Abstract
Asn-linked glycans, or the glycan code, carry crucial information for protein folding, transport, sorting, and degradation. The biochemical pathway for generating such a code is highly conserved in eukaryotic organisms and consists of ordered assembly of a lipid-linked tetradeccasaccharide. Most of our current knowledge on glycan biosynthesis was obtained from studies of yeast asparagine-linked glycosylation (alg) mutants. By contrast, little is known about biosynthesis and biological functions of N-glycans in plants. Here, we show that loss-of-function mutations in the Arabidopsis thaliana homolog of the yeast ALG12 result in transfer of incompletely assembled glycans to polypeptides. This metabolic defect significantly compromises the endoplasmic reticulum–associated degradation of bri1-9 and bri1-5, two defective transmembrane receptors for brassinosteroids. Consequently, overaccumulated bri1-9 or bri1-5 proteins saturate the quality control systems that retain the two mutated receptors in the endoplasmic reticulum and can thus leak out of the folding compartment, resulting in phenotypic suppression of the two bri1 mutants. Our results strongly suggest that the complete assembly of the lipid-linked glycans is essential for successful quality control of defective glycoproteins in Arabidopsis.
INTRODUCTION
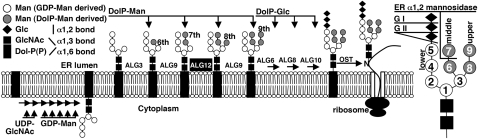
Asn (or N)-linked glycosylation is an important posttranslational protein modification process in eukaryotes (Banerjee et al., 2007). Through interactions with carbohydrate binding proteins, N-glycans regulate protein folding, transport, sorting, degradation, and intracellular signaling (Kato and Kamiya, 2007; Molinari, 2007). N-glycosylation is catalyzed in the endoplasmic reticulum (ER) by oligosaccharyltransferase (OST) that transfers a preassembled Glc3Man9GlcNAc2 glycan (Glc for glucose, Man for mannose, and GlcNAc for N-acetylglucosamine) from the dolichylpyrophosphate (Dol-PP) carrier to Asn residue in the Asn-Xaa-Ser/The motif (Xaa represents any amino acid except Pro) on nascent polypeptides (Kelleher and Gilmore, 2006). The assembly of Dol-PP-Glc3Man9GlcNAc2 starts on the cytoplasmic face of the ER membrane with the addition of GlcNAc-UDP to dolichylphosphate followed by addition of another GlcNAc and five Man residues, generating Dol-PP-Man5GlcNAc2 (Figure 1). This septasaccharide is then flipped over into the ER lumen where four more Man and three Glc residues are sequentially added to form Dol-PP-Glc3Man9GlcNAc2 with three mannose branches. The addition of 6th and 7th Man residues, catalyzed by ALG3 and ALG9, respectively, forms the middle α1,3-α1,2-dimannose branch (Aebi et al., 1996; Burda et al., 1996), while the addition of 8th and 9th Man residues, catalyzed by ALG12 and ALG9, respectively, generates the upper α1,6-α1,2-dimannose branch (Burda et al., 1999; Frank and Aebi, 2005). Adding three Glc residues to the lower branch, catalyzed sequentially by ALG6, ALG8, and ALG10, is thought to be necessary for recognition by OST (Burda et al., 1999).
Figure 1.
Proposed Scheme of Biosynthesis of Dol-PP–Linked Glc3Man9GlcNAc2 in Arabidopsis.
Dol-PP-Man5GlcNAc2 is formed at the cytoplasmic side of the ER using cytoplasmic UDP-GlcNAc and GDP-Man as substrates and is flipped over into the ER lumen. Four Man and three Glc residues are sequentially added from Dol-P-Man and Dol-P-Glc, respectively, to form Dol-PP-Glc3Man9GlcNAc2 that is transferred to nascent proteins OST. The high-mannose (H)-type glycans produced here are further processed in the Golgi into complex (C)-type glycans. The added glycoresidues, the corresponding glycosyltransferases, and three types of mannosyl bonds are shown. Three mannose branches are indicated on the enlarged Glc3Man9GlcNAc2 structure with circled numbers representing the order by which each Man is added. Glycosidases involved in trimming Glc and Man residues (GI, GII, and α-1,2 mannosidase) are also shown.
Further processing of the N-linked Glc3Man9GlcNAc2 determines the fate of nascent proteins. Immediately after glycan transfer, the first two Glc residues are sequentially removed by glucosidase I and II (GI and GII) (Trombetta, 2003; Figure 1). The resulting Glc1Man9GlcNAc2 is specifically recognized by two ER lectins, calnexin (CNX) and its soluble homolog calreticulin (CRT), which recruit other ER chaperones to assist protein folding (Helenius and Aebi, 2004). Further removal of the 3rd Glc by GII liberates glycoproteins from CNX/CRT. A fully folded glycoprotein leaves the ER and transits into the Golgi for further glycan modifications. By contrast, an incompletely/misfolded glycoprotein is recognized and reglucosylated by UDP-glucose:glycoprotein glucosyltransferase (UGGT), an ER-resident protein-folding sensor (Caramelo and Parodi, 2007). As a result, the glycoprotein reassociates with CNX/CRT for additional folding. The alternating activities of GII and UGGT drive the CNX/CRT cycle until the glycoprotein attains its native conformation (Caramelo and Parodi, 2008). A protein that fails to obtain its native structure within a time window undergoes Man trimming from Man9GlcNAc2 to Man5-8GlcNAc2 catalyzed by α1,2 mannosidases (Molinari, 2007) and is subsequently retrotranslocated into cytosol for ER-associated degradation (ERAD) (Vembar and Brodsky, 2008).
Our knowledge of glycan biosynthesis, ER quality control (ERQC), and ERAD was mainly obtained from studies of yeast mutants and mammalian cell cultures (Burda and Aebi, 1999; Molinari, 2007), but little is known about biosynthesis and biological functions of N-glycans in plants (Pattison and Amtmann, 2009). The Arabidopsis thaliana bri1-9 and bri1-5 mutants are excellent tools to study ERQC and ERAD in plants (Jin et al., 2007; Hong et al., 2008). BRI1 is a leucine-rich-repeat receptor-like kinase that functions as a cell surface receptor for brassinosteroids (BRs) (Li and Chory, 1997; Kinoshita et al., 2005). Arabidopsis mutants defective in BR biosynthesis/signaling exhibit a characteristic set of phenotypes, including dwarf stature, short hypocotyls in the dark, and delayed flowering. Studies in the past decade have uncovered a linear signaling pathway that relies on protein phosphorylation to transmit the BR signal into the nucleus (Li and Jin, 2007). Recently, we discovered that the mutant phenotypes of bri1-9 and bri1-5 are caused by failure of the two mutated BR receptors, which carry the Ser662Phe and Cys69Tyr mutations, respectively, to reach the cell surface. This failure is caused by operation of overzealous ERQC systems in Arabidopsis that retain the mutated receptors in the ER (Jin et al., 2007, 2009; Hong et al., 2008). Loss-of-function mutations in EMS-mutagenized Bri1 Suppressor1 (EBS1) and EBS2, encoding the Arabidopsis UGGT homolog and the Arabidopsis CRT3, respectively, significantly compromise the ERQC of bri1-9 to allow some mutated receptors to be correctly targeted to the cell surface. By contrast, loss of UGGT function fails to suppress but instead enhances the other ER-retained bri1 allele, bri1-5, due to involvement of other retention mechanisms to keep the Cys-69–mutated BR receptor in the ER (Hong et al., 2008).
To identify other factors that affect protein quality control in the ER, we isolated additional ebs mutants. Genetic and biochemical analyses of these mutants led to identification of several allelic ebs4 mutants that contain more bri1-9 proteins than the parental bri1-9. Using a candidate gene approach, we found that EBS4 encodes the Arabidopsis ortholog of the yeast ALG12 that catalyzes addition of the 8th Man in the assembly of Dol-PP-Glc3Man9GlcNAc2. This metabolic defect interferes with ERAD of bri1-9 and bri1-5 and is responsible for increased export of two defective receptors out of the ER. We conclude that transfer of the fully assembled glycan precursor to nascent polypeptides is critical to ensure successful ER quality control in Arabidopsis.
