 13.5%) from wild-type and tic62 thylakoids with the major photosynthetic protein complexes indicated and immunoblots with αTic62 and αFNR antibodies are shown. The approximate position of BN size markers is indicated, and the HMW Tic62/FNR complexes are marked by an asterisk.
(B) The attachment of Tic62 and FNR to thylakoids is dependent on the stromal pH. Isolated pea thylakoids were mildly solubilized and incubated in sodium phosphate buffer with either pH 6.0, 7.0, or 8.0. After separation of soluble and membrane-bound proteins, pellets (P) and supernatants (S) were analyzed by immunoblotting with antibodies against Tic62 and FNR. A typical result of four independent experiments is shown. Furthermore, the amount of Tic62 and FNR in the pellet (gray) and supernatant (white) of all experiments (n = 4) was quantified (including se bars) and is depicted as fraction of total sample (100%).
[See online article for color version of this figure.]
These data demonstrate that (1) the expression of the genes encoding LFNR is not affected in the tic62 mutants, and (2) both FNR preproteins do not seem to be imported in a strictly Tic62-dependent manner. Therefore, differences in the turnover of the FNRs inside the chloroplast are likely to cause the observed reduction in FNR amount.
HMW Tic62/FNR Complexes Are Dependent on the Presence of Tic62 and Both Leaf FNR Isoforms
Since the overall amount of FNR was reduced in tic62 chloroplasts, the converse situation was investigated, testing for the fate of Tic62 in absence of one of the LFNR isoforms. Accordingly, protein extracts from wild-type, lfnr1, and lfnr2 leaves were analyzed by immunoblotting and compared with wild-type and tic62 samples (Figure 4C). In contrast with the wild type, the amount of Tic62 was found to be drastically reduced in both lfnr knockout lines. This reciprocal phenotype indicates that the absence of either Tic62 or FNR affects the accumulation of the remaining interaction partner(s) in the chloroplasts.
This observation on the level of total chloroplast protein raised the question how this effect might be reflected more specifically at the thylakoids. In two recent reports (Lintala et al., 2007, 2009) it was demonstrated that in lfnr1 and lfnr2 mutants, thylakoid binding of the respective other isoform was affected. Similar to the tic62 phenotype, several HMW complexes were missing in lfnr2 thylakoids. Moreover, in lfnr1 knockout lines, binding of LFNR2 to the thylakoids was completely abolished. To examine these apparent similarities in parallel, the Tic62/FNR complex assembly in the thylakoids was compared in tic62, lfnr1, and lfnr2 mutants using BN-PAGE (Figure 4D). Comparison of FNR and Tic62 protein pattern in the wild-type samples corroborated the comigration in the HMW range (cf. both first lanes). FNR was lost from these complexes in tic62 and lfnr2 plants, and absolutely no FNR could be detected in lfnr1 thylakoids. Similarly, Tic62 was absent from the HMW complexes comigrating with FNR in any of the mutant lines. This fact indicates that the thylakoid-bound HMW complexes are dependent on Tic62 and both FNR isoforms, LFNR1 and LFNR2. The complexes therefore most probably represent hetero-oligomers comprised of all three components in varying composition or stoichiometry.
Thylakoidal Tic62/FNR Complexes Are Involved in Light-Regulated Processes but Do Not Participate in Photosynthetic Electron Transport
Since light regulation is common for thylakoidal protein complexes, in particular those involved in photosynthesis, the behavior of the Tic62/FNR complexes in response to light was investigated. Thylakoids prepared from dark-adapted plants were thus compared with light-treated samples and analyzed by BN-PAGE (Figure 5A). Visualization of the Tic62/FNR complexes by immunoblotting revealed that the Tic62-dependent FNR complexes decreased markedly in abundance upon irradiation (growth light). The effect was proportional to the light intensity and led to an almost complete loss of FNR from the HMW complexes under high light. The amount of Tic62 at the thylakoids likewise diminished in irradiated samples, but interestingly a fraction remained attached to the membranes even under high light. These observations confirm that Tic62 and FNR are not immobilized at the thylakoid membranes but react dynamically, yet differentially, in response to light-dependent stimuli.
Illumination of chloroplasts results in acidification of the thylakoid lumen and a concomitant alkalized stromal pH. Since energization of thylakoids and changes in the pH were described to affect FNR (Carrillo et al., 1981; Grzyb et al., 2007), we now investigated whether the observed detachment from the thylakoids might be due to this change in the environmental condition and whether Tic62 reacts accordingly. Thylakoids were isolated from dark-adapted pea leaves using a slightly acidified pH (pH 6) to mimic the nightly stromal environment. Subsequently, the membranes were either further incubated at pH 6.0 or subjected to alkalized pH (pH 7 or 8), simulating medium to strong daily changes in the stromal pH following illumination. After the incubation, solubilized proteins were separated from the membrane fraction by centrifugation and both fractions were analyzed by immunoblotting (Figure 5B). At pH 6.0, Tic62 and FNR remained mostly membrane bound. Alkalized pH, however, resulted in a strong solubilizing affect, which was slightly stronger for pH 8.0 than for pH 7. Interestingly, reminiscent of the light treatment, FNR was again found to be easier solubilized from the thylakoids than Tic62.
The ability of the thylakoidal Tic62/FNR complexes to react to changes in light and the stromal pH prompted us to investigate whether the loss of Tic62 had any influence on the light-driven electron transport. Accepting electrons from Fd and using those to produce reduction equivalents, FNR represents the link between photosynthetic electron transport and the Calvin-Benson cycle. A decreased amount of FNR could thus lead to a holdup of electrons at some point of the electron transport chain as had been seen, for example, by Lintala et al. (2007) in their lfnr1 mutants.
PSII performance of intact leaves from wild-type and tic62 mutant plants was measured using a PAM fluorometer (see Supplemental Table 2 online). However, no differences could be detected as deduced from the ratio of variable to maximum fluorescence (Fv/Fm), the quantum yield of PSII (ΦPSII), the degree of nonphotochemical quenching, and the excitation pressure of PSII (1-qP) using either growth light (90 μmol photons m−2 s−1) or high light (1100 μmol photons m−2 s−1) irradiation as actinic light. In addition to these measurements of PSII activity, we monitored the postillumination rise in chlorophyll fluorescence (F0 rise), which has been used as an indication for CEF mediated by the NDH complex (see Supplemental Figure 6A online; Shikanai et al., 1998; Endo et al., 2008; Lintala et al., 2009). Furthermore, the kinetics of the dark-induced rereduction of P700+ was determined as a parameter for PSI-driven CEF (see Supplemental Figure 6B online; Lintala et al., 2007, 2009). Both analyses did not result in any obvious differences between wild-type and mutant plants; thus, we conclude that Tic62 itself as well as the Tic62-dependent pools of FNR do not directly participate in photosynthetic linear or cyclic electron transport processes.
Tic62 Binds FNR in a High-Salt-Insensitive Manner and Does Not Distinguish between the Arabidopsis FNR Isoforms in Vitro
Thylakoid-bound FNR can be subdivided into fractions with differential binding affinities. Washing with salt readily releases a fraction of the enzyme, whereas another, stronger bound subpool is not affected by such a treatment (Matthijs et al., 1986). Comparison of the dissociation of FNR from isolated thylakoids of wild-type and tic62 mutant plants using high ionic strength buffer was employed to investigate which of those fractions contains the Tic62-bound FNR, in order to gain information about the binding mode (Figures 6A and 6B). For quantification of the amount of solubilized enzyme, a fraction of the supernatant after high-salt wash was used in cytochrome c (Cyt c) reduction assays (Figure 6B). In parallel, supernatant and pellet were immunoblotted and probed with antibodies against FNR (Figure 6A). Provided that FNR was equally well solubilized from the thylakoids in all samples, the resulting activity in the supernatant should match the difference in total quantity present in the membrane.
Figure 6.
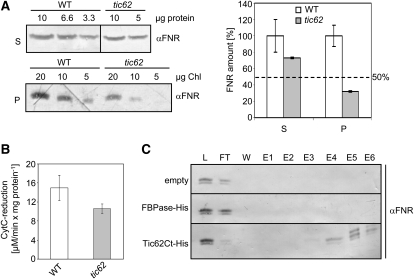
Tic62 Can Bind Both Arabidopsis FNR Isoforms in Vitro, and the Interaction Is Stable under High Salt Concentrations.
(A) FNR is more loosely bound to thylakoids from tic62 than from wild-type plants. Isolated thylakoids of wild-type and tic62 plants were washed with high ionic strength buffer (0.5 M NaCl), and membrane and soluble fractions were separated by centrifugation. Shown is a representative immunoblot of supernatant (S) and pellet (P) fractions obtained from wild-type and tic62 thylakoids, using a dilution series of protein and chlorophyll concentrations and probed with FNR antibody. Additionally, the quantification of the FNR amount in wild-type (white; 100%) and tic62 (gray) supernatant and pellet signals from the immunoblots is shown. The dotted line represents the amount of FNR detected in native tic62 thylakoids. Standard error bars are included; the experiment was performed in triplicate.
