Abstract
Cytoplasmic actin cables are the most prominent actin structures in plant cells, but the molecular mechanism underlying their formation is unknown. The function of these actin cables, which are proposed to modulate cytoplasmic streaming and intracellular movement of many organelles in plants, has not been studied by genetic means. Here, we show that Arabidopsis thaliana formin3 (AFH3) is an actin nucleation factor responsible for the formation of longitudinal actin cables in pollen tubes. The Arabidopsis AFH3 gene encodes a 785–amino acid polypeptide, which contains a formin homology 1 (FH1) and a FH2 domain. In vitro analysis revealed that the AFH3 FH1FH2 domains interact with the barbed end of actin filaments and have actin nucleation activity in the presence of G-actin or G actin-profilin. Overexpression of AFH3 in tobacco (Nicotiana tabacum) pollen tubes induced excessive actin cables, which extended into the tubes' apices. Specific downregulation of AFH3 eliminated actin cables in Arabidopsis pollen tubes and reduced the level of actin polymers in pollen grains. This led to the disruption of the reverse fountain streaming pattern in pollen tubes, confirming a role for actin cables in the regulation of cytoplasmic streaming. Furthermore, these tubes became wide and short and swelled at their tips, suggesting that actin cables may regulate growth polarity in pollen tubes. Thus, AFH3 regulates the formation of actin cables, which are important for cytoplasmic streaming and polarized growth in pollen tubes.
INTRODUCTION
The actin cytoskeleton is a highly organized but complex and dynamic structure in eukaryotic cells. The actin cytoskeleton is formed by the assembly (or polymerization) of G-actin monomers into filamentous actin (F-actin or actin microfilaments) and is generally known to participate in cell growth, polarization, morphogenesis and movement, intracellular mobility, nuclear segregation, cytokinesis, vesicle trafficking, and organelle biogenesis (Higaki et al., 2007). Each of these functions is performed by a specific configuration (or pool) of F-actin and/or its dynamics. The organization and dynamics of specific actin microfilaments are regulated by various actin binding proteins that have the ability to bind either to monomeric or to filamentous actin or to both. As polar structures, actin filaments elongate mainly at their barbed end in the cell (Casella et al., 1981; Fox and Phillips, 1981; Tellam and Frieden, 1982; Yahara et al., 1982; Symons and Mitchison, 1991; Redmond et al., 1994). A key mechanism regulating actin assembly is the creation of new barbed ends by the de novo nucleation of new actin filaments (Blanchoin et al., 2000; Wear et al., 2000; Pollard et al., 2001).
The formation of actin nuclei is a rate-limiting step during spontaneous filament assembly. Within the cell, actin nucleation factors are responsible for the generation of actin nuclei, providing a mechanism for the cell to regulate when and where to assemble actin filaments. Several actin-nucleating factors are known to date (Welch et al., 1997; Pruyne et al., 2002; Quinlan et al., 2005; Chereau et al., 2008), among which the Arp2/3 complex and formins are two well-studied actin nucleators. The Arp2/3 complex initiates actin assembly from preexisting actin filaments to generate a branched network of actin filaments (Mullins et al., 1998), whereas formins induce polymerization of parallel actin filaments for the formation of actin bundles or cables. Following the initial identification of the mouse formin limb deformity (Kleinebrecht et al., 1982), formins have been identified from various eukaryotic kingdoms, including fungi, animals, and plants. The in vitro biochemical and biophysical studies show that formin nucleates actin assembly de novo and interacts with the barbed end of actin filaments, allowing actin polymerization to occur at the barbed end while attaching to it. This behavior led to the establishment of a model in which formin was proposed to act as a processive actin-promoting motor (Kovar and Pollard, 2004; Romero et al., 2004; Kovar et al., 2006). Formins are characterized by the presence of conserved formin homologous domains, FH1 and FH2, which are required and sufficient for actin nucleation. The FH1 domain contains a poly-proline stretch, which binds to profilin (actin monomer binding protein) and profilin-actin, while the number of poly-proline repeats is variable among different formins (Kovar et al., 2006). Provided with sufficient amounts of profilin, the rate of barbed-end elongation in the presence of formin increases with the increasing number of poly-proline tracks in the FH1 domain (Paul and Pollard, 2008). The FH2 domain contains actin binding sites and acts as a dimer to nucleate new actin filaments (Xu et al., 2004). Mouse FRL (formin-related gene in leukocytes) and AFH1 (Arabidopsis thaliana formin1) were shown to have bundling activity and directly participate in organizing actin filaments into bundles (Harris et al., 2004; Michelot et al., 2005, 2006). Formins play an important role in diverse cellular and developmental processes. For example, yeast formin, which is responsible for actin cable formation, regulates cell polarity and cytokinesis (Feierbach and Chang, 2001; Evangelista et al., 2002a; Sagot et al., 2002).