RESULTS
The ER-Retained bri1-9 Is Degraded by a Proteasome-Mediated ERAD Process
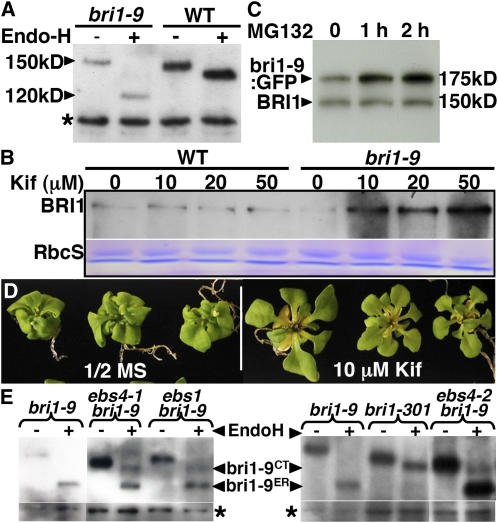
Our previous studies revealed that a mutated BR receptor bri1-9 carrying a single amino acid change (Ser662Phe) in the extracellular ligand binding domain is retained in the ER due to an overzealous protein quality control system (Jin et al., 2007, 2009). We suspected that the mutated BR receptor is cleared by ERAD to prevent blockage of the secretory pathway. Indeed, immunoblot analysis using an anti-BRI1 antibody revealed that the abundance of bri1-9 is much lower than that of the wild-type BRI1 (Figure 2A). Consistent with their subcellular localizations (Jin et al., 2007), the two BR receptors were differentially sensitive to endoglycosidase H (Endo H) that cleaves high mannose (H)-type glycans on ER-localized glycoproteins but can't remove Golgi-processed complex (C)-type N-glycans (Figure 2A). Thus, an ER-localized protein with H-type glycans will show a much greater mobility shift than a processed protein with C-type glycans. The small mobility shift of the Endo H–digested BRI1 is likely due to the presence of a few incompletely processed H-type N-glycans on the plasma membrane–localized BR receptor. This simple assay was used throughout this study to determine whether or not ER-retained BR receptors move out of the folding compartment since passage through the Golgi network converts their H-type N-glycans to C-type N-glycans. We also treated bri1-9 and the wild-type control seedlings with kifunensine (Kif), a widely used inhibitor of α1,2 mannosidases that generate the glycan signal for ERAD (Tokunaga et al., 2000). As shown in Figure 2B, Kif treatment significantly increased the bri1-9 abundance but had little effect on the BRI1 stability. We thus concluded that ER-retained bri1-9 undergoes ERAD.
Figure 2.
bri1-9 Undergoes a Proteasome-Mediated ERAD.
(A) Immunoblot analysis of bri1-9 and BRI1. The asterisk denotes a nonspecific band for loading control. Molecular masses of bri1-9 and BRI1 are indicated on the left.
(B) Immunoblot analysis of bri1-9 and BRI1 abundance in 3-week-old seedlings treated with or without Kif for 24 h. Coomassie blue staining of the small subunit of ribulose-1,5-bis-phosphate carboxylase/oxygenase (RbcS) serves as the loading control.
(C) Immunoblot analysis of bri1-9:GFP and endogenous BRI1 proteins in 3-week-old seedlings treated with or without 20 μM MG132. Molecular masses of BRI1 and bri1-9:GFP are shown on the right. The abundance of the endogenous wild-type BRI1 serves as a loading control.
(D) Longer Kif treatment partially suppresses the dwarf phenotype of bri1-9. Shown here are 4-week-old seedlings grown on regular half-strength Murashige and Skoog (MS) medium for 3 weeks followed by 1-week growth on half-strength MS medium containing no or 10 μM Kif.
(E) Immunoblot analysis of bri1-9 abundance in 3-week-old soil-grown seedlings. bri1-9CT denotes bri1-9 carrying C-type N-glycans, while bri1-9ER indicates the ER-retained bri1-9. The asterisks denote a nonspecific band for loading control.
For (A) and (E), equal amounts of total proteins extracted in 1× SDS buffer from 3-week-old seedlings were treated with 1000 units of Endo H for 1 h at 37°C, separated by SDS-PAGE, and analyzed by immunoblotting with anti-BRI1 antibody. For (B) and (C), 3-week-old seedlings were incubated in liquid half-strength MS medium supplemented with or without Kif or MG132 for a suitable length of time and extracted with 1× SDS buffer. Equal amounts of protein extracts of each sample were separated by SDS-PAGE and analyzed by immunoblot with anti-BRI1 antibody. [See online article for color version of this figure.]
We previously showed that another ER-retained BR receptor, bri1-5, is degraded by a proteasome-independent ERAD process (Hong et al., 2008). To examine if bri1-9 is similarly degraded, we treated 3-week-old seedlings of a pBRI1-bri1-9:GFP transgenic line with 20 μM MG132, a widely used proteasome inhibitor that can prevent degradation of many ERAD substrates (Schmitz and Herzog, 2004). Such a transgenic line expresses both the green fluorescent protein (GFP)-tagged bri1-9 and the endogenous BRI1 that was known to be degraded by proteasome (Hong et al., 2008). Figure 2C shows that bri1-9:GFP was more stabilized by MG132 than the wild-type BRI1, suggesting that ERAD of bri1-9 involves proteasomes. Similar to what we observed with the bri1-5 mutant (Hong et al., 2008), longer Kif treatment could suppress the bri1-9 phenotype (Figure 2D), likely due to leakage of some BR receptors as a result of saturating the bri1-9 retention mechanism by overaccumulated bri1-9 in the ER. Consistently, overexpression of bri1-9:GFP driven by its native promoter could also suppress the bri1-9 dwarf phenotype (see Supplemental Figure 1 online).
Identification of ebs Mutants That Accumulate bri1-9
Previously, we identified ∼80 ebs mutants (Jin et al., 2007). Our Kif rescue and bri1-9:GFP overexpression experiments suggested that mutations inhibiting ERAD of bri1-9 should suppress the bri1-9 mutation and that genetic studies of these mutants might uncover components or regulators of the Arabidopsis ERAD machinery. We thus performed immunoblot analysis of some ebs mutants using an anti-BRI1 antibody coupled with the Endo H assay, which can reveal if an ebs mutation results in increased bri1-9 abundance and/or escape of bri1-9 from the ER. Figures 2E shows that two such mutants contain more bri1-9 proteins than the parental bri1-9, some of which acquire Endo H–resistant N-glycans similar to bri1-9 in the ebs1 bri1-9 mutant that contains plasma membrane–localized bri1-9 due to compromised ERQC (Jin et al., 2007). These two potential ERAD mutants were later found to be allelic to each other and were named ebs4-1 and ebs4-2.
ebs4 Mutations Partially Suppress bri1-9 and Restore Its BR Sensitivity
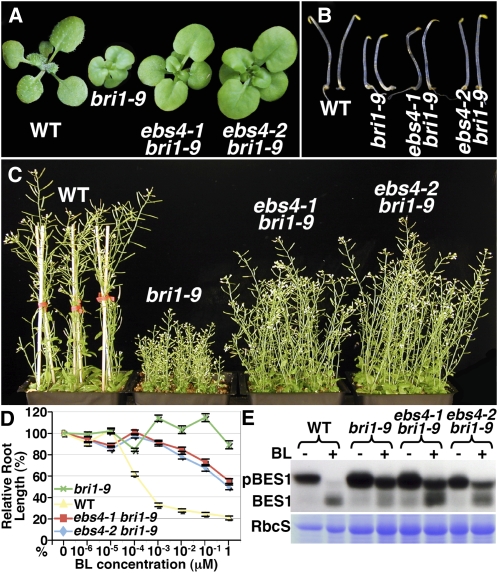
As shown in Figure 3, both ebs4 bri1-9 mutants are moderate bri1-9 suppressors. They have bigger rosette leaves with noticeable petioles (Figure 3A), exhibit longer hypocotyls in the dark (Figure 3B), and are much taller at maturity (Figure 3C) than bri1-9. Consistent with these morphological phenotypes and detection of a slower-moving Endo H–digested bri1-9 band on immunoblots (Figure 2E), the two ebs4 mutants regain partial sensitivity to brassinolide (BL), the most active BR, as measured by both the root growth inhibition and BL-induced BES1 dephosphorylation assays (Clouse et al., 1996; Mora-Garcia et al., 2004). As shown in Figure 3D, BL treatment inhibited root growth of wild-type and ebs4 bri1-9 seedlings but had little effect on that of bri1-9. The regained BR sensitivity in ebs4 bri1-9 was also observed at the biochemical level. It was known that BR treatment results in rapid dephosphorylation of BES1, an important transcription factor that regulates expression of many known BR-responsive genes (Yin et al., 2002, 2005). As shown in Figure 3E, 1-h treatment of BL resulted in nearly complete dephosphorylation of BES1 in the wild type and partial dephosphorylation of BES1 in the two ebs4 mutants but had a marginal effect on BES1 phosphorylation in bri1-9. These results thus suggested that accumulated bri1-9 proteins in ebs4 mutants might saturate the bri1-9 ERQC machinery and leak out of the ER to the cell surface where they activate BR signaling to promote plant growth.