(B) Cyt c reduction activity in the supernatant of high salt washes of tic62 thylakoids is ∼70% of wild-type activity. Activity was determined by Fd-dependent Cyt c reduction, monitored with a spectrophotometer at 550 nm. The experiment was performed in triplicate; se bars are included.
(C) Tic62 binds both LFNR1 and LFNR2 equally well. LFNR1/LFNR2 binding assay on Tic62 Ct-His affinity matrix. Overexpressed and purified Tic62 C terminus and FBPase were bound via a (His)6-tag to Ni2+ beads and used as an affinity matrix for LFNR1 and LFNR2 from tic62 Arabidopsis stroma. An empty column without the addition of His-tagged protein was used as additional negative control. After incubation, the matrix was washed (W; last wash), and bound proteins were eluted by addition of 750 mM NaCl (E1), 1 M NaCl (E2), 4 M urea (E3), 8 M urea (E4), 200 mM imidazole (E5), and 400 mM imidazole (E6). The resulting samples including 1/70 of load (L) and flow-through (FT) were subjected to urea/SDS-PAGE and immunoblotting with FNR antibody.
The Cyt c reduction activity of the high-salt washes prepared from tic62 thylakoids was ∼70% of the wild-type level, consistent with the amount of FNR in the supernatant as determined by immunoblotting. At the same time, the amount of FNR in the pellet fraction was only ∼30% of the wild type. Since the amount of FNR in tic62 thylakoids was estimated to ∼50% in both mutant lines compared with the wild type (Figure 3D), FNR was overrepresented in the supernatant fraction of the mutants. Thus, FNR is more readily washed from the thylakoid membranes in presence of high ionic strength when Tic62 is not present, indicating a strong hydrophobic binding between the two proteins in the wild type.
In the following, it was investigated whether Tic62 preferentially binds to one leaf FNR isoform. For this purpose, heterologously expressed and purified Tic62-Ct, containing the FNR binding repeats, was bound via its (His)6-tag to Ni2+ beads. These were used as an affinity matrix for LFNR1 and LFNR2 from Arabidopsis tic62 stroma, being devoid of endogenous Tic62 (Figure 6C; see Supplemental Figure 7 online). Elution was performed by increasing first the ionic strength of the solution and subsequent addition of 4 or 8 M urea, respectively, to denature the proteins still bound to the matrix. Finally, the column was stripped with imidazole. Analysis of the resulting fractions by immunoblotting revealed that both FNR isoforms eluted equally well from the Tic62-Ct matrix (Figure 6C, bottom lane), indicating a similar affinity for Tic62. In addition, only little FNR could be eluted with salt, in line with the previous findings. For a major fraction of bound FNR, denaturation with urea was necessary to release the protein from Tic62. As controls, stroma was also incubated with equal amounts of immobilized FBPase or with Ni2+-sepharose only (empty). No (unspecific) FNR binding was detected in either sample.
FNR-Tic62 Complex Formation: Tic62 Binds to the Back Side of FNR in a Defined Stoichiometry
Chemical shift mapping by nuclear magnetic resonance (NMR) spectroscopy has been successfully used to study protein–protein interactions of large flavoprotein complexes (Maeda et al., 2005; Lee et al., 2007), which is why we applied this method to investigate the interaction between FNR and Tic62. Changes in the peak positions (resonances) or in the signal intensities after titration of a ligand molecule are referred to as chemical shift perturbations, which are suggested to correspond to the interaction sites with the other molecule. In NMR experiments, however, the HMW of molecules of interest often hampers the resolution and sensitivity of resonances. We thus chose the R1 peptide, comprising a 30–amino acid repeat of the Tic62-Ct (Küchler et al., 2002), instead of the whole C-terminal part of Tic62 for the analysis.
Chemical shift perturbations of 2D 1H-15N TROSY-HSQC NMR spectra of a uniformly [2H/15N]-labeled FNR were detected. Supplemental Figure 8A online shows an overlay of the recorded spectra in the absence (green) and presence (red) of peptide. Comparison of the spectra indicated a number of cross-peaks exhibiting significant changes upon the addition of R1. Strikingly, many of the peaks of free FNR disappeared, and at the same time new peaks corresponding to a complex appeared at different resonance positions according to the titration. All of these changes were thus classified as being in a slow exchange regime in terms of the NMR time scale, indicating a strong binding mode. In addition, the intensities of the overall peaks were found to decrease significantly.
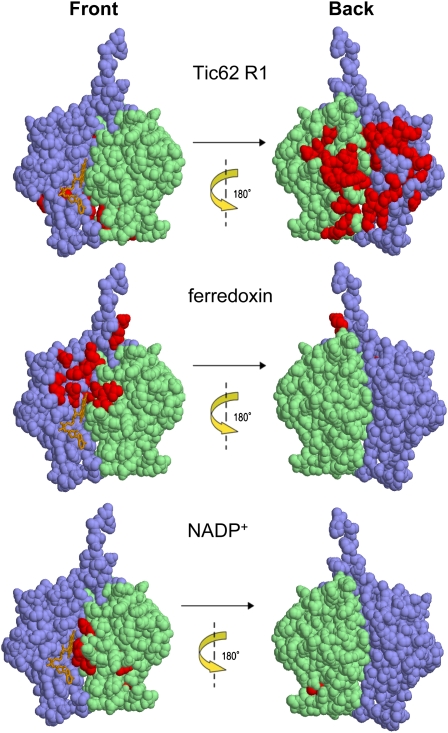
On the basis of the known resonance assignments of free FNR, the chemical shift perturbations were mapped onto the tertiary structure. Largely perturbed residues are likely to be involved in the interaction with R1. Figure 7 shows the mapping of the markedly perturbed residues on the x-ray structure of free FNR. The affected residues belong to a rather large area on only one side of FNR. Interestingly, when the binding site of FNR for R1 was compared with those for Fd and NADP+, it was found to be located on the surface opposite to that important for the catalytic activity. To further investigate the complex formation between FNR and Tic62, analytical ultracentrifugation analyses at sedimentation equilibrium (AUC-SE) in the absence and presence of R1 peptide were employed (see Supplemental Figure 8B online). The resulting molecular weights for FNR were estimated as 35,468 and 73,768 D in the absence and presence of R1 peptide, respectively, indicating that FNR and R1 peptide indeed form a complex with a stoichiometry of ∼2:1 (see Supplemental Figure 8B online, top two panels).
Figure 7.
Tic62 Binds to a Novel Binding Site at the Back Side of FNR.
Mapping of chemical shift perturbations caused by the binding of the Tic62-R1 peptide, Fd, and NADP+ onto the tertiary structure of FNR. Residues which exhibited chemical shift perturbations upon addition of the factors are shown in red. The FAD binding domain is highlighted in blue and the NADP+ binding domain in green. The front side and the back side of the FNR are displayed.
Since the AUC-SE analyses demonstrated that two FNR molecules can bind to one R1 peptide, this result suggested that at most six FNRs can form a complex with the pea homolog of the Tic62 protein, containing three repeats. To test this possibility, we next performed AUC-SE experiments of FNR in the absence and presence of the C-terminal part of Tic62 from pea (see Supplemental Figure 8B online, two bottom panels). The Tic62-Ct (19,732 D) and FNR (see above) were both found to exist as a monomer in solution when analyzed alone. After joint centrifugation, however, a complex with an estimated molecular mass of 127,782 D was formed under the condition applied. Being significantly smaller than expected, the complex was thus probably comprised of only three FNRs binding to one Tic62-Ct, indicating a steric hindrance not allowing a full saturation of binding sites by parallel attachment of six FNRs.
Tic62 Exerts a Stabilizing Effect on FNR but Does Not Act as an Activity Modulator
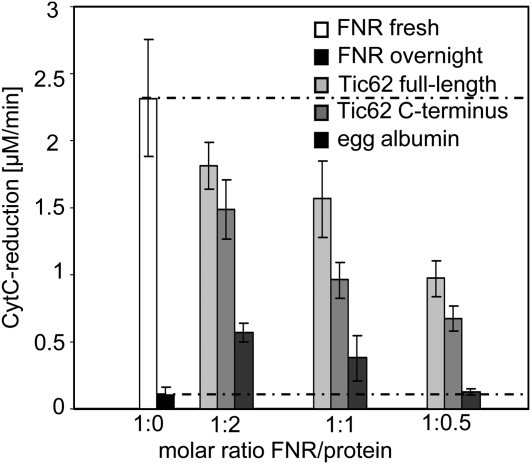
The activity of many enzymes is modulated by conformational changes upon binding of effector molecules or proteins to allosteric sites or by blocking of the active site directly. To investigate potential enzymatic effects of the Tic62/FNR interaction, the catalytic activity of heterologously expressed and purified FNR was measured in vitro using a Cyt c reduction assay in the presence or absence of varying amounts of recombinant Tic62 protein (full-length or Ct). An overnight preincubation period at +4°C was used to allow both proteins to reach a binding equilibrium. When the Cyt c reduction activity was then measured, it was discovered that the samples containing FNR only had lost most of their catalytic activity compared with fresh sample (<5% residual activity; Figure 8). In contrast with this, samples with saturating amounts of Tic62 present (ratio Tic62:FNR = 2:1) were much more active, still showing ∼80% of the original Cyt c reduction capacity. Similarly, in the presence of Tic62-Ct, ∼64% of FNR activity was retained. To test whether the observed effect was specific for the Tic62/FNR interaction or simply due to an increased protein concentration in the overnight reaction, FNR was additionally incubated with equal molar amounts of egg albumin as a control protein using otherwise identical conditions. In this case, a portion of FNR stayed active, but to a much lower extend than seen for the Tic62 constructs (<25% original activity). When the amount of added protein was decreased to a ratio of 1:0.5 (FNR:protein), activity in the presence of egg albumin was indistinguishable to the values without protein. Under the same conditions, however, the Tic62 constructs still had a considerable effect on FNR activity (∼42 or ∼29% activity, respectively). The observable effect of Tic62 (full-length or Ct) on FNR activity implies that the interaction helps to stabilize FNR.
