Abstract
During compatible pollination of the angiosperms, pollen tubes grow in the pistil transmitting tract (TT) and are guided to the ovule for fertilization. Lily (Lilium longiflorum) stigma/style Cys-rich adhesin (SCA), a plant lipid transfer protein (LTP), is a small, secreted peptide involved in pollen tube adhesion-mediated guidance. Here, we used a reverse genetic approach to study biological roles of Arabidopsis thaliana LTP5, a SCA-like LTP. The T-DNA insertional gain-of-function mutant plant for LTP5 (ltp5-1) exhibited ballooned pollen tubes, delayed pollen tube growth, and decreased numbers of fertilized eggs. Our reciprocal cross-pollination study revealed that ltp5-1 results in both male and female partial sterility. RT-PCR and β-glucuronidase analyses showed that LTP5 is present in pollen and the pistil TT in low levels. Pollen-targeted overexpression of either ltp5-1 or wild-type LTP5 resulted in defects in polar tip growth of pollen tubes and thereby decreased seed set, suggesting that mutant ltp5-1 acts as a dominant-active form of wild-type LTP5 in pollen tube growth. The ltp5-1 protein has additional hydrophobic C-terminal sequences, compared with LTP5. In our structural homology/molecular dynamics modeling, Tyr-91 in ltp5-1, replacing Val-91 in LTP5, was predicted to interact with Arg-45 and Tyr-81, which are known to interact with a lipid ligand in maize (Zea mays) LTP. Thus, Arabidopsis LTP5 plays a significant role in reproduction.
INTRODUCTION
In angiosperms, pollination is a key regulatory step in plant sexual reproduction and crop yield. Growing pollen tubes traverse a series of interspecies or intraspecies barriers in the pistil transmitting tract (TT) during a journey to reach the ovule for fertilization (Franklin-Tong, 1999, 2002; Lord and Russell, 2002). Cell–cell communications are known to be involved in regulating polar pollen tube growth in floral reproductive tissues (Franklin-Tong, 1999; Johnson and Preuss, 2002; Lord, 2003).
The lily (Lilium longiflorum) plant has a large flower with a 15-cm pistil. Pollen germinate on the wide stigmatic surface, and pollen tubes grow and are guided to enter the long hollow style (Lord and Russell, 2002; Lord, 2003). This guidance on the stigma is mediated by chemotrophic activity of a small (∼9.8 kD) extracellular protein, chemocyanin (Kim et al., 2003). When passing into the style, pollen tubes grow adhering to each other and to the extracellular matrix of the pistil TT epidermis and deliver two sperm cells to each ovule (Lord, 2000). Lily SCA (for stigma/style Cys-rich adhesin), a small (∼9.4 kD), basic (pI ∼9.0), secreted protein, is involved in this haptotactic (adhesion-mediated) pollen tube guidance via an ionic interaction with a stylar pectic polysaccharide in the pistil TT cell walls (Mollet et al., 2000; Park et al., 2000). SCA may act as a lectin-like molecule on the pollen tube cell wall, back from the tip, where adhesive pectins are mainly found (Mollet et al., 2000, 2007). However, SCA was shown to be localized as well at the tip of the pollen tube and to enter the in vitro and in vivo growing pollen tubes via an endocytotic pathway (Kim et al., 2004, 2006). A previous study showed that SCA can facilitate the activity of chemocyanin for chemotrophic guidance of tip growth as well (Kim et al., 2003). SCA, as a pistil factor (Park and Lord, 2003), may be involved in cell–cell communication both in pollen tube adhesion and tip growth (Chae et al., 2007).
SCA is a plant nonspecific lipid transfer protein (LTP) (Park et al., 2000). Plant nonspecific LTPs are small (9 to 10 kD), basic (pI 8.8 to 10), and secreted proteins (Kader, 1996). They contain four conserved disulfide bonds with eight Cys residues and two consensus pentapeptide motifs (Thr/Ser-X1-X2-Asp-Arg/Lys and Pro-Tyr-X-Ile-Ser) (Douliez et al., 2000b). These features were also identified in SCA (Chae et al., 2007). Computational modeling indicates that SCA has an LTP-like structure with a globular shape of the orthogonal four-helix bundle architecture and a hydrophobic core (Chae et al., 2007). The hydrophobic cavity of several plant LTPs is known to interact with lipids and fatty acids in vitro without ligand specificity (Zachowski et al., 1998; Charvolin et al., 1999; Douliez et al., 2000a, 2001; Hamilton, 2004). Only a few plant LTPs related to lily SCA have been functionally characterized, including a few molecules involved in plant defense (Phillippe et al., 1995; Molina and Garcia-Olmedo, 1997) and one in cell wall loosening (Nieuwland et al., 2005).
Due to the difficulties of using lily for a genetic approach, we have been hindered in our attempts to further describe the biological function of SCA. Here, we used Arabidopsis thaliana to confirm a biological role for a SCA-like LTP, LTP5, in pollination. Our reverse genetic approach, using a T-DNA insertional gain-of-function mutant (ltp5-1) and pollen-targeted overexpression of ltp5-1 or wild-type LTP5 gene, suggests that Arabidopsis LTP5, a small secreted peptide from both pollen and the pistil, plays a role in pollen tube tip growth and in pistil function.
RESULTS
Isolation of ltp5-1, a Gain-of-Function Mutant for Arabidopsis LTP5
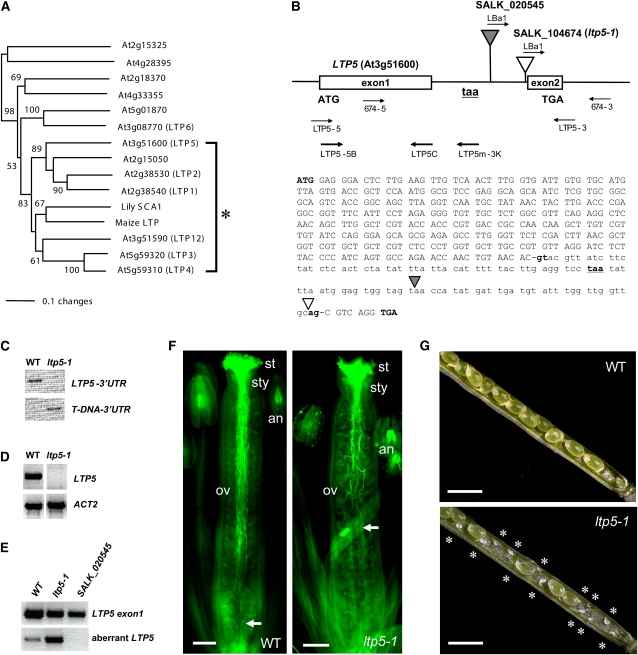
The Arabidopsis genome has a large LTP family (Kader, 1997; Arondel et al., 2000). As the first attempt to narrow down to SCA-like Arabidopsis LTPs, we obtained all LTP or LTP-like protein sequences from The Arabidopsis Information Resource databank (Altschul et al., 1990). Our phylogenetic analysis revealed 13 Arabidopsis LTPs closely related to lily SCA (see Supplemental Figure 1 and Supplemental Data Set 1 online). Based upon bootstrap percentage (83%), we further narrowed down seven of them as SCA-like Arabidopsis LTPs: LTP1 to 4, 5, 12, and a putative LTP (At2g15050) (Figure 1A). To study biological functions of SCA-like LTPs from Arabidopsis, one or two T-DNA insertion alleles per LTP gene were obtained from the ABRC (Alonso et al., 2003). We isolated homozygous SALK T-DNA insertion lines using PCR-based genotyping analysis (see Supplemental Table 1 online). RT-PCR analysis using Arabidopsis LTP gene-specific primer sets (see Supplemental Table 2 online) showed that almost all homozygous T-DNA insertion mutants were not knockouts of gene expression (see Supplemental Figure 2 online) nor did they show any phenotype, except for one allele for LTP5 (ltp5-1, SALK_104674) (Figure 1).
Figure 1.
The ltp5-1 Plant Has Defects in Pollen Tube Growth and Fertilization.
(A) A portion of the unrooted neighbor-joining tree presented in Supplemental Figure 1 online showing phylogenetic relationships of SCA and SCA-like LTPs in Arabidopsis. Asterisk indicates lily SCA, maize LTP, and seven closely related Arabidopsis SCA-like LTPs. The values on the branches indicate the number of bootstrap replicates supporting the branch. Only bootstrap replication values >50 are shown.