Figure 3.
ebs4-1 and ebs4-2 Are Moderate Suppressors of bri1-9.
(A) Two-week-old soil-grown seedlings of wild-type, bri1-9, and two allelic ebs4 mutants.
(B) Five-day-old dark-grown seedlings of wild-type, bri1-9, and two allelic ebs4 mutants.
(C) Seven-week-old soil-grown mature plants of wild-type, bri1-9, and two allelic ebs4 mutants.
(D) The root growth-inhibition assay. Root lengths of 7-d-old seedlings grown on BL-containing half-strength MS medium under a 16-h-light/8-h-dark growth condition in a 22°C growth chamber were measured and presented as the relative value of the average root length of BL-treated seedlings to that of untreated seedlings of the same genotype. Each data point represents the average of ∼40 seedlings of duplicated experiments. Error bars denote se.
(E) Immunoblot analysis of the BL-induced BES1 dephosphorylation. Total proteins were extracted in 1× SDS buffer from 3-week-old seedlings treated with or without 1 μM BL for 1 h in liquid half-strength MS medium, separated by 10% SDS-PAGE, and analyzed by immunoblotting with anti-BES1 antibody. Coomassie blue staining of RbcS serves as the loading control.
The ebs4 Mutations Likely Affect Assembly of Lipid-Linked Glycans
In addition to stabilizing bri1-9, the ebs4 mutations also reduce its molecular weight. As shown in Figure 2E, the bri1-9 mobility in both ebs4 bri1-9 mutants is slightly faster than that in the bri1-9 single mutant. Endo H assay revealed that most of the bri1-9 proteins are still retained in the ER and that the mobility of the fast-moving band of Endo H–digested bri1-9 in the two ebs4 mutants is the same as that in the parental bri1-9, indicating that the ebs4 mutations likely affect glycoforms on bri1-9. The bri1-9 is either hypoglycosylated with Glc1Man9GlcNAc2 on fewer glycosylation sites or fully glycosylated on all 14 predicted glycosylation sites with incompletely assembled glycans. Since hypoglycosylation often results in discrete bands each with different numbers of glycans, the presence of a single faster-moving bri1-9 band on immunoblots suggested that the ebs4 mutations most likely affect the assembly of the lipid-linked tetradeccasaccharide. This hypothesis was supported by our finding that the mobility of the wild-type BRI1 in ebs4-2 is the same as that in the EBS4+ background (see Supplemental Figure 2 online) since hypoglycosylation (on fewer sites) should result in a fast-moving BRI1 band on immunoblots, whereas full glycosylation with incompletely assembled glycans has no effect on BRI1 mobility due to Golgi-mediated further glycan modifications (Henquet et al., 2008).
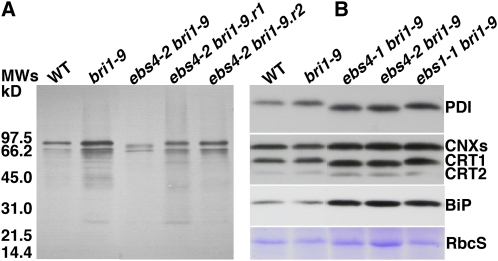
The effect of ebs4 mutation on N-glycosylation was also investigated by both immunoblotting and lectin affinoblotting. The immunoblotting was performed using antibodies against α1,3-fucose or β1,2-xylose residues characteristic of plant-specific C-type N-glycans (Faye et al., 1993), while the affinoblotting was performed with the concanavalin A (Con A)/peroxidase system specific for H-type N-glycans (Faye and Chrispeels, 1985). Whereas no significant difference in C-type N-glycans between bri1-9 and ebs4-2 bri1-9 was detected by either antibody (see Supplemental Figure 3 online), noticeable differences in H-type N-glycans between the two mutants were detected by affinoblotting. As shown in Figure 4A, not only is the intensity of Con A–positive signals lower in ebs4-2 bri1-9 than in bri1-9, but also the mobility of two major Con A–positive bands is faster in ebs4-2 bri1-9 than in bri1-9, most likely due to presence of H-type N-glycans with fewer Man residues in the double mutant. These results further support our conclusion that ebs4 mutations result in transfer of incompletely assembled glycan precursor to nascent glycoproteins in the ER.
Figure 4.
ebs4 Mutations Affect Mobility of Several Glycoproteins Carrying H-Type N-Glycans and Lead to UPR.
(A) Affinoblotting analysis of glycoproteins. Proteins were extracted by the phenol method (Fitchette et al., 1999) from leaves of 4-week-old soil-grown plants of wild-type, bri1-9, ebs4-2 bri1-9, and two independently rescued ebs4-2 bri1-9 lines, resolved in 1× SDS buffer, separated by SDS-PAGE, and analyzed by the Con A/peroxidase system (Faye and Chrispeels, 1985). Equal amounts of proteins were loaded in each lane. The sizes and positions of molecular mass markers are shown on the left.
(B) Immunoblot analysis of PDI, CNX/CRT, and BiP. Equal amounts of total protein extracts in 1× SDS sample buffer from 3-week-old seedlings of wild-type, bri1-9, two ebs4 bri1-9 mutants, and ebs1-1 bri1-9 were separated by SDS-PAGE and analyzed by immunoblotting with antibodies made against the Arabidopsis PDIL1-1, maize (Zea mays) CRTs, or a spinach (Spinacia oleracea) BiP. Coomassie blue staining of RbcS serves as the loading control. [See online article for color version of this figure.]
ebs4 Mutations Activate the Unfolded Protein Response
We also examined the effect of ebs4 mutations on N-glycosylation of an important ER-localized folding enzyme, protein disulfide isomerase (PDI), which catalyzes the thiol-disulfide exchange reaction. The Arabidopsis genome encodes at least nine PDIs (named PDIL1-1 to 1-6 and PDIL2-1 to 2-3) (Houston et al., 2005). Using an anti-PDIL1 antibody, we repeatedly detected a faster-moving PDI band in two ebs4 mutants but not in the ebs1 mutant defective in UGGT that catalyzes reglucosylation of bri1-9 (Jin et al., 2007). In addition to the mobility change, all three ebs mutations also increase the abundance of PDI (Figure 4B), likely caused by unfolded protein response (UPR), a highly conserved ER stress response that stimulates production of ER chaperones/folding enzymes (Bernales et al., 2006). Further support for the UPR induction by ebs4 mutations was provided by additional immunoblot analyses using anti-spinach BiP antibody and anti-maize CRT antibody that detects two CRTs and two CNXs in Arabidopsis (Persson et al., 2003). As shown in Figure 4B, accumulation of BiP, CNXs, and at least one CRT were substantially increased in the three ebs mutants. It is worthwhile to note that the mobility of both CRT1 and CRT2 is also altered in the ebs4 mutants (Figure 4B), consistent with previous prediction that CRT1 and CRT2 carry three and one N-glycans, respectively (Persson et al., 2003).