Figure 8.
Tic62 Has a Stabilizing Effect on FNR in Vitro.
Catalytic activity of overexpressed and purified FNR was measured in vitro using the Fd-dependent Cyt c reduction assay. FNR was either used fresh (white), or activity was determined after overnight incubation (black). Both values are additionally represented by dotted lines for comparison with the other results. Various amounts (molar ratio of FNR:used protein was 1:2, 1:1, and 1:0.5) of full-length Tic62 (light gray), Tic62 C terminus (gray), or egg albumin as control (dark gray) were added to FNR before overnight incubation, and activity was measured the next day. None of the interacting proteins displayed any detectable Cyt c reductase activity independently of FNR. The mean values of triplicate experiments with se bars are depicted.
DISCUSSION
Tic62 Is a Major FNR Interaction Partner at the Thylakoids
Evaluation of Tic62 coexpression clusters as well as GUS reporter gene analysis demonstrate a clear link to processes such as protein turnover in the chloroplast and photosynthesis, suggesting a role in the regulation of the fate of chloroplastic proteins involved in general photosynthetic functions (see Supplemental Figure 1A online). The experimental results fit surprisingly well to these initial in silico predictions, since Tic62 is obviously closely connected to the fate of FNR, one of the main photosynthetic proteins present in the chloroplast.
Chloroplast subfractionation (Figure 1) as well as the localization of GFP-tagged constructs (see Supplemental Figure 2 online) demonstrate that Tic62 and FNR display a very similar localization pattern within the chloroplast as they are both found in the same compartments. Notably, the thylakoid pool of Tic62 is obviously present in addition to the known pools found at the envelope and in the stroma. However, in contrast with FNR, Tic62 seems to be preferentially (but not exclusively) membrane bound in Arabidopsis (Figure 1). It is interesting to note that this feature differs from the situation in pea, where an ∼50:50 distribution between soluble and membrane-bound form was described (Stengel et al., 2008). Although the reason for this difference is not clear, it might be based on differences in the Tic62 sequence (Balsera et al., 2007) or varying growth conditions of the plants. Since Tic62 seems to react sensibly to changes in the redox status, it is possible that these differences are reflected by a shift in the balance between membrane-bound and soluble protein.
The presented results indicate a strong preference for Tic62 localization at the stroma lamellae (Figure 3F). In addition, localization of GFP-tagged constructs corroborates the hypothesis that the N-terminal half of Tic62 with its hydrophobic patch is responsible for the attachment to membranes (see Supplemental Figure 2 online; Balsera et al., 2007), while the Tic62-Ct was found to be completely soluble in the chloroplast stroma.
For FNR, the subthylakoidal distribution was less well defined (see Supplemental Figure 2 online; Figure 3F). However, as the FNR-red fluorescent protein signal was not very strong and found to be dispersed throughout the thylakoid system, the possibility of an unspecific aggregation of the constructs was excluded. Rather, the punctate (dot-like) pattern represents an accumulation at or around their main site of action, which is clearly located at the thylakoid-to-stroma border, where FNR mediates the transfer of electrons from PSI/Fd to the reduction equivalents used for the metabolic processes.
We conclude that Tic62 is almost exclusively present in the stroma thylakoids. FNR, on the other hand, is not restricted to one location within the thylakoid system. It apparently resides with Tic62 at the stroma lamellae, but seems to be additionally associated to unknown other factors, either at the margins or even at the grana stacks.
Tic62 Acts as a Membrane Anchor of FNR
In tic62 knockout plants, we found the amount and distribution of FNR to be specifically altered (Figures 3D and 3F). In particular, the membrane-bound pools of FNR are drastically reduced: ∼50% of the thylakoid-bound FNR is lost and the envelope fraction is almost completely depleted (Figure 3D). Transcript analysis and in vitro import assays indicate that neither expression nor import of the FNR precursors is affected (under standard conditions) but that the incorporation into the HMW protein complexes of the thylakoid membrane is defective (Figure 4B). It was furthermore shown by BN-PAGE that Tic62 and FNR comigrate perfectly in the HMW range (Figure 2A). These results demonstrate that Tic62 acts as a major FNR binding protein at the thylakoids. Moreover, it is most likely the sole FNR binding factor at the inner envelope membrane. However, some degree of caution has to be exercised regarding the import assays since the impact of Tic62 on preprotein import has not been studied in detail so far. The fact that Tic62 is a dynamically associated regulatory Tic subunit, as well as the high viability of tic62 knockout plants, indicates that Tic62 might only affect the translocation of a small subset of preproteins under certain redox conditions. Further analyses to identify these have to be performed but are complicated by the complex molecular processes that Tic62 is apparently involved in.
As mentioned above, the presence of a thylakoid-bound form of an otherwise soluble protein like FNR has stimulated a number of studies aimed at the identification of a factor providing a docking station for the protein (Vallejos et al., 1984; Shin et al., 1985; Matthijs et al., 1986; Chan et al., 1987; Soncini and Vallejos, 1989; Guedeney et al., 1996; Zhang et al., 2001). However, all these results describing potential FNR binding proteins were to a large extend not followed up, are still disputed, and often the real physiological significance is not well understood. Moreover, the interactions are likely very short lived and dynamic, and it is also possible that reduced Fd first detaches from the thylakoids and subsequently interacts with FNR in the stroma. The ability of soluble FNR to sustain the main electron flow is clearly demonstrated by the high viability of lfnr1 knockout plants, which do not contain any thylakoid-bound FNR (Lintala et al., 2007). Moreover, substrate affinity and catalytic activity of FNR were demonstrated to be enhanced by energization of thylakoids following illumination (Carrillo et al., 1981). Since illumination clearly results in solubilization of the Tic62/FNR complex (Figure 5A), the membrane-bound state might indeed represent the more inactive form of the enzyme. It is therefore quite reasonable to suggest that the soluble pool of FNR in the stroma is the most responsible for the photosynthetic electron transport and that binding to the thylakoids might serve a different purpose, possibly in redox sensing or regulation. In addition, since FNR stability seems to be lowered at more acidic pH values (Lee et al., 2007), assembly into Tic62 complexes could therefore stabilize the enzyme during the hour of (photosynthetic) inactivity.
The results presented now allow a more detailed understanding of the thylakoid-localized pool of FNR. As indicated by BN-PAGE and thylakoid subfractionation, the HMW Tic62/FNR complexes are most likely present in the stroma lamellae. In absence of Tic62, the remaining FNR complexes are small in size and probably located either in the grana or at the margins of thylakoids (Figures 3F and 4D). No indications could be found that Tic62 or FNR stably associate with any other thylakoid protein complexes, since none (of those tested) displayed a clear comigration behavior (Figure 2). By contrast, comparison of the Tic62/FNR complex assembly in tic62, lfnr1, and lfnr2 plants by BN-PAGE revealed that the HMW complexes depend on the presence of all three components (Figure 4D). The amount and migration behavior of FNR, on the other hand, was shown not to be affected in the absence of a functional NDH complex (Burrows et al., 1998). We thus propose that the HMW Tic62/FNR complexes are composed mainly, if not exclusively, of Tic62 and FNR. This would extend the hypothesis of an LFNR1/LFNR2 dimer by Lintala et al. (2007), suggesting that it is actually a hetero-oligomer, composed of both leaf isoforms of FNR together with Tic62, that is required for the attachment of FNR to the envelope and thylakoid membranes.
Another hint for the existence of a hetero-oligomeric complex derives from the binding experiments performed with stromal FNRs and a Tic62-Ct affinity matrix (Figure 6C). Even though it cannot be ruled out that Tic62 could bind homodimers (or homo-oligomers) of both kind, Tic62 obviously does not distinguish between the two FNR isoforms and binds both equally well. A hetero-oligomeric complex therefore seems likely. The observation of several distinct complexes of varying size in the thylakoids would then be due to different oligomerization states of Tic62 and FNR, although the participation of other proteins cannot be excluded solely based on these facts. Other proteins might also assist in the membrane binding of FNR complexes that can be found also in absence of Tic62. Most recently, for example, the identification of a so far unknown thylakoid protein (TROL) was reported that seems to be involved in the formation of one of the smaller FNR complexes (Juric et al., 2009), while another study demonstrated that FNR is able to bind to artificial membranes directly, independent of proteinaceous factors (Grzyb et al., 2007).
