Abstract
Transcription-related chromatin decondensation has been studied in mammals for clusters of structurally and/or functionally related genes that are coordinately regulated (e.g., the homeobox locus in mice and the major histocompatability complex locus in humans). Plant genes have generally been considered to be randomly distributed throughout the genome, although several examples of metabolic gene clusters for synthesis of plant defense compounds have recently been discovered. Clustering provides for genetic linkage of genes that together confer a selective advantage and may also facilitate coordinate regulation of gene expression by enabling localized changes in chromatin structure. Here, we use cytological methods to investigate components of a metabolic gene cluster for synthesis of developmentally regulated defense compounds (avenacins) in diploid oat (Avena strigosa). Our experiments reveal that expression of the avenacin gene cluster is associated with cell type–specific chromatin decondensation, providing new insights into regulation of gene clusters in plants. Importantly, chromatin decondensation could be visualized not only at the large-scale level but down to the single gene level. We further show that the avenacin and sterol pathways are likely to be inversely regulated at the level of transcription.
INTRODUCTION
In mammalian systems, transcription-related chromatin decondensation has been observed in a number of artificial gene clusters, in highly transcribed chromosomal regions and for clusters of functionally related genes that are coordinately regulated during development or between different cells types, such as the Hoxb locus (Tumbar et al., 1999; Tsukamoto et al., 2000; Volpi et al., 2000; Williams et al., 2002; Chambeyron and Bickmore, 2004; Janicki et al., 2004; Chambeyron et al., 2005; Sproul et al., 2005; Morey et al., 2007). In plants, genes are normally considered to be randomly distributed throughout the genome, with the obvious exception of groups of physically linked genes that share sequence homology and that have arisen through tandem gene duplication (e.g., gene-for-gene type disease resistance genes) (Friedman and Baker, 2007). Recently, however, several examples of clusters of genes for synthesis of plant defense compounds have been found (Frey et al., 1997; Qi et al., 2004; Wilderman et al., 2004; Shimura et al., 2007; Field and Osbourn, 2008; Swaminathan et al., 2009). The identification of groups of genes that do not share sequence relatedness and that contribute to a common pathway raises some intriguing questions about the evolutionary pressures responsible for such self-organization (Amoutzias and Van de Peer, 2008). Clustering will facilitate the inheritance of beneficial combinations of genes that together confer a selective advantage (Gierl and Frey, 2001; Qi et al., 2004; Wong and Wolfe, 2005; Field and Osbourn, 2008), while disruption of such metabolic gene clusters may lead not only to loss of ability to synthesize a beneficial pathway end product but also to accumulation of toxic pathway intermediates (Field and Osbourn, 2008; Mylona et al., 2008). Physical clustering may also facilitate coordinate regulation of expression of pathway genes by enabling localized changes in chromatin structure (Sproul et al., 2005; Osbourn and Field, 2009).
Our investigations of the role of natural products in plant defense have led us to identify an operon-like gene cluster for the synthesis of antimicrobial terpenes (avenacins) in diploid oat (Avena strigosa) (Papadopoulou et al., 1999; Qi et al., 2004). These defense compounds accumulate in the roots, where they confer broad-spectrum resistance to attack by soil-borne pathogens (Papadopoulou et al., 1999). The ability to produce avenacins is restricted to the genus Avena and has evolved since the divergence of oats from other cereals and grasses. To date, we have cloned a total of five genes in this cluster and have defined a further two loci that show complete genetic cosegregation with these five genes (Papadopoulou et al., 1999; Haralampidis et al., 2001; Qi et al., 2004, 2006; Mugford et al., 2009). The expression of the avenacin biosynthesis genes is tightly regulated and is restricted to the epidermal cells of the root meristem, which is the site of accumulation of avenacins (Haralampidis et al., 2001; Qi et al., 2006; Mugford et al., 2009).
Here, we have selected the avenacin biosynthetic genes Sad1 and Sad2 for cytological investigation of chromatin decondensation in oat. Sad1 and Sad2 are adjacent genes that lie ∼60 kb apart from each other. The two genes are separated by an intervening intergenic sequence that contains repetitive elements but no obvious open reading frames (Qi et al., 2006). Sad1 encodes the enzyme that catalyzes the first committed step in the avenacin pathway (the oxidosqualene cyclase, β-amyrin synthase) (Haralampidis et al., 2001), and Sad2 encodes a cytochrome P450 that is required for further modification of the product of SAD1 action, β-amyrin (Qi et al., 2006). Sad1 and Sad2 are particularly amenable for study because, in addition to encoding key early pathway enzymes, these genes are longer than the other genes in the cluster and so are particularly amenable to cytological analysis. Here, we use mRNA and DNA in situ hybridization, focusing on Sad1 and Sad2, to investigate the relationship between cell type–specific gene expression and chromatin decondensation within the oat gene cluster.
RESULTS
Sad1 and Sad2 Are Cotranscribed in Root Epidermis Cells and Are Transcriptionally Silent in the Cortex
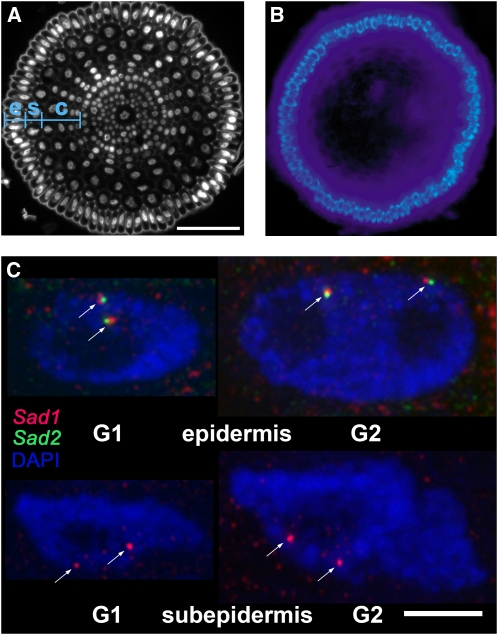
Avenacin A-1 is the major UV fluorescent compound present in oat roots and accumulates in the epidermal cells of the root tips, which are the site of synthesis (Hostettmann and Marston, 1995; Haralampidis et al., 2001; Qi et al., 2006; Mugford et al., 2009) (Figures 1A and 1B). We used mRNA fluorescence in situ hybridization (FISH) to examine the expression of the Sad1 and Sad2 genes in root cells. All experiments were performed on wax-embedded root sections of 3-d-old seedlings of A. strigosa. Sad1 and Sad2 were simultaneously transcriptionally active in the nuclei of over 80% of root tip epidermal cells (Figure 1C, Table 1). Sense probes for these genes did not give signals, indicating that the signals that we observed were derived from mRNA and not DNA (see Supplemental Figure 1 online). Sad1 was additionally expressed in ∼50% of the cells of the outermost cortex layer, referred to here as the subepidermis (Figures 1A and 1C), while Sad2 transcripts were seldom detected in this cell layer. Sad1 and Sad2 transcripts were not observed in other cell types within the root (see Supplemental Figure 1 online). There were no obvious differences between G1 and G2 nuclei in coexpression of Sad1 and Sad2 in root tip epidermal cells (Figure 1C), implying that expression of Sad1 and Sad2 is not cell cycle dependent.