ebs4 Mutations Also Suppress the bri1-5 Mutation
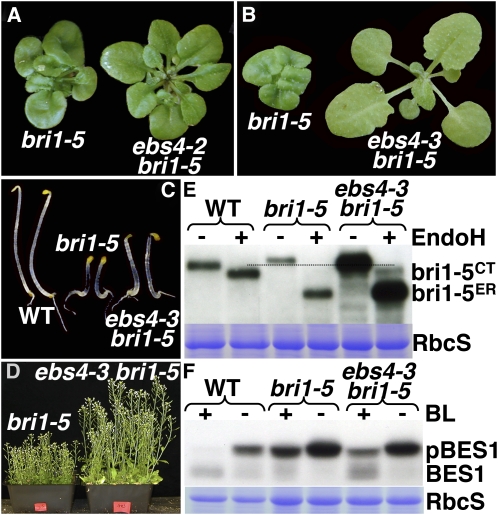
Since Kif treatment prevented ERAD of bri1-5 and suppressed the bri1-5 mutation (Hong et al., 2008), we suspected that the ebs4 mutations should also be able to suppress bri1-5. Indeed, when crossed into bri1-5, ebs4-2 was able to partially suppress the rosette phenotype of bri1-5 (Figure 5A). This was in sharp contrast with the ebs1 mutations that fail to suppress but instead enhance the bri1-5 mutation (Hong et al., 2008). In addition, a genetic screen for extragenic bri1-5 suppressors resulted in identification of the third ethyl methanesulfonate–generated ebs4 allele that suppresses many of the bri1-5 mutant phenotypes, including small rosette, short hypocotyls in the dark, and short inflorescence stems of mature plants (Figures 5B to 5D). Figure 5E shows that the ebs4-3 bri1-5 mutant accumulated more BR receptors than bri1-5 or even the wild-type control. Consistent with the detection of C-type N-glycan–containing bri1-5 suggestive of cell surface localization (Figure 5E), a BR-induced BES1 dephosphorylation assay confirmed that ebs4-3 bri1-5 regained partial sensitivity to the plant steroid hormone. As shown in Figure 5F, 1-h treatment with 1 μM BL resulted in almost complete, marginal, and partial BES1 dephosphorylation in wild-type, bri1-5, and ebs4-3 bri1-5, respectively.
Figure 5.
ebs4 Can Suppress the bri1-5 Mutation.
(A) Four-week-old soil-grown plants of bri1-5 and ebs4-2 bri1-5.
(B) Two-week-old soil-grown plants of bri1-5 and ebs4-3 bri1-5.
(C) Five-day-old dark-grown seedlings of bri1-5 and ebs4-3 bri1-5.
(D) Seven-week-old soil-grown mature plants of bri1-5 and ebs4-3 bri1-5.
(E) Immunoblot analysis of BRI1 and bri1-5 with or without Endo H treatment in 3-week-old seedlings. Equal amounts of total proteins extracted in 1× SDS buffer from 3-week-old seedlings were treated with or without 1000 units of Endo H for 1 h at 37°C, separated by SDS-PAGE, and analyzed by immunoblotting with anti-BRI1 antibody. bri1-5ER is the ER-localized form, while bri1-5CT indicates bri1-5 carrying C-type N-glycans.
(F) Immunoblot analysis of the BES1 phosphorylation status in 3-week-old seedlings treated with or without 1 μM BL. Total proteins were extracted in 1× SDS buffer from 3-week-old seedlings treated with or without 1 μM BL for 1 h in liquid half-strength MS medium, separated by 10% SDS-PAGE, and analyzed by immunoblotting with anti-BES1 antibody. pBES1 is the phosphorylated form of BES1. In both (E) and (F), Coomassie blue staining of RbcS serves as the loading control. [See online article for color version of this figure.]
EBS4 Encodes the Arabidopsis Homolog of the Yeast ALG12 Enzyme
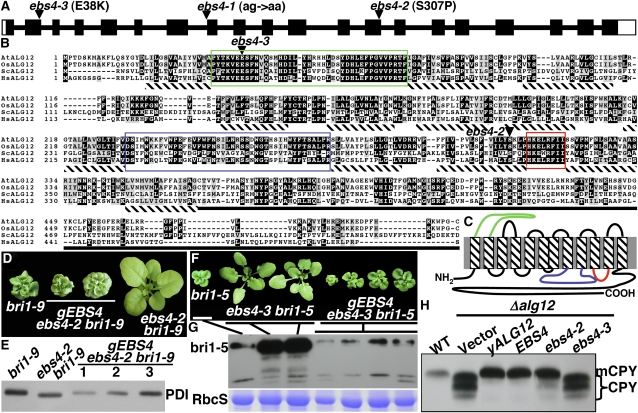
Our discovery that the ebs4 mutations affect the size of H-type N-glycans and inhibit ERAD of both bri1-5 and bri1-9 but have no effect on biosynthesis of C-type N-glycans suggested that EBS4 might encode one of the three ER-localized mannosyltransferases catalyzing the assembly of the Dol-PP-Glc3Man9GlcNAc2 (Figure 1). We thus sequenced the two Arabidopsis genes At2g47760 and At1g16900 annotated to encode homologs of the yeast ALG3 and ALG9, respectively, from the ebs4 mutants but failed to identify any nucleotide change. At the time of sequencing, there was no annotated gene encoding an ALG12 homolog. Using the yeast ALG12 (Burda et al., 1999) as a query, a BLASTX search against the entire Arabidopsis genome did identify a region between At1g02140 and At1g02150 that encodes a potential ALG12 homolog (annotated later in GenBank as At1g02145). As shown in Figures 6A and 6B, At1g02145 contains 20 exons and 19 introns, and its predicted 497–amino acid polypeptide displays high sequence homology with ALG12s of yeast and human and a predicted rice (Oryza sativa) ALG12 homolog. Based on previous sequence analysis of ALG12 proteins and our bioinformatic analysis, we predicted that the Arabidopsis ALG12 also contains 12 transmembrane segments with both N and C termini exposed to the cytosol and the highly conserved 1st loop facing the ER lumen (Figure 6C).
Figure 6.
EBS4 Is the likely Arabidopsis Ortholog of the Yeast ALG12.
(A) Schematic presentation of EBS4 gene structure. Black bars denote exons, with open bars denoting untranslated regions, and the thin lines represent introns. The positions and molecular nature of the three ebs4 mutations are indicated.
(B) Sequence alignment of EBS4 with the yeast ALG12 and ALG12 homologs of rice and human. Alignment of EBS4 (accession number NP_001077448) with the yeast ALG12 (ScALG12, NP_014427) and ALG12 homologs from rice (OsALG12, NP_001053463) and human (HsALG12, NP_077010) was performed at the Tcoffee server (http://tcoffee.vital-it.ch/cgi-bin/Tcoffee/tcoffee_cgi/index.cgi). Identical residues in ≥3 sequences are shaded black, while similar residues are shaded in gray using the BoxShade server at http://www.ch.embnet.org/software/BOX_form.html. Two largest loops and a highly conserved cytoplasmic loop are color-boxed, and potential transmembrane domains are underlined with hatched bars. Triangles indicate the residues mutated in ebs4-2 and ebs4-3.
(C) EBS4 contains 12 predicted transmembrane segments. Prediction of potential transmembrane domains was performed at the DAS server (http://www.sbc.su.se/∼miklos/DAS/) and was later adjusted based on the predicted topology of other ALG12 homologs (Oriol et al., 2002). The two largest loops and the highly conserved 5th cytoplasmic loop are shown in colors corresponding to the boxes in (B).
(D) Four-week-old soil-grown plants of bri1-9, ebs4-2 bri1-9, and two EBS4-complemented ebs4-2 bri1-9 transgenic lines carrying a genomic EBS4 transgene (gEBS4) that contains its native promoter and 3′-terminator.
(E) Immunoblot analysis of a PDI in bri1-9, ebs4-2 bri1-9, and three independent gEBS4-complemented ebs4-2 bri1-9 lines.
(F) Three-week-old soil-grown plants of one bri1-5 mutant, two ebs4-3 bri1-5 mutants, and four independent gEBS4-rescued ebs4-3 bri1-5 transgenic lines.
(G) Immunoblot analysis of bri1-5 abundance in plants shown in (F). For both (E) and (G), equal amounts of total proteins extracted in 1× SDS sample buffer from 3-week-old seedlings were separated by SDS-PAGE and analyzed by immunoblots using anti-PDI (E) or anti-BRI1 (G) antibody. Coomassie blue staining of RbcS serves as a loading control (G).