(B) Structures of two T-DNA insertion alleles for Arabidopsis LTP5. The white and gray triangles indicate T-DNA insertion sites in SALK_104674 (ltp5-1) and SALK_020545 plants, respectively. 674-5, 674-3, and LBa1 are PCR primers for genotyping analysis in (C). LTP5-5 and LTP5-3 are LTP5 gene-specific PCR primers for evaluating gene transcript levels in (D). LTP5-5B and LTP5C are forward and reverse primers for the first exon of LTP5 shown in (E). LTP5m-3K is the reverse primer for ltp5-1 whose putative translation termination codon (taa, underlined, bold, lowercase letters) was found in frame in the intron. Translation start (ATG) and termination codons (TGA) for LTP5 are shown in bold uppercase letters. Bold lowercase letters indicate both 5′- and 3′-splicing recognition sites.
(C) PCR-based genotyping analysis for ltp5-1 plants. The LTP5 gene locus was not amplified in ltp5-1 when 674-5 and 674-3 primers were used for 35 cycles of PCR (LTP5-3′UTR) (top panel). This is due to a homozygous T-DNA insertion, identified by use of LBa1 and 674-3 primers (T-DNA-3′UTR) (bottom panel).
(D) RT-PCR analysis for ltp5-1 plants from three replicates. LTP5 gene expression was not found in 35 cycles of PCR using the LTP5 gene-specific primer set (top panel). ACT2 is the PCR control (bottom panel).
(E) Identification of aberrant LTP5 transcript in the ltp5-1 plant. Three replicates of RT-PCR using LTP5-5B and LTP5C primers showed the presence of the transcript of LTP5 exon1 in the wild type, ltp5-1, and SALK_020545. LTP5-5B and LTP5m-3K primers were used to amplify the transcript of aberrant LTP5. Thirty-five PCR cycles were performed.
(F) Wild-type and ltp5-1 flowers at stage 14 (Smyth et al., 1990) were stained with aniline blue to visualize in vivo pollen tube growth in the pistil (n = 20). Arrow, pollen tube front; st, stigma; sty, style; ov, ovary; an, anther. Bars = 200 μm.
(G) Mature siliques of wild-type and ltp5-1 plants were dissected to examine ovules (n = 10). Asterisks indicate unfertilized ovules in the ltp5-1 silique. Bars = 1 mm.
The ltp5-1 plant harbored the T-DNA next to the 3′-splicing recognition site (ag) in the intron of the LTP5 gene locus (Figures 1B). This insertion probably abrogated a splicing process to generate mature mRNA of LTP5. So, we did not identify the full LTP5 (Figure 1D). This failure in splicing may also allow an alternative translation termination codon (taa), found in the intron, to be used to generate an aberrant LTP5 transcript (Figure 1B). To see if this aberrant, unspliced transcript is accumulated in ltp5-1 plants, we designed specific primer sets for aberrant LTP5 and LTP5 exon1 (control), respectively (Figure 1B; see Supplemental Table 2 online). In RT-PCR analysis, we found that a high level of the aberrant LTP5 transcript is present in the ltp5-1 plant (Figure 1E). A weak level of PCR product for the aberrant LTP5 transcript was also amplified in the wild-type control, probably from a certain level of unspliced RNA precursors.
In the presence of the aberrant LTP5 transcript, ltp5-1 plants have severe defects in pollination and seed formation (Figures 1F and 1G). Aniline blue staining visualized in vivo pollen tubes growing to the base of the wild-type pistil (Figure 1F). However, the majority of ltp5-1 pollen tubes were shown to reach only the middle of the ovary. The ltp5-1 siliques were shown to harbor significant numbers of unfertilized ovules mainly at the base of the ovary (Figure 1G). Average sizes of ltp5-1 and wild-type siliques were 1.15 ± 0.16 and 1.53 ± 0.08 cm, respectively (n = 100). Another T-DNA insertion allele for LTP5 (SALK_020545), which has the T-DNA in the middle of the intron (Figure 1B), did not show any aberrant LTP5 transcript (Figure 1E) nor did it show any mutant phenotype. This suggests that the presence of aberrant LTP5 (ltp5-1) in the ltp5-1 plant may contribute to the examined mutant phenotypes as a gain-of-function mutation. Indeed, our efforts to complement the mutant phenotypes of ltp5-1 plants were not successful. Due to the nature of the gain-of-function mutant, we cannot exclude the possibility that ltp5-1 may act as a dominant-active mutant for LTP5 or as a neomorph (i.e., dominant gain of function different from normal gene function).
Decreased Seed Set in ltp5-1 Is Caused by Both Abnormal Pollen Tube Growth and Disturbed Pistil Function
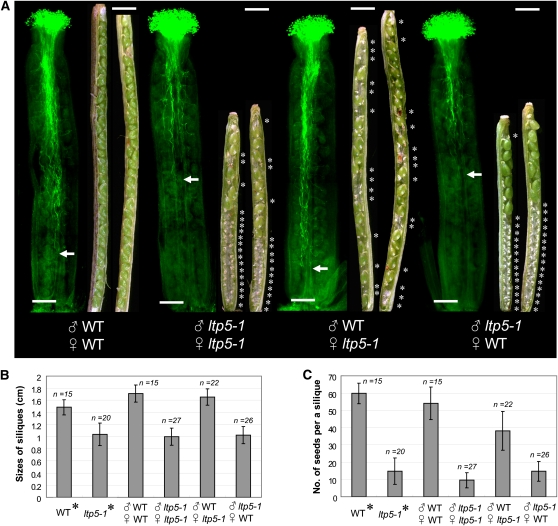
To clarify the effect of ltp5-1 on Arabidopsis fertilization, we performed reciprocal cross-pollinations between wild-type and ltp5-1 plants (Figure 2). Arabidopsis pollen germinated on the stigma within 1 h and penetrated the pistil TT in 3 h (see Supplemental Figure 3 online). A 12-h period was sufficient for the growing pollen tube front to arrive at the base of the ovary (see Supplemental Figure 3 online). In reciprocal cross-pollinations, wild-type pollen showed active pollen tube growth to the base of the wild-type pistil in 12 h, and the mature siliques were shown to be completely filled with fertilized ovules (Figure 2A). The 2qaverage size of mature siliques and number of seeds per silique from this cross were equivalent to those of self-pollinated wild-type plants (Figures 2B and 2C). By contrast, ltp5-1 pollen did not show normal pollen tube growth in either ltp5-1 or wild-type pistils, resulting in unfertilized ovules mainly at the base of the mature siliques (Figure 2A). This caused short siliques and small numbers of seeds per silique, similar to self-pollinated ltp5-1 plants (Figures 2B and 2C). Cross-pollination of wild-type pollen onto the ltp5-1 pistil resulted in normal pollen tube growth and silique sizes (Figures 2A and 2B). However, the dissected siliques harbored many unfertilized ovules, dispersed randomly from the top to the base (Figure 2A), so the average number of seeds per silique was decreased in this cross, compared with the wild type (Figure 2C). Our reciprocal cross-pollination study showed that ltp5-1 has a subtle effect on pistil function for seed formation as well as a more pronounced effect on pollen tube growth.
Figure 2.
In Vivo Reciprocal Cross-Pollination of ltp5-1 to Wild-Type Plants.
(A) Flowers at stage 12 (Smyth et al., 1990) were emasculated a day before each cross-pollination (n = 15 per cross). At 12 h after pollination, six to seven pistils were fixed, and pollen tube growth was examined by aniline blue staining. The remaining pollinated pistils ripened into mature siliques in 8 d. Siliques were then dissected for examination of fertilized ovules. Bars = 200 μm. Arrows indicate the pollen tube front in the pistil. Asterisks designate unfertilized ovules in the silique. Bars = 1 mm.
(B) and (C) In another set of reciprocal cross-pollinations, sizes of mature siliques (B) and numbers of seeds per silique (C) were examined 8 d after pollination. Control flowers were allowed to self-pollinate (asterisks). Data are shown as mean ± sd.
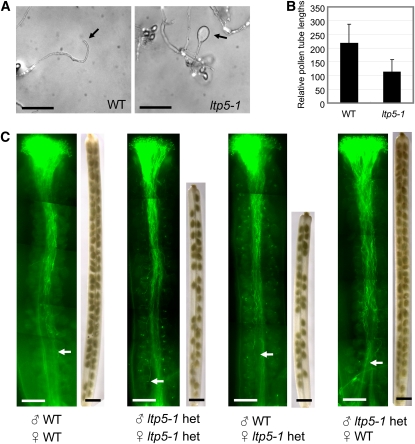
The ltp5-1 Plant Has Defects in Both Tip Growth of Pollen Tubes in Vitro and in Pistil Function
To better define the underlying cause of male sterility of ltp5-1, we performed in vitro pollen tube growth assays at room temperature. Over 50% of ltp5-1 pollen tubes exhibited abnormal tip morphology in 6 h: the tube tip was obviously swollen (Figure 3A). These abnormal pollen tubes were arrested in their growth within 6 h of germination, and their relative tube length was shorter than that of the wild type by about half (Figure 3B). In addition, ltp5-1 pollen precociously germinated in vitro (see Supplemental Figure 4D online).