The Strong Interaction between Tic62 and FNR Involves a Novel Binding Mode and Has a Stabilizing Function for the Complex
Several lines of evidence suggest that the association between Tic62 and FNR is not only very specific, but also involves a strong binding mode. The resistance to high salt concentrations (Figure 6) indicates the participation of mainly hydrophobic interactions in the binding of FNR and Tic62 as opposed to extensive hydrogen bonds or ionic interactions, which would be destabilized in the presence of salts.
Further insight into the binding mode of FNR and Tic62 was obtained by NMR spectroscopy. The 30–amino acid R1 peptide of Tic62 interacted with FNR, thereby confirming the interaction between FNR and Tic62 on a molecular basis (see Supplemental Figure 8A online). Interestingly, the data revealed that FNR binds to the Tic62 Ct via its back side (Figure 7). To our knowledge, this binding mode between FNR and Tic62 is novel because no other flavoprotein has been found to have such a binding pattern. Further investigation of this interaction might lead to finding of a new function of flavoproteins.
The suggested binding pattern of Tic62 and FNR should not block the active site but could be an indication for an allosteric regulation of the enzyme. However, from our results (Figure 8) we conclude that Tic62 is not acting as a modulator of FNR activity but rather as a stabilizing factor, thereby increasing the half-life of FNR. Instability and subsequent degradation might also be accountable for the observation that FNR that is not correctly assembled into thylakoid-bound complexes in tic62 plants, initially accumulates in the stroma after import, and is not visible in the steady state situation (Figure 4B versus 3D). Interestingly, the same holds true for Tic62 in lfnr1 and lfnr2 plants (Figure 4C). Surplus protein that cannot be integrated into the membrane-bound complexes in either mutant thus seems to be prone to proteolytical degradation, further supporting the notion of a reciprocal and interdependent stabilization of the components.
The Basic Tic62/FNR Complex Adopts a 1:3 Stoichiometry
In sedimentation velocity experiments with FNR in the presence or absence of Tic62 peptide, we determined the molecular masses of the resultant complexes (see Supplemental Figure 8B online). The finding of a 2:1 stoichiometry between FNR and the R1 peptide suggests that two FNR molecules dimerize upon addition of R1, thereby sandwiching it between them. This formation of a rather large heterotrimer could also explain the general decrease of NMR signal intensities, which tend to get weaker with an increasing target size. At the same time, the recorded chemical shift perturbations (on the back side of FNR) are therefore induced by binding of R1 and a second FNR molecule, making the clear assignment of the binding site difficult.
Interestingly, the complete Tic62 Ct from pea (containing three repeats) was found to associate with only three FNRs, suggesting that a steric hindrance within the complex does not allow the full theoretical set of six FNR molecules to bind at the same time. Further evidence supporting this in vitro result comes from a semiquantitative proteomic analysis of pea inner envelope, where the ratio of FNR:Tic62 was found to be 3:1 (Bräutigam et al., 2008; A. Weber, personal communication). As mentioned before, FNR is almost completely depleted from the envelopes in tic62 plants, suggesting that all the FNR found at this membrane is complexed by Tic62. Hence, the observed 3:1 ratio can be considered a measure of the complex at the envelope in vivo and fits surprisingly well to the values derived from the AUC experiments. However, this ratio may vary since the affinity of Tic62 to FNR (and the Tic complex) was demonstrated to be dependent on the redox status of the organelle (Stengel et al., 2008). This gives room to speculate that the number of FNR molecules bound to the Tic62 Ct can likewise vary dependent on the chloroplast redox conditions, which remains to be tested.
The Thylakoidal Tic62/FNR Complexes Are Dynamically Regulated by Light and the Stromal pH
The dissociation of the thylakoidal HMW complexes under light exposure (Figure 5A) indicates that those are subject to a light-dependent regulation. Interestingly, however, no photosynthetic phenotype could be detected in tic62 plants (see Supplemental Table 2 online), although about half of the thylakoid-bound FNR is missing in these mutants (Figure 3D), supporting the notion that the thylakoidal Tic62/FNR hetero-oligomers are not associated to photosynthetic complexes.
Biochemical assays with isolated wild-type thylakoids from pea revealed that the solubilization of the membrane-attached Tic62/FNR complexes might be the result of an alkalized stromal environment. This result is consistent with results from in vitro assays demonstrating that FNR binding to artificial membranes is enhanced under acidified pH conditions, possibly as a result of exposure of hydrophobic structures on the protein surface (Grzyb et al., 2007). However, the fact that Tic62 and FNR are both identically regulated by the metabolic redox status (Stengel et al., 2008) and react extremely similar to changes in the pH or light (Figure 5) indicates that the correct relocation of FNR in vivo depends on Tic62.
Because Tic62 is a redox-sensing protein that is not involved in LET or CET, the Tic62-dependent relocation of FNR between the thylakoids and the stroma can be envisioned to be part of a regulatory step in the adaptation to changing redox conditions. Since FNR competes with the stromal enzyme FTR for electrons from Fd, a compartmentation of FNR could thus optimize electron flows in the chloroplast. Preliminary experiments with tic62 mutants as well as an altered redox state in lfnr1 plants (Lintala et al., 2007) indicate that this might be the case, but more data need to be acquired to resolve this question.
In summary of the presented data, it can therefore be concluded that Tic62 performs at least two important functions in the chloroplast: (1) stabilizing FNR, probably in the form of HMW heterotrimeric complexes (e.g. in phases of prolonged inactivity), and (2) regulating the allocation of FNR between stroma and membranes (thylakoids and envelope) by providing a membrane anchor and a platform for efficient redox sensing, which is in line with the proposed function of Tic62 as a redox sensor protein (Küchler et al., 2002; Stengel et al., 2008).
It is likely that Tic62 is engaged in distinct functions dependent on its localization within the chloroplast, with some role being a more ancient trait and another being a rather young evolutionary modification (Gross and Bhattacharya, 2009). However, the subpools could be closely connected by the redox-dependent shuttling behavior. This would allow the transport of electrons from the photosynthetic machinery via the Tic62/FNR complex directly to the Tic translocon. It will be an interesting and challenging task to address this question in the future.
METHODS
Coexpression Analysis
Coexpression analysis with the A. thaliana Co-Response Database (AthCoR@CSB.DB; http://csbdb.mpimp-golm.mpg.de/csbdb/dbcor/ath.html) was performed with the following settings: single gene query, Matrix: Developmental Series (only wild type); ATH1 chip; AtGenExpress; 12,200 genes, coefficient: nonparametric Spearman's Rho rank correlation, output: positive, significant coresponding genes (Bonferroni correction).
Coexpression analysis by hierarchical clustering of microarray data (Eisen et al., 1998) was performed as follows: gcRMA normalized (Irizarry et al., 2003), log2-transformed Arabidopsis thaliana microarray data of the AtGenExpress Developmental Series (Schmid et al., 2005) were downloaded from the AtGenExpress website (http://www.weigelworld.org/resources/microarray/AtGenExpress/), and arithmetic means were calculated for each of the triplicate values of the 79 tested conditions provided by AtGenExpress. The data were loaded into the program Cluster 3.0 (de Hoon et al., 2004) and were then adjusted by median-centering rows (genes) and columns (arrays) (in this order) for five consecutive rounds each. After data adjustment, genes and arrays were hierarchically clustered using a Spearman Rank Correlation similarity metric and Average Linkage as the clustering method. The clustered data file and the tree files were loaded into the program Java Treeview (Saldanha, 2004) for visualization and data mining.
TIC62 was found to be part of a large, well coexpressed cluster of genes (394 and 193 genes coexpressed with a Spearman rank ρ ≥ 0.9, respectively). A total of 142 genes were present in the cross section of both analyses. Only those genes were used for further analysis and grouped into bins based on MapMan (Thimm et al., 2004).
Plant Material and Growth Conditions
All experiments were performed with Arabidopsis Columbia-0 (Lehle Seeds). The T-DNA insertion lines used for At-TIC62 (At3g18890) and At-LFNR1 (At5g66190) were SAIL_124G04 (tic62-1), GABI_439H04 (tic62-2), and SALK_067668 (lfnr1) (Alonso et al., 2003; Rosso et al., 2003) and were purchased from the Nottingham Arabidopsis Stock Centre (University of Nottingham) and GABI-Kat (Max Planck Institute for Plant Breeding Research). Plants depleted for LFNR2 (At1g20020) by RNA interference were line AGRIKOLA-N204598 (lfnr2) (Hilson et al., 2004). To synchronize germination, all seeds were subjected to vernalization at 4°C for 2 d. Plants were grown on soil or on 0.3% Gelrite medium (Serva) containing 1% d-sucrose and 0.5× Murashige and Skoog salts at pH 5.7. Plant growth occurred in growth chambers with a 16-h light (21°C; 100 μmol photons m−2 s−1) and 8-h dark (16°C) cycle. If not stated otherwise, plants were harvested from the dark or early in the day from growth light. Material was usually used immediately and fresh. If not possible, leaf material was shock-frozen in liquid nitrogen and stored at −80°C until use.