(H) Immunoblot analysis of CPY glycosylation in wild-type or transformed Δalg12 yeast cells containing the vector plasmid, the yeast ALG12 gene, the wild-type EBS4, or a mutated EBS4 gene carrying the ebs4-2 or ebs4-3 mutation. See Methods for experimental details.
Sequencing At1g02145 of ebs4-1 bri1-9 identified a single nucleotide substitution (G to A) at the end of the predicted 7th intron (Figure 6A), which likely results in aberrant RNA splicing leading to premature translational termination. The identity of At1g02145 as EBS4 was confirmed by two different experiments. First, sequencing of two other ebs4 alleles identified additional single nucleotide changes in At1g02145, mutating the predicted Ser-307 to Phe in ebs4-2 and Glu-38 to Lys in ebs4-3 (Figures 6A and 6B). We also conducted complementation experiments by transforming a genomic transgene (gEBS4), which contains the entire At1g02145 gene with its native promoter and terminator, into the ebs4-2 bri1-9 or ebs4-3 bri1-5 mutants. As shown in Figures 6D and 6F, expression of the At1g02145 gene in two ebs4 mutants inhibited the suppression activity of the ebs4 mutations on the two bri1 mutants. In addition, At1g02145 expression rescued several biochemical defects of the ebs4 mutations, including the faster mobility of the two major Con A–positive bands (Figure 3A) and PDI (Figure 6E) on immunoblots as well as compromised ERAD of bri1-5 (Figure 6G).
The EBS4 Gene Can Complement the Yeast Δalg12 Mutation
To directly test if the EBS4 gene can replace the function of the yeast ALG12, we generated yeast expression plasmids pYEp352-EBS4, pYEp352-ebs4-2, and pYEp352-ebs4-3, each containing the promoter and terminator of the yeast ALG12 gene, and transformed them individually into the yeast Δalg12 cells that exhibit hypoglycosylation of an ER-localized carboxypeptidase Y (CPY) (Burda et al., 1999). Total yeast proteins were extracted from the wild-type and transformed Δalg12 cells, and the CPY glycosylation pattern was analyzed by immunoblotting. As shown in Figure 6H, a single band of mCPY (m indicates mature) was detected in wild-type cells, and three discrete faster-moving CPY* bands (* denotes abnormal CPY) were detected in Δalg12 cells transformed with an empty vector. Expression of the yeast ALG12 gene or the wild-type EBS4 cDNA was able to rescue the glycosylation defect of CPY, converting the three CPY* bands to a single mCPY band on immunoblots, whereas the ebs4-3–mutated construct failed to complement the Δalg12 mutation. Interestingly, although the Arabidopsis ebs4-2 mutant exhibits almost identical phenotypes to ebs4-1 that carries a splicing-defective mutation (Figure 3), the ebs4-2 mutation only slightly reduces the EBS4 ability to complement the Δalg12 mutation since the majority of detected CPY in pYEp352-ebs4-2 Δalg12 yeast cells exhibited the same mobility as the single mCPY band in wild-type cells (Figure 6H). We thus conclude that EBS4 is the likely Arabidopsis ortholog of the yeast ALG12 that catalyzes the addition of 8th Man in assembling the Dol-PP-Glc3Man9GlcNAc2 glycan.
Expression of the EBS2 Gene Rescues the ebs4 bri1-9 Phenotype
There are two possible reasons that the ebs4 mutations suppress the bri1-9 mutant phenotype. The first reason is that reduced numbers of Man residues of N-glycans on bri1-9 might reduce its affinity to interact with ER lectins; the second reason is that overaccumulated bri1-9 saturates its ER retention machinery. Our Kif rescue and bri1-9 overexpression experiments (Figure 2D; see Supplemental Figure 1 online) seemed to support the second explanation, which was further supported by our discovery that the ebs4-2 mutation did not reduce the bri1-9-CNX interaction (see Supplemental Figure 4 online). Our previous studies discovered two major components of this ERQC machinery, EBS1, the Arabidopsis UGGT homolog, and EBS2, a plant-specific CRT3, which act together to keep bri1-9 in the ER (Jin et al., 2007, 2009). To test which component is likely saturated by overaccumulated bri1-9, we introduced an EBS1 or EBS2 transgene into ebs4-2 bri1-9. As shown in Figures 7C to 7E, overexpression of EBS1 driven by the 35S promoter had little effect on ebs4-2 bri1-9, but expression of EBS2 driven by its native promoter resulted in transgenic plants that are similar to or even smaller than the gEBS4-complemented ebs4-2 bri1-9 mutants. Thus, EBS2 is likely a limiting factor that can be easily saturated by overaccumulated bri1-9 proteins.
Figure 7.
The Morphological Effect of Transgene Expression on the ebs4-2 bri1-9 Mutant.
Shown here are images of 4-week-old soil-grown transgenic ebs4-2 bri1-9 plants carrying the empty pPZP212 vector (A), a genomic EBS4 transgene (gEBS4) (B), an EBS1 overexpression transgene driven by the 35S promoter (p35S-EBS1) (C), and a genomic EBS2 transgene (gEBS2) (D). [See online article for color version of this figure.]
DISCUSSION
bri1-9 Is Degraded by a Proteasome-Dependent ERAD Pathway
In this study, we have shown using chemical inhibitors that the ER-retained bri1-9 is degraded by a proteasome-mediated ERAD process. We demonstrated that treatment of bri1-9 seedlings with Kif resulted in a strong increase in bri1-9 abundance and phenotypic suppression of its dwarf phenotype. It was this result that prompted us to conduct a secondary screen with previously isolated ebs mutants for mutations that compromise the ERAD of bri1-9. What was a bit surprising was our discovery that the degradation of bri1-9 seems to be dependent on proteasome since treatment with MG132 was able to stabilize bri1-9. This is in sharp contrast with our previous finding that ERAD of bri1-5 does not involve a proteasome-mediated process. It will be interesting to know why the two ER-retained BR receptors are degraded differently and to determine the factors that drive them into different degradation pathways.
EBS4 Is the likely Arabidopsis Ortholog of the Yeast ALG12
The assembly of lipid-linked glycans is a highly conserved pathway in higher eukaryotes and was elucidated mainly through genetic and biochemical analysis of the yeast alg mutants (Burda and Aebi, 1999). Mutations in human ALG genes result in type I congenital disorders of glycosylation largely due to hypoglycosylation of important glycoproteins (Freeze and Aebi, 2005). However, little is known about the assembly of lipid-linked glycan in plants (Pattison and Amtmann, 2009). A recent study reported the first loss-of-function Arabidopsis alg mutant through a reverse genetic approach and revealed that the incompletely assembled Man5GlcNAc2 glycan can be efficiently transferred to glycoproteins and processed in the Golgi to produce normal C-type N-glycans in Arabidopsis (Henquet et al., 2008).
In this study, we identified another Arabidopsis ALG gene through a forward genetic screen looking for second site mutations that suppress bri1-5 or bri1-9. We present strong evidence that EBS4 encodes a putative Arabidopsis ortholog of the yeast ALG12, an α1,6 mannosyltransferase that catalyzes the addition of the 8th Man during the assembly of the lipid-linked Glc3Man9GlcNAc2 glycan (Figure 1). First, the EBS4 candidate gene was initially discovered by BLASTX search against the entire Arabidopsis genome using the yeast ALG12 as query. Second, ebs4 mutations, while having no effect on C-type N-glycan biosynthesis, result in transfer of incompletely assembled glycans onto proteins, as shown by slower mobility of bri1-9, PDI, CRTs, and two major Con A–positive bands on immunoblots (Figures 2E, 4A, and 4B). Third, ebs4 mutations inhibit ERAD of two ER-retained BR receptors, consistent with the effect of the yeast Δalg12 mutation on a model yeast ERAD substrate CPY (Jakob et al., 1998). Finally, the wild-type EBS4 gene but not the mutated EBS4 plasmid carrying the E38K mutation (ebs4-3) was able to complement the yeast Δalg12 mutation.