Figure 3.
The ltp5-1 Plants Showed Abnormal Pollen Tube Tip Growth in Vitro and Disturbed Pistil Function in Seed Set Formation.
(A) and (B) In vitro pollen tube growth assay.
(A) Pollen from mature flowers was grown on solid germination medium in vitro for 6 h at room temperature. Arrows indicate pollen tube tips. Bars = 100 μm.
(B) Relative pollen tube lengths were measured at 6 h in vitro germination (n = 100). Data are shown as mean ± sd. Student's t test showed a significant difference in the comparison (P = 0.0001, 95% confidence interval for mean: 206 to 229 for the wild type and 101 to 124 for ltp5-1).
(C) In vivo reciprocal cross-pollination of ltp5-1 heterozygote to the wild type. Pollination was allowed to grow for 12 h on the emasculated (previous day) pistils (n = 10). Aniline blue staining shows in vivo pollen tube growth. Arrows indicate the pollen tube front. Bars = 200 μm. Mature siliques at 8 d after pollination were decolorized with 100% ethanol to visualize seeds. Bars = 1 mm.
We also examined heterozygous ltp5-1 plants (ltp5-1 het) for mutant phenotypes in pollen tube growth and in pistil function. The ltp5-1 het, with half of the pollen normal, showed typical in vivo pollen tube growth (see Supplemental Figure 4A online). However, significant numbers of unfertilized seeds were found dispersed overall in ltp5-1 het siliques, regardless of pollen tube growth behavior in vivo (see Supplemental Figure 4A online). The average longitudinal silique size (1.27 ± 0.1 cm, n = 34) was shorter, compared with the wild type (LTP5/LTP5 obtained from backcrossing, 1.52 ± 0.12 cm, n = 23). Average seed numbers per silique (33 ± 2.1, n = 9) were around half that of the wild type (59 ± 4.3, n = 5).
In reciprocal cross-pollination of ltp5-1 het to the wild type (Figure 3C), ltp5-1 het pollen tubes were able to grow to the base of either wild-type or ltp5-1 het pistils in 12 h. However, when ltp5-1 het pistils were used as pollen acceptors, resulting siliques were shorter, regardless of the pollen donor was of the wild type (1.3 ± 0.1 cm, n = 5) or ltp5-1 het (1.4 ± 0.1 cm, n = 5). Their average seed numbers per silique were 28 ± 2.5 and 32 ± 3.8, respectively. This shows that ltp5-1 het pistils are partially defective in seed formation, as was the ltp5-1 pistil shown in Figure 2.
In vitro pollen tube growth assays showed that significant numbers of ltp5-1 het pollen, similar to ltp5-1, germinated precociously at 2 h, which is rare for wild-type pollen (see Supplemental Figures 4B to 4D online). Some pollen tubes started to show abnormal pollen tube tips by 6 h after germination (see Supplemental Figures 4F and 4G online) and were arrested with an obviously swollen tip (∼16% for ltp5-1 het and ∼59% for ltp5-1) by 8 h (see Supplemental Figures 4I and 4J online).
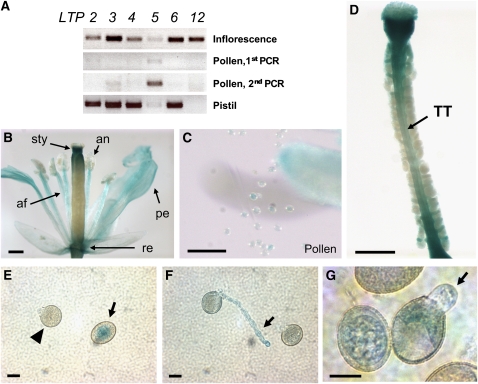
Expression Patterns of LTP5 in the Arabidopsis Flower
In RT-PCR analysis (Figure 4A), LTP5 was found to be the most weakly expressed gene in the inflorescence among SCA-like LTPs examined. However, we found that LTP5 is specifically present in pollen, though at a low level. Only the LTP5 transcript became obvious when a second round of PCR was performed using the first PCR products as templates. LTP5 transcript levels also were very low in the pistil compared with other LTPs.
Figure 4.
Arabidopsis LTP5 Is Present in Pollen, Pollen Tubes, and the Pistil TT.
(A) Gene expression levels of some SCA-like Arabidopsis LTPs were evaluated by two replicates of RT-PCR using the gene-specific primer sets. Thirty cycles of PCRs were performed for gene expression in the inflorescence apex and pistil tissues and 35 cycles for pollen. Among the LTP genes examined, LTP5 gene transcripts were shown to be present in both pollen and pistil at a low level. Only the second PCR using the first PCR products as templates was able to show a significant level of LTP5 transcript in pollen.
(B) to (D) GUS assay of LTP5pro:GUS flower. GUS signals were developed for 5 d to make the weak LTP5 gene level more visible.
(B) Gene expression was shown in the style (sty), anthers (an), anther filaments (af), petals (pe), and the receptacle (re). Bar = 400 μm.
(C) Weak GUS signals were shown in pollen grains. Bar = 100 μm.
(D) A dissected pistil showed a low level of LTP5 gene expression in the pistil TT (arrow) Bar = 400 μm.
(E) to (G) GUS assay of LTP5pro:GUS pollen. Pollen tubes were grown on the solid medium in vitro for 6 h. GUS signals were developed for 3 d. Arrowhead indicates pollen grain right before germination; arrows indicate GUS signals in pollen grain and pollen tubes. Bars = 10 μm.
To examine LTP5 gene expression in planta, an ∼2-kb upstream genomic DNA sequence of the LTP5 gene was fused with a β-glucuronidase (GUS) reporter gene. In GUS assays of LTP5pro:GUS flowers, overall signals were very low in pollen and the pistil, similar to RT-PCR results. So, we incubated the flower samples in GUS reaction for up to 5 d to clearly visualize LTP5 gene expression patterns. We found that LTP5 gene expression was obvious in the style, the top surface of the ovary, the receptacle, petals, anthers, and anther filaments (Figure 4B). When we removed pollen from the anthers, we could identify a weak GUS signal in pollen (Figure 4C). We also identified gene expression in the pistil TT, where pollen tubes grow (Figure 4D).
To verify LTP5 expression in growing pollen tubes, we germinated LTP5pro:GUS pollen in vitro for 6 h and incubated them in GUS reaction for 3 d. GUS signals were clearly identified in LTP5pro:GUS pollen grains (Figure 4E, arrow). When pollen grains were about to germinate, the signals became very weak (Figure 4E, triangle), but then the weak GUS signals were clearly identified in the growing tubes and at the tip (Figures 5F and 5G, arrows).
Figure 5.
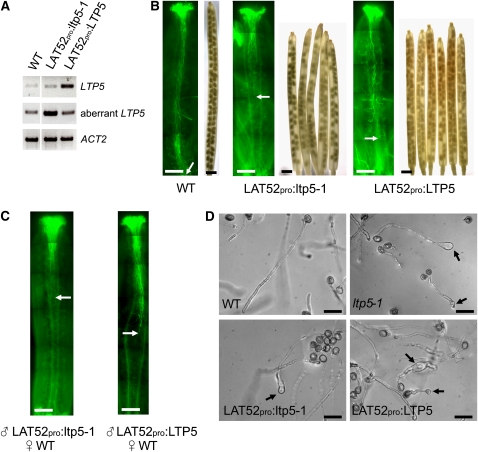
Pollen-Targeted Overexpression of ltp5-1 or LTP5 Gene in Arabidopsis Plants.
(A) RT-PCR analysis for LAT52pro:ltp5-1 or LAT52pro:LTP5 plants from two replicates. LTP5-5 and LTP5-3 primers shown in Figure 1B were used to examine LTP5 levels. LTP5-5B and LTP5m-3K primers were used to examine aberrant LTP5 levels. PCR was performed in 30 cycles. The ACT2 primer set was used as the PCR control.
(B) In vivo pollen tube growth and silique examination for LAT52pro:ltp5-1 and LAT52pro:LTP5. Mature flowers were stained with aniline blue to visualize pollen tube growth in the pistil (n = 10). Arrows indicate the pollen tube front. Bars = 200 μm. Mature siliques were destained with 100% ethanol to examine seed sets (n = 20). Bars = 1 mm.