GUS Reporter Gene Detection
The ProTIC62-GUS construct was made by cloning the 1.55-kb upstream promoter region of At3g18890 as a translational fusion into the pBI101-GUS vector (Clontech).
Plants were transformed with Agrobacterium tumefaciens, strain UIA143 (kind gift of C. Bolle, Ludwig-Maximilians-Universität München), using the floral dip procedure (Clough and Bent, 1998) and subsequently selected for positive transformation events on kanamycin plates. Single insertion events were selected (3 Kanr:1 Kans) and homozygous lines (all descendants Kanr) from those plants analyzed.
Histochemical localization of GUS was performed as described by Guo et al. (2001).
Isolation and Fractionation of Chloroplasts
Intact Arabidopsis chloroplasts were prepared from ∼150 g fresh weight leaf material of 4-week-old plants from the dark grown on soil essentially as described by Seigneurin-Berny et al. (2008). Chloroplasts were subsequently resuspended in 15 mL of 10 mM HEPES-KOH, pH 7.6, 5 mM MgCl2, and lysed using 50 strokes in a small (15 mL) Dounce homogenizer (Wheaton). Further separation in stroma, thylakoids, and envelopes was done according to Li et al. (1991).
For high-ionic-strength washes of Arabidopsis thylakoids, chloroplasts were isolated with the above protocol, ruptured by incubation in 10 mM HEPES-KOH, pH 7.6, and 5 mM MgCl2 for 20 min on ice and separated into membranes and supernatant by centrifugation at 5000g for 5 min at 4°C. The membrane fraction was washed several times (in HEPES/Mg buffer without additional salt) to get rid of stroma proteins and then incubated with rotation for 30 min (at 4°C in the dark) with HEPES/Mg buffer + 500 mM NaCl. The supernatant was then used in sprectrophotometric Cyt c reduction assays and for immunoblotting. The pellet was resuspended in HEPES/Mg buffer and likewise used for immunoblotting.
Protoplast Transfection
Arabidopsis mesophyll protoplasts were isolated from leaves of 4-week-old plants and transiently transfected according to the protocol of Jen Sheen (available online at http://genetics.mgh.harvard.edu/sheenweb/protocols_reg.html). GFP fluorescence was observed with a Leica TCS SP5 confocal laser scanning microscope (Leica Microsystems).
BN Gel Electrophoresis, 2D BN/SDS-PAGE, Native PAGE, and Immunoblotting
BN gel electrophoresis (Figures 2 and 4B; see Supplemental Figure 5 online) was performed essentially as described by Schagger and von Jagow (1991) and Küchler et al. (2002) with minor modifications. BN-PAGE as shown in Figures 4D and 5A was performed as described by Sirpiö et al. (2007). For 2D BN/SDS-PAGE, the lanes were cut out after the run and incubated in 1% SDS, 1 mM β-mercaptoethanol (β-ME) for 15 min, followed by 15 min in 1% SDS without β-ME and 15 min in SDS-PAGE electrophoresis buffer (25 mM Tris, 192 mM glycine, and 0.1% SDS) at room temperature. Single lanes were then placed on top of SDS-PAGE gels (10 or 12.5% polyacrylamide), and the individual complexes were separated into their constituent subunits by electrophoresis. Native PAGE (Figure 4C) was performed as described by Lintala et al. (2009). For immunoblotting, proteins were transferred onto polyvinylidenfluoride membrane using a semidry protein gel blotting system. Labeling with protein-specific antibodies was performed by standard techniques, and bound antibodies were visualized with alkaline phosphatase or using a chemiluminescence detection system (Pierce).
Characterization of the T-DNA Mutants tic62-1 and tic62-2
Genomic DNA of the T-DNA insertion lines inside TIC62 was screened by PCR genotyping. TIC62 gene-specific primers in combination with T-DNA–specific left border primers generated fragments on heterozygous and homozygous plants. To identify plants with the T-DNA insertion in both alleles of TIC62, we used gene-specific primers flanking the predicted T-DNA insertion sites. The following primers were used: for tic62-1, Ex5fwd (5′-GATCTCCGATATTACCGGTCCTTAC-3′) and Ex8rev (5′-AGTTTCTTTGTATGCATCAGTCG-3′); for tic62-2, Ex1fwd (5′-ATGGAAGGAACTTGTTTTCTCCGTGGACAACC-3′) and Ex2rev (5′-TTGCTTCTGTTACTACAGAGCTTG-3′); as well as SAIL LB1 (5′-GCCTTTTCAGAAATGGATAAATAGCCTTGCTTCC 3′) and T-DNA-GABI (5′-GGACGTGAATGTAGACACGTCG-3′). For positions and orientations of the T-DNA inserts and oligonucleotide primers, see Figure 3A. To verify PCR products and T-DNA insertion sites, amplified DNA fragments were sequenced.
RT-PCR
Total RNA from leaves of 4-week-old Arabidopsis plants from the end of the dark period was isolated using the plant RNeasy extraction kit (Qiagen). The RNA was digested with DNase and reverse transcribed into cDNA. Detection and quantification of transcripts were performed as described previously (Philippar et al., 2004) using a LightCycler (Roche). For TIC62, we constructed the gene-specific primers Ex3fwd (5′-CTGGGATTTCGGGTTAGAG-3′) and Ex7rev (5′-CGTAATTAAGACCGCTTTCA-3′), amplifying a product of 416 bp, spanning both sites of the T-DNA insertion of tic62-1.
The same RNA was also used for Affymetrix experiments.
Thylakoid Subfractionation
Arabidopsis leaves harvested from plants at the end of the dark period were homogenized in 25 mL isolation buffer (0.4 M sorbitol, 0.1 M Tricine-KOH, pH 7.8, and 0.3 mM PMSF). After two rounds of filtration through a layer of gauze, the homogenate was pelleted at 1400g for 10 min at 4°C, resuspended, and washed once in isolation buffer. Chloroplasts were resuspended in lysis buffer (25 mM HEPES-KOH, pH 7.8, 5 mM MgCl2, and 0.3 mM PMSF) and incubated for 15 min on ice in the dark. After centrifugation at 10,000g for 10 min at 4°C, thylakoids were resuspended in buffer B (15 mM Tricine-KOH, pH 7.9, 0.1 M sorbitol, 10 mM NaCl, and 5 mM MgCl2).
To separate stroma thylakoids from grana thylakoids, the protocol of Ossenbühl et al. (2002) was followed. Fractionation was obtained by four rounds of centrifugation at 4°C, beginning with 1000g for 30 min (1K, unlysed chloroplasts), 10,000g for 30 min (K10, grana), 40,000g for 1 h (40K, margins), and at 140,000g for 1.5 h (140K, stroma lamellae). The final supernatant was precipitated with trichloroacetic acid. Subfractions were used for immunoblots.
In Vitro Transcription and Translation
The coding regions including the transit peptides of the analyzed preproteins were cloned into the vector pSP65 (Promega) under the control of the SP6 promoter. Transcription of linearized plasmids was carried as previously described (Firlej-Kwoka et al., 2008). Translation was performed using the Wheat Germ Extract Translation Kit (Promega) or the Flexi Rabbit Reticulocyte Lysate System (only pLFNR1 import for Figure 4B; Promega) in the presence of [35S]-Met for radioactive labeling.
Chloroplast Isolation and Protein Import
Chloroplasts were isolated from 17- to 18-d-old Arabidopsis plants (grown on plates) from the dark according to the protocol by Aronsson and Jarvis (2002) with the following exceptions: all buffers were supplied with 0.4 M sorbitol, and NaHCO3 and gluconic acid were omitted. An import reaction (containing chloroplasts equivalent to 7.5 μg chlorophyll) was subsequently performed in 100 μL volume containing 3 mM ATP and 1 to 5% (v/v) [35S]-labeled translation products. Import reactions were initiated by the addition of translation product to the import/chloroplast mix and performed for the indicated time at 25°C. Reactions were terminated by the addition of two volumes ice-cold washing buffer. Chloroplasts were washed twice in wash medium (0.4 M sorbitol, 50 mM HEPES-KOH, pH 8.0, and 3 mM MgSO4) and finally resuspended in Laemmli buffer (50 mM Tris-HCl, pH 6.8, 100 mM β-ME, 2% [w/v] SDS, 0.1% bromophenol blue [w/v], and 10% [v/v] glycerol). For BN-PAGE separation of thylakoid proteins, the import was performed as above (30 min import time), and the chloroplasts lysed after the second wash in 25 μL of shock buffer (10 mM HEPES-KOH, pH 8.0, and 5 mM MgCl2) for 10 min on ice. Stroma was separated from the membranes by ultracentrifugation (10 min 256,000g at 4°C). The supernatant was treated with BN loading buffer and the pellet solubilized using standard procedures (see above).
Import products were separated by SDS-PAGE, and radiolabeled proteins were analyzed by a phosphor imager or by exposure on x-ray films.