Figure 1.
Visualization of Nascent Sad1 and Sad2 Transcripts in Nuclei of Oat Root Tip Cells.
(A) Cross section of an oat root tip with nuclei stained with 4',6-diamidino-2-phenylindole (DAPI) showing the different cell types (e, epidermis; s, subepidermis; c, cortex). Bar = 50 μm.
(B) Fluorescence of avenacin A-1 in the root tip epidermal cells.
(C) Detection of Sad1 (red) and Sad2 (green) transcripts in nuclei of root tip cells; blue, chromatin stained with DAPI. Representative G1 and G2 nuclei are shown for the epidermal and subepidermal calls. The images are overlays of 27 optical sections with section spacing of 0.2 μm. Arrows indicate the nuclear sites of nascent transcripts. In each panel of (C), a single nucleus is shown. Bar = 5 μm.
Table 1.
Detection of Nascent Transcripts of Sad1 and Sad2 in the Nuclei of Cells within the Oat Root Tip
| Percentage of Alleles Expressing: |
|||||
|---|---|---|---|---|---|
| Cell Type | Sad1 + Sad2 | Sad1 | Sad2 | Neither | Total |
| Epidermis | 81.4 | 1.8 | 9.5 | 7.3 | 100 |
| Subepidermis | 2.1 | 50.7 | 0.7 | 46.4 | 100 |
Total alleles counted: Epidermis, 220; subepidermis, 140.
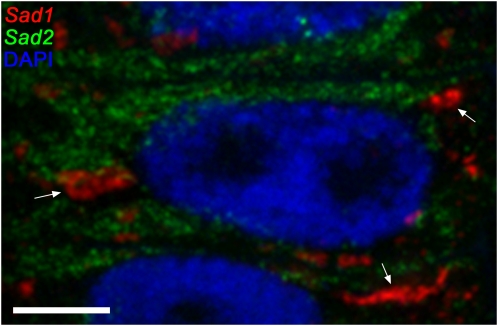
Cytoplasmic proteins and nuclear-encoded plastid and mitochondrial proteins are generally translated on free ribosomes in the cytoplasm, while integral membrane proteins and those that are secreted are translated on ribosomes bound to the ER. SAD1 and SAD2 are not predicted to be integral membrane proteins, but based on structural predictions and by analogy with their sterol biosynthetic counterparts, both are likely to be membrane associated (Haralampidis et al., 2001; Qi et al., 2004, 2006). In addition to the nascent transcripts that we detected in the nuclei (Figure 1), cytoplasmic transcripts derived from Sad1 and Sad2 were readily detectable in the root tip epidermal cells (Figure 2). Cytoplasmic Sad1 transcript was also evident at lower levels in the cytoplasm of the cells of the subepidermis (see Supplemental Figure 2 online). The spatial pattern of the Sad2 transcript resembled a typical reticulate endoplasmic reticulum (ER) distribution in the cytoplasm. By contrast, the Sad1 transcript was localized in foci that may represent branched strands of a membranous system, possibly specialized subdomains of the ER. These experiments indicate that the patterns of localization of the Sad1 and Sad2 mRNAs in the cytoplasm are distinct and rarely overlap, suggesting that the translation of these two mRNA species into proteins is unlikely to be spatially coordinated. A few examples of targeting of transcripts to specific parts of the cytoplasm in plant cells have been reported, including transcripts for expansins and seed storage proteins (Choi et al., 2000; Okita and Choi, 2002; Crofts et al., 2004; Washida et al., 2004). Avenacins accumulate in the cell vacuole (Mylona et al., 2008), but the route by which pathway intermediates are transported from the ER to this final destination is unknown. It is possible that this process could be mediated via zip-coded regions of the ER (Grotewold and Davies, 2008).
Figure 2.
Localization of Cytoplasmic Transcripts Derived from Sad1 and Sad2.
Cytoplasmic localization of Sad1 and Sad2 transcripts in oat root epidermal cells visualized by mRNA FISH. The Sad1 transcript (red) is localized in discrete foci (examples shown by the arrows), while the Sad2 transcript (green) shows a reticulate distribution suggestive of localization to the reticulate ER. Blue, chromatin stained with DAPI. Bar = 5 μm.
The Genomic Region Spanning Sad1 and Sad2 Is Decondensed in the Root Epidermis
We then used DNA FISH to determine whether expression of genes within the avenacin gene cluster is accompanied by chromatin decondensation. Previously, we constructed and sequenced a BAC contig spanning the Sad1/Sad2 region and showed that these two genes lie within 58 kb of each other in the A. strigosa genome (Figure 3A) (Qi et al., 2004, 2006). We generated probes for the Sad1 and Sad2 genes (15.2 and 5.8 kb, respectively) (Figure 3A). Using these probes, we were able to detect the Sad1 and Sad2 loci in nuclei of the epidermal, subepidermal, and cortical cells of A. strigosa root tips by DNA FISH (Figures 3B and 3C). The hybridization signal for Sad1 in the nuclei of epidermal cells frequently comprised more than one fluorescent focus. In several cases, this was also true for Sad2. However, in the subepidermal and cortical cells, each locus was almost always visualized as a discrete focus. The nuclei that we examined in these experiments were all G1 nuclei (see Methods) and so these multiple foci are not attributable to signals derived from sister chromatids. The multiple foci that we have observed for Sad1 (and to a lesser extent Sad2) in the nuclei of the root tip epidermal cells are therefore likely to reflect decondensation of the respective loci. Beaded signals have also been seen in much larger, transcriptionally active chromosome regions spanning groups of major histocompatibility complex and other genes in mammalian cells (Volpi et al., 2000; Müller et al., 2004).