(C) In vivo reciprocal cross-pollination of LAT52pro:ltp5-1 or LAT52pro:LTP5 pollen to wild-type pistils. Pollination was allowed for 12 h on the emasculated (previous day) wild-type pistils (n = 10). Aniline blue staining shows in vivo pollen tube growth. Arrows indicate the pollen tube front. Bars = 200 μm.
(D) In vitro pollen tube growth assay. Pollen tubes were grown on the solid medium for 16 h. Bars = 50 μm.
Pollen-Targeted Overexpression of Wild-Type LTP5 or ltp5-1 Results in Adverse Effects on Pollen Tube Tip Growth and Seed Set
Our study of LTP5 gene expression indicates that ltp5-1, the aberrant LTP5, is also present in the gain-of-function mutant pollen. The ltp5-1 gene in the pollen might express into the ltp5-1 protein, which eventually could cause abnormal pollen tube tip growth as a dominant active form of wild-type LTP5. If so, one would expect that overexpression of wild-type LTP5 or ltp5-1 in wild-type pollen may cause a similar defect as in ltp5-1 pollen. To understand this dominant defect of ltp5-1 on pollen tube growth, we generated transgenic Arabidopsis plants overexpressing either ltp5-1 or LTP5 genes under the control of the pollen-specific LAT52 promoter (LAT52pro:ltp5-1 and LAT52pro:LTP5, respectively) (see Supplemental Figure 5A online). The majority of LAT52pro:ltp5-1 (86%) and LAT52pro:LTP5 (73.6%) plants in the T1 generation exhibited abnormally swollen pollen tube tip morphology in in vitro pollen tube growth assays (see Supplemental Figure 5B and Supplemental Table 3 online). About 44% of LAT52pro:ltp5-1 and 28% of LAT52pro:LTP5 plants had short siliques, <1.3 cm in average length, with many unfertilized eggs (see Supplemental Figure 5C and Supplemental Table 3 online). Overall, the effects of ltp5-1 overexpression on mutant phenotypes in pollen tube growth and seed formation were slightly more severe than those of LTP5 overexpression.
We obtained homozygous transgenic plants with a single transgene copy for further analysis (Figure 5A). We first examined in vivo pollen tube growth and siliques of LAT52pro:ltp5-1 and LAT52pro:LTP5 plants (Figure 5B). The majority of LAT52pro:ltp5-1 pollen tubes were arrested in the middle of the ovary. By contrast, LAT52pro:LTP5 pollen tubes grew to the base of the ovary. However, both produced shorter siliques (∼1.3 cm) with many unfertilized ovules. Average seed numbers per silique were 33 ± 2.4 (n = 9) for LAT52pro:ltp5-1 and 46 ± 2.1 (n = 10) for LAT52pro:LTP5 plants. In cross-pollination to the wild-type pistil, the majority of both LAT52pro:ltp5-1 and LAT52pro:LTP5 pollen tubes were shown to be unable to grow to the base of the ovary in 12 h (Figure 5C). We also evaluated in vitro pollen tube growth for the transgenic lines (Figure 5D; see Supplemental Figure 6 online). Unlike ltp5-1, both LAT52pro:ltp5-1 and LAT52pro:LTP5 pollen did not show precocious germination nor did they show abnormal tip morphology in 8 h. However, their tips eventually swelled by 16 h of germination.
The overall mutant phenotypes examined in both LAT52pro:ltp5-1 and LAT52pro:LTP5 lines were not identical to those in ltp5-1 plants. Their pollen tubes were shown to have swollen tips at 16 h in vitro, while ltp5-1 pollen tubes displayed obvious tip swelling at 6 h. Their effect on silique size and seed set formation was milder, more similar to those of ltp5-1 heterozygous plants. These discrepancies may be due to the different promoters used in our study. LTP5 or ltp5-1 gene overexpressing plant lines had the transgene under the control of the LAT52 promoter, while ltp5-1 plants expressed the mutant gene by LTP5 promoter activity. Nonetheless, our work suggests that ltp5-1 protein causes abnormal pollen tube growth behavior and thereby a decreased seed set in ltp5-1 plants as a dominant mutant of LTP5.
Three-Dimensional Structures of Arabidopsis LTP5 and ltp5-1
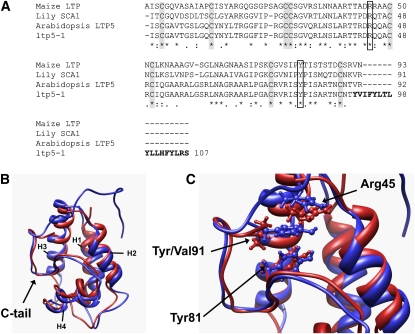
We sequenced the PCR product of the aberrant LTP5 transcript shown in Figure 1E to deduce amino acid sequences of ltp5-1. With elimination of the predicted N-terminal signal sequences, our sequence alignment showed that mature LTP5 protein has 49% amino acid identity to maize (Zea mays) LTP (Shin et al., 1995), 50% to lily SCA1 (Chae et al., 2007), and 97% to ltp5-1 (Figure 6A). Conserved sequence motifs of the plant LTP family were also found in Arabidopsis LTP5: eight conserved Cys residues and two pentapeptide motifs (Thr/Ser-X1-X2-Asp-Arg/Lys and Pro-Tyr-X-Ile-Ser) (Douliez et al., 2000b). Two amino acids (Val-91 and Arg-92) that were translated from the second exon (Figure 1B) in the C-tail of LTP5 were replaced with a long, hydrophobic sequence in ltp5-1.
Figure 6.
The ltp5-1 Protein Has an Additional Hydrophobic C-Terminal Tail.
(A) Protein sequence alignment. Gray boxes indicate the eight Cys residues conserved in the plant LTP family. White boxes indicate Arg-45 and Tyr-81 in the conserved consensus pentapeptide motifs (Thr/Ser-X1-X2-Asp-Arg/Lys and Pro-Tyr-X-Ile-Ser) (Douliez et al., 2000b). Bold letters are additional C-terminal tail sequences in ltp5-1.
(B) Superposition of ribbon representations of the structures of LTP5 and ltp5-1. The structures were generated using homology modeling and 1-ns molecular dynamics simulations. The additional, predominantly hydrophobic, C-terminal tail of ltp5-1 is shown to cap one side of the protein, which is known to be an entrance for a putative ligand to the internal hydrophobic cavity in maize LTP (Han et al., 2001). Red, LTP5; blue, ltp5-1; H1 to 4, helix 1 to 4.
(C) Tyr-91 in ltp5-1, replacing Val-91 of LTP5, is shown to interact with Arg-45 and Tyr-81. A focused view of the superposition of (B) is shown, with residues of interest (Arg-45, Tyr-81, Val-91, and Tyr-91) depicted in ball and stick representations. Replacement of Val-91 with Tyr-91 results in stabilizing π-cation interactions with Arg-45 and π-stacking interactions with Tyr-81 (also see Supplemental Figure 7 online). The coloring scheme is the same as in (B).
To examine the structural difference between LTP5 and ltp5-1, we predicted three-dimensional structures using structural homology/molecular dynamics modeling (Figures 6B and 6C; see Supplemental Figure 7 online). Both LTP5 and ltp5-1 showed a typical plant LTP structure with a globular shape of the orthogonal four-helix bundle architecture. However, the additional C-terminal tail of ltp5-1 was predicted to lie down in the vicinity of the first and second helices (Figure 6B). Interestingly, Tyr-91 in ltp5-1, replacing Val-91 in LTP5, was predicted to localize in between Arg-45 and Tyr-81, which are known to be crucial for the interaction with a lipid molecule in maize LTP in vitro (Han et al., 2001). This localization is favored by the π-cation interaction between the aromatic group of Tyr-91 and the guanidinium group of Arg-45 and by the π-stacking interaction between the aromatic groups of Tyr-91 and Tyr-81 (see Supplemental Figure 7 online).
DISCUSSION
A biological function of plant LTPs has long been explored since they were first proposed to function as lipid carriers between intracellular organelles (Kader et al., 1984). The finding that LTPs are extracellular matrix proteins (Thoma et al., 1993) shifted our attention to their extracellular role. Lily LTP, SCA, is secreted from the pistil TT epidermis and functions in forming an adhesive matrix with pectin that guides pollen tubes to the ovules (Mollet et al., 2000; Park et al., 2000). SCA was also shown to be involved in pollen tube chemotropism by synergistically enhancing the activity of lily chemocyanin on the stigma (Kim et al., 2003). Further work revealed that lily SCA, a secreted pistil factor, is endocytosed through the pollen tube tip and may function in tip growth as a signal transducer (Kim et al., 2006; Chae et al., 2007). LTPs related to SCA in other plant species have a variety of proposed biological roles. Purified Ace-AMP1 from onion (Allium cepa) seeds showed antifungal activity (Phillippe et al., 1995). Transgenic tobacco (Nicotiana tabacum) expressing barley (Hordeum vulgare) LTP4 had enhanced resistance against a bacterial pathogen (Molina and Garcia-Olmedo, 1997). More recently, it was shown that tobacco LTP2 mediates cell wall loosening in vitro (Nieuwland et al., 2005). However, no genetic evidence has yet been provided for these proposed functions.
