Abstract
To examine the role of the tonoplast in plant salt tolerance and identify proteins involved in the regulation of transporters for vacuolar Na+ sequestration, we exploited a targeted quantitative proteomics approach. Two-dimensional differential in-gel electrophoresis analysis of free flow zonal electrophoresis separated tonoplast fractions from control, and salt-treated Mesembryanthemum crystallinum plants revealed the membrane association of glycolytic enzymes aldolase and enolase, along with subunits of the vacuolar H+-ATPase V-ATPase. Protein blot analysis confirmed coordinated salt regulation of these proteins, and chaotrope treatment indicated a strong tonoplast association. Reciprocal coimmunoprecipitation studies revealed that the glycolytic enzymes interacted with the V-ATPase subunit B VHA-B, and aldolase was shown to stimulate V-ATPase activity in vitro by increasing the affinity for ATP. To investigate a physiological role for this association, the Arabidopsis thaliana cytoplasmic enolase mutant, los2, was characterized. These plants were salt sensitive, and there was a specific reduction in enolase abundance in the tonoplast from salt-treated plants. Moreover, tonoplast isolated from mutant plants showed an impaired ability for aldolase stimulation of V-ATPase hydrolytic activity. The association of glycolytic proteins with the tonoplast may not only channel ATP to the V-ATPase, but also directly upregulate H+-pump activity.
INTRODUCTION
The vacuole plays an important role in a plant's tolerance to salinity. Low cytoplasmic sodium concentrations are maintained partially through active sequestration of sodium into the vacuole lumen, serving to compartmentalize this toxic ion away from the cytoplasm. This also provides solutes for osmotic adjustment, facilitating water uptake. Transport of sodium across the vacuolar membrane (tonoplast) is attributed to members of the family of Na+/H+ exchangers (NHXs) and is driven by the inside acidic pH gradient generated by the vacuolar H+-ATPase (V-ATPase). While NHX proteins are encoded by single polypeptides, the V-ATPase is a multisubunit enzyme composed of at least 13 subunits, organized to form two distinct sectors: a peripheral domain (V1) and a membrane domain (Vo) (Cipriano et al., 2008). VHA subunits A, B, C, D, E, F, G, and H make up the V1 sector, and subunits a, b, c, d, and e compose the Vo sector (Cipriano et al., 2008). In both salt-tolerant halophytes and salt-sensitive glycophytes, sodium regulates the expression at the transcript and protein levels for the tonoplast NHXs (Shi and Zhu, 2002; Yokoi et al., 2002) and different subunits of the V-ATPase (Löw et al., 1996; Tsiantis et al., 1996; Dietz et al., 2001; Vera-Estrella et al., 2005). In addition, their transport activity has been shown to increase under salt stress (Reuveni et al., 1990; Barkla et al., 1995; Qiu et al., 2004; Vera-Estrella et al., 2004). In Arabidopsis thaliana, mutants of NHX family members displayed enhanced salt sensitivity (Shi et al., 2000; Apse et al., 2003), as do mutants in subunits of the V-ATPase. Specifically, det3, a VHA-C subunit mutant (Batelli et al., 2007), and vha-c3, a double-stranded RNA interference mutant of the VHA-c subunit (Padmanaban et al., 2004), are salt sensitive, confirming the role of these proteins in plant salt tolerance. Despite the large number of studies of the function of these transporters in plant salt tolerance, information on how these processes are regulated and the signaling molecules involved is just beginning to emerge.
Accumulating evidence implicates components of the salt overly sensitive (SOS) pathway, and, specifically, the calcineurin B-like interacting protein kinase-interacting protein kinase, SOS2/CIPK24, has recently been revealed to regulate both the activity of the tonoplast Na+/H+ exchanger, NHX1 (Qiu et al., 2004), and that of the V-ATPase (Batelli et al., 2007), through a possible calcineurin B-like protein-calcineurin B-like interacting protein kinase network. In Arabidopsis, SOS2 mutants show a 60% reduction in Na+/H+ exchange and a 30% reduction in V-ATPase H+ transport activity. However, only Na+/H+ exchange activity was returned to wild-type levels in the sos2 mutant by incubation with a constitutively activated SOS2 protein (Qiu et al., 2004). Neither transporter appeared to be directly phosphorylated by SOS2, and, in the case of the V-ATPase, regulation appears to be via direct interaction of the SOS2 protein with VHA-B (Batelli et al., 2007), although how this regulation is achieved was not addressed.
Other possible mechanisms for regulation of the V-ATPase include in vitro evidence that WNK8, a member of the Arabidopsis WNK family of protein kinases, binds to and phosphorylates VHA-C of the V-ATPase (Hong-Hermesdorf et al., 2006); however, the involvement of this kinase in salt regulation of the transporters is not known. It has also been proposed that regulation of the V-ATPase may result from changes in assembly brought about by alterations in subunit availability or expression, as well as reversible dissociation of the complex into its component V1 and V0 domains (Qi et al., 2007), although this has not yet been investigated in plants.
In this study, we exploit a quantitative proteomics approach with the aim to identify regulatory proteins involved in salt tolerance in the halophyte Mesembryanthemum crystallinum, employing an organelle-centered study to enable the characterization of a specific subset of proteins targeted to the tonoplast. Free flow zonal electrophoresis (FFZE) purified tonoplast was subjected to two-dimensional differential in-gel electrophoresis (2D-DIGE), a powerful approach for the comparative analysis of protein abundance due to its high degree of accuracy and reproducibility. This technique allowed us to detect statistically significant changes in tonoplast protein abundance between untreated and salt-treated M. crystallinum plants. Analysis of gels using Decyder Software V.6.5 highlighted a small number of tonoplast proteins that showed significant changes in expression level in the presence of NaCl, and these were selected for identification by mass spectroscopy and further characterization.
RESULTS
FFZE
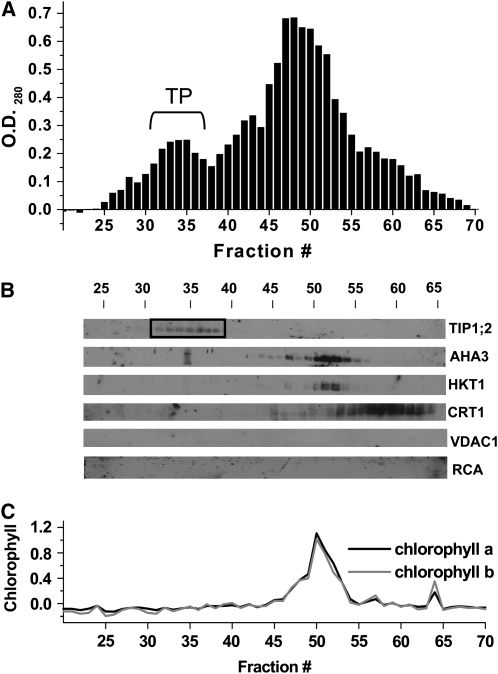
One of the difficulties with subproteome or directed proteome analysis is the presence of contaminating proteins from other cellular membranes that can be erroneously allocated to a particular subcellular structure or endomembrane (Millar, 2004). In this study, we avoided using traditional fractionation techniques, which are known to result in the presence of contaminating membranes and subsequent identification of nontonoplast proteins (Carter et al., 2004; Shimaoka et al., 2004; Endler et al., 2006) by using FFZE. This technique separates tonoplast from other membranes based on surface charge by laminar flow through a thin aqueous layer (Heidrich and Hannig, 1989; Moritz and Simpson, 2005). Previously, we have shown that addition of 3 mM ATP to microsomal membranes prior to FFZE results in a shift in tonoplast toward the positive electrode, most likely due to a screening of positive surface charges by the negatively charged ATP4− (Figure 1A; Barkla et al., 2007). To confirm the origin and purity of this population of membranes for this study, FFZE fractions of M. crystallinum microsomal membranes were collected and subjected to protein blot analysis (Figure 1B). Based on membrane protein marker analysis for different membrane compartments, including the tonoplast aquaporin TIP1;2 (Kirch et al., 2000), the plasma membrane H+-ATPase AHA3 (Parets-Soler et al., 1990), the plasma membrane Na+/K+ cotransporter HKT1 (Su et al., 2003), the endoplasmic reticulum Ca2+ binding protein calreticulin (CRT1; Nelson et al., 1997), the mitochondrial voltage-dependent anion channel VDAC1 (Clausen et al., 2004), and chloroplast ribulose-1,5-bis-phosphate carboxylase/oxygenase activase (RCA; Vargas-Suárez et al., 2004), as well as direct chlorophyll measurements (Figure 1C), we found that the ATP-dependent peak of membranes between fractions 31 to 37 corresponded to tonoplast (Figure 1A), in agreement to our previous results (Barkla et al., 2007).
Figure 1.
Purification of M. crystallinum Tonoplast by FFZE.
M. crystallinum microsomal membranes were separated by FFZE in the presence of 3 mM ATP.
(A) Protein profile of FFZE fractions showing a positive OD280. The bracket indicates the location of the ATP-dependent peak of tonoplast (TP).
(B) Immunological detection in the respective fractions of (from top to bottom) the tonoplast marker TIP1;2 (26 kD), the plasma membrane marker AHA3 (100 kD), the plasma membrane marker HKT1 (56 kD), the endoplasmic reticulum marker CRT1 (57 kD), the mitochondrial marker VDAC1 (29 kD), and the chloroplast marker RCA (43 and 41 kD). The fractions corresponding to pure tonoplast are enclosed in the box.
(C) Measurement of chlorophyll a and b in FFZE fractions.
DIGE of Salt-Regulated Tonoplast Proteins
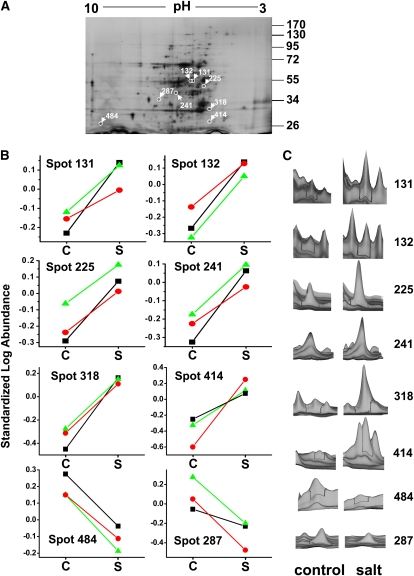
To identify tonoplast proteins with altered abundance in salt-treated M. crystallinum plants compared with control plants, we performed 2D-DIGE using CyDye fluorescent labeling. Tonoplast proteins from three independent biological replicates (control and salt-treated) were prepared, minimally labeled with Cy2, Cy3, and Cy5, and processed for DIGE analysis as described in Methods and shown in Table 1. The Cy2 dye was used to label an internal standard consisting of an equal amount of protein from control and salt-treated membranes from all experiments; the internal standard is included on all gels, allowing for normalization of spot volume ratios in order to overcome intergel variability (Alban et al., 2003). This eliminates error due to protein loading, gel polymerization artifacts, and experimental variation. Protein spots were detected automatically using the DIA module of the software DeCyder 6.5 (GE LifeSciences) with an initial estimation of 2000 spots/gel. A representative 2D preparative gel of tonoplast protein from salt-treated plants is shown in Figure 2A. Following spot matching and filtering, within-gel analysis yielded normalized spot ratio values for all 471 included spots. These values were used to generate histograms that showed a normal distribution with an average 2 sd, corresponding to a threshold volume ratio of ±1.49198. Normalized spot volume ratios falling outside this threshold value can be reasonably expected to represent biological changes at a confidence of 95%, with only a 5% false positive chance of it being an artifact of system variation. Matching between the different gels was done by means of landmarking spots in the Cy2 internal standard images from each gel using the BVA module of the Decyder 6.5 software. Statistical analysis was then performed on matched spots, and those that were 1.5-fold up- or downregulated in salt-treated compared with control tonoplasts across all three gels (representing three independent experiments) with a Student's t test probability score of ≤0.03 were considered differentially expressed (Figures 2B and 2C). Under these criteria, eight spots exhibited statistically significant expression changes (Figure 2). The spot ratios for all the protein spots detected are presented in Supplemental Data Set 1 online.