Pea Thylakoid Isolation and pH Treatment
Chloroplasts from pea were isolated from dark-adapted leaves of 9- to 11-d-old pea seedlings (Pisum sativum var Arvica) and purified through Percoll density gradients as previously described (Keegstra and Youssif, 1986; Waegemann and Soll, 1995). Chloroplasts were subsequently lysed by incubation in 5 mM MES-KOH, pH 6.0, 5 mM MgCl2 (100 μg chlorophyll/1 mL) for 20 min on ice and thylakoids separated from the stroma by centrifugation (5 min 5000g at 4°C).
For the solubilization assay, thylakoids (10 μg chlorophyll) were rebuffered in 100 mM sodium phosphate buffer (either pH 6.0, 7.0, or 8.0), 10 mM MgCl2, 1 mM EDTA, and 0.1% n-decyl β-d-maltoside and incubated for 10 min on ice and an additional 30 min at 25°C followed by centrifugation (50,000g, 4°C for 20 min) to separate soluble from insoluble proteins.
Chlorophyll Fluorescence Measurements
In vivo chlorophyll a fluorescence of PSII from single leaves was measured using a PAM 101/103 fluorometer (Walz). Plants were dark adapted for 30 min and minimal fluorescence (F0) was measured. Then, pulses (0.8 s) of saturating white light (5000 μmol photons m−2 s−1) were applied to determine maximal fluorescence (Fm) and calculate the ratio Fv/Fm = (Fm – F0)/Fm (maximum quantum yield of PSII). A 15-min illumination with actinic light of 90 or 1100 μmol, respectively, was supplied to drive e− transport between PSII and PSI. Then, the steady state fluorescence (Fs) and by a further saturating pulse the max fluorescence in the light (Fm′) were determined, and the effective quantum yield of PSII (ΦPSII) was calculated as (Fm′ − Fs)/Fm′. Additionally, the photosynthetic parameters 1-qP [excitation pressure of PSII; qP = (Fm′ − Fs)/(Fm′ − F0)] and nonphotochemical quenching [(Fm − Fm′)/Fm′] were determined.
The redox state of PSI reaction center chlorophyll P700 was basically measured as by Lintala et al. (2009) with small modifications. The transient postillumination increase in chlorophyll fluorescence (F0 rise) was measured basically as described by Allahverdiyeva et al. (2005).
Enzymatic Assays
Fd-dependent Cyt c reductase activity was determined by an assay consisting of 20 μM Cyt c (horse heart), 0.1 μM Fd (spinach [Spinacia oleracea]), and 100 μM NADPH in 1 mL of a 50 mM Tris-HCl, pH 7.5, 100 mM NaCl, and 1 mM MgCl2 reaction buffer. The reduction of Cyt c was monitored with a spectrophotometer (Ultrospec 3100pro; Amersham Biosciences) at 550 nm in kinetic mode over a course of 120 s. The amount of reduced Cyt c was calculated from the extinction coefficient ε = 21.1 mM−1 cm−1.
Protein Expression and Purification
The constructs PsTic62-IA3 and LeTic62-fl were as described by Stengel et al. (2008). Leaf isoform FNR from pea was cloned from cDNA, and the mature part (PsmFNR-L; Glu-53 to Tyr-360) was inserted into pET21d (Novagen). FBPase from Arabidopsis (At3g54050) and AtTic62-Ct (Pro-334 to His-641) were cloned from leaf cDNA and also inserted into pET21d. For heterologous expression, the clones were transformed into Escherichia coli BL21 (DE3) cells and were grown at 37°C in the presence of 100 μg/mL ampicillin to an A600 of 0.5. Expression was induced by addition of 1 mM isopropyl-1-thio-β-d-galactopyranoside, and cells were grown for 3 h at 37°C (PsTic62-IA1 and FBPase) or at 12°C overnight (LeTic62-fl, AtTic62-Ct, and PsmFNR-L), respectively. All proteins were purified via their C-terminal polyhistidine tags using Ni-NTA-Sepharose (GE Healthcare) under native conditions and eluted with 100 to 400 mM imidazole. The proteins were always used fresh and concentrated, and buffer was exchanged for 50 mM Tris-HCl, pH 8.0, and 150 mM NaCl prior to analysis.
In preparation for the Cyt c reduction assay, 10 μg of purified FNR was incubated overnight at 4°C in a rotary shaker in 1 mL of reaction buffer, supplied with or without the indicated molar amounts of Tic62 or control (egg albumin) protein.
Protein Interaction Study
AtTic62-Ct was expressed and purified as described above with addition of a subsequent size exclusion chromatography using a Superdex 75 column (GE Healthcare) in 50 mM sodium phosphate buffer, pH 8, and 150 mM NaCl. The purest fractions were again bound to Ni-NTA-Sepharose beads and washed three times with 30 bead volumes each (50 mM sodium phosphate buffer, pH 8, and 300 mM NaCl), followed by three washes with 1% egg albumin in the same buffer to saturate unspecific binding sites and five washes without albumin. An empty column without the addition of His-tagged protein was used as negative control. The columns were then incubated with concentrated stroma from tic62-1 Arabidopsis plants (lacking endogenous Tic62 protein) for 1 h at 4°C followed by eight washes and elution by sequential addition of 750 mM NaCl (E1), 1 M NaCl (E2), 4 M urea (E3), 8 M urea (E4), 200 mM imidazole (E5), and 400 mM imidazole (E6). Proteins were subsequently separated on a 12% 4 M urea-SDS-PAGE, blotted, and probed for FNR using an antibody raised against AtFNR-L1 (reacting with both leaf isoforms).
NMR Spectroscopy
Samples of uniformly 2H/15N-labeled maize (Zea mays) FNR were prepared as described previously (Maeda et al., 2005; Lee et al., 2007), and the R1 peptide (amino acid sequence: VAKTEQPLSPYTAYDDLKPPSSPSPTKPSE) was purchased from Toray Research Center. The NMR samples were dissolved in 25 mM sodium phosphate buffer, pH 8.0, 50 mM NaCl, and 90% H2O/10% D2O. The NMR titration experiments were performed by acquiring 2D 1H-15N TROSY-HSQC spectra of 0.1 mM [U-2H; U-15N]-FNR, while adding the R1 peptide. The data matrix of each 2D spectrum was comprised of 128 and 1024 complex points for 15N and 1H dimension corresponding to acquisition times of 49.0 and 63.8 ms. The indirect 15N dimension was doubled by linear prediction, and the data matrix was zero-filled to 512 and 2048 complex points prior to Fourier transformation. All the NMR experiments were performed on a Bruker DRX-800 spectrometer equipped with a cryogenic probe with a z-axis gradient coil at 313 K, and all of the data were processed with the program NMRPipe (Delaglio et al., 1995) and analyzed using Sparky (developed by T.D. Goddard and D.G. Kneller at the University of California, San Francisco).
AUC
The sedimentation equilibrium measurements for analytical ultracentrifugation were performed using a Beckman-Coulter Optima XL-1 analytical ultracentrifuge and by atomic force microscopy and electron microscopy images. The samples were centrifuged at 10,000 rpm (1050g) for the Tic62 Ct-FNR complex, 13,000 rpm (1360g) for the free FNR and the FNR-R1 complex, and 23,000 rpm (2400g) for the free Tic62 Ct. The concentrations of polypeptides were 10 μM for FNR, 15 μM for R1, and 60 μM for the Tic62 Ct. Equilibrium concentration profiles were recorded by monitoring absorbance at 280 nm for the Tic62 Ct and 450 nm for FNR across the centrifugation cell with a radial increment of 0.001 cm in the continuous scanning mode. All measurements were performed at a constant temperature of 37°C with a radial increment of 0.003 cm in the continuous scanning mode. For the analysis of sedimentation equilibrium data, the experimental data were fitted by the following equation:
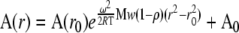 |
where A is absorbance, r is the distance from the rotation center, r0 is the distance of the meniscus, ω is angular velocity,  is the partial specific volume of proteins, ρ is the density of the solvent, A0 is baseline absorbance, T is temperature in Kelvin, and R is the gas constant. The values of ρ were obtained with the software UltraScan 8.0 (www.ultrascan.uthscsa.edu), and the partial specific volume of FNR determined in our previous study was used (Lee et al., 2009).
is the partial specific volume of proteins, ρ is the density of the solvent, A0 is baseline absorbance, T is temperature in Kelvin, and R is the gas constant. The values of ρ were obtained with the software UltraScan 8.0 (www.ultrascan.uthscsa.edu), and the partial specific volume of FNR determined in our previous study was used (Lee et al., 2009).
Accession Numbers
Sequence data from this article can be found under the respective Arabidopsis gene index (Arabidopsis Genome Initiative locus identifier): Arabidopsis Tic62 (At3g18890), LFNR1 (At5g66190), and LFNR2 (At1g20020); P. sativum Tic62 (CAC87810) and LFNR (CAA67796). A list with all Arabidopsis Genome Initiative codes of the 142 genes derived from the coexpression analysis is provided in Supplemental Table 1 online. T-DNA insertion lines used were SAIL_124G04/CS871343 (tic62-1), GABI_439H04/N442136 (tic62-2), and SALK_085403/N585403 (lfnr1). lfnr2 was Agricola RNA interference line CATMA1a19020/N204598.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Tic62 (Co-)Expression Analysis.