Some Arabidopsis LTPs were studied using a genetic approach and found to have extracellular functions. Defective in induced resistance 1 (DIR1) is involved in systemic acquired resistance (Maldonado et al., 2002). Azelaic acid induced 1 (AZI1) is involved in salicylic acid–mediated plant defense (Jung et al., 2009). Arabidopsis glycosylphosphatidylinositol-anchored LTP1 (LTPG1) is involved in cuticular wax deposition (DeBono et al., 2009). However, their homology to SCA-like LTPs is very low. SCA-like LTPs are small (∼10 kD) and basic (pI ∼9) molecules (Chae et al., 2007). DIR1 is ∼10 kD but is an acidic (pI ∼4.5) molecule. AZI1 and LTPG are relatively basic (pIs 7.7 for AZI1 and 9.2 for LTPG), but they are predicted to be much bigger than SCA-like LTPs (∼15 kD for AZI1 and ∼19 kD for LTPG). Their amino acid identities to SCA are 20% for DIR1, 7% for AZI1, and 16% for LTPG. In this study, a SCA-like Arabidopsis LTP group, shown in Figure 1A, has 40 to 50% amino acid identity to lily SCA. Our work reveals a biological function for a SCA-like LTP using a genetic approach.
We examined T-DNA mutants of SCA-like Arabidopsis LTPs, focusing on silique size and seed set, which are direct indicators of successful fertilization. The gain-of-function mutant plant for LTP5 (ltp5-1) was shown to have small siliques with a significantly decreased seed set compared with wild-type plants or other mutants. We initially thought this defect would be caused only by a lack of pistil function because SCA was known as a pistil factor for lily pollen tube growth and guidance. However, our reciprocal cross-pollinations and in vitro pollen tube growth assays revealed an unexpected potential male function for LTP5 as well. A majority of ltp5-1 pollen tubes were unable to grow further than the middle of the ovary, resulting in failure to fertilize the ovules at the base of the silique. In addition, in vitro–grown ltp5-1 pollen showed abnormal growth behavior and a swollen tube tip. Our gene expression study demonstrates that LTP5 is present in pollen and in growing pollen tubes at a low level, as well as in the pistil TT. To date, several Arabidopsis LTPs and lily SCA have been shown to be expressed in various tissues, but not in pollen (Thoma et al., 1994; Arondel et al., 2000; Park and Lord, 2003). Arabidopsis LTP5 is the first LTP found in pollen.
Our initial attempt to complement the abnormal pollen tube growth and seed formation of ltp5-1 plants was not successful. Soon after, we revealed the presence of aberrant LTP5 transcript in ltp5-1 plants by RT-PCR analysis. Our study using pollen-targeted gene overexpression suggests that the examined male sterility of ltp5-1 plants could be due to activity of aberrant LTP5. The effects of wild-type LTP5 overexpression was similar to, though slightly milder than, those of ltp5-1 overexpression. This suggests that ltp5-1 may act as a dominant-active mutant of LTP5 in pollen tube tip growth and fertilization, though neomorphic activity cannot be excluded yet.
Arabidopsis LTP1/2, LTP3/4, and LTP5/12 genes are thought to be pairs of duplicated genes. Two genes in each pair were found to be located right next to each other in tandem orientation (Arondel et al., 2000). However, LTP5 has only a 45% cDNA sequence identity with LTP12, while other pairs showed above 80% identity. In addition, LTP12 transcripts were not found in either pollen or the pistil (Figure 4A). Arabidopsis LTP5 may not be a duplicated gene. Nonetheless, our examination of T-DNA insertional mutants for seven SCA-like Arabidopsis LTPs showed that a downmodulation or knockout of a single gene was not sufficient to cause any mutant phenotype. This suggests that functional redundancy occurs in the SCA-like LTP group. The examined phenotypes of ltp5-1 plants in this study may reflect the disturbance in the redundant function by the gain-of-function mutation.
Our structural analysis showed that Arabidopsis LTP5 has typical features of plant LTPs. It has the eight conserved Cys residues, the consensus motifs, and a globular shape of the orthogonal four-helix bundle architecture. The mutant protein, ltp5-1, was shown to have an additional hydrophobic C-terminal tail, compared with LTP5. Structural homology/MD modeling allowed us to predict that Tyr-91, in the ltp5-1 C-terminal tail, interacts strongly with both Tyr-81 and Arg-45, both of which are highly conserved in plant LTPs and responsible for interaction with the carboxylate group of a fatty acid (Han et al., 2001). Analysis of the MD trajectories indicated that Tyr-91–Arg-45 and Tyr-91–Tyr-81 form interactions through π-cation and π-stacking, respectively. These potential interactions in ltp5-1 are yet to be examined. So far, we have no evidence that a SCA-like LTP has a binding partner such as a lipid.
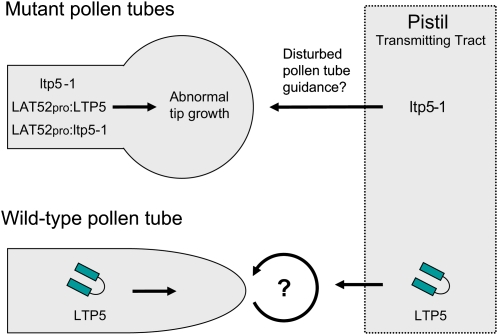
Our study suggests that putative ltp5-1 may disturb a signaling pathway that regulates polarity of pollen tube cells, either in an autocrine manner or as a neomorph (Figure 7). The abnormal tip morphology of the mutant pollen tubes resembles that of pollen tubes overexpressing Rop1 GTPase or its activator RopGEFs, which regulate cellular responses for polar tip growth (Li et al., 1999; Fu et al., 2001; Gu et al., 2006). A putative upstream activator of the Rop signaling pathway in pollen is a putative cell surface receptor kinase, PRK2, whose overexpression also causes a swollen tip (Zhang and McCormick, 2007).
Figure 7.
Model for the Role of LTP5 in Arabidopsis Sexual Reproduction.
LTP5, present in pollen, may be secreted from the pollen tube tip and function in establishing or maintaining cell polarity at the tip of pollen tubes. Mutant pollen tubes showed a phenotype similar to that of mutants for Rop signaling (Fu et al., 2001; Gu et al., 2006; Zhang and McCormick, 2007), suggestive of a role of LTP5 in signaling for polar pollen tube tip growth. LTP5 was also found in the pistil TT, where pollen tubes grow. Our study showed a subtle effect of ltp5-1 on normal pistil function for seed formation. In the pistil, it may be both endocytosed into the pollen tube and interact with pectin in an adhesion-mediated guidance mechanism, as SCA does in lily (Kim et al., 2006). Other LTPs may also be involved in pollen tube guidance to the ovule for fertilization since several LTPs are known to be present in the Arabidopsis pistil TT (Thoma et al., 1994; Arondel et al., 2000; Tung et al., 2005).
[See online article for color version of this figure.]
Polarized cell growth shares many fundamental features across species (Madden and Snyder, 1998; Momany, 2002; Baker et al., 2006; Cole and Fowler, 2006; Stumm and Hollt, 2007; Cheung and Wu, 2008; Lee and Yang, 2008). Small secreted peptides play a crucial role in cell–cell communication to trigger and maintain cell polarization. Yeast mating pair cells use small secreted peptides (a/α factors) to initiate polarized cell growth toward each partner (Madden and Snyder, 1998). The CXC motif chemokine stromal cell-derived factor-1 (SDF-1) and netrins interact with receptors to regulate polar axonal extension of neurons (Baker et al., 2006; Stumm and Hollt, 2007). A challenge will be to determine whether Arabidopsis LTP5, or other SCA-like LTPs, act as an extracellular cue to regulate pollen tube cell polarity through a plasma membrane receptor, as do their functional counterparts in yeast and neurons.