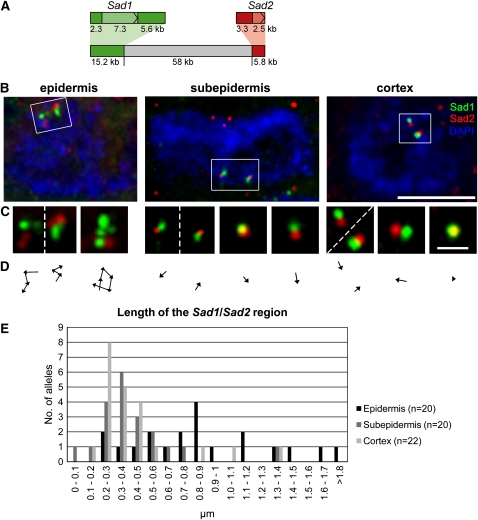
Figure 3.
Chromatin Decondensation in the Sad1/Sad2 Region.
(A) Diagram showing the Sad1 (green) and Sad2 (red) probes used for DNA in situ hybridization (coding regions in lighter shades of green and red).
(B) The Sad1/Sad2 region is decondensed in the nuclei of epidermal cells (left) compared with those of the subepidermis (center) and the cortex (right). In each panel, a single nucleus is shown. Bar = 5 μm.
(C) Detailed views of individual gene regions. Dashed lines separate loci on two adjacent chromosomes. Bar = 1 μm. The images shown in (B) and (C) are overlays of several optical sections, section spacing 0.2 μm. Blue, chromatin stained with DAPI.
(D) Line drawings showing the minimum paths measured in (C), starting from Sad1.
(E) Length distributions of the Sad1/Sad2 region in nuclei of epidermal, subepidermal, and cortical cells.
We then examined the length of the region encompassing Sad1 and Sad2 in the nuclei of the epidermal, subepidermal, and cortical cells. To do this, we measured a path consisting of the shortest distance in three dimensions traversing the intensity maxima of all fluorescent foci, starting with all Sad1 foci (green) and ending with all Sad2 foci (red), as shown by Figure 3C and the accompanying path outlines (Figure 3D). These data are summarized in Figure 3E. We calculated a mean length for the whole region of 0.97 μm for epidermis nuclei and of 0.41 and 0.42 μm for subepidermis and cortex nuclei, respectively. The length differences between epidermis and subepidermis/cortex are significant (P = 0.0001 for both, two-tailed Mann-Whitney U-test) and correspond to a chromatin compaction of 28-fold for the epidermis and 66- and 65-fold for subepidermis and cortical cells, compared with naked B-DNA (Watson and Crick, 1953). The length differences between subepidermal and cortical cells are not significant (P = 0.7, two-tailed Mann-Whitney U-test). Thus, the region shown in Figure 3A is more extended in nuclei of cells expressing both Sad1 and Sad2 (the epidermal cells) than in those cells that do not express these genes (the cortical cells) or in the subepidermal cell layer where Sad1 is expressed more sporadically and at lower levels. This suggests that coordinate expression of Sad1 and Sad2 in the root epidermal cells may be associated with chromatin decondensation. However, lower-level expression of Sad1 in the subepidermal cell layer is not associated with significant chromatin decondensation as assessed using these methods. In order to distinguish the contributions of the two genes to chromatin decondensation from that made by the intergenic sequence, we compared the distances between the closest Sad1 and Sad2 foci for each chromosome that we examined, as an estimated lower bound for the intergenic distance. The mean lengths obtained were 0.47 μm for epidermis nuclei and 0.31 and 0.34 μm for subepidermal and cortex nuclei, respectively. These differences between epidermis and subepidermis/cortex nuclei were significant (P = 0.03 for the difference between epidermis and cortex and P = 0.02 for the difference between epidermis and subepidermis, two-tailed Mann-Whitney U-test). There was no significant difference in the length of the intergenic regions when the values for subepidermis and cortex were compared (P = 0.9, two-tailed Mann-Whitney U-test). Our strategy for defining the paths that we have measured is based on the assumption that the closest foci that we observe for each gene are nearest neighbors in terms of the linear organization of the gene and that the Sad1 and Sad2 foci that are closest to each other represent the shortest intergenic distance. Clearly, this need not always be the case, and so we may have underestimated the degree of chromatin decondensation for both of these parameters.