Interestingly, E38 is absolutely conserved not only among ALG12 homologs but also among members of three α2-mannosyltransferase families that include ALG9, PIG-B, and SMP3 (Oriol et al., 2002). ALG9 catalyzes the addition of 7th and 9th Man resides in the assembly of Dol-PP-Glc3Man9GlcNAc2, while the other two are involved in the synthesis of the phosphatidyl-inositol glycan anchor, better known as PIG (Ferguson, 1992). Sequence analysis indicated that this acidic residue is located within the highly conserved long loop between the first two transmembrane segments (Figure 6C). We predicted that this Glu residue might be directly involved in catalyzing the mannosyltransferase reaction. It should also be interesting to note that the Ser-307 residue mutated in ebs4-2 is located in the 10th transmembrane segment, three amino acids away from a highly conserved small loop linking the 10th and 11th transmembrane domains. Surprisingly, while this mutation results in similar phenotypes as ebs4-1 that carries a 7th intron/8th exon junction mutation that likely causes a splicing defect and early translational termination (Figure 3), it only slightly reduces the ALG12 activity in yeast cells (Figure 6H). Based on its location, the Ser307Pro mutation in EBS4 might behave similarly to the Ser662Phe mutation in BRI1 by affecting protein folding with a marginal effect on catalytic activity. It is possible that the Ser307Pro-mutated EBS4 protein is not efficiently recognized by the yeast quality control system but is recognized and degraded by a high-fidelity ER quality control system and its associated ERAD machinery in Arabidopsis. Further studies using an anti-EBS4 antibody or a GFP-tagged EBS4 protein could shed light on this interesting mutation.
The Likely Reasons for ebs4 Mutations to Inhibit ERAD of bri1-5 and bri1-9
One likely explanation for the ebs4 mutations' ability to inhibit ERAD of both bri1-5 and bri1-9 is lack of the ERAD glycan signal on the two defective BR receptors due to their glycosylation with incompletely assembled glycans. Although the removal of the 7th Man residue was previously considered as the ERAD signal (Lederkremer and Glickman, 2005), two recent studies presented convincing evidence that the exposure of an α1,6 mannose on N-glycans is a true ERAD signal to degrade misfolded glycoproteins (Quan et al., 2008; Clerc et al., 2009). These two studies showed that the yeast HTM1 protein, which was previously thought to function as an ERAD receptor (Jakob et al., 2001), is responsible for removing the 9th Man from the upper dimannose branch. Further investigation, such as overexpression of EBS4 in an alg9 mutant or construction of a triple mutant of alg3 ebs4 bri1-9, could tell if a similar mechanism is used in plants to generate the ERAD glycan signal on a misfolded glycoprotein. The former approach will generate an Man7GlcNAc2 glycan with an exposed α1,3 Man on the middle arm (6th Man) and an exposed α1,6 Man (8th Man) on the upper arm, while the latter approach will generate an Man5GlcNAc2 glycan exposing a different α1,6 Man (3rd Man) that can also be recognized by an ERAD lectin (Clerc et al., 2009).
The Role of N-Glycosylation in Plant Development and Plant Stress Response
N-glycosylation is a complex process involving the assembly of Glc3Man9GlcNAc2 and its subsequent transfer to nascent polypeptide in the ER and extensive N-glycan remodeling in the Golgi. Recent studies using Arabidopsis mutants indicated that N-glycosylation plays an essential role in plant development as mutations in most enzymes involved in the assembly of Glc3Man9GlcNAc2 cause severe developmental defect including embryo lethality (reviewed in Pattison and Amtmann, 2009). By contrast, mutations of a Golgi-localized N-glycan–modifying enzyme have little effect on plant development but result in reduced stress tolerance (Strasser et al., 2005; Frank et al., 2008; Kang et al., 2008). It seems that mutations affecting the formation of a monoglucosylated lower mannose branch (Figure 1) often lead to severe developmental defects since the terminal Glc residue is essential for a nascent protein to acquire its native conformation in the ER by interacting with CNX/CRT. This study and the previous report on the Arabidopsis alg3 mutants revealed that mutations in enzymes involved in the ER luminal addition of Man residues to assemble Glc3Man9GlcNAc2 have no detectable effect on plant growth and development or the formation of C-type N-glycan, raising a question why plants need these enzymes in the first place. Our discovery that ebs4 mutations block ERAD of a mutated cell surface receptor provided a possible answer to this question. The assembly of a complete Glc3Man9GlcNAc2 N-glycan precursor is required for generating an ERAD signal that diverts a misfolded protein into the ERAD pathway and might therefore play an important role in plant stress tolerance since stressful conditions could reduce protein folding efficiency and lead to accumulation of misfolded proteins in the ER. Further experiments using alg3, alg9, and alg12/ebs4 mutants will be needed to test our hypothesis.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana ecotypes Columbia (Col-0) and Wassileskija-2 (Ws-2) were used as wild-type controls. The ebs4-1, ebs4-2, and other ebs mutants were discovered in the genetic screen for extragenic suppressors of bri1-9 (Jin et al., 2007), while ebs4-3 was identified as a suppressor for bri1-5. Ethyl methanesulfonate mutagenesis of bri1-5 was performed as previously described (Jin et al., 2007). Seed sterilization and plant growth conditions were described previously (Li et al., 2001), and a root growth inhibition assay was performed as described (Clouse et al., 1996).
Isolation of the Full-Length EBS4 cDNA
A partial EBS4 cDNA clone DQ492199 was obtained from the ABRC. The missing 5′ 130-bp fragment was obtained by RT-PCR from total RNAs of 2-week-old wild-type seedlings using the EBS4-5′RT primer set: 5′-CGAAGCTTGAGACGATGCCGACGGATTC-3′ and 5′-TACCATATGCCAGATTGACTAATCC-3′ (the underlined sequences are restriction sites for HindIII and NdeI, respectively) and the SuperScript first-strand synthesis system for RT-PCR (Invitrogen). The resulting PCR fragment was double digested with HindIII and NdeI and cloned into a HindIII/XbaI-digested pBluescript KS− (Stratagene) plasmid along with an NdeI/XbaI-cut DQ492199 cDNA fragment to create a full-length EBS4 cDNA plasmid pBS-EBS4.
Plasmid Construction and Plant Transformation
The construction of a pPZP222-gEBS2 was previously described (Jin et al., 2009). The full-length EBS1 cDNA was PCR amplified and cloned into the binary vector pCHF1 that carries the 35S promoter and the pea (Pisum sativum) RbcS-E9 terminator (Fankhauser et al., 1999). A 6.6-kb genomic fragment, including 1.6-kb promoter region and the entire EBS4 coding region, was PCR amplified from BAC T7I23 and cloned into the binary vector pPZP212 (Hajdukiewicz et al., 1994). Each constructed plasmid involving PCR was fully sequenced to ensure that there were no PCR-introduced errors. These plasmids were transformed individually into ebs4-2 bri1-9 or ebs4-3 bri1-5 mutants via the Agrobacterium tumefaciens–mediated vacuum infiltration method (Clough and Bent, 1998).
Yeast Complementation Assay
For the yeast complementation assay, the yeast pYEp352-ALG12 was used as the positive control and to make the pYEp352-EBS4 plasmid by replacing the entire open reading frame of the yeast ALG12 with that of the EBS4 gene. Site-directed mutagenesis using the QuickChange XL site-directed mutagenesis kit (Stratagene) was performed to generate pYEp352-ebs4-2 and pYEp352-ebs4-3 plasmids, while a pYEp352 plasmid lacking only the ALG12 open reading frame was used as the negative control. Each plasmid was fully sequenced to ensure that there were no PCR-generated errors and was transformed into Δalg12 yeast cell using a rapid yeast transformation protocol (Gietz and Woods, 2002).
Protein Extraction and Immunoblot Analysis
Arabidopsis seedlings harvested from agar, soil, or liquid half-strength MS medium supplemented with or without BL (Chemiclones), Kif (Toronto Research Chemicals), or MG132 (Sigma-Aldrich) were grounded in liquid nitrogen, dissolved in 1× SDS sample buffer, and boiled for 10 min. After centrifugation to remove tissue debris, supernatants were used for immunoblot analyses or subjected to Endo H assay (Jin et al., 2007). Ninety microliters of supernatant were mixed with 10 μL 10× G5 buffer and incubated with or without 1000 units of Endo Hf (New England Biolabs) at 37°C for 1 h. Protein samples extracted from the same amount of seedlings were separated on 7% or 10% SDS-PAGE gel and stained with Coomassie Brilliant Blue to determine the relative amount of total proteins among different samples. Proteins from duplicated gels were transferred onto immobilon-P membranes (Millipore) and analyzed by immunoblot with antibodies made against BRI1 (Mora-Garcia et al., 2004), BES1 (Mora-Garcia et al., 2004), PDI (Rose Biotechnology), BiP (SPA-818; Stressgen), and maize (Zea mays) CRTs (Pagny et al., 2000).