In addition to the effect of ltp5-1 on pollen tube growth, we showed that ltp5-1 has a subtle effect on normal pistil function for seed formation in reciprocal cross-pollinations using the ltp5-1 or ltp5-1 het pistil. In spite of normal in vivo pollen tube growth, the mutant pistils did not generate fertilized seed set properly. Our GUS analysis showed an obvious LTP5 gene expression in the pistil TT. Several Arabidopsis LTPs are known to exist in the pistil TT where pollen tubes grow (Thoma et al., 1994; Arondel et al., 2000; Tung et al., 2005). In addition, lily SCA synergistically enhances the pollen tube guidance activity of chemocyanin, a small secreted peptide in the pistil (Kim et al., 2003). Arabidopsis LTP5 and other SCA-like molecules may be involved in a signaling hierarchy of pollen tube guidance in the pistil TT (Figure 7), through which pollen tubes become competent to receive a guidance signal from the ovule (Lord and Russell, 2002).
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana (ecotype Columbia) plants were grown in a growth chamber in the Department of Botany and Plant Sciences at the University of California, Riverside. SALK T-DNA insertion lines were obtained from the ABRC at Ohio State University (Alonso et al., 2003). Seeds were grown to 6-week-old plants in soil (Sunshine Mix 1; Sun Gro Horticulture) at 22°C under a 16-h-light/8-h-dark photoperiod (200 μE m−2 s−1).
Phylogenetic Analysis
Amino acid sequences of 107 Arabidopsis LTPs, SCA1 (Chae et al., 2007), and maize (Zea mays) LTP (Shin et al., 1995) were aligned using default parameters of ClustalX 2.0.10 (Thompson et al., 1997), and the aligned sequences were analyzed using PAUP* 4.0 (Swofford, 2003). Pairwise amino acid divergence was calculated, and a neighbor-joining (Saitou and Nei, 1987) tree was constructed using PAUP*. Support for groups was examined by 1000 bootstrap replicates (Felsenstein, 1985). Parsimony analysis was also performed using the heuristic search option with tree-bisection-reconnection branch swapping and the multiple parsimony (MULPARS) option on. The heuristic search found two equally parsimonious trees (tree length = 6804, consistency index = 0.4705, retention index = 0.4899), and tree topologies were very similar to the neighbor-joining tree. Therefore, we presented the neighbor-joining tree in Figure 1 for further discussion.
PCR-Based Genotyping Analysis
For PCR-based genotyping analysis, genomic DNAs were purified from an inflorescence. The forward primer (674-5) and the reverse primer (674-3) were designed to amplify an ∼1.5-kb genomic DNA sequence including LTP5 from a genomic DNA strand, in which the T-DNA was not inserted (see Supplemental Table 2 online). The LBa1 and 674-3 primers were used for PCR to amplify about a 1-kb genomic DNA sequence from the T-DNA inserted genomic DNA strand.
RT-PCR Analysis
For purification of Arabidopsis pollen RNAs, ∼200 Arabidopsis flowers were collected in liquid pollen germination medium [18% sucrose, 0.01% boric acid, 1 mM CaCl2, 1 mM Ca(NO3)2, and 1 mM MgSO4] containing diethylpyrocarbonate and vortexed to harvest pollen. The pollen was filtered through an 80-μm nylon mesh (NITEX; SEFAR America). The filtered pollen was rinsed with liquid germination medium three times via centrifugation at 1200 rpm. For purification of pistil RNAs, ∼20 pistils were dissected out from mature flowers. Except for the gene expression study using pollen and pistil RNAs shown in Figure 4A, all others were performed using RNAs purified from one inflorescence apex. Total RNAs were extracted using the RNeasy Mini Kit (Qiagen). The cDNA synthesis was performed using 0.5 μg of total RNA and the iScript cDNA synthesis kit (Bio-Rad).
Forward and reverse gene-specific primers for SCA-like Arabidopsis LTPs were designed to include several 5′- and 3′-untranslated region sequences, respectively (see Supplemental Table 2 online). The PCR products, obtained using each primer set, were fully sequenced, confirming the gene specificity of each primer set.
For aberrant LTP5 transcripts in ltp5-1 plants, the forward primer containing the translation start codon (ATG; LTP5-5B) and the reverse primer containing the putative termination codon from the intron (TAA; LTP5m-3K) were used for PCR (see Supplemental Table 2 online). The LTP5-5B primer and another reverse primer (LTP5C) were used to amplify the first exon of LTP5 as the control (see Supplemental Table 2 online).
In Vitro Pollen Tube Growth Assay
Plants were removed from the growth chamber for 2 h before pollen was removed from flowers. Pollen was grown on solid germination medium [18% sucrose, 0.01% boric acid, 1 mM CaCl2, 1 mM Ca(NO3)2, 1 mM MgSO4, and 0.5% Noble agar] at room temperature. Pollen tube length and tip morphology were examined at various time points (2 to 16 h) using a Leica dissecting microscope or an Eclipse inverted microscope (TE300; Nikon) for higher magnification. The relative length of pollen tubes was measured at 6 h using ImageJ (Abramoff et al., 2004).
Study of in Vivo Pollen Tube Growth and Seed Formation in Siliques
To examine in vivo pollen tube growth, about 10 mature flowers at stage 14 (Smyth et al., 1990) were fixed in acetic acid/ethanol (1:3) solution. Fixed floral tissues were cleared in 8 M NaOH and stained with aniline blue following a previously published method (Mori et al., 2006). Pollen tube growth in the pistil was examined using a fluorescent compound microscope (Nikon Microphot FXA), and digital images were captured using a Spot Insight camera (Diagnostic Instruments).
To evaluate fertilization, mature siliques were measured for their lengths and dissected to identify unfertilized ovules. Siliques were also decolorized by incubation in 100% ethanol at 37°C overnight to visualize the seed set.
For in vivo reciprocal cross-pollination, ∼20 floral buds at stage 12 (Smyth et al., 1990) were emasculated per cross a day before hand-pollination. Fresh pollen at flower stage 13 (Smyth et al., 1990) was fully applied to the stigma of the emasculated flower. After a 12-h pollination, the pollinated pistil was fixed, stained with aniline blue, and examined as described above. For the fertilization study, half of the pollinated flowers were further grown in the growth chamber for 8 to 10 d. Siliques were dissected or decolorized at maturity to examine seed set.
Generation of Transgenic Plants (LAT52pro:ltp5-1, LAT52pro:LTP5, and LTP5pro:GUS)
The 35S promoter of Cauliflower mosaic virus in a binary vector pCL0011 (Lin et al., 2003) was replaced by the PCR-amplified EcoRI-BamHI fragment (see Supplemental Table 2 online) containing the tomato (Solanum lycopersicum) LAT52 promoter (Twell et al., 1990) to generate the plasmid construct pCL0011-LAT52pro. The PCR-amplified BamHI-XbaI fragment (see Supplemental Table 2 online) containing ltp5-1 or wild-type LTP5 coding sequence was cloned into pCL0011-LAT52pro. The binary plasmid constructs, pCL0011-LAT52pro:ltp5-1 and pCL0011-LAT52pro:LTP5, were introduced into Arabidopsis plants using Agrobacterium tumefaciens strain GV3101 with a floral dip method (Clough and Bent, 1998). LAT52pro:ltp5-1 or LAT52pro:LTP5 transgenic plants were selected on soil by spraying with a 1000-fold dilution of Finale (AgrEvo Environmental Health; BASTA). Spraying was initiated at 10 d after germination and was performed three times every 2 d.
For the LTP5pro:GUS fusion, a 2-kb-upstream region of LTP5 was amplified from genomic DNA by PCR (see Supplemental Table 2 online), and cloned into a Gateway Binary Vector pGWB3 (Invitrogen). This plasmid construct was used for transformation of Arabidopsis plants. LTP5pro:GUS transgenic plants were selected on solid MS medium containing 30 μg/ml kanamycin.
GUS Assay
Mature LTP5:GUS flowers were incubated for 5 d at 37°C in a GUS reaction buffer (10 mM EDTA, 100 mM sodium phosphate, pH 7.0, 0.5 mM potassium ferrocyanide, 0.5 mM potassium ferricyanide) and 0.1% Triton X-100 with 1 mM 5-bromo-4-chloro-β-d-glucuronide. GUS-stained tissues were decolorized with 70% ethanol three times in a 12-h incubation at 37°C. The developed GUS signals were examined using a Leica dissecting microscope. For examination of gene expression in pollen, LTP5pro:GUS pollen were grown in vitro on solid medium for 6 h and incubated for 3 d in a GUS buffer. GUS signals were examined using a fluorescent compound microscope (Nikon Microphot FXA) in the RGB color mode.