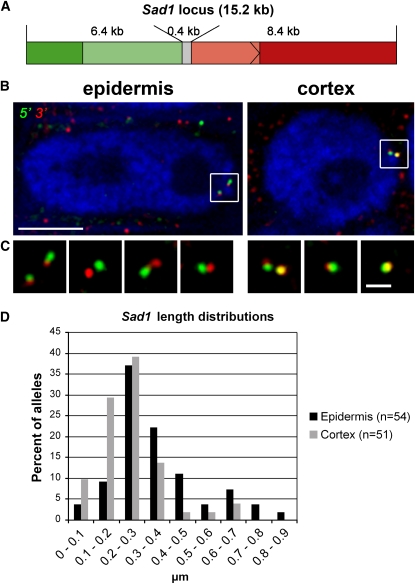
Sad1 Is Decondensed in the Root Epidermis
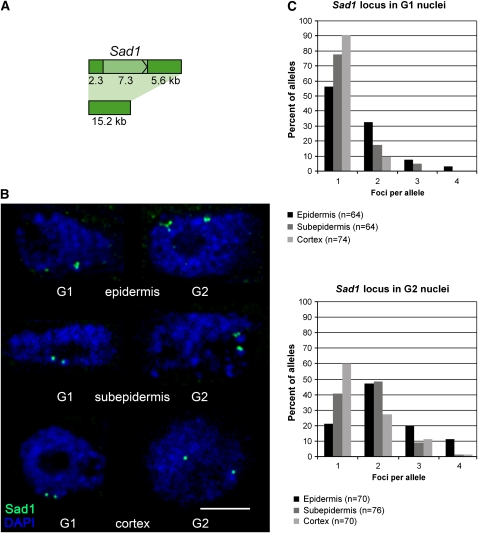
Next, we examined decondensation at the single gene level. First, we performed DNA FISH experiments using the same 15.2-kb Sad1 probe as in our previous experiments (Figures 3A and 4A) and analyzed the number of fluorescent foci in G1 and G2 nuclei of cells in the epidermis, subepidermis, and cortex of the root tips (Figures 4B and 4C) by three-dimensional optical sectioning through the nuclei. There were more fluorescent foci for Sad1 in the nuclei of epidermal cells when compared with the nuclei of subepidermal and cortical cells, consistent with decondensation of Sad1 in the nuclei of the epidermal cells. These differences were significant for both G1 and G2 nuclei (G1, P = 0.001; G2, P = 0.002, for epidermis compared with subepidermis; G1, P = 0.000004; G2, P = 0.00002 for epidermis compared with cortex; Figure 4C). There also appeared to be more Sad1 foci in the subepidermis compared with the cortex, although these differences were significant for G2 nuclei (P = 0.03) but not for G1 nuclei (P = 0.2). These results are consistent with our expression data (Figure 1C, Table 1), which indicate that Sad1 is expressed in the nuclei of root epidermal cells and to a lesser extent in nuclei of the subepidermal cells but not in the cortex. Sad1 shows an intermediate degree of decondensation when G2 nuclei of the cortex, subepidermis, and epidermis are compared (Figure 4), although cell cycle–related differences in Sad1 expression were not observed (Figure 1).
Figure 4.
The Sad1 Gene Locus Is Decondensed in Nuclei of Root Tip Epidermal Cells as Shown Using a 15.2-kb Probe Spanning Sad1.
(A) Diagram showing the 15.2-kb probe spanning the Sad1 gene locus used for DNA in situ hybridization (coding region in light green).
(B) Representative examples of Sad1 loci in different cell types. The images are overlays of several optical sections, section spacing 0.2 μm. Blue, chromatin stained with DAPI. Bar = 5 μm.
(C) Distributions of numbers of fluorescent foci in G1 and G2 nuclei in the three different cell types.
We then labeled the 5′ and 3′ parts of the Sad1 locus with different fluorochromes (Figure 5). In this case, we did not differentiate between G1 and G2 nuclei. We restricted our analysis to those loci that showed one green and one red focus (and so represented either G1 nuclei or overlapping chromatids in G2 nuclei) or that showed two red and two green foci that could be identified as belonging to two different chromatids (Figures 5B and 5C). We then measured the distances in three dimensions between the intensity maxima of the two halves for nuclei of root epidermal and cortical cells (Figure 5D). Based on these measurements, the mean length of Sad1 was calculated to be 0.35 μm in the epidermis and 0.23 μm in the cortex. This corresponds to a chromatin compaction of 15-fold in the epidermis and 22-fold in the cortex compared with naked B-DNA (Watson and Crick, 1953). These length differences are highly significant (P = 0.0004).
Figure 5.
Decondensation of the Sad1 Gene Locus Can Be Visualized Using Probes for the 5′ and 3′ Ends of the Gene.
(A) Diagram showing the probes for the 5′ and 3′ ends of the Sad1 gene used for DNA in situ hybridization. The two probes split the locus in half. The parts of the coding region encompassed by each probe are shown in lighter shades of green or red.
(B) G2 nuclei in epidermal and cortical root cells each showing two Sad1 loci: 5′ probe in green and 3′ probe in red. Bar = 5 μm.
(C) Detailed views of individual gene loci (with the framed loci shown in [B] on the left of each set of panels). Bar = 1 μm. The images shown in (B) and (C) are overlays of several optical sections, section spacing 0.2 μm. Blue, chromatin stained with DAPI.
(D) Length distributions of the Sad1 locus in nuclei of epidermal and cortical cells. The length of each locus was determined by measuring the distance in three dimensions between the intensity maxima of the two halves.
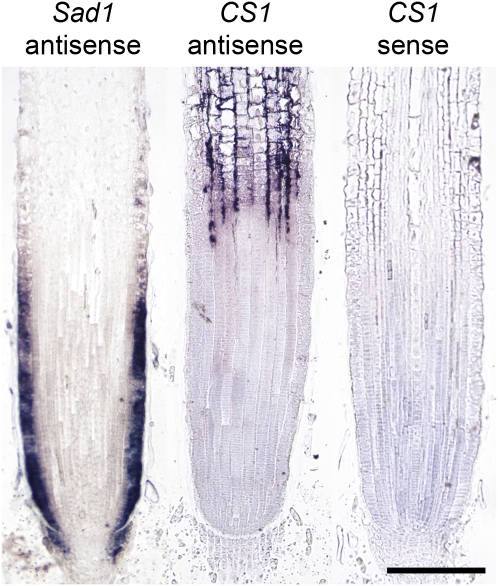
CS1, the Sad1 Homolog in Sterol Biosynthesis, Is Expressed in the Root Cortex but Not in the Epidermis of the Root Tip
Sad1 is likely to have been recruited from primary sterol metabolism by duplication of the cycloartenol synthase gene CS1, followed by acquisition of new function (Haralampidis et al., 2001; Qi et al., 2004). Sterols are essential for plant growth and development, and the genes for sterol biosynthesis are normally expressed throughout the plant (Ohyama et al., 2009). Previously, we cloned and characterized the A. strigosa CS1 gene and showed by RNA gel blot analysis that this gene (which shares 42% nucleotide sequence identity with Sad1) is expressed in the roots, stems, leaves, and flowers, as expected (Haralampidis et al., 2001; Qi et al., 2004). Here, we have performed mRNA in situ hybridization using alkaline phosphatase signal amplification to examine the spatial expression pattern of CS1 at higher resolution. Surprisingly, we found that CS1 transcripts were not detectable in the root tip. While Sad1 transcripts were detected primarily in the epidermal cells of the meristematic zone of the root tips of 3-d-old A. strigosa seedlings (and more weakly in the subepidermal cell layer and in parts of the columella), CS1 transcripts were only evident in the cortical cells behind this region (Figure 6). This suggests that the two pathways are likely to be inversely regulated at the level of transcription. Inverse coordinate regulation of the two pathways may be important because the two pathways compete for a common substrate (2,3-oxidosqualene) (Chappell, 2002). It could also represent a self-protection mechanism, since amphipathic triterpene glycosides, such as avenacins, exert their toxic effects on cells by complexing with sterols, causing membrane permeabilization (Morrissey and Osbourn, 1999). A cycloartenol synthase-independent pathway to plant sterols that involves synthesis of low levels of sterols via lanosterol has recently been described (Ohyama et al., 2009). It is possible that the cells of the root meristem synthesize essential sterols via this route.