Table 1.
DIGE Experimental Design
| CyDye |
||||
|---|---|---|---|---|
| Gel No. | Cy3 | Cy5 | Cy2 (Internal Standard Pool) | Protein/IEF Gel Strip |
| 1 | 50 μg C1 | 50 μg S1 | 8.333 μg each of C1+C2+C3+S1+S2+S3 | 150 μg |
| 2 | 50 μg S2 | 50 μg C2 | 8.333 μg each of C1+C2+C3+S1+S2+S3 | 150 μg |
| 3 | 50 μg C3 | 50 μg S3 | 8.333 μg each of C1+C2+C3+S1+S2+S3 | 150 μg |
C1, control experiment 1; C2, control experiment 2; C3, control experiment 3; S1, salt-treated experiment 1; S2, salt-treated experiment 2; S3, salt-treated experiment 3.
Figure 2.
2D-DIGE of FFZE Separated Tonoplast from M. crystallinum.
(A) A representative preparative silver-stained gel of tonoplast fractions from salt-treated plants. Protein (200 μg) was separated by isoelectric focusing on 3 to 10 linear immobilized pH gradient strips for the first dimension and by SDS-PAGE on a 10% linear acrylamide gel for the second dimension. Eight protein spots that showed significant changes in abundance between the control and salt-treated tonoplast samples after analysis with Decyder Software (>1.5-fold change, P ≤ 0.05; Student's t test [P ≤ 0.03]; n = 3) are circled and labeled with the software-derived spot number. The positions of PAGE molecular mass markers are shown in kilodaltons on the right of the gel image.
(B) Graphical representation of the standardized log abundance (i.e., log abundance of Cy3- or Cy5-labeled spot over log abundance of Cy2-labeled standard spot). Individual lines show each of the three biological replicates from control (C) and salt (S)-treated tonoplast. Triangles, values from gel 1; circles, values from gel 2; squares, values from gel 3.
(C) The three-dimensional fluorescence intensity profiles of the individual spots shown for one of the biological replicates comparing control and salt-treated profiles of each of the eight protein spots that showed significant changes.
[See online article for color version of this figure.]
Identification of Differentially Regulated Tonoplast Proteins by Mass Spectrometry
Following spot picking and tryptic digestion, protein identification was performed using nano-liquid chromatography–tandem mass spectrometry (MS/MS). Of the eight differentially regulated spots that were present in all the gels from three independent experiments, four unique proteins were identified that all showed increases in abundance in the salt-treated M. crystallinum tonoplast compared with tonoplast isolated from control plants (Table 2; see Supplemental Table 1 online). These included two peripheral subunits of the V-ATPase, VHA-d and VHA-B. In the case of subunit VHA-B, two spots were identified (spots 318 and 414) that differed in molecular mass (32 and 26 kD, respectively). While the deduced molecular mass of VHA-B is 55 kD, it is known that this subunit can under certain conditions undergo in vivo proteolytic processing into two subunits with apparent molecular masses of 31 to 32 kD and 27 to 28 kD (Zhigang et al., 1996; Krisch et al., 2000; Ratajczak, 2000), matching the size of the proteins identified in this study. This phenomenon is not limited to M. crystallinum, as the V-ATPase B subunit of yeast, Vma2p, was also susceptible to proteolytic processing with the appearance of a 30-kD protein (Landolt-Marticorena et al., 1999). It has been suggested that this posttranslational modification may be a means of stabilizing the holoenzyme complex or for regulating function (Ratajczak, 2000). Both VHA-B protein spots showed a similar fold increase in abundance in tonoplast from salt-treated plants.
Table 2.
Identification of Salt-Responsive Tonoplast Proteins in M. crystallinum
| Spot No. | DIGE Ave. Ratioa | DIGE t Testb | Gene Name Description | Accessionc | No. of Identified Peptidesd | Sequence of Identified Peptidese | zf | Xcorrg | ΔCnh | Molecular Function |
|---|---|---|---|---|---|---|---|---|---|---|
| 131 | 1.80 | 0.011 | PGH1- Enolase | Q43130/S79242 | 3 | K.VNQIGSVTESIEAVK.M K.NVNEIIGPALVGK.D R.AAVPSGASTGVYEALELR.D | 2 | 3.94 | 0.66 | Glycolysis, vacuolar fusion, and trafficking |
| 2 | 3.36 | 0.66 | ||||||||
| 2 | 4.41 | 0.81 | ||||||||
| 132 | 2.19 | 0.0046 | PGH1- Enolase | Q43130/S79242 | 2 | K.VQIVGDDLLVTNPK.R K.VNQIGSVTESIEAVK.M | 2 | 3.99 | 0.77 | Glycolysis, vacuolar fusion, and trafficking |
| 2 | 2.98 | 0.71 | ||||||||
| 225 | 1.93 | 0.026 | VHA-d - V-ATPase subunit d | Q8GUB0/AJ439342 | 2 | R.DVQELLEK.C K.AYLEDFYR.F | 2 | 2.77 | 0.38 | Primary proton pump |
| 1 | 2.04 | 0.30 | ||||||||
| 241 | 1.92 | 0.0084 | ALF1 - Fructose bisphosphate aldolase | O04975/AF003124 | 5 | K.TAAGKPFVEVLK.E R.FAGINVENVESNR.R K.VAPEVIAEYTVR.A K.GVVELAGTNGETTT QGLDGLGAR.C K.YADELIANAAYIGTPGK.G | 2 | 3.53 | 0.60 | Glycolysis |
| 2 | 3.52 | 0.66 | ||||||||
| 2 | 3.34 | 0.71 | ||||||||
| 2 | 3.89 | 0.68 | ||||||||
| 2 | 4.63 | 0.71 | ||||||||
| 318 | 3.02 | 0.0010 | VHA-B - V-ATPase subunit B | Q8GUB5/AJ438590 | 2 | R.QIYPPINVLPSLSR.L R.VTLFLNLANDPTIER.I | 2 | 2.87 | 0.64 | Primary proton pump |
| 2 | 3.71 | 0.67 | ||||||||
| 414 | 3.32 | 0.0099 | VHA-B - V-ATPase subunit B | Q8GUB5/AJ438590 | 3 | K.AVVQVFEGTSGIDNK.Y K.TPVSLDMLGR.I R.TYPEEM*IQTGISTIDVM *NSIAR.G | 2 | 5.38 | 0.70 | Primary proton pump |
| 2 | 2.85 | 0.70 | ||||||||
| 3 | 5.10 | 0.51 |
Protein spots chosen for MS/MS analysis met the following criteria: >1.5-fold change (P ≤ 0.05); n = 3; t test (P ≤ 0.03).
Average ratios of abundance of salt-treated tonoplast relative to the untreated control represent data from three separate experiments.
Student's t test P values are given as a measure of confidence for the ratio of each spot measured.
UniProtKB/GenBank accession numbers.
Number of matched peptides from MS/MS. Proteins were identified by two or more unique peptides.
The amino acid residues appearing before and after the periods correspond to the residues proceeding and following the peptide in the protein sequence, and the asterisks within the peptide sequence indicate a differential modification on the preceding amino acid.
The charge state of the candidate peptide.
For data validation, we accepted spectra with SEQUEST cross-correlation scores (Xcorr) of at least 2.5 for doubly and 3.5 for triply charged ions.
SEQUEST ΔCn value gives the difference of the cross-correlation scores between the best hit and the following hits.
Two enzymes most commonly associated with glycolysis and categorized as cytoplasmic soluble proteins were also identified. One of these, 2-phosphoglycerate dehydratase (enolase), was also identified from two distinct spots, 131 and 132. These spots had the same molecular mass of ∼50 kD, but their pI values differed slightly (between ∼5.6 and 5.7). This suggested posttranslational modification of the protein and was supported by previous evidence of reversible phosphorylation of the enzyme (Dannelly et al., 1989; Forsthoefel et al., 1995). The other enzyme, fructose bis-phosphate aldolase (aldolase), was identified from spot 241 with molecular mass of 38 kD and pI 6.5. The remaining two spots (287 and 484) were unidentifiable, as no spectra were obtained for these proteins, indicating there were most likely insufficient quantities in the excised spots. These two were the only proteins found to be significantly downregulated in the salt-treated tonoplast.
It is not surprising that we did not identify any highly hydrophobic integral membrane proteins in this analysis, as these proteins are known to precipitate out of solution during isoelectric focusing; this is one of the inherent problems of gel-based proteomics approaches for hydrophobic membrane proteins (Henningsen et al., 2002).
Confirmation of 2D-DIGE Results by Protein Blot Analysis and Enzymatic Activity
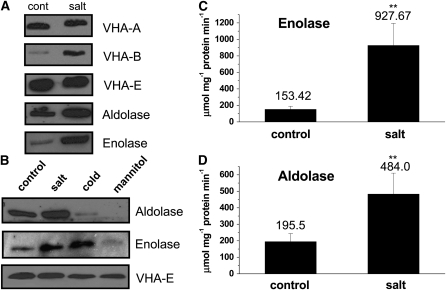
Direct protein blot analysis of purified tonoplast from control and salt-treated M. crystallinum supported the localization of the glycolytic proteins to tonoplast fractions and corroborated the differential regulation of the proteins identified in the DIGE gels under our growth and treatment regimes. Aldolase and enolase were both detected in the purified tonoplast fractions of leaves of control plants and showed increased abundance in the salt-treated plants, validating the DIGE experimental results (Figure 3A). V-ATPase subunit VHA-B was also confirmed to be salt regulated, while subunits VHA-A and VHA-E showed no change in abundance between control and salt-treated plants (Figure 3A). Differential expression of V-ATPase subunits is thought to be important for the regulation of the enzyme under stress conditions (Qi et al., 2007). In addition to regulation by salt treatment, the effect of cold and mannitol treatment on the levels of the glycolytic enzymes at the tonoplast were also examined (Figure 3B), as enolase has previously been shown to be regulated at the level of the transcript by low temperature stress (Lee et al., 2002). Tonoplast aldolase levels were significantly lower when plants were treated with mannitol or exposed to low temperatures (4°C). By contrast, enolase showed an increase in protein amount in tonoplast fractions over control values following cold exposure, similar to the increase observed in the presence of NaCl. However, mannitol treatment resulted in decreased enolase levels (Figure 3B). These results indicate that only salt treatment results in the coordinate upregulation of both glycolytic enzymes at the tonoplast.