Homology Modeling/Molecular Dynamics Simulations
To generate homology models for LTP5 and ltp5-1, we first generated amino acid sequence alignments between our target sequences and the maize LTP sequence using ClustalW (Chenna et al., 2003). The obtained amino acid sequence alignments and the template structure, maize LTP (PDB Code: 1MZL) (Shin et al., 1995), were imported into the Automodel module of the homology modeling package Modeler 9v5 (Fiser and Sali, 2003). After generation, models were visualized using Chimera (Pettersen et al., 2004) and inspected for van der Waals clashes, secondary structure quality, and disulfide bond correctness.
Following the homology modeling procedure, molecular dynamics (MD) simulations were performed to relax the structures through the optimization of local contacts and geometries under the influence of the protein's force field. MD trajectories were obtained using an explicit water sphere and spherical boundary conditions. The simulations were allowed to run for 500,000 iterative steps with a 2-fs time step, resulting in trajectories of 1-ns durations. The scalable molecular dynamics package, Nanoscale Molecular Dynamics (Phillips et al., 2005), was used in conjunction with the CHARMM22 (MacKerell et al., 1998) force field. The setup and analysis of the generated MD trajectories was performed using visual molecular dynamics (Humphrey et al., 1996).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: LTP1, At2g38540; LTP2, At2g38530; LTP3, At5g59320; LTP4, At5g59310; LTP5, At3g51600; a putative SCA-like LTP, At2g15050; LTP12, At3g51590; DIR1, At5g48485; AZI1, At4g12470; and LTPG1, At1g27950.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. A Genome-Wide Search for SCA-Like Arabidopsis LTPs.
Supplemental Figure 2. Gene Expression Levels in T-DNA Insertion Lines of SCA-Like Arabidopsis LTPs.
Supplemental Figure 3. In Vivo Arabidopsis Pollen Tube Growth.
Supplemental Figure 4. Heterozygous ltp5-1 Showed Disturbed Pistil Function in Seed Formation and Abnormal in Vitro Pollen Tube Morphology.
Supplemental Figure 5. Pollen-Targeted ltp5-1 or LTP5 Overexpression Lines at T1 Generation Resulted in Abnormal Pollen Tube Tip Morphology in Vitro and a Defect in Seed Formation.
Supplemental Figure 6. In Vitro Pollen Tube Growth of LAT52pro:ltp5-1 or LAT52pro:LTP5 at T3 Generation.
Supplemental Figure 7. Comparison of Molecular Dynamics Snapshots for Maize LTP, Arabidopsis LTP5, and ltp5-1.
Supplemental Table 1. PCR-based Genotyping Analysis for SALK T-DNA Insertion Alleles of SCA-Like Arabidopsis LTPs.
Supplemental Table 2. PCR Primer Sequences.
Supplemental Table 3. Transgenic Plants with Pollen-Specific Overexpression of LTP5 or ltp5-1 Gene in T1 Generation.
Supplemental Data Set 1. Sequences and Alignment Used to Generate the Phylogenetic Trees Presented in Supplemental Figure 1 and Figure 1.
Supplemental References.
Supplementary Material
Acknowledgments
We thank Zhenbiao Yang for critical advice and review of this manuscript, Shelia McCormick and Ravi Palanivelu for helpful discussions, Yong Jik Lee and Jae-Ung Hwang for valuable technical comments, and Benedict J. Gonong for technical assistance. This study was supported by the National Science Foundation (Grant IBM0420445 to E.M.L.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Elizabeth M. Lord (lord@ucr.edu).
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Abramoff, M.D., Magelhaes, P.J., and Ram, S.J. (2004). Image processing with ImageJ. Biophotonics International 11 36–42. [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215 403–410. [DOI] [PubMed] [Google Scholar]
- Arondel, V., Vergnolle, C., Cantrel, C., and Kader, J.C. (2000). Lipid transfer proteins are encoded by a small multigene family in Arabidopsis thaliana. Plant Sci. 157 1–12. [DOI] [PubMed] [Google Scholar]
- Baker, K.A., Moore, S.W., Jarjour, A.A., and Kennedy, T.E. (2006). When a diffusible axon guidance cue stops diffusing: roles for netrins in adhesion and morphogenesis. Curr. Opin. Neurobiol. 16 529–534. [DOI] [PubMed] [Google Scholar]
- Chae, K., Li, Z., Li, K., Morikis, D., Kim, S.T., Mollet, J.C., de la Rosa, N., Tan, K., and Lord, E.M. (2007). Two SCA (stigma/style cysteine-rich adhesin) isoforms show structural differences that correlate with their levels of in vitro pollen tube adhesion activity. J. Biol. Chem. 282 33845–33858. [DOI] [PubMed] [Google Scholar]
- Charvolin, D., Douliez, J.P., Marion, D., Cohen-Addad, C., and Pebay-Peyroula, E. (1999). The crystal structure of a wheat nonspecific lipid transfer protein (nsLTP1) complexed with two molecules of phospholipid at 2.1 angstrom resolution. Eur. J. Biochem. 264 562–568. [DOI] [PubMed] [Google Scholar]
- Chenna, R., Sugawara, H., Koike, T., Lopez, R., Gibson, T.J., Higgins, D.G., and Thompson, J.D. (2003). Multiple sequence alignment with the clustal series of programs. Nucleic Acids Res. 31 3497–3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung, A.Y., and Wu, H.M. (2008). Structural and signaling networks for the polar cell growth machinery in pollen tubes. Annu. Rev. Plant Biol. 59 547–572. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Cole, R.A., and Fowler, J.E. (2006). Polarized growth: Maintaining focus on the tip. Curr. Opin. Plant Biol. 9 579–588. [DOI] [PubMed] [Google Scholar]
- DeBono, A., Yeats, T.H., Rose, J.K.C., Bird, D., Jetter, R., Kunst, L., and Samuelsa, L. (2009). Arabidopsis LTPG is a glycosylphosphatidylinositol-anchored lipid transfer protein required for export of lipids to the plant surface. Plant Cell 21 1230–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douliez, J.P., Michon, T., Elmorjani, K., and Marion, D. (2000. b). Structure, biological and technological functions of lipid transfer proteins and indolines, the major lipid binding proteins from cereal kernels. J. Cereal Sci. 32 1–20. [Google Scholar]
- Douliez, J.P., Michon, T., and Marion, D. (2000. a). Steady-state tyrosine fluorescence to study the lipid-binding properties of a wheat non-specific lipid-transfer protein (nsLTP1). Biochim. Biophys. Acta 1467 65–72. [PubMed] [Google Scholar]
- Douliez, J.P., Pato, C., Rabesona, H., Molle, D., and Marion, D. (2001). Disulfide bond assignment, lipid transfer activity and secondary structure of a 7-kDa plant lipid transfer protein, LTP2. Eur. J. Biochem. 268 1400–1403. [DOI] [PubMed] [Google Scholar]
- Felsenstein, J. (1985). Confidence-limits on phylogenies - An approach using the bootstrap. Evolution 39 783–791. [DOI] [PubMed] [Google Scholar]
- Fiser, A.S., and Sali, A. (2003). MODELLER: Generation and refinement of homology-based protein structure models. Methods Enzymol. 374 461–491. [DOI] [PubMed] [Google Scholar]
- Franklin-Tong, V.E. (1999). Signaling and the modulation of pollen tube growth. Plant Cell 11 727–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin-Tong, V.E. (2002). The difficult question of sex: The mating game. Curr. Opin. Plant Biol. 5 14–18. [DOI] [PubMed] [Google Scholar]
- Fu, Y., Wu, G., and Yang, Z.B. (2001). Rop GTPase-dependent dynamics of tip-localized F-actin controls tip growth in pollen tubes. J. Cell Biol. 152 1019–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, Y., Li, S.D., Lord, E.M., and Yang, Z.B. (2006). Members of a novel class of Arabidopsis Rho guanine nucleotide exchange factors control rho GTPase-dependent polar growth. Plant Cell 18 366–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton, J.A. (2004). Fatty acid interactions with proteins: What X-ray crystal and NMR solution structures tell us. Prog. Lipid Res. 43 177–199. [DOI] [PubMed] [Google Scholar]
- Han, G.W., et al. (2001). Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography. J. Mol. Biol. 308 263–278. [DOI] [PubMed] [Google Scholar]
- Humphrey, W., Dalke, A., and Schulten, K. (1996). VMD: Visual molecular dynamics. J. Mol. Graph. 14 33–38. [DOI] [PubMed] [Google Scholar]
- Johnson, M.A., and Preuss, D. (2002). Plotting a course: Multiple signals guide pollen tubes to their targets. Dev. Cell 2 273–281. [DOI] [PubMed] [Google Scholar]