Supplemental Figure 2. Tic62 and FNR Colocalize at the (Stroma-) Thylakoids.
Supplemental Figure 3. Ultrastructural Analysis of Wild-Type and tic62 Mesophyll Chloroplasts by Transmission Electron Microscopy.
Supplemental Figure 4. Tic62 Knockout Lines Display a Specific Reduction of FNR in the Thylakoids.
Supplemental Figure 5. FNR Is Absent from HMW Complexes in tic62 Mutants.
Supplemental Figure 6. Tic62 Knockout Lines Display No Differences in Cyclic Electron Transfer Processes.
Supplemental Figure 7. Coomassie Control Gel of Tic62-FNR Binding Assay.
Supplemental Figure 8. Structural Analyses of the Tic62/FNR Complex.
Supplemental Table 1. Result of Combined Tic62 Coexpression Analysis.
Supplemental Table 2. Photosynthetic Properties of Arabidopsis Wild-Type and tic62 Plants.
Supplemental Methods. Transmission Electron Microscopy and 2D Isoelectric Focusing/SDS-PAGE.
Supplemental References.
Supplementary Material
Acknowledgments
We thank Eike Petersen and Yvonne Seidel for excellent technical assistance. Eva-Mari Aro, Dario Leister, and Ute Armbruster are gratefully acknowledged for their help with chlorophyll fluorescence measurements. We also thank Cordelia Bolle for providing the Agrobacterium strain UIA143, Nigel M. Crawford for the GUS vector pBI101, Manuela Baumgartner for the construct AtTic110-GFP, and Ute C. Vothknecht for critical reading of the manuscript. This work was supported by Deutsche Forschungsgemeinschaft (SFB594) (B.B., J.P.B., A.S., and J.S.), Elitenetzwerk Bayern (J.P.B.), International Max-Planck Research School for Life Sciences (A.S.), Deutscher Akadmischer Austausch Dienst (J.P.B. and M.L.), and Studienstiftung des Deutschen Volkes (A.S.). A.W. acknowledges support from Deutsche Forschungsgemeinschaft Grant WE 2231/4-1. P.M. and M.L. acknowledge funding from the Academy of Finland (110099, 130075, and 126649).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: B. Bölter (boelter@lrz.uni-muenchen.de).
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Allahverdiyeva, Y., Mamedov, F., Maenpaa, P., Vass, I., and Aro, E.M. (2005). Modulation of photosynthetic electron transport in the absence of terminal electron acceptors: Characterization of the rbcL deletion mutant of tobacco. Biochim. Biophys. Acta 1709 69–83. [DOI] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide Insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Andersen, B., Scheller, H.V., and Moller, B.L. (1992). The PSI-E subunit of photosystem I binds ferredoxin:NADP+ oxidoreductase. FEBS Lett. 311 169–173. [DOI] [PubMed] [Google Scholar]
- Aro, E.M., Suorsa, M., Rokka, A., Allahverdiyeva, Y., Paakkarinen, V., Saleem, A., Battchikova, N., and Rintamaki, E. (2005). Dynamics of photosystem II: A proteomic approach to thylakoid protein complexes. J. Exp. Bot. 56 347–356. [DOI] [PubMed] [Google Scholar]
- Aronsson, H., and Jarvis, P. (2002). A simple method for isolating import-competent Arabidopsis chloroplasts. FEBS Lett. 529 215–220. [DOI] [PubMed] [Google Scholar]
- Balsera, M., Stengel, A., Soll, J., and Bölter, B. (2007). Tic62: A protein family from metabolism to protein translocation. BMC Evol. Biol. 7 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biehl, A., Richly, E., Noutsos, C., Salamini, F., and Leister, D. (2005). Analysis of 101 nuclear transcriptomes reveals 23 distinct regulons and their relationship to metabolism, chromosomal gene distribution and co-ordination of nuclear and plastid gene expression. Gene 344 33–41. [DOI] [PubMed] [Google Scholar]
- Bräutigam, A., Hofmann-Benning, S., and Weber, A.P.M. (2008). Comparative proteomics of chloroplast envelopes from C-3 and C-4 plants reveals specific adaptations of the plastid envelope to C-4 photosynthesis and candidate proteins required for maintaining C-4 metabolite fluxes. Plant Physiol. 148 568–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breyton, C., Nandha, B., Johnson, G.N., Joliot, P., and Finazzi, G. (2006). Redox modulation of cyclic electron flow around photosystem I in C3 plants. Biochemistry 45 13465–13475. [DOI] [PubMed] [Google Scholar]
- Burrows, P.A., Sazanov, L.A., Svab, Z., Maliga, P., and Nixon, P.J. (1998). Identification of a functional respiratory complex in chloroplasts through analysis of tobacco mutants containing disrupted plastid ndh genes. EMBO J. 17 868–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo, N., Lucero, H.A., and Vallejos, R.H. (1981). Light-modulation of chloroplast membrane-bound ferredoxin-Nadp+ oxidoreductase. J. Biol. Chem. 256 1058–1059. [PubMed] [Google Scholar]
- Chan, R.L., Ceccarelli, E.A., and Vallejos, R.H. (1987). Immunological studies of the binding protein for chloroplast ferredoxin-NADP+ reductase. Arch. Biochem. Biophys. 253 56–61. [DOI] [PubMed] [Google Scholar]
- Chigri, F., Hörmann, F., Stamp, A., Stammers, D.K., Bölter, B., Soll, J., and Vothknecht, U.C. (2006). Calcium regulation of chloroplast protein translocation is mediated by calmodulin binding to Tic32. Proc. Natl. Acad. Sci. USA 103 16051–16056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Darie, C.C., Biniossek, M.L., Winter, V., Mutschler, B., and Haehnel, W. (2005). Isolation and structural characterization of the Ndh complex from mesophyll and bundle sheath chloroplasts of Zea mays. FEBS J. 272 2705–2716. [DOI] [PubMed] [Google Scholar]
- de Hoon, M.J., Imoto, S., Nolan, J., and Miyano, S. (2004). Open source clustering software. Bioinformatics 20 1453–1454. [DOI] [PubMed] [Google Scholar]
- Dekker, J.P., and Boekema, E.J. (2005). Supramolecular organization of thylakoid membrane proteins in green plants. Biochim. Biophys. Acta 1706 12–39. [DOI] [PubMed] [Google Scholar]
- Delaglio, F., Grzesiek, S., Vuister, G.W., Zhu, G., Pfeifer, J., and Bax, A. (1995). NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6 277–293. [DOI] [PubMed] [Google Scholar]
- Eisen, M.B., Spellman, P.T., Brown, P.O., and Botstein, D. (1998). Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo, T., Ishida, S., Ishikawa, N., and Sato, F. (2008). Chloroplastic NAD(P)H dehydrogenase complex and cyclic electron transport around photosystem I. Mol. Cells 25 158–162. [PubMed] [Google Scholar]
- Firlej-Kwoka, E., Strittmatter, P., Soll, J., and Bölter, B. (2008). Import of preproteins into the chloroplast inner envelope membrane. Plant Mol. Biol. 68 505–519. [DOI] [PubMed] [Google Scholar]
- Gross, J., and Bhattacharya, D. (2009). Revaluating the evolution of the Toc and Tic protein translocons. Trends Plant Sci. 14 13–20. [DOI] [PubMed] [Google Scholar]
- Grzyb, J., Gagos, M., Gruszecki, W.I., Bojko, M., and Strzalka, K. (2007). Interaction of ferredoxin: NADP(+) oxidoreductase with model membranes. Biochim. Biophys. Acta 1778 133–142. [DOI] [PubMed] [Google Scholar]
- Guedeney, G., Corneille, S., Cuine, S., and Peltier, G. (1996). Evidence for an association of ndh B, ndh J gene products and ferredoxin-NADP-reductase as components of a chloroplastic NAD(P)H dehydrogenase complex. FEBS Lett. 378 277–280. [DOI] [PubMed] [Google Scholar]
- Guo, F.Q., Wang, R., Chen, M., and Crawford, N.M. (2001). The Arabidopsis dual-affinity nitrate transporter gene AtNRT1.1 (CHL1) is activated and functions in nascent organ development during vegetative and reproductive growth. Plant Cell 13 1761–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilson, P., et al. (2004). Versatile gene-specific sequence tags for Arabidopsis functional genomics: transcript profiling and reverse genetics applications. Genome Res. 14 2176–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hörmann, F., Küchler, M., Sveshnikov, D., Oppermann, U., Li, Y., and Soll, J. (2004). Tic32, an essential component in chloroplast biogenesis. J. Biol. Chem. 279 34756–34762. [DOI] [PubMed] [Google Scholar]
- Irizarry, R.A., Hobbs, B., Collin, F., Beazer-Barclay, Y.D., Antonellis, K.J., Scherf, U., and Speed, T.P. (2003). Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4 249–264. [DOI] [PubMed] [Google Scholar]
- Ishihara, S., Takabayashi, A., Ido, K., Endo, T., Ifuku, K., and Sato, F. (2007). Distinct functions for the two PsbP-like proteins PPL1 and PPL2 in the chloroplast thylakoid lumen of Arabidopsis. Plant Physiol. 145 668–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juric, S., Hazler-Pilepic, K., Tomasic, A., Lepedus, H., Jelicic, B., Puthiyaveetil, S., Bionda, T., Vojta, L., Allen, J.F., Schleiff, E., and Fulgosi, H. (2009). Tethering of ferredoxin:NADP(+) oxidoreductase to thylakoid membranes is mediated by novel chloroplast protein TROL. Plant J. 60 783–794. [DOI] [PubMed] [Google Scholar]
- Keegstra, K., and Youssif, A.E. (1986). Isolation and characterization of chloroplast envelope membranes. In Methods Enzymology - Plant Molecular Biology, A. Weissbach and H. Weissbach, eds (Orlando, FL: Academic Press), pp. 316–325.