Figure 3.
Aldolase and Enolase Are Salt-Regulated Proteins Detected in Tonoplast Fractions of M. crystallinum.
(A) Protein blots of isolated tonoplast from plants treated for 1 week in the absence (cont) or presence (salt) of 200 mM NaCl. SDS-PAGE separated tonoplast protein was probed with polyclonal antibodies individually raised against VHA-A, VHA-B, and VHA-E subunits of the V-ATPase, or the glycolytic enzymes aldolase and enolase, which recognized proteins of 72, 55, 29, 38, and 50 kD polypeptides, respectively. Blots are representative of three independent experiments.
(B) Protein blots of isolated tonoplast from plants treated for 1 week in the absence (control) or presence of 200 mM NaCl (salt) or at 4°C (cold) in the presence of 400 mM mannitol. Blots are representative of three independent experiments.
(C) Aldolase and enolase enzymatic activity in tonoplast isolated from control and salt-treated plants. Results are presented as mean ± se (n = 3). Statistical significance was evaluated using Student's t test for pairwise comparison and analysis of variance for comparison of data from several groups. A probability level of <0.01 (indicated by asterisks) was considered highly significant.
Measurement of enzyme activity was also used to confirm the presence of aldolase and enolase in the purified tonoplast fraction and the upregulation by salinity treatment (Figure 3C). Activities of both enzymes were measured as the utilization of NADH in coupled reactions. The aldolase activity assay employed the substrate fructose 1,6 bisphosphate and the enzymes glycerophosphate dehydrogenase and triosephosphate isomerase (Hodgson and Plaxton, 1998), whereas the enolase activity assay used the substrate 2-phosphoglycerate and the enzymes pyruvate kinase and lactate dehydrogenase (Hüther et al., 1990). Aldolase and enolase activities were both detected in tonoplast from control plants with averaged measured activities of 195 μmol NADH mg−1 protein min−1 and 153 μmol NADH mg−1 protein min−1, respectively. Both enzymes showed significant increases in activity in tonoplast from salt-treated plants: a 6.0-fold increase in activity for enolase and a 2.6-fold increase for aldolase (Figure 3C).
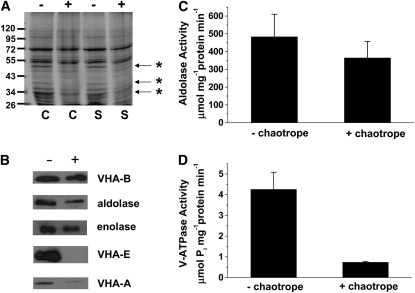
Chaotrope Treatment of Tonoplast Demonstrates Membrane Association of Aldolase and Enolase
Chaotropes are routinely used to destabilize protein–protein and protein–lipid interactions that mediate the peripheral association of proteins to a membrane by disrupting hydrogen bonds, van der Waals forces, and hydrophobic effects and as such allow for the determination of the strength of that association (Hatefi and Hanstein, 1974). Addition of these compounds has been used successfully to remove nonintegral proteins from membranes, including subunits of the yeast and plant V-ATPase that constitute the V1 sector of the holoenzyme (Kane et al., 1989; Ward et al., 1992). In this study, 200 mM sodium carbonate buffer, pH 11.4, in the presence of 3 mM MgATP, was used to determine the membrane association of aldolase and enolase in purified tonoplast, compared with specific peripheral subunits of the V-ATPase. The addition of MgATP is known to facilitate the removal of V1 sector subunits into the soluble fraction in the presence of the chaotrope (Ward et al., 1992). Visualization of Coomassie blue–stained gels of tonoplast proteins showed that several polypeptides ranging in molecular mass from 25 to 100 kD were absent in the chaotrope-treated membranes (Figure 4A). Subsequent protein blot analysis of tonoplast isolated from salt-treated plants demonstrated that while chaotropic treatment was able to successfully remove most of VHA-E and VHA-A from the tonoplast fraction, only a small decrease in aldolase or enolase abundance was observed and little to no decrease in VHA-B subunit was detected (Figure 4B), suggesting a strong association of these proteins, including the glycolytic enzymes, with the tonoplast. When aldolase activity was measured in chaotrope-incubated membranes isolated from salt-treated plants, there was only a slight decrease compared with values measured in the absence of chaotrope (484 μmol NADH mg−1 protein min−1 compared with 365 μmol NADH mg−1 protein min−1 in the presence of chaotrope) (Figure 4C). These data confirmed that the majority of these enzymes remained attached to the tonoplast. By contrast, V-ATPase hydrolytic activity was severely reduced in the chaotrope-treated tonoplast (Figure 4D), most likely due to the almost complete loss of the catalytic subunit, VHA-A (Figure 4B).
Figure 4.
Protein Blot Analysis and Effect of Chaotrope Treatment on Enzyme Activities in Tonoplast of M. crystallinum.
(A) Coomassie blue–stained gel of tonoplast isolated from control (C) and salt (S)-treated plants (200 mM NaCl for 1 week) incubated in the presence (+) or absence (−) of 200 mM Na2CO3, pH 11.4, and 3 mM MgATP. Proteins that are clearly absent in the chaotrope treated lanes are marked (asterisks).
(B) Protein blot analysis of tonoplast from salt-treated plants incubated in the presence (+) or absence (−) of 200 mM Na2CO3, pH 11.4, and 3 mM MgATP and probed with antibodies against V-ATPase subunits VHA-B, VHA-E, and VHA-A and the enzymes aldolase and enolase.
(C) Aldolase activity in tonoplast from salt-treated plants incubated in the presence (+ chaotrope) or absence (− chaotrope) of 200 mM Na2CO3, pH 11.4, and 3 mM MgATP. Values are means ± se from three experiments.
(D) V-ATPase hydrolytic activity in tonoplast from salt-treated plants incubated in the presence (+ chaotrope) or absence (− chaotrope) of 200 mM Na2CO3, pH 11.4, and 3 mM MgATP. Values are means ± se from three experiments.
Immunoprecipitation Reveals Interaction between Glycolytic Enzymes and Subunits of the V-ATPase
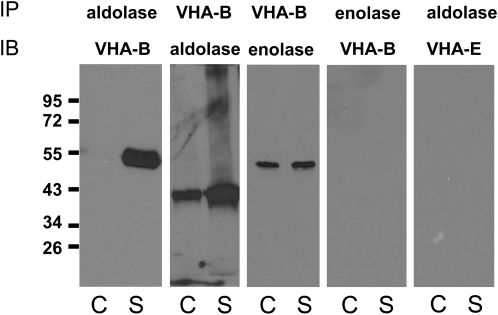
Enzymes of glycolysis are increasingly being assigned roles in other nonmetabolic processes, suggesting so-called moonlighting functions (Gancedo and Flores, 2008). With particular relevance to this study, yeast aldolase has been shown to bind to subunits of the yeast vacuolar V-ATPase, including subunits E and B, mediating assembly, and regulating expression and activity of the proton pump (Lu et al., 2001, 2004). This in vivo interaction did not require aldolase catalytic activity, as mutants lacking the catalytic site were still capable of interacting (Lu et al., 2007), although the exact mechanism of regulation remains unclear, and it has not been related to a specific stress condition. Enolase also appears to bind to yeast vacuoles through a peripheral membrane association, but the target protein on the membrane remains unknown (Decker and Wickner, 2006; Wiederhold et al., 2009). In order to determine if in M. crystallinum these enzymes are associated to the tonoplast by means of physical interaction with subunits of the V-ATPase, we performed reciprocal immunoprecipitation assays on tonoplast isolated from control and salt-treated plants using antibodies against aldolase, enolase, VHA-B, and VHA-E subunits (Figure 5). Immunoprecipitation of aldolase followed by immunoblotting with anti-VHA-B antibodies showed association between the two proteins, which was confirmed by reciprocal experiments in which VHA-B was immunoprecipitated and interacting proteins were probed with anti-aldolase antibodies (Figure 5). In both cases, more of the interacting protein was detected in salt-treated tonoplast fractions. Immunoprecipitation of VHA-B followed by probing against enolase also suggested an interaction; however, in this case, no VHA-B was detected in reciprocal experiments wherein tonoplast enolase was immunoprecipitated (Figure 5). This could result from failure of the enolase peptide-specific polyclonal antibody to recognize the nondenatured native form of the protein or obstruction of the epitope by the protein interaction but may also suggest the detection of VHA-B/enolase interaction was nonspecific. Immunoprecipitation experiments using other V-ATPase subunits (VHA-A and -E) failed to detect protein associations with either enolase or aldolase, indicating specificity for VHA-B (Figure 5).
Figure 5.
Aldolase Interacts with the VHA-B Subunit of the V-ATPase.
Leaf tonoplast protein (15 μg) from control (C) and salt-treated (S) M. crystallinum plants was analyzed by reciprocal immunoprecipitation (IP), SDS-PAGE, and immunoblotting (IB) as described in Methods, using the indicated antibodies (top antibody was used for immunoprecipitation; bottom antibody was used to probe blots). The results shown are representative of experiments that were repeated three times, which yielded identical results. The positions of PAGE molecular mass markers are shown in kilodaltons on the left of the panels.
Aldolase Stimulates V-ATPase Hydrolytic Activity in Vitro in Tonoplast from M. crystallinum by Increasing the Affinity for ATP
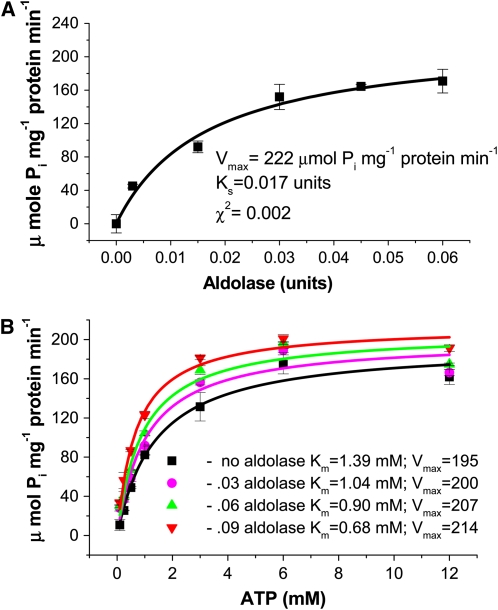
To investigate the functional significance of the reciprocal association of aldolase with the V-ATPase subunit VHA-B, we examined whether the presence of purified spinach (Spinacia oleracea) aldolase was able to regulate the activity of the V-ATPase in vitro, in tonoplast fractions from M. crystallinum leaf tissue. As demonstrated in Figure 6A, the addition of increasing concentrations of aldolase resulted in a concomitant increase in bafilomycin-sensitive and azide- and vanadate-insensitive V-ATPase hydrolytic activity. The stimulation of activity followed Michaelis Menten kinetics with a Ks for aldolase stimulation of 0.017 units of aldolase and a Vmax of 1.11 μmol Pi min−1 mg−1 protein. To determine if the regulation by aldolase of the V-ATPase was unique to M. crystallinum tonoplast or a phenomenon also present in other plants, we isolated tonoplast from leaves of pineapple (Ananas comosus) and measured V-ATPase activity in the presence of aldolase. Similar to values obtained for M. crystallinum, aldolase stimulation of V-ATPase hydrolytic activity gave a Ks of 0.015 units of aldolase and a Vmax of 1.31 μmol Pi mg−1 protein min−1 (see Supplemental Figure 1 online), demonstrating that aldolase regulation was not species specific.