Equal amounts of yeast cells of mid-log phase grown at 30°C were collected by centrifugation, resuspended in 1× bead buffer (0.3 M sorbitol, 0.1 M NaCl, 5 mM MgCl2, and 10 mM Tris, pH 7.4), lysed by vortexing with glass beads, and mixed with equal volumes of 2× SDS buffer. After 5 min boiling and 5 min centrifugation, the resulting supernatants were separated by SDS-PAGE and analyzed by immunoblotting with a monoclonal anti-CPY antibody (10A5; Invitrogen).
For all immunoblot experiments, horseradish peroxidase–conjugated goat anti-mouse (for anti-BiP or anti-CPY antibody), goat anti-rat (for anti-BES1 antibody), or goat anti-rabbit (for anti-BRI1, anti-PDI, or anti-maize-CRT antibody) IgG secondary antibodies and an Immobilon Western Chemiluminescent HRP substrate (ECL) kit (Millipore) were used for detection. The chemiluminescent signals were recorded on x-ray films (Blue Basic Autorad Film; ISC BioExpress) with multiple exposures to obtain nonsaturated signals for each protein of interest.
N-Linked Glycan Assays
Total proteins were extracted from lyophilized shoot tissues of 4-week-old soil-grown plants by the phenol method (Fitchette et al., 1999), resolved in 1× SDS sample buffer. Twenty-five milligrams of proteins/each extract were separated on 15% SDS-PAGE gels and visualized by Coomassie Brilliant Blue staining. After transferring to nitrocellulose membranes, glycoproteins were analyzed by lectin affinoblotting using the Con A/peroxidase system specific for H-type N-glycans (Faye and Chrispeels, 1985) or immunoblotting with antibodies made against β1,2-xylose or α1,3-fucose characteristic of C-type N-glycans in plants (Faye et al., 1993). Briefly, affinodetection of blots was performed by saturation in TBS buffer (20 mM Tris-HCl, pH 7.4, containing 0.5M NaCl) containing 0.1% (v/v) Tween 20 (TTBS) for 1 h, followed by two successive incubations at room temperature in 25 μg/mL Con A (Sigma-Aldrich) in TTBS containing 1 mM MgCl2 and 1mM CaCl2 for 1.5 h and in 50 μg/mL horseradish peroxidase (Sigma-Aldrich) in TTBS for 1 h. For immunodetection, blots were first saturated in 3% (w/v) gelatin in TBS for at least 1 h and incubated with anti-glycan antibodies (1/1000 dilution) in TBS with 1% (v/v) gelatin for 2 h and with horseradish peroxidase–conjugated goat anti-rabbit IgG antibodies (Bio-Rad) (1/3000 dilution) in TBS with 1% (v/v) gelatin for 1 h at room temperature. All nitrocellulose membranes were incubated with 4-chloro-1-naphtol and hydrogen peroxide with gentle agitation at room temperature until signals reached desired intensities.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: NP_001077448 (EBS4, At1g01245), NP_014427 (yeast ALG12), NP_077010 (human ALG12), and NP_001053463 (a rice ALG12 homolog).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Overexpression of bri1-9:GFP Can Suppress the Dwarf Phenotype of bri1-9.
Supplemental Figure 2. The ebs4-2 Mutation Has Little Effect on the Molecular Weight of the Wild-Type BRI1.
Supplemental Figure 3. The ebs4-2 Mutation Has No Effect on the C-Type N-Glycan Biosynthesis.
Supplemental Figure 4. The ebs4-2 Mutation Does Not Inhibit the bri1-9–CNX Interaction.
Supplementary Material
Acknowledgments
We thank the ABRC for providing BAC and cDNA clones, M. Aebi for providing the yeast SS328 and Δalg12 (YG840) strains and the pYEp352-ALG12 plasmid, F. Tax for bri1-5 seeds, A. Chang for the monoclonal anti-CPY antibody, R. Boston for anti-maize CRT antibody, Y. Yin for anti-BES1 antibody, and J. Chory for anti-BRI1 antiserum. This work was supported in part by grants from the National Institutes of Health (GM60519) and the Department of Energy (ER15672) to J.L. and by grants from Centre National de la Recherche Scientifique and the Agence Nationale de la Recherche to L.F.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Jianming Li (jian@umich.edu).
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Aebi, M., Gassenhuber, J., Domdey, H., and te Heesen, S. (1996). Cloning and characterization of the ALG3 gene of Saccharomyces cerevisiae. Glycobiology 6 439–444. [DOI] [PubMed] [Google Scholar]
- Banerjee, S., Vishwanath, P., Cui, J., Kelleher, D.J., Gilmore, R., Robbins, P.W., and Samuelson, J. (2007). The evolution of N-glycan-dependent endoplasmic reticulum quality control factors for glycoprotein folding and degradation. Proc. Natl. Acad. Sci. USA 104 11676–11681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernales, S., Papa, F.R., and Walter, P. (2006). Intracellular signaling by the unfolded protein response. Annu. Rev. Cell Dev. Biol. 22 487–508. [DOI] [PubMed] [Google Scholar]
- Burda, P., and Aebi, M. (1999). The dolichol pathway of N-linked glycosylation. Biochim. Biophys. Acta 1426 239–257. [DOI] [PubMed] [Google Scholar]
- Burda, P., Jakob, C.A., Beinhauer, J., Hegemann, J.H., and Aebi, M. (1999). Ordered assembly of the asymmetrically branched lipid-linked oligosaccharide in the endoplasmic reticulum is ensured by the substrate specificity of the individual glycosyltransferases. Glycobiology 9 617–625. [DOI] [PubMed] [Google Scholar]
- Burda, P., te Heesen, S., Brachat, A., Wach, A., Dusterhoft, A., and Aebi, M. (1996). Stepwise assembly of the lipid-linked oligosaccharide in the endoplasmic reticulum of Saccharomyces cerevisiae: Identification of the ALG9 gene encoding a putative mannosyl transferase. Proc. Natl. Acad. Sci. USA 93 7160–7165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caramelo, J.J., and Parodi, A.J. (2007). How sugars convey information on protein conformation in the endoplasmic reticulum. Semin. Cell Dev. Biol. 18 732–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caramelo, J.J., and Parodi, A.J. (2008). Getting in and out from calnexin/calreticulin cycles. J. Biol. Chem. 283 10221–10225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerc, S., Hirsch, C., Oggier, D.M., Deprez, P., Jakob, C., Sommer, T., and Aebi, M. (2009). Htm1 protein generates the N-glycan signal for glycoprotein degradation in the endoplasmic reticulum. J. Cell Biol. 184 159–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Clouse, S.D., Langford, M., and McMorris, T.C. (1996). A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 111 671–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser, C., Yeh, K.C., Lagarias, J.C., Zhang, H., Elich, T.D., and Chory, J. (1999). PKS1, a substrate phosphorylated by phytochrome that modulates light signaling in Arabidopsis. Science 284 1539–1541. [DOI] [PubMed] [Google Scholar]
- Faye, L., and Chrispeels, M.J. (1985). Characterization of N-linked oligosaccharides by affinoblotting with concanavalin A-peroxidase and treatment of the blots with glycosidases. Anal. Biochem. 149 218–224. [DOI] [PubMed] [Google Scholar]
- Faye, L., Gomord, V., Fitchette-Laine, A.C., and Chrispeels, M.J. (1993). Affinity purification of antibodies specific for Asn-linked glycans containing alpha 1→3 fucose or beta 1→2 xylose. Anal. Biochem. 209 104–108. [DOI] [PubMed] [Google Scholar]
- Ferguson, M.A. (1992). Colworth Medal Lecture. Glycosyl-phosphatidylinositol membrane anchors: The tale of a tail. Biochem. Soc. Trans. 20 243–256. [DOI] [PubMed] [Google Scholar]
- Fitchette, A.C., Cabanes-Macheteau, M., Marvin, L., Martin, B., Satiat-Jeunemaitre, B., Gomord, V., Crooks, K., Lerouge, P., Faye, L., and Hawes, C. (1999). Biosynthesis and immunolocalization of Lewis a-containing N-glycans in the plant cell. Plant Physiol. 121 333–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank, C.G., and Aebi, M. (2005). ALG9 mannosyltransferase is involved in two different steps of lipid-linked oligosaccharide biosynthesis. Glycobiology 15 1156–1163. [DOI] [PubMed] [Google Scholar]