- Küchler, M., Decker, S., Hörmann, F., Soll, J., and Heins, L. (2002). Protein import into chloroplasts involves redox-regulated proteins. EMBO J. 21 6136–6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y.H., Chatani, E., Sasahara, K., Naiki, H., and Goto, Y. (2009). A comprehensive model for packing and hydration for amyloid fibrils of beta2-microglobulin. J. Biol. Chem. 284 2169–2175. [DOI] [PubMed] [Google Scholar]
- Lee, Y.H., Tamura, K., Maeda, M., Hoshino, M., Sakurai, K., Takahashi, S., Ikegami, T., Hase, T., and Goto, Y. (2007). Cores and pH-dependent dynamics of ferredoxin-NADP(+) reductase revealed by hydrogen/deuterium exchange. J. Biol. Chem. 282 5959–5967. [DOI] [PubMed] [Google Scholar]
- Li, H.M., Moore, T., and Keegstra, K. (1991). Targeting of proteins to the outer envelope membrane uses a different pathway than transport into chloroplasts. Plant Cell 3 709–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lintala, M., Allahverdiyeva, Y., Kangasjarvi, S., Lehtimaki, N., Keranen, M., Rintamaki, E., Aro, E.M., and Mulo, P. (2009). Comparative analysis of leaf-type ferredoxin-NADP oxidoreductase isoforms in Arabidopsis thaliana. Plant J. 57 1103–1115. [DOI] [PubMed] [Google Scholar]
- Lintala, M., Allahverdiyeva, Y., Kidron, H., Piippo, M., Battchikova, N., Suorsa, M., Rintamaki, E., Salminen, T.A., Aro, E.M., and Mulo, P. (2007). Structural and functional characterization of ferredoxin-NADP+-oxidoreductase using knock-out mutants of Arabidopsis. Plant J. 49 1041–1052. [DOI] [PubMed] [Google Scholar]
- Maeda, M., Lee, Y.H., Ikegami, T., Tamura, K., Hoshino, M., Yamazaki, T., Nakayama, M., Hase, T., and Goto, Y. (2005). Identification of the n- and c-terminal substrate binding segments of Ferredoxin-NADP plus reductase by NMR. Biochemistry 44 10644–10653. [DOI] [PubMed] [Google Scholar]
- Matthijs, H.C.P., Coughlan, S.J., and Hind, G. (1986). Removal of ferredoxin - NADP+ oxidoreductase from thylakoid membranes, rebinding to depleted membranes, and identification of the binding site. J. Biol. Chem. 261 2154–2158. [PubMed] [Google Scholar]
- Okutani, S., Hanke, G.T., Satomi, Y., Takao, T., Kurisu, G., Suzuki, A., and Hase, T. (2005). Three maize leaf ferredoxin:NADPH oxidoreductases vary in subchloroplast location, expression, and interaction with ferredoxin. Plant Physiol. 139 1451–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossenbühl, F., Hartmann, K., and Nickelsen, J. (2002). A chloroplast RNA binding protein from stromal thylakoid membranes specifically binds to the 5′ untranslated region of the psbA mRNA. Eur. J. Biochem. 269 3912–3919. [DOI] [PubMed] [Google Scholar]
- Peltier, J.B., Ytterberg, A.J., Sun, Q., and van Wijk, K.J. (2004). New functions of the thylakoid membrane proteome of Arabidopsis thaliana revealed by a simple, fast, and versatile fractionation strategy. J. Biol. Chem. 279 49367–49383. [DOI] [PubMed] [Google Scholar]
- Philippar, K., Ivashikina, N., Ache, P., Christian, M., Luthen, H., Palme, K., and Hedrich, R. (2004). Auxin activates KAT1 and KAT2, two K+-channel genes expressed in seedlings of Arabidopsis thaliana. Plant J. 37 815–827. [DOI] [PubMed] [Google Scholar]
- Quiles, M.J., and Cuello, J. (1998). Association of ferredoxin-NADP oxidoreductase with the chloroplastic pyridine nucleotide dehydrogenase complex in barley leaves. Plant Physiol. 117 235–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiles, M.J., Garcia, A., and Cuello, J. (2000). Separation by blue-native PAGE and identification of the whole NAD(P)H dehydrogenase complex from barley stroma thylakoids. Plant Physiol. Biochem. 38 225–232. [Google Scholar]
- Romano, P., Gray, J., Horton, P., and Luan, S. (2005). Plant immunophilins: Functional versatility beyond protein maturation. New Phytol. 166 753–769. [DOI] [PubMed] [Google Scholar]
- Rosso, M.G., Li, Y., Strizhov, N., Reiss, B., Dekker, K., and Weisshaar, B. (2003). An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 53 247–259. [DOI] [PubMed] [Google Scholar]
- Rumeau, D., Peltier, G., and Cournac, L. (2007). Chlororespiration and cyclic electron flow around PSI during photosynthesis and plant stress response. Plant Cell Environ. 30 1041–1051. [DOI] [PubMed] [Google Scholar]
- Saldanha, A.J. (2004). Java Treeview–Extensible visualization of microarray data. Bioinformatics 20 3246–3248. [DOI] [PubMed] [Google Scholar]
- Schagger, H., and von Jagow, G. (1991). Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 199 223–231. [DOI] [PubMed] [Google Scholar]
- Schmid, M., Davison, T.S., Henz, S.R., Pape, U.J., Demar, M., Vingron, M., Scholkopf, B., Weigel, D., and Lohmann, J.U. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37 501–506. [DOI] [PubMed] [Google Scholar]
- Seigneurin-Berny, D., Salvi, D., Dorne, A.J., Joyard, J., and Rolland, N. (2008). Percoll-purified and photosynthetically active chloroplasts from Arabidopsis thaliana leaves. Plant Physiol. Biochem. 46 951–955. [DOI] [PubMed] [Google Scholar]
- Shikanai, T., Endo, T., Hashimoto, T., Yamada, Y., Asada, K., and Yokota, A. (1998). Directed disruption of the tobacco ndhB gene impairs cyclic electron flow around photosystem I. Proc. Natl. Acad. Sci. USA 95 9705–9709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin, M., Ishida, H., and Nozaki, Y. (1985). A new protein factor, connectein, as a constituent of the large form of ferredoxin-NADP reductase. Plant Cell Physiol. 26 559–563. [Google Scholar]
- Sirpiö, S., Allahverdiyeva, Y., Suorsa, M., Paakkarinen, V., Vainonen, J., Battchikova, N., and Aro, E.M. (2007). TLP18.3, a novel thylakoid lumen protein regulating photosystem II repair cycle. Biochem. J. 406 415–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soncini, F.C., and Vallejos, R.H. (1989). The chloroplast reductase-binding protein is identical to the 16.5-kDa polypeptide described as a component of the oxygen-evolving complex. J. Biol. Chem. 264 21112–21115. [PubMed] [Google Scholar]
- Stengel, A., Benz, P., Balsera, M., Soll, J., and Bölter, B. (2008). TIC62 redox-regulated translocon composition and dynamics. J. Biol. Chem. 283 6656–6667. [DOI] [PubMed] [Google Scholar]
- Thimm, O., Blasing, O., Gibon, Y., Nagel, A., Meyer, S., Kruger, P., Selbig, J., Muller, L.A., Rhee, S.Y., and Stitt, M. (2004). MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37 914–939. [DOI] [PubMed] [Google Scholar]
- Vallejos, R.H., Ceccarelli, E., and Chan, R. (1984). Evidence for the existence of a thylakoid intrinsic protein that binds ferredoxin-NADP+ oxidoreductase. J. Biol. Chem. 259 8048–8051. [PubMed] [Google Scholar]
- Waegemann, K., and Soll, J. (1995). Characterization and isolation of the chloroplast protein import machinery. Methods Cell Biol. 50 255–267. [DOI] [PubMed] [Google Scholar]
- Zhang, H., Whitelegge, J.P., and Cramer, W.A. (2001). Ferredoxin:NADP+ oxidoreductase is a subunit of the chloroplast cytochrome b6f complex. J. Biol. Chem. 276 38159–38165. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.