Figure 6.
Distributions of Sad1 and Cycloartenol Synthase Transcripts in the Root Tip.
In situ mRNA hybridization of young root tips. Sad1 is expressed in the epidermis and weakly in parts of the subepidermis of the root tip and some columella cells, and CS1 expression is restricted to the cortex and starts in the elongation zone above the root tip. Bar = 200 μm.
[See online article for color version of this figure.]
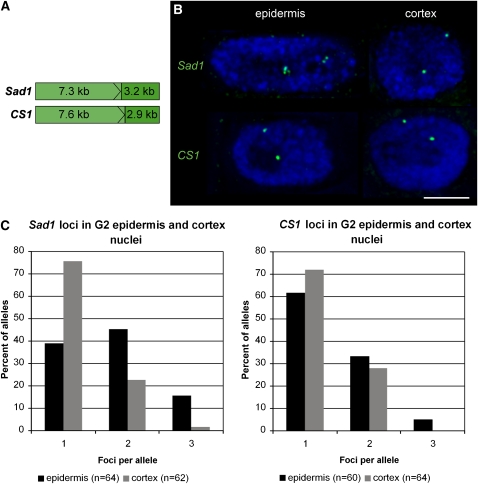
The coding regions (including introns) of Sad1 and CS1 are of similar length (7.3 and 7.6 kb, respectively). CS1 is not expressed at detectable levels in the root meristem and is unlinked to Sad1 (Qi et al., 2004). We labeled 10.5-kb sequences comprising the coding regions and 3′ flanks for each of the two genes (Figure 7A) and used these in DNA FISH experiments to compare the behavior of the Sad1 and CS1 loci in the nuclei of epidermal and cortical cells within the root tip (Figure 7B). We selected G2 nuclei for these experiments because the hybridization signals are more readily visible. Analysis of the number of fluorescent foci that were detected for the two loci in the nuclei of epidermis and cortical cells revealed that Sad1 is significantly more decondensed than CS1 in epidermal cells (P = 0.01). This confirms our previous findings that expression of Sad1 in the nuclei of epidermal cells is associated with chromatin decondensation at the Sad1 locus. By contrast, both the Sad1 and CS1 loci are condensed in the nuclei of cortical cells, and there is no significant difference in the number of foci observed for each (P = 0.6). Equally, there is no significant difference in the number of foci observed for CS1 loci in the nuclei of cortical and epidermal cells (P = 0.2). These experiments indicate that the differences in the degree of chromatin decondensation of the Sad1 locus in the epidermal and cortical cells of the root are unlikely to be due to general differences in chromatin conformation or compaction between the two cell types.
Figure 7.
Sad1 and CS1 Loci in Epidermis and Cortex Nuclei.
(A) After in situ hybridization with 10.5-kb probes spanning the coding regions and 3′ flanks of Sad1 or CS1, the number of fluorescent foci per allele was determined in G2 root epidermis and cortex nuclei.
(B) Representative epidermis and cortex nuclei with Sad1 or CS1 labeling. Overlay of several optical sections; section spacing, 0.15 μm. Blue, chromatin stained with DAPI. Bar = 5 μm.
(C) Distributions of fluorescent foci in epidermis and cortex nuclei (Sad1 appears more condensed in both the epidermis and the cortex than in Fig. 4 due to differences in probe lengths).
DISCUSSION
Previously, we showed that a multicopy transgene cluster in wheat (Triticum aestivum) containing endogenous genes for seed storage proteins under the control of their own promoters undergoes large-scale chromatin decondensation upon transcriptional activation during seed development (Wegel et al., 2005). Here, we have shown that a naturally occurring gene cluster for synthesis of developmentally regulated plant defense compounds decondenses in nuclei of root epidermal cells as assessed by cytological analysis of the region of the genome encompassing Sad1 and Sad2. Sad1 and Sad2 are the longest genes in the cluster that we have so far identified, and this enabled us to image them, while the sequences of the other genes were below the detection limit. We were able to correlate the expression of Sad1 and Sad2 with an increase in both the number of foci of in situ labeling and with the minimum path length necessary to join the respective foci of the genes. The latter provides a minimum estimate for decondensation of the cluster and corresponds to 28- and 65-fold compaction compared with B-DNA for the epidermal and cortical cells, respectively. This is likely to be an underestimate since the actual length may be longer than the shortest path and also because the signal for the most decondensed regions may be too faint to be detected. In other systems, the appearance of multiple foci instead of continuous signal has been attributed to the underlying chromatin structure, with less densely packed (invisible) regions linking more densely packed (visible) regions (Volpi et al., 2000; Müller et al., 2004). The murine HoxB gene cluster has been shown to decondense from a DNA compaction of around 300- to ∼34-fold after activation (Chambeyron and Bickmore, 2004), while a 375-kb region of the human major histocompatibility complex (MHCII) has a DNA compaction of ∼100-fold before induction with interferon and ∼60-fold after induction (Müller et al., 2004; Table 2). In both these cases, the values are averaged over the whole region, and the compaction of the actual transcribed genes compared with the intergenic regions cannot be deduced. Our experiments suggest that the Sad1 gene itself decondenses on activation, with a 15-fold compaction in the epidermis compared with 22-fold compaction in the cortex. This assumes a linear gene organization. If the active gene adopts a more complex structure, the compaction values are likely to be lower. The condensation state of the Sad1 gene also seems to depend on its level of activity. Judging by the number of fluorescent foci, Sad1 was more condensed in the subepidermis than in the epidermis. This is likely to be due to a combination of condensed, nonexpressing loci and also loci that are expressing at a lower level than in the epidermis. Transcriptional activity can precede any visible loosening of the chromatin structure (Janicki et al., 2004; Wegel et al., 2005), but as more genes within a colinear region decondense at the same time, the region itself becomes more decondensed, and this is likely to further facilitate expression. The fact that the intergenic region is only visibly decondensed in the nuclei of epidermal cells is consistent with this scenario.