Figure 6.
Aldolase Stimulates V-ATPase Hydrolytic Activity by Increasing Affinity for ATP.
(A) V-ATPase hydrolytic activity (bafilomycin-sensitive and azide- and vanadate-insensitive) was estimated by spectrophotometric measurement of inorganic phosphate release as described in Methods. Activity was measured in tonoplast vesicles (15 μg protein) isolated from M. crystallinum over a range of aldolase concentrations. Data represent means ± se of three replicate experiments. Each replicate experiment was performed using independent membrane preparations. The solid lines show the fit of the kinetic data with the Michaelis-Menten equation, and from this the rate constants Ks and Vmax were calculated. Ks refers to the concentration of aldolase that gives half the maximal velocity, and Vmax refers to the velocity of the enzyme catalyzed reaction at saturating aldolase concentrations. The χ2 value indicates the goodness of fit and confirmed that the data fitted the equation at a probability level of at least P < 0.01.
(B) V-ATPase hydrolytic activity was measured over a range of ATP concentrations in the presence of increasing amounts of aldolase. Data represent means ± se of three replicate experiments performed using independent membrane preparations. The solid lines show the fit of the data with the Michaelis-Menten equation. Units for Vmax are μmol Pi mg−1 protein min−1. The χ2 values, indicating the goodness of fit of the data to the equation, gave probabilities of at least P < 0.05.
[See online article for color version of this figure.]
In order to understand the mechanism underlying aldolase stimulation of the V-ATPase, we measured hydrolytic activity in the presence of increasing ATP concentrations at different concentrations of aldolase to determine the effect on the kinetic properties of the V-ATPase enzyme. As aldolase concentration was increased, there was a concomitant decrease in the Km value for ATP with an increase in Vmax, indicating an increased affinity for ATP in the presence of aldolase and an allosteric regulation of V-ATPase by the glycolytic enzyme (Figure 6B).
The ability of enolase to stimulate in vitro V-ATPase hydrolytic activity was also investigated. The addition of increasing concentrations of purified yeast enolase did not result in a change in V-ATPase hydrolytic activity (see Supplemental Figure 2A online). Enolase (0.04 units) was also added in the presence of aldolase (0.03 units) to determine if there was a synergistic or additive effect of the two glycolytic enzymes (see Supplemental Figure 2B online); however, the resulting stimulation of V-ATPase activity was similar to levels obtained in the presence of aldolase alone (Figure 6A). The absence of an effect of yeast enolase on the stimulation of the M. crystallinum V-ATPase activity may be attributed to differences between the yeast enzyme and plant enolases (only 54% similarity or less between the sequences). Previously, it has been shown that structural differences between yeast and a rabbit enolase resulted in the inability of the yeast enzyme to substitute for the rabbit enzyme in stimulation of immunoglobulin production (Sugahara et al., 1998) and highlights the need to use plant-specific enzymes.
Arabidopsis Enolase Mutants Are Salt Sensitive, Have Reduced Levels of Enolase at the Tonoplast, and Show a Reduction in Aldolase Stimulated V-ATPase Activity
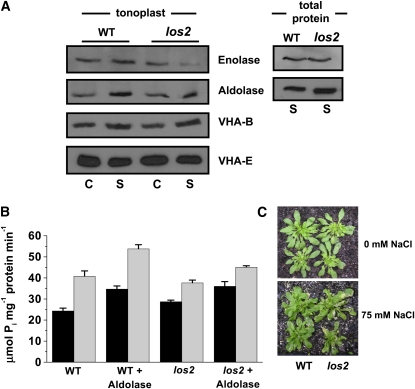
Similar to results obtained using M. crystallinum, Arabidopsis shows tonoplast glycolytic enzyme association that is upregulated by salinity (Figure 7A, WT), and the addition of aldolase is observed to stimulate in vitro the hydrolytic activity of the V-ATPase (Figure 7B, WT), suggesting it is an appropriate model for further studies on the in vivo role of the salinity-induced association of glycolytic enzymes with the tonoplast and their regulation of the V-ATPase. To do this, we obtained an Arabidopsis enolase mutant, los2, which has been shown to have severely reduced cytoplasmic enolase activity and a reduction in enolase transcript specifically under cold stress, as well as alterations in cold-responsive gene expression, which suggested another role for enolase as a cold-specific transcriptional repressor (Lee et al., 2002).
Figure 7.
The los2 Arabidopsis Enolase Mutant Shows a Salinity-Dependent Reduction in Enolase Abundance at the Tonoplast and a Salinity-Dependent Reduction in Aldolase Stimulation of V-ATPase Hydrolytic Activity and Is Salt Sensitive.
(A) Immunodetection of enolase, aldolase, and VHA subunits in tonoplast (left) and enolase and aldolase in total protein fractions (right), isolated from wild-type (Col-0) and los2 Arabidopsis plants grown in the absence (C) or presence (S) of 75 mM NaCl for 4 d as indicated. Blots are representative of three independent experiments.
(B) V-ATPase hydrolytic activity in the presence or absence of 0.03 units of aldolase was estimated by spectrophotometric measurement of inorganic phosphate release as described in Methods. Activity was measured in tonoplast vesicles (15 ug protein) isolated from wild-type (Col-0) or los2 plants grown in the absence (black bars) or presence (thatched bars) of 75 mM NaCl for 4 d. Data represent means ± se of three replicate experiments.
(C) Response of wild-type (Col-0) and los2 plants to salinity. Plants were grown in the absence (top) or presence (bottom) of 75 mM NaCl for 4 d. Visual phenotype of leaves is shown with noticeable wilting and chlorotic lesions on the mutant plant.
In this study, under control growth conditions, both wild-type and los2 plants showed similar levels of enolase associated with the tonoplast (Figure 7A, left panel). However, in membranes isolated from salt-stressed los2 plants, there was a noticeable reduction in enolase protein levels at the tonoplast (Figure 7A, left panel). At the same time, there was no apparent reduction of the enzyme in total protein extracts from salt-treated plants (Figure 7A, right panel). We also investigated the protein levels of aldolase and several of the VHA subunits in the los2 enolase mutant (Figure 7A). There was no change in abundance of aldolase in total protein extracts from wild-type and los2 control or salt-treated plants, and the salt-induced accumulation of the protein was maintained in tonoplast fractions as well. As expected for membrane-associated proteins, no VHA subunits were detected in the total protein extracts (data not shown), whereas in tonoplast fractions, VHA-B maintained its salt regulation, while there were no detectable changes in VHA-E (Figure 7A).
This specific reduction in enolase protein at the tonoplast appeared to affect directly the ability of exogenous aldolase to stimulate V-ATPase activity from salt-treated los2 mutant plants. In the salt-treated los2 mutant plants, V-ATPase stimulation by aldolase was significantly less than that measured in wild-type salt-treated plants (45.15 μmol Pi mg−1 protein min−1 and 53.84 μmol Pi mg−1 protein min−1, respectively; P < 0.05, Figure 7B). Moreover, salt-treated los2 enolase mutant plants were considerably more salt sensitive than wild-type salt-treated plants (Figure 7C), showing severe wilting and chlorotic lesions on leaves.
DISCUSSION
The identification of aldolase and enolase as associated with the plant tonoplast is not as surprising as it may first appear, and it may be that we have to rethink our fundamental view that soluble glycolytic enzymes diffuse freely within the cytoplasmic volume. Increasingly, proteins involved in glycolysis are being assigned to membrane fractions in organisms ranging from mammals to yeast, as well as in plants. It is argued that binding to different organelles/membranes may concentrate glycolytic complexes in regions of high demand for ATP or pyruvate, directly channeling these substrates to specific transporters or proton pumps by forming functionally compartmentalized energy networks (Dhar- Chowdhury et al., 2007), and this glycolysis-derived ATP is preferentially used to drive rapid biological processes, including membrane transporters (Ikemoto et al., 2003). In plants, an Arabidopsis mitochondrial proteomics study identified the presence of seven glycolytic enzymes, including aldolase and enolase, associated with the outer mitochondrial membrane (Giegé et al., 2003), and mitochondrial membrane-associated enzyme activities for all 10 of the glycolytic enzymes were confirmed (Giegé et al., 2003). The mitochondrial association of the enzymes increased with higher respiratory demand, and pull-down experiments suggested protein interactions with the outer membrane channel VDAC, which anchors the glycolytic complex to the mitochondrial surface via direct and strong interaction with aldolase (Graham et al., 2007).
Evidence for tonoplast localization of glycolytic enzymes in plants has come from a number of independent proteomic studies, although it has been widely ignored as having no functional significance, being attributed to contaminating fractions (Carter et al., 2004) or characterized as soluble cytoplasmic proteins that have no role at the tonoplast (Schmidt et al., 2007; Endler et al., 2009). Triosephosphate isomerase was identified in a proteomic study of barley (Hordeum vulgare) tonoplast (Endler et al., 2006), as well as in the vegetative vacuole proteome of Arabidopsis of both whole vacuoles and tonoplast (Carter et al., 2004). The latter study also identified hexokinase in the tonoplast fraction (Carter et al., 2004). Glyceraldehyde 3 phosphate dehydrogenase was present in a proteomic study of vacuoles purified from cauliflower (Brassica oleracea) buds (Schmidt et al., 2007) and also in a barley tonoplast phosphoproteomic study (Endler et al., 2009). Enolase was identified as a protein in highly purified vacuoles from Arabidopsis cell suspensions (Shimaoka et al., 2004). Aldolase has been identified in several studies, including a quantitative proteomics analysis of rice (Oryza sativa) root tonoplast proteins induced by gibberellin treatment (Tanaka et al., 2004), in the proteome of vacuoles from cauliflower buds (Schmidt et al., 2007), and as a phosphopeptide in barley tonoplast (Endler et al., 2009). Jaquinod et al. (2007), in a proteomic study of vacuoles from Arabidopsis cell suspensions, identified five glycolytic enzymes associated with the chaotrope-treated membrane fraction, namely, hexokinase, glyceraldehyde-3-phosphate dehydrogenase, phosphoglycerate kinase, aldolase, and triosephosphate isomerase. In specific studies, the glycolytic proteins were associated with the membrane fractions following chaotrope treatment (Schmidt et al., 2007).