Plant formins contain the core FH1 and FH2 domains located in their C-terminal halves (Deeks et al., 2002; Cvrckova et al., 2004; Blanchoin and Staiger, 2008). However, plant formins lack autoinhibition and Rho GTPase activation domains found in animal and fungal formins (Cvrckova et al., 2004; Blanchoin and Staiger, 2008). Plant formins have evolved into two groups, referred to as group I and group II. Group I formins are characterized by the presence of a signal peptide and a transmembrane domain at its N terminus, which are presumably involved in their targeting to the plasma membrane (PM). No conserved domains were identified beside FH1 and FH2 in group II formins. Bioinformatic analysis reveals 21 formin genes in the Arabidopsis genome with 11 members in group I and 10 members in group II (Cvrckova et al., 2004). AFH1 and At FH5 have been shown to nucleate actin in vitro (Ingouff et al., 2005; Michelot et al., 2005). Overexpression of AFH1 (from group I) induced microfilament bundling and PM deformation in pollen tubes (Cheung and Wu, 2004). The localization of group I formins to the PM implies their potential roles in the regulation of cortical actin organization (Cheung and Wu, 2004; Michelot et al., 2005). Overexpression of At FH8 (group I) perturbs root hair tip growth (Deeks et al., 2005; Yi et al., 2005). At FH6 is upregulated during nematode infection and partially complemented the loss of BNI1p and BNR1p in Saccharomyces cerevisiae (Favery et al., 2004). At FH5 (group I) is localized to the cell plate, and its loss-of-function mutations reduce the rate of cell wall formation (Ingouff et al., 2005). This is the only example of functional analysis of plant formins by loss-of-function genetic studies to date. However, the cellular and developmental functions for most plant formins remain unclear. In this study, we integrated biochemical and loss- and gain- of-function genetic approaches to determine the function of pollen-specific AFH3 (a group I formin) in Arabidopsis pollen.
The pollen tube has been a favorite model system for the investigation of polarized cell growth and of its relation to the actin cytoskeleton (Vidali and Hepler, 2001; Sagot et al., 2002; Cole and Fowler, 2006; Lee and Yang, 2008; Yalovsky et al., 2008; Yang, 2008). Pollen tubes extend rapidly via tip growth, searching for the ovule to deliver the sperm cells to the embryo sac for fertilization. This polarized tip growth is known to require the actin cytoskeleton (Gibbon et al., 1999; Cole and Fowler, 2006). The growing pollen tube apparently contains multiple forms of actin microfilaments: fine, less abundant but highly dynamic actin microfilaments in the extreme apex, a dense cortical fringe or collar of microfilaments just behind the extreme apex, and the abundant longitudinal actin cables in the shank (Hepler et al., 2001; Sagot et al., 2002; Higashida et al., 2004). Evidence suggests that the apical F-actin modulates the apical accumulation of exocytic vesicles and their exocytosis (Sagot et al., 2002; Lee et al., 2008). The role of the actin fringe is not clear, although it is proposed to mediate endocytosis to retrieve excess materials deposited by exocytosis and/or the cytoplasmic organization of the apical region (Lovy-Wheeler et al., 2005; Cardenas et al., 2008). The actin cables, generally staying away from the apical region, have been proposed to regulate the cytoplasmic streaming, which occurs in the reverse fountain pattern (i.e., moving down toward the apex along the tube cortex and back in the center of the cell once reaching the subapical region). However, the function of these cables has not been studied by genetic means.
In this report, we demonstrate that the Arabidopsis formin AFH3 is required for the assembly of actin cables in pollen tubes and that disruption of these cables caused by genetic downregulation of AFH3 was associated with the inhibition of cytoplasmic streaming as well as growth depolarization, providing direct evidence that these cables are involved in the regulation of cytoplasmic streaming and polarized pollen tube growth.