Table 2.
DNA Compaction
| DNA | Size (kb) | Untranscribed Length Transcribed Length (μm) | kb/μm | Compaction | Reference |
|---|---|---|---|---|---|
| B-DNA | – | – | 2.9 | 1.0 | Watson and Crick (1953) |
| 10-nm fiber | – | – | 20.3 | 7.0 | Woodcock and Dimitrov (2001) |
| 30-nm fiber | – | – | 116–145 | 40–50 | Woodcock and Dimitrov (2001) |
| Hox1b-Hox9 | 90 | 0.1 | 900 | 310 | Chambeyron and Bickmore (2004) |
| 0.9 | 100 | 34 | |||
| MHCII | 375 | 1.2 | 312 | 107 | Müller et al. (2004) |
| 2.1 | 178 | 61 | |||
| Sad1-Sad2 | 79 | 0.42 (cortex) | 178 | 65 | This study |
| 0.97 (epidermis) | 81 | 28 | |||
| Sad1 | 15 | 0.23 (cortex)) | 65 | 22 | This study |
| 0.35 (epidermis) | 43 | 15 | |||
| Pea rDNA (transcribed) | 6.6 | 0.2-0.3 | 22–33 | 7.6–11.4 | Gonzalez-Melendi et al. (2001) |
| Chironomus BR2 (transcribed) | 37 | 3.5 | 10.4 | 3.6 | Daneholt et al. (1982) |
| Wheat glutenin array (20 copies) | 180 | 2.17 | 83 | 29 | Wegel et al. (2005) |
| 13.66 | 13 | 4.5 |
The higher-order structure of chromatin is still a matter of considerable debate. There is good evidence that nucleosomes are organized into 10-nm fibers, visualized as beads on a string under the electron microscope, which would have a compaction of sevenfold (Table 2) (Olins and Olins, 1974; Oudet et al., 1975; Goodrich and Tweedie, 2002). Thus, our data for Sad1 suggest that on average this gene is more compact than a 10-nm fiber, although it is likely that decondensation varies along the gene and may be greater where the polymerases are bound. There is very little data so far available about the condensation status of active genes. The Balbiani rings (giant chromosomal puffs) of the Dipteran midge Chironomus have been shown by electron microscopy to have a compaction of 3.6, well below that of the 10-nm fiber (Daneholt et al., 1982), while the rRNA genes in pea (Pisum sativum) show a greater degree of compaction (8- to 11-fold), which corresponds well with that of the 10-nm fiber (Gonzalez-Melendi et al., 2001) (Table 2). The Chironomus and pea genes studied in these investigations are very highly transcribed and have many polymerases attached, which may be expected to lead to a higher degree of decondensation. Most active genes are likely to have only one or two polymerases attached, so our value of 15 for compaction of active Sad1 seems reasonable.
The proposed levels of structural organization above the 10-nm fiber are highly controversial. A well-publicized secondary structure of chromatin is the 30-nm fiber, which is 40- to 50-fold more condensed than naked B-DNA (Goodrich and Tweedie, 2002). Such 30-nm fibers have been observed in vitro (Robinson et al., 2006), but as yet there is no clear evidence for these in vivo or, indeed, for more condensed higher-order structures (Branco and Pombo, 2007; Fakan and van Driel, 2007). Our mean chromatin compaction values indicate that in both the epidermis and the cortex, Sad1 is packaged into a structure that has a higher packing order than the 10-nm fiber and a lower one than the 30-nm fiber. For the Sad1/Sad2 region with its repeat-rich intergenic region (Qi et al., 2006), the packing order increases but is still less than the 30-nm fiber level in the epidermis, while it is above this level in the cortex (Table 2).
Our data provide cytological evidence to link cell type–specific chromatin decondensation with expression of the avenacin gene cluster in nuclei of oat root epidermal cells, providing new insights into regulation of gene clusters in plants. It has been suggested that opening up domains of secondary chromatin structure may place genes in a transcriptionally permissive environment, where their expression can then be triggered by transcription factors (Sproul et al., 2005). Further experiments are required to establish the cause and effect of this phenomenon and to identify the factors required for pathway regulation. The most satisfactory way to achieve this will be through characterization of regulatory mutants. We have isolated a resource of ∼100 avenacin-deficient mutants of A. strigosa (Papadopoulou et al., 1999; Qi et al., 2006; Mugford et al., 2009). Analysis of this mutant collection is expected to lead to the identification of loci that are required for regulation of expression of the gene cluster. These may include genes for pathway-specific transcription factors and also for regulation at the chromatin level.
METHODS
Plant Material
The diploid oat (Avena strigosa) accession number S75 (from the Institute of Grasslands and Environmental Research, Aberystwyth, Wales, UK) was used for all experiments.
RNA and DNA in Situ Hybridization
Oat seeds were imbibed in water for 24 h at 4°C and germinated on wet filter paper for 3 d at 22°C. Root tips were fixed in 4% (w/v) formaldehyde in PBS and embedded in wax and longitudinal sections (12 μm) prepared. Probes for in situ hybridization were prepared by in vitro transcription (for RNA probes) or by nick translation (for DNA probes) and labeled with either digoxigenin-11-UTP (Roche) or dinitrophenol-11-UTP (Perkin-Elmer) (see Supplemental Methods online for further details of probes, hybridization, and immunodetection methods).