The increased abundance and activity of aldolase and enolase in the tonoplast in M. crystallinum under conditions of salt stress (Figure 3) may provide a means for localized increases in ATP generation via glycolysis to meet demands of the V-ATPase for increased proton-driven transport, which would in turn facilitate the accumulation of Na+ into the vacuole. Evidence that aldolase and enolase are interacting with the tonoplast through direct association with V-ATPase subunits, specifically the regulatory subunit VHA-B, supports this view (Figure 5) and is not unprecedented. In both bovine kidney and yeast, aldolase has been shown to interact with the VHA-B of the V-ATPase (Lu et al., 2001, 2004), with the possible involvement of other subunits including VHA-E. Furthermore, this interaction modulates V-ATPase activity and assembly (Lu et al., 2007). Yeast aldolase mutants that maintained enzyme catalytic activity but were impaired in binding to VHA-B resulted in decreased V-ATPase activity (Lu et al., 2007). In yeast, the physiological significance of this association has not been determined, and studies have yet to decipher under what conditions the interaction takes place. In plants, an association between aldolase and V-ATPase subunits was noted in rice roots following the identification of aldolase in the tonoplast fraction (Tanaka et al., 2004); VHA-A, VHA-B, and VHA-a coimmunoprecipitated with aldolase, but regulation of activity was not demonstrated (Konishi et al., 2004, 2005), and physiological relevance was also lacking for this association.
In this study, increases in both aldolase and enolase protein at the tonoplast are paralleled by increases in amount of the VHA-B subunit, but not by changes in abundance of either VHA-E or VHA-A (Figure 3). Similar nonstoichiometric changes in subunit abundance have been reported in yeast and in mammalian renal tissue, where it is thought that changes in specific subunits of the protein complex can regulate assembly and/or coupling efficiency (Valles et al., 2005; Cipriano et al., 2008), without changing the final subunit stoichiometry. In plants, noncoordinated regulation of VHA subunits in tonoplast fractions has been suggested to be important for stress tolerance (Dietz et al., 2001), although how this is achieved is not addressed. An increase in VHA-B and not VHA-A could also indicate a previously unknown, V-ATPase–independent function for a particular VHA-B isoform, of which there are three expressed in plants (Sze et al., 2002). Cho et al., (2006) showed VHA-B1 interactions with hexokinase and transcription factors in the nucleus were involved in glucose sensing, which was independent of its function in V-ATPase vacuole acidification. However, in our study, an unconventional role for the VHA-B subunit appears less likely, as aldolase is shown to have direct effects on modulating the activity of the V-ATPase on the tonoplast and may have a role in regulation of salt accumulation, particularly as the association is induced by salinity. Hydrolytic activity in the presence of aldolase was increased in a concentration-dependent manner and was reflected as an increase in the affinity of the proton pump for ATP (Figure 6). This possible steric effect may be due to the association of aldolase with the VHA-B subunit (Figure 5) and seems to imply that the presence of the glycolytic enzymes on the tonoplast has an additional, more direct role to that of merely a localized supply of ATP. Further work will need to be performed to identify the isoform(s) of the VHA-B that is involved in the interaction, as antibodies used in this study were not isoform specific.
The Arabidopsis enolase mutant los2 provides more direct evidence that the association of glycolytic enzymes with the tonoplast, and in particular the regulation of the V-ATPase, plays a role in plant salinity tolerance (Figure 7). Enolase mutant plants show a salinity-induced, tonoplast-specific reduction in enolase protein that is not observed in whole-cell protein extracts or in plants that are grown under control conditions. This decreased abundance of enolase at the tonoplast results in a reduction in the ability of aldolase to stimulate V-ATPase activity in vitro, suggesting that a protein complex comprising aldolase, subunits of the V-ATPase, and enolase may be required for successful regulation. Moreover, the los2 plants show a salt-sensitive phenotype, highlighting the importance of this association in plant salt tolerance.
Interestingly, in a meta-analysis of 2D proteomics data, enolase is one of the top 15 identified differentially expressed proteins in human quantitative proteomic studies (Petrak et al., 2008), raising the concern that its frequent identification results from a technical artifact. However, a reexamination of compiled transcriptomic data confirmed that enolase is also frequently identified as a differentially expressed transcript, and the observed transcriptional changes for the gene correlated surprisingly well with the frequencies calculated in the proteomic meta-analysis (Petrak et al., 2008). This finding, along with the growing evidence that enolase is a multifunctional protein across species, implies that it may play a regulatory or sensor role in multiple stress situations in diverse cellular locations. This would fit well with a role in salt tolerance as suggested by our study.
METHODS
Plant Materials and Growth Conditions
Mesembryanthemum crystallinum plants were grown from seed in soil (MetroMix 500; Sun Gro Horticulture) in a propagation tray. Three weeks following germination, individual seedlings were transplanted to pots containing the soil mixture, with two plants per 15-cm-diameter pot. Plants were watered daily and one-half strength Hoagland medium (Hoagland and Arnon, 1938) was supplied weekly. NaCl (200 mM) or mannitol (400 mM) treatment was initiated 6 weeks after germination for a period of 7 d. For cold treatment, plants were placed at 4°C under a normal photoperiod for a period of 4 d.
Arabidopsis thaliana wild-type (Columbia-0 [Col-0]) and los2 (enolase mutant) (Lee et al., 2002) plants were grown from seed in soil in a propagation tray and then transplanted to hydroponic trays containing one-half strength Hoagland medium at 3 weeks (Hoagland and Arnon, 1938). NaCl-treatment (75 mM for 4 d) was initiated 8 weeks after germination. Ananas comosus was propagated vegetatively from the crowns of commercially obtained fruits. All plants were grown in a glasshouse under natural irradiation and photoperiod. Temperature was maintained at 25°C ± 3°C.
The enolase (los2) mutant seeds were generously provided by J.-K. Zhu (University of California, Riverside) and confirmed by PCR sequencing analysis. Primers derived from the LOS2 sequence (LP, 5′-CCAACTCCTCCTCAATACGCAA-3′; and RP, 5′-GTTGGTGATGAAGGTGGGTTTG-3′) were used to amplify the corresponding region from genomic DNA of the wild type and los2. Sequence analysis of two independent PCR products of Col-0 and los2 detected a single G-to-A substitution at position 326 in the los2 enolase mutant as previously described (Lee et al., 2002).
Microsomal Membrane Isolation
Leaf material (30 g) from M. crystallinum, A. comosus, and Arabidopsis was harvested and sliced into small pieces (following the removal of major veins for M. crystallinum). Tissue was placed directly into 300 mL of ice-cold homogenization medium in a prechilled Waring blender. All subsequent operations were performed at 4°C. The homogenization medium consisted of 400 mM mannitol, 10% (w/v) glycerol, 5% (w/v) PVP-10, 0.5% (w/v) BSA, 1 mM PMSF, 30 mM Tris, 2 mM DTT, 5 mM EGTA, 5 mM MgSO4, 0.5 mM butylated hydroxytoluene, 0.25 mM dibucaine, 1 mM benzamidine, and 26 mM K+-metabisulfite, adjusted to pH 8.0 with H2SO4. Microsomal membranes and tonoplast were isolated as previously described (Barkla et al., 1995). Membranes were frozen directly in liquid N2 and stored at −80°C.
Purification of Tonoplast by FFZE
Microsomal membranes were fractionated by FFZE using the BD FFE system (BD Proteomics). Prior to fractionation, the microsomal sample was diluted 2:1 (v:v) in separation medium (10 mM triethanol amine [TEA], 10 mM acetic acid, 2 mM KCl, and 250 mM sucrose) and centrifuged at 14,000g for 20 min at 4°C. The sample (3 mg/mL protein) was injected continuously via a peristaltic pump at a rate of 1.2 mL/h using the anodic sample inlet. The media inlets of the chamber had the following buffer compositions: inlets 2 to 6, separation medium (10 mM TEA, 10 mM acetic acid, 2 mM KCl, and 250 mM sucrose); inlets 1 and 7, stabilization medium (40 mM TEA, 40 mM acetic acid, 8 mM KCl, and 180 mM sucrose). The cathodic and anodic circuit electrolyte solutions consisted of 100 mM TEA, 100 mM acetic acid, and 20 mM KCl adjusted to pH 7.4 with NaOH, with 0.4% formaldehyde added to the anodic solution to prevent loss of chloride by anodic oxidation. The counter flow medium for inlets C1, C2, and C3 was the same as the separation medium.
FFZE was performed in horizontal mode at a constant voltage of 750 V (118 mA), with a media and counter flow rate of 250 mL/h. The temperature during the run was maintained at 5°C by the continual flow of coolant, below the glass separation plate, from a circulating water bath. Following separation in the chamber, membrane fractions were collected continually in 96 deep well microtiter plates (4 mL/well; Sunergia Medical). Fractions from sequential runs were pooled and membranes concentrated by centrifugation in a Beckman 55.2 Ti rotor in an L8-M ultracentrifuge at 100,000g for 50 min at 4°C. Membrane pellets were resuspended in 50 to 100 μL of suspension buffer containing 250 mM mannitol, 10% glycerol (w/v), 10 mM Tris/MES, pH 8.0, and 2 mM DTT and frozen in liquid N2 for storage at −80°C. Separation by FFZE was monitored by collecting microtiter plates (250 μL/well) at several time points during the run and measuring protein (O.D.280) using a microplate scanning spectrophotometer (Power WaveX; Bio-Tek Instruments).
Protein and Chlorophyll Determination in Samples
Protein in microsomal and FFZE fractions was measured by a modification of the Bradford method (Bradford, 1976), in which membrane protein was partially solubilized with 0.5% (v/v) Triton X-100 for 5 min before dilution and the addition of the dye reagent concentrate; the final concentration of Triton X-100 in the assay was 0.015%. Protein in samples prepared for 2D-DIGE analysis was measured by the RCDC protein assay kit (Bio-Rad) according to the manufacturer's instructions. BSA was employed as the protein standard.
Chlorophyll in FFZE fractions was measured spectrophotometrically according to the method of Arnon (1949) using a microplate scanning spectrophotometer (Power WaveX). The absorbance was measured at 645 and 663 nm, and calculations were made according to the following equations. Chlorophyll a (μg/mL) = 12.7 (A663) to 2.69 (A645). Chlorophyll b (μg/mL) = 22.9 (A645) to 4.68 (A663).