- Jung, H.W., Tschaplinski, T.J., Wang, L., Glazebrook, J., and Greenberg, J.T. (2009). Priming in systemic plant immunity. Science 324 89–91. [DOI] [PubMed] [Google Scholar]
- Kader, J.C. (1996). Lipid transfer proteins in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47 627–654. [DOI] [PubMed] [Google Scholar]
- Kader, J.C. (1997). Lipid transfer proteins: A puzzling family of plant proteins. Trends Plant Sci. 2 66–70. [Google Scholar]
- Kader, J.C., Julienne, M., and Vergnolle, C. (1984). Purification and characterization of a spinach leaf protein capable of transferring phospholipids from liposomes to mitochondria or chloroplasts. Eur. J. Biochem. 139 411–416. [DOI] [PubMed] [Google Scholar]
- Kim, S., Mollet, J.C., Dong, J., Zhang, K.L., Park, S.Y., and Lord, E.M. (2003). Chemocyanin, a small basic protein from the lily stigma, induces pollen tube chemotropism. Proc. Natl. Acad. Sci. USA 100 16125–16130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S.R., Dong, J., and Lord, E.M. (2004). Pollen tube guidance: The role of adhesion and chemotropic molecules. Curr. Top. Dev. Biol. 61 61–79. [DOI] [PubMed] [Google Scholar]
- Kim, S.T., Zhang, K.L., Dong, J., and Lord, E.M. (2006). Exogenous free ubiquitin enhances lily pollen tube adhesion to an in vitro stylar matrix and may facilitate endocytosis of SCA. Plant Physiol. 142 1397–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y.J., and Yang, Z. (2008). Tip growth: Signaling in the apical dome. Curr. Opin. Plant Biol. 11 662–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., Lin, Y.K., Heath, R.M., Zhu, M.X., and Yang, Z.B. (1999). Control of pollen tube tip growth by a Rop GTPase-dependent pathway that leads to tip-localized calcium influx. Plant Cell 11 1731–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, W.C., Shuai, B., and Springer, P.S. (2003). The Arabidopsis LATERAL ORGAN BOUNDARIES domain gene ASYMMETRIC LEAVES2 functions in the repression of KNOX gene expression and in adaxial-abaxial patterning. Plant Cell 15 2241–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord, E. (2000). Adhesion and cell movement during pollination: Cherchez la femme. Trends Plant Sci. 5 368–373. [DOI] [PubMed] [Google Scholar]
- Lord, E.M. (2003). Adhesion and guidance in compatible pollination. J. Exp. Bot. 54 47–54. [DOI] [PubMed] [Google Scholar]
- Lord, E.M., and Russell, S.D. (2002). The mechanisms of pollination and fertilization in plants. Annu. Rev. Cell Dev. Biol. 18 81–105. [DOI] [PubMed] [Google Scholar]
- MacKerell, A.D., et al. (1998). An all-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 102 3586–3616. [DOI] [PubMed] [Google Scholar]
- Madden, K., and Snyder, M. (1998). Cell polarity and morphogenesis in budding yeast. Annu. Rev. Microbiol. 52 687–744. [DOI] [PubMed] [Google Scholar]
- Maldonado, A.M., Doerner, P., Dixon, R.A., Lamb, C.J., and Cameron, R.K. (2002). A putative lipid transfer protein involved in systemic resistance signalling in Arabidopsis. Nature 419 399–403. [DOI] [PubMed] [Google Scholar]
- Molina, A., and García-Olmedo, F. (1997). Enhanced tolerance to bacterial pathogens caused by the transgenic expression of barley lipid transfer protein LTP2. Plant J. 12 669–675. [DOI] [PubMed] [Google Scholar]
- Mollet, J.C., Faugeron, C., and Morvan, H. (2007). Cell adhesion, separation and guidance in compatible plant reproduction. Annu. Plant Rev. 25 69–90. [Google Scholar]
- Mollet, J.C., Park, S.Y., Nothnagel, E.A., and Lord, E.M. (2000). A lily stylar pectin is necessary for pollen tube adhesion to an in vitro stylar matrix. Plant Cell 12 1737–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momany, M. (2002). Polarity in filamentous fungi: establishment, maintenance and new axes. Curr. Opin. Microbiol. 5 580–585. [DOI] [PubMed] [Google Scholar]
- Mori, T., Kuroiwa, H., Higashiyama, T., and Kuroiwa, T. (2006). GENERATIVE CELL SPECIFIC 1 is essential for angiosperm fertilization. Nat. Cell Biol. 8 64–71. [DOI] [PubMed] [Google Scholar]
- Nieuwland, J., Feron, R., Huisman, B.A.H., Fasolino, A., Hilbers, C.W., Derksen, J., and Mariani, C. (2005). Lipid transfer proteins enhance cell wall extension in tobacco. Plant Cell 17 2009–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, S.Y., Jauh, G.Y., Mollet, J.C., Eckard, K.J., Nothnagel, E.A., Walling, L.L., and Lord, E.M. (2000). A lipid transfer-like protein is necessary for lily pollen tube adhesion to an in vitro stylar matrix. Plant Cell 12 151–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, S.Y., and Lord, E.M. (2003). Expression studies of SCA in lily and confirmation of its role in pollen tube adhesion. Plant Mol. Biol. 51 183–189. [DOI] [PubMed] [Google Scholar]
- Pettersen, E.F., Goddard, T.D., Huang, C.C., Couch, G.S., Greenblatt, D.M., Meng, E.C., and Ferrin, T.E. (2004). UCSF chimera - A visualization system for exploratory research and analysis. J. Comput. Chem. 25 1605–1612. [DOI] [PubMed] [Google Scholar]
- Phillippe, B., Cammue, B.P.A., Thevissen, K., Hendriks, M., Eggermont, K., Goderis, I.J., Proost, P., Vandamme, J., Osborn, R.W., Guerbette, F., Kader, J.C., and Broekaert, W.F. (1995). A potent antimicrobial protein from onion seeds showing sequence homology to plant lipid transfer proteins. Plant Physiol. 109 445–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips, J.C., Braun, R., Wang, W., Gumbart, J., Tajkhorshid, E., Villa, E., Chipot, C., Skeel, R.D., Kale, L., and Schulten, K. (2005). Scalable molecular dynamics with NAMD. J. Comput. Chem. 26 1781–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou, N., and Nei, M. (1987). The neighbor joining method - A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4 406–425. [DOI] [PubMed] [Google Scholar]
- Shin, D.H., Lee, J.Y., Hwang, K.Y., Kim, K.K., and Suh, S.W. (1995). High-resolution crystal structure of the nonspecific lipid transfer protein from maize seedlings. Structure 3 189–199. [DOI] [PubMed] [Google Scholar]
- Smyth, D.R., Bowman, J.L., and Meyerowitz, E.M. (1990). Early flower development in Arabidopsis. Plant Cell 2 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumm, R., and Hollt, V. (2007). CXC chemokine regulator receptor 4 regulates neuronal migration and axonal pathfinding in the developing nervous system: implications for neuronal regeneration in the adult brain. J. Mol. Endocrinol. 38 377–382. [DOI] [PubMed] [Google Scholar]
- Swofford, D.L. (2003). PAUP*: Phylogenetic Analysis Using Parsimony (and Other Methods), Version 4. (Sunderland, MA: Sinauer Associate).
- Thoma, S., Hecht, U., Kippers, A., Botella, J., Devries, S., and Somerville, C. (1994). Tissue specific expression of a gene encoding a cell wall localized lipid transfer protein from Arabidopsis. Plant Physiol. 105 35–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma, S., Kaneko, Y., and Somerville, C. (1993). A nonspecific lipid transfer protein from Arabidopsis is a cell wall protein. Plant J. 3 427–436. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. (1997). The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung, C.W., Dwyer, K.G., Nasrallah, M.E., and Nasrallah, J.B. (2005). Genome-wide identification of genes expressed in Arabidopsis pistils specifically along the path of pollen tube growth. Plant Physiol. 138 977–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twell, D., Yamaguchi, J., and McCormick, S. (1990). Pollen-specific gene expression in transgenic plants: Coordinate regulation of 2 different tomato gene promoters during microsporogenesis. Development 109 705–713. [DOI] [PubMed] [Google Scholar]
- Zachowski, A., Guerbette, F., Grosbois, M., Jolliot-Croquin, A., and Kader, J.C. (1998). Characterisation of acyl binding by a plant lipid transfer protein. Eur. J. Biochem. 257 443–448. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., and McCormick, S. (2007). A distinct mechanism regulating a pollen-specific guanine nucleotide exchange factor for the small GTPase Rop in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 104 18830–18835. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.