Image Acquisition, Analysis, and Measurements
RNA in situ experiments with enzymatic detection were analyzed with a Nikon Eclipse 800 microscope using a ×10 objective with a numerical aperture of 0.3. Images were taken on a Pixera Pro 600 digital camera. For the FISH experiment, root sections were analyzed using a ×60 objective (numerical aperture 1.4, oil) on a Nikon Eclipse 600 microscope equipped with a Hamamatsu Orca ER cooled CCD digital camera, a motorized xy stage, and a z-focus drive. Further details concerning filters, correction of chromatic aberration, deconvolution of raw data, differentiation between G1 and G2 nuclei, length measurements, and preparation of images are given in Supplemental Methods online.
Accession Numbers
Sequence data for the genes referred to in this article can be found in the GenBank/EMBL data libraries under accession numbers DQ680849 (Sad1 and Sad2) and AY618693 (CS1).
Supplemental Data
The following materials are available in the online version of this article:
Supplemental Figure 1. Control RNA in Situ Hybridization Experiments for Sad1 and Sad2.
Supplemental Figure 2. Transcripts of Sad1 (red) and Sad2 (green) in Oat Root Tips.
Supplemental Methods.
Acknowledgments
This project was funded by the Biotechnology and Biological Sciences Research Council, UK (Grant BB/C504435/1).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Anne Osbourn (anne.osbourn@bbsrc.ac.uk).
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Amoutzias, G., and Van de Peer, Y. (2008). Together we stand: Genes cluster to coordinate regulation. Dev. Cell 14 640–642. [DOI] [PubMed] [Google Scholar]
- Branco, M.R., and Pombo, A. (2007). Chromosome organization: New facts, new models. Trends Cell Biol. 17 127–134. [DOI] [PubMed] [Google Scholar]
- Chambeyron, S., and Bickmore, W.A. (2004). Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev. 18 1119–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambeyron, S., Da Silva, N.R., Lawson, K.A., and Bickmore, W.A. (2005). Nuclear re-organisation of the Hoxb complex during mouse embryonic development. Development 132 2215–2223. [DOI] [PubMed] [Google Scholar]
- Chappell, J. (2002). The genetics and molecular genetics of terpene and sterol origami. Curr. Opin. Plant Biol. 5 151–157. [DOI] [PubMed] [Google Scholar]
- Choi, S.B., Wang, C., Muench, D.G., Ozawa, K., Franceschi, V.R., Wu, Y., and Okita, T.W. (2000). Messenger RNA targeting of rice seed storage proteins to specific ER subdomains. Nature 407 765–767. [DOI] [PubMed] [Google Scholar]
- Crofts, A.J., Washida, H., Okita, T.W., Ogawa, M., Kumamaru, T., and Satoh, H. (2004). Targeting of proteins to endoplasmic reticulum-derived compartments in plants. The importance of RNA localization. Plant Physiol. 136 3414–3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daneholt, B., Andersson, K., Bjorkroth, B., and Lamb, M.M. (1982). Visualization of active 75 S RNA genes in the Balbiani rings of Chironomus tentans. Eur. J. Cell Biol. 26 325–332. [PubMed] [Google Scholar]
- Fakan, S., and van Driel, R. (2007). The perichromatin region: A functional compartment in the nucleus that determines large-scale chromatin folding. Semin. Cell Dev. Biol. 18 676–681. [DOI] [PubMed] [Google Scholar]
- Field, B., and Osbourn, A.E. (2008). Metabolic diversification - Independent assembly of operon-like gene clusters in different plants. Science 320 543–547. [DOI] [PubMed] [Google Scholar]
- Frey, M., Chomet, P., Glawischnig, E., Stettner, C., Grun, S., Winklmair, A., Eisenreich, W., Bacher, A., Meeley, R.B., Briggs, S.P., Simcox, K., and Gierl, A. (1997). Analysis of a chemical plant defense mechanism in grasses. Science 277 696–699. [DOI] [PubMed] [Google Scholar]
- Friedman, A.R., and Baker, B.J. (2007). The evolution of resistance genes in multi-protein plant resistance systems. Curr. Opin. Genet. Dev. 17 493–499. [DOI] [PubMed] [Google Scholar]
- Gierl, A., and Frey, M. (2001). Evolution of benzoxazinone biosynthesis and indole production in maize. Planta 213 493–498. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Melendi, P., Wells, B., Beven, A.F., and Shaw, P.J. (2001). Single ribosomal transcription units are linear, compacted Christmas trees in plant nucleoli. Plant J. 27 223–233. [DOI] [PubMed] [Google Scholar]
- Goodrich, J., and Tweedie, S. (2002). Remembrance of things past: Chromatin remodeling in plant development. Annu. Rev. Cell Dev. Biol. 18 707–746. [DOI] [PubMed] [Google Scholar]
- Grotewold, E., and Davies, K. (2008). Trafficking and sequestration of anthocyanins. Nat. Prod. Comms. 3 1251–1258. [Google Scholar]
- Haralampidis, K., Bryan, G., Qi, X., Papadopoulou, K., Bakht, S., Melton, R., and Osbourn, A. (2001). A new class of oxidosqualene cyclases directs synthesis of antimicrobial phytoprotectants in monocots. Proc. Natl. Acad. Sci. USA 98 13431–13436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hostettmann, K., and Marston, A. (1995). Chemistry and Pharmacology of Natural Products: Saponins. (Cambridge, UK: Cambridge University Press).