SDS-PAGE, Staining, and Immunoblotting
FFZE fractions and tonoplast samples were precipitated by dilution of the samples 50-fold in 1:1 (v/v) ethanol/acetone and incubated overnight at −30°C according to the method of Parry et al. (1989). Samples were then centrifuged at 13,000g for 20 min at 4°C using an F2402 rotor in a GS-15R table-top centrifuge (Beckman). Pellets were air dried, resuspended with sample buffer (2.5% SDS), and heated at 60°C for 2 min before loading (15 μg of protein per lane) onto 10% (w/v) linear acrylamide extra wide mini-gels (Scie-Plas). After electrophoresis, SDS-PAGE separated proteins were either fixed and stained with Coomassie Brilliant Blue R 250 or electrophoretically transferred onto nitrocellulose membranes (ECL; GE Lifesciences) for immunoblot analysis, as previously described (Vera-Estrella et al., 2004). Digital images were captured using a Hewlett Packard flatbed scanner (Scan Jet 8250). Primary antibodies and dilutions used in this study are as follows: castor bean (Ricinus communis) anti-aldolase (1/2000) (Hodgson and Plaxton, 1998); human anti-enolase (1/1000) (purchased from Santa Cruz Biotechnology); Kalanchoe anti-VHA-B (1/1000) (Long et al., 1995); Kalanchoe anti-VHA-A (1/1000) (Long et al., 1995); anti-VHA-E (1/2000) (purchased from Agrisera; Reuveni et al., 2001); M. crystallinum anti-TIP1;2 (1/2000) (Kirch et al., 2000); Arabidopsis anti-AHA3 (1/2000) (Parets-Soler et al., 1990); M. crystallinum anti-HKT1 (1/2000) (Su et al., 2003); M. crystallinum anti-CRT1 (1/2000) (Nelson et al., 1997); anti-VDAC1 (1/2000) (purchased from Agrisera; Clausen et al., 2004); and Zea mays anti-RCA (Vargas-Suárez et al., 2004). With the exception of anti-TIP1;2, the antibodies used did not distinguish between specific isoforms of the various proteins detected.
Sample Preparation and CyDye Fluorescent Labeling
FFZE fractions (33 to 37), representing pure tonoplast, were pooled, and 75 μg protein was then desalted/cleaned with the ReadyPrep 2D Cleanup kit (Bio-Rad) according to the manufacturer's instructions. The final protein pellet was resuspended in labeling buffer (30 mM Tris-HCl, pH 8.5, 7 M urea, 2 M thiourea, 2% CHAPS, and 2% amidosulfobetaine-14. Samples were labeled with the appropriate CyDye (Cy2, Cy3, or Cy5) according to the three dye strategy for minimal labeling as instructed by the manufacturer (GE Lifesciences). Briefly, 300 pmol dye/50 μg protein was added and samples were incubated in the dark on ice for 30 min. The labeling reaction was stopped by the addition of 1 μL of 10 mM lysine and incubated on ice for a further 10 min. This was repeated for control and treated samples from all three independent experiments as described in the experimental design (Table 1). Dye swapping between experimental samples was performed to control for any dye-specific artifacts resulting from preferential labeling or variable fluorescence characteristics of the gel or glass plates at the different excitation wavelengths of Cy2, Cy3, and Cy5. Following labeling, equal volumes of rehydration buffer containing 2× DTT and ampholytes was added to each reaction (7 M urea, 2 M thiourea, 2% amidosulfobetaine-14, 2% CHAPS, 100 mM DTT, and 1% Bio-Lyte 3-10 ampholytes [Bio-Rad]), giving a final concentration of 50 mM DTT and 0.5% Bio-Lyte 3-10 ampholytes. The three different CyDye samples for each gel were pooled and brought to a final volume of 300 μL with 1× rehydration buffer.
2D Gel Electrophoresis, Gel Imaging, and Image Analysis
Ready Strip IPG strips (17 cm, linear pH 3 to 10; Bio-Rad) were layered gel side down onto samples placed in the Protean IEF tray (Bio-Rad). Strips were covered with 2 mL of mineral oil, and active rehydration was performed overnight in a Protean IEF Cell (Bio-Rad) at 50 V and 20°C. Following strip rehydration, isoelectric focusing (IEF) was performed with a three-step ramping protocol for a total of 40,000 V/h at 20°C and a maximum current setting of 50 μA per strip. After IEF, the IPG strips were first equilibrated for 15 min in DTT equilibration buffer (6 M urea, 0.375 M Tris-HCl, pH 8.8, 2% SDS, 20% glycerol, and 2% DTT) on a shaker, followed by an additional 15 min in iodoacetamide equilibration buffer (6 M urea, 0.375 M Tris-HCl, pH 8.8, 2% SDS, 20% glycerol, and 2.5% iodoacetamide). IEF gel strips were loaded onto 10% acrylamide gels cast between low fluorescence glass plates coated on one side with bind silane solution (80% ethanol, 2% glacial acetic acid, and 0.001% bind silane). Two reference markers were included on the glass plate that contained the bind silane to facilitate robotic spot picking. Strips were overlayed with 0.5% low melting point agarose in SDS running buffer (25 mM Tris-HCl, pH 8.3, 192 mM glycine, 0.1% SDS, and bromophenol blue), and SDS-PAGE was performed using the Ettan Daltsix electrophoresis system (GE Lifesciences) at 10 mA/gel for 1 h followed by 12 mA/gel for a total of 17 to 20 h at 25°C.
Individual gels were scanned three times using a Typhoon Variable Mode TM 9410 imager (GE Lifesciences) to obtain the images for each of the three CyDyes according to the acquisition conditions outlined in Supplemental Table 2 online. Image analysis was performed using the DeCyder 2D Software V6.5 following the manufacturer's instructions (GE Lifesciences) and as described (Casasoli et al., 2007).
Spot Picking and Protein Identification by Electrospray Ionization LTQ-Orbitrap MS
Protein spots of interest were excised from the gels using the Ettan Spot Picker robot (GE Lifesciences) and the spot pick map generated by the Decyder software, according to the instrument handbook and user manual. Digestion of protein was performed according to the in-gel method (Kinter and Sherman, 2000) with some modifications. Excised gel spots were dehydrated in 100 μL of acetonitrile (ACN) for 5 min at room temperature, the supernatant was removed, and gel fragments were air dried. Samples were rehydrated in 20 μL of trypsin (25 ng/μL) until the solution was absorbed (10 min). Ammonium bicarbonate (BICAM; 50 mM) was added to completely cover the rehydrated gel, and the sample was incubated overnight at 37°C. Following overnight incubation, an additional 20 μL of 50 mM BICAM was added to the sample and incubated at room temperature for 10 min. The sample was then centrifuged, and the extract was transferred into an empty 0.5-mL eppendorf tube. The gel spot was reextracted with 20 μL of extraction solution (5% formic acid in 50% ACN) and incubated for 10 min at room temperature. The sample was then vortexed and centrifuged for 30 s. The supernatant was collected without disturbing the gel piece and combined with the previous extract. Protein digests were desalted by solid-phase extraction by means of a modified protocol for C18-ZipTips from Millipore. For peptide binding and washing, the sample was aspirated 60 times, and peptides were eluted in 15 μL of 1% formic acid in 50% ACN.
Peptide mixtures were analyzed by nano-liquid chromatography–MS/MS using a Finnigan MicroAS autosampler and a Surveyor MS pump system coupled to an LTQ-Orbitrap (Thermo). Forty microliters of each mixture was loaded on a C18 precolumn (Symmetry300 C18 5 μm; NanoEase Trap Column, Waters) at 3 μL/min for 15 min in 0.1% formic acid in 5% ACN. Peptides were eluted using a 5 to 35% gradient of solvent B (0.1% formic acid in 100% ACN) during 60 min at a flow rate of 300 nL/min with a BioBasic C18 picofrit column (PFC7515/BI/10; New Objective). Data-dependent acquisition mode was performed with the Xcalibur software. A fourier transformed full scan was acquired by means of the Orbitrap, from 300 to 1800 m/z with resolving power set at 30,000 (400 m/z). The five most intense peaks were sequentially isolated for the MS/MS experiments in the linear ion trap (LTQ) using collisionally induced dissociation. To prevent duplication of MS/MS data for the same peptide, dynamic exclusion was set to 2, and selected ions were placed on the exclusion list for 45 s. The MS/MS raw spectra data were converted to DTA files using ThermoElectron Bioworks 3.2 and analyzed by means of Turbo SEQUEST (ThermoFisher Scientific). From a general Uniprot100 database, two decoy databases were generated for M. crystallinum and Arabidopsis. Independent searching was performed using rigorous parameters (Xcorr z = 1: 1.90, z = 2: 2.70, z = 3: 3.50, z = 4: 3.75 and Delta Cn >0.1). The search parameters were set for dynamic modifications for Cys alkylation with iodoacetamide and Met oxidation.
Glycolytic Enzyme Activity
Aldolase and enolase activities were measured spectrophotometrically by monitoring the initial rate of NADH utilization at 340 nm according to the coupled methods as described by Hodgson and Plaxton (1998) for aldolase and Hüther et al. (1990) for enolase, using a Hewlett Packard 8452A Diode-array spectrophotometer. The rate of NADH oxidation in the presence of 30 μg total protein was calculated using the NADH molar extinction coefficient of 6220 mol cm−1 and expressed as μmol mg−1 protein min−1.
Immunoprecipitation
Tonoplast protein (30 μg) was incubated with 500 μL of NET-gel buffer (50 mM Tris/HCl, pH 7.5, 150 mM NaCl, 0.1% NP-40, 1 mM EDTA, and 0.25% gelatin) containing the appropriate antibodies at the above mentioned dilutions for 1 h at 4°C on a rocking table. Protein A Sepharose CL-4B (20 μL; GE Lifesciences) was added and the slurry rocked for a further 2 h at 4°C. Samples were centrifuged at 12,000g for 30 s, and the pellet was washed twice with NET-gel buffer and once with wash buffer (10 mM Tris/HCl, pH 7.5, and 0.1% Nonidet P-40). The pellet was resuspended with 20 μL of 2.5% Laemmli sample buffer and heated at 95°C for 3 min. Samples were centrifuged at 10,000g prior to loading onto a 10% linear acrylamide gel for SDS-PAGE and subsequent protein blotting as described.
V-ATPase Hydrolytic Activity
Hydrolytic activity of V-ATPase was measured by the release of Pi according to the method of Ames (1966), as previously described (Vera-Estrella et al., 2004). Tonoplast protein (15 μg) was incubated in 300 μL of solution containing 50 mM KCl, 1 mM sodium molybdate, 3 mM Tris/ATP, 6 mM MgSO4, and 30 mM Tris/MES, pH 8.0. Activity was measured in the presence or absence of 100 nM bafilomycin, 10 mM azide, and 100 μM vanadate to inhibit the activities of the V-ATPase, F-ATPase, and the P-ATPase, respectively, when necessary. Lyopholized and purified spinach (Spinacia oleracea) aldolase (E.C. 4.1.2.13) and rabbit enolase (E.C. 4.2.1.11), purchased from Sigma-Aldrich, were resuspended in sterile milli-Q water, and the indicated units were added to the assay. The reaction medium was incubated at 37°C for 30 min and stopped by adding Ames solution (1 volume of 10% ascorbic acid to 6 volumes of 0.42% ammonium molybdate in H2SO4), and the A820 was measured using a Hewlett Packard 8452A diode array spectrophotometer. Bafilomycin-sensitive and vanadate- and azide-resistant activity is presented. The values were calculated and represented as μmol of Pi released mg−1 protein min−1.