- Frank, J., Kaulfurst-Soboll, H., Rips, S., Koiwa, H., and von Schaewen, A. (2008). Comparative analyses of Arabidopsis complex glycan1 mutants and genetic interaction with staurosporin and temperature sensitive3a. Plant Physiol. 148 1354–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeze, H.H., and Aebi, M. (2005). Altered glycan structures: the molecular basis of congenital disorders of glycosylation. Curr. Opin. Struct. Biol. 15 490–498. [DOI] [PubMed] [Google Scholar]
- Gietz, R.D., and Woods, R.A. (2002). Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350 87–96. [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz, P., Svab, Z., and Maliga, P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25 989–994. [DOI] [PubMed] [Google Scholar]
- Helenius, A., and Aebi, M. (2004). Roles of N-linked glycans in the endoplasmic reticulum. Annu. Rev. Biochem. 73 1019–1049. [DOI] [PubMed] [Google Scholar]
- Henquet, M., Lehle, L., Schreuder, M., Rouwendal, G., Molthoff, J., Helsper, J., van der Krol, S., and Bosch, D. (2008). Identification of the gene encoding the alpha1,3-mannosyltransferase (ALG3) in Arabidopsis and characterization of downstream N-glycan processing. Plant Cell 20 1652–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, Z., Jin, H., Tzfira, T., and Li, J. (2008). Multiple mechanism-mediated retention of a defective brassinosteroid receptor in the endoplasmic reticulum of Arabidopsis. Plant Cell 20 3418–3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houston, N.L., Fan, C., Xiang, J.Q., Schulze, J.M., Jung, R., and Boston, R.S. (2005). Phylogenetic analyses identify 10 classes of the protein disulfide isomerase family in plants, including single-domain protein disulfide isomerase-related proteins. Plant Physiol. 137 762–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob, C.A., Bodmer, D., Spirig, U., Battig, P., Marcil, A., Dignard, D., Bergeron, J.J., Thomas, D.Y., and Aebi, M. (2001). Htm1p, a mannosidase-like protein, is involved in glycoprotein degradation in yeast. EMBO Rep. 2 423–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob, C.A., Burda, P., Roth, J., and Aebi, M. (1998). Degradation of misfolded endoplasmic reticulum glycoproteins in Saccharomyces cerevisiae is determined by a specific oligosaccharide structure. J. Cell Biol. 142 1223–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, H., Hong, Z., Su, W., and Li, J. (2009). A plant-specific calreticulin is a key retention factor for a defective brassinosteroid receptor in the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 106 13612–13617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, H., Yan, Z., Nam, K.H., and Li, J. (2007). Allele-specific suppression of a defective brassinosteroid receptor reveals a physiological role of UGGT in ER quality control. Mol. Cell 26 821–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, J.S., et al. (2008). Salt tolerance of Arabidopsis thaliana requires maturation of N-glycosylated proteins in the Golgi apparatus. Proc. Natl. Acad. Sci. USA 105 5933–5938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, K., and Kamiya, Y. (2007). Structural views of glycoprotein-fate determination in cells. Glycobiology 17 1031–1044. [DOI] [PubMed] [Google Scholar]
- Kelleher, D.J., and Gilmore, R. (2006). An evolving view of the eukaryotic oligosaccharyltransferase. Glycobiology 16 47R–62R. [DOI] [PubMed] [Google Scholar]
- Kinoshita, T., Cano-Delgado, A., Seto, H., Hiranuma, S., Fujioka, S., Yoshida, S., and Chory, J. (2005). Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature 433 167–171. [DOI] [PubMed] [Google Scholar]
- Lederkremer, G.Z., and Glickman, M.H. (2005). A window of opportunity: Timing protein degradation by trimming of sugars and ubiquitins. Trends Biochem. Sci. 30 297–303. [DOI] [PubMed] [Google Scholar]
- Li, J., and Chory, J. (1997). A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90 929–938. [DOI] [PubMed] [Google Scholar]
- Li, J., and Jin, H. (2007). Regulation of brassinosteroid signaling. Trends Plant Sci. 12 37–41. [DOI] [PubMed] [Google Scholar]
- Li, J., Nam, K.H., Vafeados, D., and Chory, J. (2001). BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol. 127 14–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinari, M. (2007). N-glycan structure dictates extension of protein folding or onset of disposal. Nat. Chem. Biol. 3 313–320. [DOI] [PubMed] [Google Scholar]
- Mora-Garcia, S., Vert, G., Yin, Y., Cano-Delgado, A., Cheong, H., and Chory, J. (2004). Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev. 18 448–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oriol, R., Martinez-Duncker, I., Chantret, I., Mollicone, R., and Codogno, P. (2002). Common origin and evolution of glycosyltransferases using Dol-P-monosaccharides as donor substrate. Mol. Biol. Evol. 19 1451–1463. [DOI] [PubMed] [Google Scholar]
- Pagny, S., Cabanes-Macheteau, M., Gillikin, J.W., Leborgne-Castel, N., Lerouge, P., Boston, R.S., Faye, L., and Gomord, V. (2000). Protein recycling from the Golgi apparatus to the endoplasmic reticulum in plants and its minor contribution to calreticulin retention. Plant Cell 12 739–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattison, R.J., and Amtmann, A. (2009). N-glycan production in the endoplasmic reticulum of plants. Trends Plant Sci. 14 92–99. [DOI] [PubMed] [Google Scholar]
- Persson, S., Rosenquist, M., Svensson, K., Galvao, R., Boss, W.F., and Sommarin, M. (2003). Phylogenetic analyses and expression studies reveal two distinct groups of calreticulin isoforms in higher plants. Plant Physiol. 133 1385–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan, E.M., Kamiya, Y., Kamiya, D., Denic, V., Weibezahn, J., Kato, K., and Weissman, J.S. (2008). Defining the glycan destruction signal for endoplasmic reticulum-associated degradation. Mol. Cell 32 870–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz, A., and Herzog, V. (2004). Endoplasmic reticulum-associated degradation: exceptions to the rule. Eur. J. Cell Biol. 83 501–509. [DOI] [PubMed] [Google Scholar]
- Strasser, R., Stadlmann, J., Svoboda, B., Altmann, F., Glossl, J., and Mach, L. (2005). Molecular basis of N-acetylglucosaminyltransferase I deficiency in Arabidopsis thaliana plants lacking complex N-glycans. Biochem. J. 387 385–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga, F., Brostrom, C., Koide, T., and Arvan, P. (2000). Endoplasmic reticulum (ER)-associated degradation of misfolded N-linked glycoproteins is suppressed upon inhibition of ER mannosidase I. J. Biol. Chem. 275 40757–40764. [DOI] [PubMed] [Google Scholar]
- Trombetta, E.S. (2003). The contribution of N-glycans and their processing in the endoplasmic reticulum to glycoprotein biosynthesis. Glycobiology 13 77R–91R. [DOI] [PubMed] [Google Scholar]
- Vembar, S.S., and Brodsky, J.L. (2008). One step at a time: Endoplasmic reticulum-associated degradation. Nat. Rev. Mol. Cell Biol. 9 944–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, Y., Vafeados, D., Tao, Y., Yoshida, S., Asami, T., and Chory, J. (2005). A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 120 249–259. [DOI] [PubMed] [Google Scholar]
- Yin, Y., Wang, Z.Y., Mora-Garcia, S., Li, J., Yoshida, S., Asami, T., and Chory, J. (2002). BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109 181–191. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.