- Janicki, S.M., Tsukamoto, T., Salghetti, S.E., Tansey, W.P., Sachidanandam, R., Prasanth, K.V., Ried, T., Shav-Tal, Y., Bertrand, E., Singer, R.H., and Spector, D.L. (2004). From silencing to gene expression: Real-time analysis in single cells. Cell 116 683–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morey, C., Da Silva, N.R., Perry, P., and Bickmore, W.A. (2007). Nuclear reorganisation and chromatin decondensation are conserved, but distinct, mechanisms linked to Hox gene activation. Development 134 909–919. [DOI] [PubMed] [Google Scholar]
- Morrissey, J.P., and Osbourn, A.E. (1999). Fungal resistance to plant antibiotics as a mechanism of pathogenesis. Microbiol. Mol. Biol. Rev. 63 708–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugford, S.T., et al. (2009). A serine carboxypeptidase-like acyltransferase is required for synthesis of antimicrobial compounds and disease resistance in oats. Plant Cell 21 2473–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, W.G., Rieder, D., Kreth, G., Cremer, C., Trajanoski, Z., and McNally, J.G. (2004). Generic features of tertiary chromatin structure as detected in natural chromosomes. Mol. Cell. Biol. 24 9359–9370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylona, P., Owatworakit, A., Papadopoulou, K., Jenner, H., Qin, B., Findlay, K., Hill, L., Qi, X., Bakht, S., Melton, R., and Osbourn, A. (2008). Sad3 and Sad4 are required for saponin biosynthesis and root development in oat. Plant Cell 20 201–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohyama, K., Suzuki, M., Kikuchi, J., Saito, K., and Muranaka, T. (2009). Dual biosynthetic pathways to phytosterol via cycloartenol and lanosterol in Arabidopsis. Proc. Natl. Acad. Sci. USA 106 725–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okita, T.W., and Choi, S.B. (2002). mRNA localization in plants: Targeting to the cell's cortical region and beyond. Curr. Opin. Plant Biol. 5 553–559. [DOI] [PubMed] [Google Scholar]
- Olins, A.L., and Olins, D.E. (1974). Spheroid chromatin units (v bodies). Science 183 330–332. [DOI] [PubMed] [Google Scholar]
- Osbourn, A.E., and Field, B. (2009). Operons. Cell. Mol. Life Sci. 66 3755-3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oudet, P., Gross-Bellard, M., and Chambon, P. (1975). Electron microscopic and biochemical evidence that chromatin structure is a repeating unit. Cell 4 281–300. [DOI] [PubMed] [Google Scholar]
- Papadopoulou, K., Melton, R.E., Leggett, M., Daniels, M.J., and Osbourn, A.E. (1999). Compromised disease resistance in saponin-deficient plants. Proc. Natl. Acad. Sci. USA 96 12923–12928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, X., Bakht, S., Leggett, M., Maxwell, C., Melton, R., and Osbourn, A. (2004). A gene cluster for secondary metabolism in oat: Implications for the evolution of metabolic diversity in plants. Proc. Natl. Acad. Sci. USA 101 8233–8238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, X., Bakht, S., Qin, B., Leggett, M., Hemmings, A., Mellon, F., Eagles, J., Werck-Reichhart, D., Schaller, H., Lesot, A., Melton, R., and Osbourn, A. (2006). A different function for a member of an ancient and highly conserved cytochrome P450 family: From essential sterols to plant defense. Proc. Natl. Acad. Sci. USA 103 18848–18853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, P.J., Fairall, L., Huynh, V.A., and Rhodes, D. (2006). EM measurements define the dimensions of the “30-nm” chromatin fiber: Evidence for a compact, interdigitated structure. Proc. Natl. Acad. Sci. USA 103 6506–6511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimura, K., et al. (2007). Identification of a biosynthetic gene cluster in rice for momilactones. J. Biol. Chem. 282 34013–34018. [DOI] [PubMed] [Google Scholar]
- Sproul, D., Gilbert, N., and Bickmore, W.A. (2005). The role of chromatin structure in regulating the expression of clustered genes. Nat. Rev. Genet. 6 775–781. [DOI] [PubMed] [Google Scholar]
- Swaminathan, S., Morrone, D., Wang, Q., Fulton, B.D., Peters, R.J. (2009). CYP76M7 is an ent-cassadiene C11α-hydroxylase defining a second multifunctional diterpenoid biosynthetic gene cluster in rice. Plant Cell 21 3315–3325.doi/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto, T., Hashiguchi, N., Janicki, S.M., Tumbar, T., Belmont, A.S., and Spector, D.L. (2000). Visualization of gene activity in living cells. Nat. Cell Biol. 2 871–878. [DOI] [PubMed] [Google Scholar]
- Tumbar, T., Sudlow, G., and Belmont, A.S. (1999). Large-scale chromatin unfolding and remodeling induced by VP16 acidic activation domain. J. Cell Biol. 145 1341–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpi, E.V., Chevret, E., Jones, T., Vatcheva, R., Williamson, J., Beck, S., Campbell, R.D., Goldsworthy, M., Powis, S.H., Ragoussis, J., Trowsdale, J., and Sheer, D. (2000). Large-scale chromatin organization of the major histocompatibility complex and other regions of human chromosome 6 and its response to interferon in interphase nuclei. J. Cell Sci. 113 1565–1576. [DOI] [PubMed] [Google Scholar]
- Washida, H., Sugino, A., Messing, J., Esen, A., and Okita, T.W. (2004). Asymmetric localization of seed storage protein RNAs to distinct subdomains of the endoplasmic reticulum in developing maize endosperm cells. Plant Cell Physiol. 45 1830–1837. [DOI] [PubMed] [Google Scholar]
- Watson, J.D., and Crick, F.H.C. (1953). The structure of DNA. Cold Spring Harb. Symp. Quant. Biol. 18 123–131. [DOI] [PubMed] [Google Scholar]
- Wegel, E., Vallejos, R.H., Christou, P., Stoger, E., and Shaw, P. (2005). Large-scale chromatin decondensation induced in a developmentally activated transgene locus. J. Cell Sci. 118 1021–1031. [DOI] [PubMed] [Google Scholar]
- Wilderman, P.R., Xu, M., Jin, Y., Coates, R.M., and Peters, R.J. (2004). Identification of syn-pimara-7,15-diene synthase reveals functional clustering of terpene synthases involved in rice phytoalexin/allelochemical biosynthesis. Plant Physiol. 135 2098–2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, R.R., Broad, S., Sheer, D., and Ragoussis, J. (2002). Subchromosomal positioning of the epidermal differentiation complex (EDC) in keratinocyte and lymphoblast interphase nuclei. Exp. Cell Res. 272 163–175. [DOI] [PubMed] [Google Scholar]
- Wong, S., and Wolfe, K.H. (2005). Birth of a metabolic gene cluster in yeast by adaptive gene relocation. Nat. Genet. 37 777–782. [DOI] [PubMed] [Google Scholar]
- Woodcock, C.L., and Dimitrov, S. (2001). Higher-order structure of chromatin and chromosomes. Curr. Opin. Genet. Dev. 11 130–135. [DOI] [PubMed] [Google Scholar]