Chaotrope Treatment of Tonoplast
Purified tonoplast (200 μg) was incubated for 30 min at 4°C in suspension buffer containing 250 mM mannitol, 10% glycerol (w/v), 10 mM Tris/MES, pH 8.0, and 2 mM DTT in the presence or absence of 200 mM Na2CO3, pH 11.4, supplemented with 3 mM MgSO4 and 3 mM BTP-ATP. Following incubation, membranes were concentrated by centrifugation in a Beckman 55.2 Ti rotor in an L8-M ultracentrifuge at 100,000g for 50 min at 4°C.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: LOS2, At2g36530; ALD, X65742; and ENO1, J01322 (see additional accession numbers in Table 2).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Aldolase Stimulates V-ATPase Hydrolytic Activity in Ananas comosus.
Supplemental Figure 2. Yeast Enolase Does Not Stimulate V-ATPase Hydrolytic Activity in Vitro.
Supplemental Table 1. Peptide Precursor Mass and Mass Accuracy of Identified Peptides.
Supplemental Table 2. Gel Scanning Parameters Used for the Typhoon Scanner.
Supplemental Data Set 1. Average Spot Ratios from DIGE Analysis.
Supplementary Material
Acknowledgments
Mass spectra were obtained at the INMEGEN Medical Proteomics Facility with the assistance of Jose Luis Gallegos Pérez. We thank Victoria Pando (Instituto de Salud Pública) for guidance with the DIGE method and Susana López (Instituto de Biotecnología) for access to the Typhoon imager. The gifts of the anti-aldolase antibody from Bill Plaxton (Queens University, Kingston, Canada), the anti-AHA3 antibody from Ramon Serrano (Universidad Politécnica de Valencia-Consejo Superior de Investigaciones Científicas, Spain), anti-RCA from Estela Sánchez-de-Jiménez (Universidad Nacional Autónoma de México), and the los2 enolase mutant seeds from Jian-Kang Zhu (University of California, Riverside, CA) are acknowledged. This work was supported by Consejo Nacional de Ciencia y Tecnología Grants 49735 to B.J.B. and 57685 to R.V.-E. and by Dirección General de Asuntos del Personal Académico IN221308.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Bronwyn J. Barkla (bronwyn@ibt.unam.mx).
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Alban, A., David, S.O., Bjorkesten, L., and Andersson, C. (2003). A novel experimental design for comparative two-dimensional gel analysis: Two-dimensional difference gel electrophoresis incorporating a pooled internal standard. Proteomics 3 36–44. [DOI] [PubMed] [Google Scholar]
- Ames, B.N. (1966). Assay of inorganic phosphate, total phosphate and phosphatases. Methods Enzymol. 8 115–118. [Google Scholar]
- Apse, M.P., Sottosanto, J.B., and Blumwald, E. (2003). Vacuolar cation/H+ exchange, ion homeostasis, and leaf development are altered in a T-DNA insertional mutant of AtNHX1, the Arabidopsis vacuolar Na+/H+ antiporter. Plant J. 36 229–239. [DOI] [PubMed] [Google Scholar]
- Arnon, D.I. (1949). Copper enzymes in isolated chloroplasts: polyphenol oxidase in Beta vulgaris. Plant Physiol. 24 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkla, B.J., Vera-Estrella, R., and Pantoja, O. (2007). Enhanced separation of membranes during free flow zonal electrophoresis in plants. Anal. Chem. 79 5181–5187. [DOI] [PubMed] [Google Scholar]
- Barkla, B.J., Zingarelli, L., Blumwald, E., and Smith, J.A.C. (1995). Tonoplast Na+/H+ antiport activity and its energization by the vacuolar H+-ATPase in the halophyte Mesembryanthemum crystallinum. Plant Physiol. 109 549–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batelli, G., Verslues, P.E., Agius, F., Qiu, Q., Fujii, H., Pan, S.Q., Schumaker, K., Grillo, S., and Zhu, J.-K. (2007). SOS2 promotes salt tolerance in part by interacting with the vacuolar H+-ATPase and upregulating its transport activity. Mol. Cell. Biol. 27 7781–7790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford, M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72 248–254. [DOI] [PubMed] [Google Scholar]
- Carter, C., Pan, S., Zouhar, J., Avila, E.L., Girke, T., and Raikhel, N.V. (2004). The vegetative vacuole proteome of Arabidopsis thaliana reveals predicted and unexpected proteins. Plant Cell 16 3285–3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casasoli, M., Meliciani, I., Cervone, F., De Lorenzo, G., and Mattei, B. (2007). Oligogalacturonide-induced changes in the nuclear proteome of Arabidopsis thaliana. Int. J. Mass Spectrom. 268 277–283. [Google Scholar]
- Cho, Y.-H., Yoo, S.-D., and Sheen, J. (2006). Regulatory functions of Nuclear Hexokinase1 complex in glucose signaling. Cell 127 579–589. [DOI] [PubMed] [Google Scholar]
- Cipriano, D.J., Wang, Y., Bond, S., Hinton, A., Jefferies, K.C., Qi, J., and Forgac, M. (2008). Structure and regulation of the vacuolar ATPases. Biochim. Biophys. Acta 1777 599–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen, C., Ilkavets, I., Thomson, R., Philippar, K., Vojta, A., Möhlmann, T., Neuhaus, E., Fulgosi, H., and Soll, J. (2004). Intracellular localization of VDAC proteins in plants. Planta 220 30–37. [DOI] [PubMed] [Google Scholar]
- Dannelly, H.C., Cortay, J.-C., Cozzone, A.J., and Reeves, H.C. (1989). Identification of phosphoserine in in vivo-labeled enolase from Escherichia coli. Curr. Microbiol. 19 237–240. [Google Scholar]
- Decker, B.L., and Wickner, W.T. (2006). Enolase activates homotypic vacuole fusion and protein transport to the vacuole in yeast. J. Biol. Chem. 281 14523–14528. [DOI] [PubMed] [Google Scholar]
- Dhar-Chowdhury, P., Malestera, B., Rajacica, P., and Coetzeea, W.A. (2007). The regulation of ion channels and transporters by glycolytically derived ATP. Cell. Mol. Life Sci. 64 3069–3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz, K.J., Tavakoli, N., Kluge, C., Mimura, T., Sharma, S.S., Harris, G.C., Chardonnens, A.N., and Golldack, D. (2001). Significance of the V-type ATPase for the adaptation to stressful growth conditions and its regulation on the molecular and biochemical level. J. Exp. Bot. 52 1969–1980. [DOI] [PubMed] [Google Scholar]
- Endler, A., Meyer, S., Schelbert, S., Schneider, T., Weschke, W., Peters, S.W., Keller, F., Baginsky, S., Martinoia, E., and Schmidt, U.G. (2006). Identification of a vacuolar sucrose transporter in barley and Arabidopsis mesophyll cells by a tonoplast proteomic approach. Plant Physiol. 141 196–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endler, A., Reiland, S., Gerrits, B., Schmidt, U.G., Baginsky, S., and Martinoia, E. (2009). In vivo phosphorylation sites of barley tonoplast proteins identified by a phosphoproteomic approach. Proteomics 9 310–321. [DOI] [PubMed] [Google Scholar]
- Forsthoefel, N., Cushman, M., and Cushman, J. (1995). Posttranscriptional and posttranslational control of enolase expression in the facultative Crassulacean acid metabolism plant Mesembryanthemum crystallinum L. Plant Physiol. 108 1185–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gancedo, C., and Flores, C.-L. (2008). Moonlighting proteins in yeasts. Microbiol. Mol. Biol. Rev. 72 197–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giegé, P., Heazlewood, J.L., Roessner-Tunali, U., Millar, A.H., Fernie, A.R., Leaver, C.J., and Sweetlove, L.J. (2003). Enzymes of glycolysis are functionally associated with the mitochondrion in Arabidopsis cells. Plant Cell 15 2140–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham, J.W.A., Williams, T.C.R., Morgan, M., Fernie, A.R., Ratcliffe, R.G., and Sweetlove, L.J. (2007). Glycolytic enzymes associate dynamically with mitochondria in response to respiratory demand and support substrate channeling. Plant Cell 19 3723–3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatefi, Y., and Hanstein, W.G. (1974). Destabilization of membranes with chaotropic ions. Methods Enzymol. 31 770–790. [DOI] [PubMed] [Google Scholar]
- Heidrich, H.G., and Hannig, K. (1989). Separation of cell populations by free-flow electrophoresis. Methods Enzymol. 171 513–531. [DOI] [PubMed] [Google Scholar]
- Henningsen, R., Gale, B.L., Straub, K.M., and DeNagel, D.C. (2002). Application of zwitterionic detergents to the solubilization of integral membrane proteins for two-dimensional gel electrophoresis and mass spectrometry. Proteomics 2 1479–1488. [DOI] [PubMed] [Google Scholar]
- Hoagland, D.R., and Arnon, D.I. (1938). The water culture method for growing plants without soil. Univ. Calif. Exp. Stn. Circ. 347 1–39. [Google Scholar]
- Hodgson, R.J., and Plaxton, W.C. (1998). Purification and characterization of cytosolic fructose-1,6-bisphosphate aldolase from endosperm of germinated castor oil seeds. Arch. Biochem. Biophys. 355 189–196. [DOI] [PubMed] [Google Scholar]
- Hong-Hermesdorf, A., Brüx, A., Grüber, A., Grüber, G., and Schumacher, K. (2006). A WNK kinase binds and phosphorylates V-ATPase subunit C. FEBS Lett. 580 932–939. [DOI] [PubMed] [Google Scholar]
- Hüther, F.-J., Psarros, N., and Duschner, H. (1990). Isolation, characterization, and inhibition kinetics of enolase from Streptococcus rattus FA-1. Infect. Immun. 58 1043–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikemoto, A., Bole, D., and Ueda, T. (2003). Glycolysis and glutamate accumulation into synaptic vesicles: Role of glyceraldehyde phosphate dehydrogenase and 3-phosphoglycerate kinase. J. Biol. Chem. 278 5929–5940. [DOI] [PubMed] [Google Scholar]
- Jaquinod, M., Villiers, F., Kieffer-Jaquinod, S., Hugouvieux, V., Bruley, C., Garin, J., and Bourguignon, J. (2007). A proteomics dissection of Arabidopsis thaliana vacuoles isolated from cell culture. Mol. Cell. Proteomics 6 394–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane, P.M., Yamashiro, C.T., and Stevens, T.H. (1989). Biochemical characterization of the yeast vacuolar H(+)-ATPase. J. Biol. Chem. 264 19236–19244. [PubMed] [Google Scholar]
- Kinter, M., and Sherman, N.E. (2000). Protein Sequencing and Identification Using Tandem Mass Spectrometry. (New York: Wiley-Interscience).
- Kirch, H.-H., Vera-Estrella, R., Golldack, D., Quigley, F., Michalowski, C.B., Barkla, B.J., and Bohnert, H.J. (2000). Expression of water channel proteins in Mesembryanthemum crystallinum. Plant Physiol. 123 111–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi, H., Maeshima, M., and Komatsu, S. (2005). Characterization of vacuolar membrane proteins changed in rice root treated with gibberellin. J. Proteome Res. 4 1775–1780. [DOI] [PubMed] [Google Scholar]
- Konishi, H., Yamane, H., Maeshima, M., and Komatsu, S. (2004). Characterization of fructose-biphosphate aldolase regulated by gibberllin in roots of rice seedling. Plant Mol. Biol. 56 839–848. [DOI] [PubMed] [Google Scholar]
- Krisch, R., Rakowski, K., and Ratajczak, R. (2000). Processing of V-ATPase Subunit B of Mesembryanthemum crystallinum L. is mediated in vitro by a protease and/or reactive oxygen species. J. Biol. Chem. 381 583–592. [DOI] [PubMed] [Google Scholar]
- Landolt-Marticorena, C., Kahr, W.H., Zawarinski, P., Correa, J., and Manolson, M.F. (1999). Substrate- and inhibitor-induced conformational changes in the yeast V-ATPase provide evidence for communication between the catalytic and proton-translocating sectors. J. Biol. Chem. 274 26057–26064. [DOI] [PubMed] [Google Scholar]
- Lee, H., Guo, Y., Ohta, M., Xiong, L., Stevenson, B., and Zhu, J.K. (2002). LOS2, a genetic locus required for cold-responsive gene transcription encodes a bi-functional enolase. EMBO J. 21 2692–2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, A.R., Williams, L.E., Nelson, S.J., and Hall, J.L. (1995). Localization of membrane pyrophosphatase activity in Ricinus communis seedlings. J. Plant Physiol. 146 629–638. [Google Scholar]
- Löw, R., Rockel, B., Kirsch, M., Ratajczak, R., Hörtensteiner, S., Martinoia, E., Lüttge, U., and Rausch, T. (1996). Early salt stress effects on the differential expression of vacuolar H+-ATPase genes in roots and leaves of Mesembryanthemum crystallinum. Plant Physiol. 110 259–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, M., Ammar, D., Ives, H., Albrecht, F., and Gluck, S.L. (2007). Physical interaction between aldolase and vacuolar H+-ATPase is essential for the assembly and activity of the proton pump. J. Biol. Chem. 282 24495–24503. [DOI] [PubMed] [Google Scholar]
- Lu, M., Holliday, L.S., Zhang, L., Dunn, W.A., Jr., and Gluck, S.L. (2001). Interaction between aldolase and vacuolar H+-ATPase. Evidence for direct coupling of glycolysis to the ATP-hydrolyzing proton pump. J. Biol. Chem. 276 30407–30413. [DOI] [PubMed] [Google Scholar]
- Lu, M., Sautin, Y.Y., Holliday, L.S., and Gluck, S.L. (2004). The glycolytic enzyme aldolase mediates assembly, expression, and activity of vacuolar H+-ATPase. J. Biol. Chem. 279 8732–8739. [DOI] [PubMed] [Google Scholar]
- Millar, A.H. (2004). Location, location, location: Surveying the intracellular real estate through proteomics in plants. Funct. Plant Biol. 31 563–571. [DOI] [PubMed] [Google Scholar]
- Moritz, R.L., and Simpson, R.J. (2005). Liquid-based free-flow electrophoresis–reversed-phase HPLC: A proteomic tool. Nat. Methods 2 863–873. [DOI] [PubMed] [Google Scholar]
- Nelson, D.E., Glaunsinger, B., and Bohnert, H.J. (1997). Abundant accumulation of the calcium-binding molecular chaperone calreticulin in specific floral tissues of Arabidopsis thaliana. Plant Physiol. 114 29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanaban, S., Lin, X., Perera, I., Kawamura, Y., and Sze, H. (2004). Differential expression of vacuolar H+-ATPase subunit c genes in tissues active in membrane trafficking and their roles in plant growth as revealed by RNAi. Plant Physiol. 134 1514–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parets-Soler, A., Pardo, J.M., and Serrano, R. (1990). Immunocytolocalization of plasma membrane H+-ATPase. Plant Physiol. 93 1654–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry, R.V., Turner, J.C., and Rea, P.A. (1989). High purity preparations of higher plant vacuolar H+-ATPase reveal additional subunits: Revised subunit composition. J. Biol. Chem. 264 20025–20032. [PubMed] [Google Scholar]
- Petrak, J., Ivanek, R., Toman, O., Cmejla, R., Cmejlova, J., Vyoral, D., Zivny, J., and Vulpe, C.D. (2008). Déjà vu in proteomics. A hit parade of repeatedly identified differentially expressed proteins. Proteomics 8 1744–1749. [DOI] [PubMed] [Google Scholar]
- Qi, J., Wang, Y., and Forgac, M. (2007). The vacuolar (H+)-ATPase: Subunit arrangement and in vivo regulation. J. Bioenerg. Biomembr. 39 423–426. [DOI] [PubMed] [Google Scholar]
- Qiu, Q.-S., Guo, F., Quintero, F.J., Pardo, J.M., Schumaker, K.S., and Zhu, J.-K. (2004). Regulation of vacuolar Na+/H+ exchange in Arabidopsis thaliana by the salt-overly-sensitive (SOS) pathway. J. Biol. Chem. 279 207–215. [DOI] [PubMed] [Google Scholar]
- Ratajczak, R. (2000). Structure, function and regulation of the plant vacuolar H+-translocating ATPase. Biochim. Biophys. Acta 1465 17–36. [DOI] [PubMed] [Google Scholar]
- Reuveni, M., Bennett, A.B., Bressan, R.A., and Hasegawa, P.M. (1990). Enhanced H+ transport capacity and atp hydrolysis activity of the tonoplast H+-ATPase after NaCl adaptation. Plant Physiol. 94 524–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuveni, M., Evenor, D., Artzi, B., Perl, A., and Erner, Y. (2001). Decrease in vacuolar pH during petunia flower opening is reflected in the activity of tonoplast H+-ATPase. J. Plant Physiol. 158 991–998. [Google Scholar]
- Schmidt, U.G., Endler, A., Schelbert, S., Brunner, A., Schnell, M., Neuhaus, H.E., Marty-Mazars, D., Marty, F., Baginsky, S., and Martinoia, E. (2007). Novel tonoplast transporters identified using a proteomic approach with vacuoles isolated from cauliflower buds. Plant Physiol. 145 216–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, H., Ishitani, M., Kim, C., and Zhu, J.-K. (2000). The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. USA 97 6896–6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, H., and Zhu, J.-K. (2002). Regulation of expression of the vacuolar Na+/H+ antiporter gene AtNHX1 by salt stress and abscisic acid. Plant Mol. Biol. 50 543–550. [DOI] [PubMed] [Google Scholar]
- Shimaoka, T., Ohnishi, M., Sazuka, T., Mitsuhashi, N., Hara-Nishimura, I., Shimazaki, K.-I., Maeshima, M., Yokota, A., Tomizawa, K.-I., and Mimura, M. (2004). Isolation of intact vacuoles and proteomic analysis of tonoplast from suspension-cultured cells of Arabidopsis thaliana. Plant Cell Physiol. 45 672–683. [DOI] [PubMed] [Google Scholar]
- Su, H., Balderas, E., Vera-Estrella, R., Golldack, D., Quigley, F., Zhao, C., Pantoja, O., and Bohnert, H.J. (2003). Expression of the cation transporter McHKT1 in a halophyte. Plant Mol. Biol. 52 967–980. [DOI] [PubMed] [Google Scholar]
- Sugahara, T., Shimizu, S., Abiru, M., Matsuoka, S., and Sasaki, T. (1998). A novel function of enolase from rabbit muscle; an immunoglobulin production stimulating factor. Biochim. Biophys. Acta 1380 163–176. [DOI] [PubMed] [Google Scholar]
- Sze, H., Schumacher, K., Muller, M.L., Padmanaban, S., and Taiz, L. (2002). A simple nomenclature for a complex proton pump: VHA genes encode the vacuolar H+-ATPase. Trends Plant Sci. 7 157–161. [DOI] [PubMed] [Google Scholar]
- Tanaka, N., Fujita, M., Handa, H., Marayama, S., Uemura, M., Kawamura, Y., Mitsui, T., Mikami, S., Tozawa, Y., Yoshinaga, T., and Komatsu, S. (2004). Proteomics of the rice cell: Systematic identification of the protein populations in subcellular compartments. Mol. Genet. Genomics 271 566–576. [DOI] [PubMed] [Google Scholar]
- Tsiantis, M.S., Bartholomew, D.M., and Smith, J.A.C. (1996). Salt regulation of transcript levels for the c subunit of a leaf vacuolar H+-ATPase in the halophyte Mesembryanthemum crystallinum. Plant J. 9 729–736. [DOI] [PubMed] [Google Scholar]
- Valles, P., Wysocki, J., Salabat, M.R., Cokic, I., Ye, M., LaPointe, M.S., and Batlle, D. (2005). Angiotensin II increases H+-ATPase B1 subunit expression in medullary collecting ducts. Hypertension 45 818–823. [DOI] [PubMed] [Google Scholar]
- Vargas-Suárez, M., Ayala-Ochoa, A., Lozano-Franco, J., García-Torres, I., Díaz-Quiñonez, A., Ortíz-Navarrete, V.F., and Sánchez-de-Jiménez, E. (2004). Rubisco activase chaperone activity is regulated by a post-translational mechanism in maize leaves. J. Exp. Bot. 55 2533–2539. [DOI] [PubMed] [Google Scholar]
- Vera-Estrella, R., Barkla, B.J., García-Ramírez, L., and Pantoja, O. (2004). Novel regulation of aquaporins during osmotic stress. Plant Physiol. 135 2318–2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward, J., Reinders, A., Hsu, H.-T., and Sze, H. (1992). Dissociation and reassembly of the vacuolar H+-ATPase complex from oat roots. Plant Physiol. 99 161–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederhold, E., Gandhi, T., Permentier, H.P., Breitling, R., Poolman, B., and Slotboom, D.J. (2009). The yeast vacuolar membrane proteome. Mol. Cell. Proteomics 8 380–392. [DOI] [PubMed] [Google Scholar]
- Yokoi, S., Quintero, F.J., Cubero, B., Ruiz, T., Bressan, R.A., Hasegawa, P.M., and Pardo, J.M. (2002). Differential expression and function of Arabidopsis thaliana NHX Na+/H+ antiporters in the salt stress response. Plant J. 30 1–12. [DOI] [PubMed] [Google Scholar]
- Zhigang, A., Löw, R., Rausch, T., Lüttge, U., and Ratajczak, R. (1996). The 32 kDa tonoplast polypeptide Di associated with the V-type H+-ATPase of Mesembryanthemum crystallinum L. in the CAM plant state: A proteolytically processed subunit B? FEBS Lett. 389 314–318. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.