RESULTS
Molecular Analysis of FH3
To investigate the molecular mechanism regulating the organization of the actin cytoskeleton in pollen tubes, we focused on AFH3, a group I formin that is preferentially expressed in Arabidopsis pollen. AFH3 deduced from the genomic sequence is predicted to encode a p140 mDial-like protein. There are two cDNA sequences encoding AFH3 reported in The Arabidopsis Information Resource (TAIR) database, one is 1938 bp encoding a 645–amino acid polypeptide, and the other is 1803 bp (lacking the N-terminal 45 amino acids) encoding a 600–amino acid polypeptide. The latter is consistent with the analysis of Cvrckova (2000). However, it contains most subdomains of FH2 but lacks the C-terminal I and J subdomains of FH2 (Cvrckova, 2000). To determine whether the AFH3 cDNA sequence annotated by TAIR is correct, we first designed a set of primers to amplify a cDNA starting at the ATG and ending at the stop codon TAA of the 1803-bp cDNA sequence predicted from TAIR (www.Arabidopsis.org). An ∼1.8-kb cDNA fragment was amplified. Sequencing of this cDNA revealed that the sequence before 1381 bp was identical to that of the TAIR prediction, except for an A replacing a G at 508 bp and an A replacing a T at 561 bp. An additional C was found at 1385 and 8 bp after this C, there was an additional 47 bp sequence, which was previously thought to be an intron. The inclusion of this sequence does not cause a frame shift, and the whole FH2 domain amino acid sequence is unchanged. These results demonstrate a sequencing error in the genome sequence, which was suggested by Cvrckova et al. (2004), who suspected that such an error could cause a missing exon within the FH2 domain (Cvrckova et al., 2004). However, our sequenced cDNA contains another G at the 1812 bp position that was not presented in the TAIR genome sequence. The presence of this G changes the reading frame so that it does not contain a TAA stop codon as predicted from the TAIR sequence. Based on this sequence analysis, we reannotated the genomic DNA sequence information and accordingly designed two 5′ primers and three downstream primers before the poly(A) signal sequence. These primers were used to amplify the full-length cDNA from Arabidopsis inflorescence mRNA. We concluded that the full-length AFH3 coding sequence is 2358 bp, which encodes a 785–amino acid protein including the sequence of a complete FH2 domain (see Supplemental Figure 1 online).
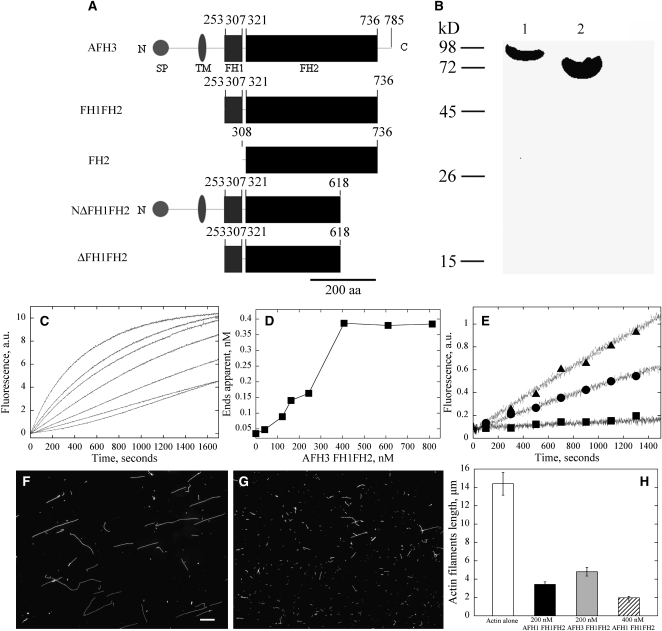
As shown in Figure 1A, AFH3 is a typical group I formin; it contains a signal peptide (1 to 20 amino acids) and a transmembrane domain (143 to 167 amino acids) at its N terminus. The FH1 domain (253 to 307 amino acids) contains a typical polyproline-rich stretch, implicated in binding to profilin (Evangelista et al., 1997; Watanabe et al., 1997; Banno and Chua, 2000). The conserved FH2 domain is located at its C-terminal halves (321 to 736 amino acids). The FH2 of AFH3 shares 34.8% amino acid similarity with that of Bni1p and 67.3% and 77.8% amino acid similarity with those of two well characterized Arabidopsis formin proteins, AFH1 and At FH5, respectively (see Supplemental Figure 2 online).
Figure 1.
Recombinant AFH3 FH1FH2 Fusion Protein Nucleates Actin Assembly
(A) Schematic representation of the predicted domain organization of AFH3. The full-length AFH3 contains 785 amino acids. The FH1 domain is from amino acids 253 to 307, and the FH2 domain is from amino acids 321 to 736. Recombinant AFH3 FH1FH2 and AFH3 FH2 fusion proteins include amino acids from 253 to 736 and 301 to 736, respectively. NΔFH1FH2 and ΔFH1FH2 are TAIR annotated full-length AFH3 and FH1FH2, respectively. SP, predicted signal peptide; TM, transmembrane domain; aa, amino acids.
(B) Purification of AFH3 FH1FH2 and AFH3 FH2 fusion proteins. Coomassie blue–stained protein gel of recombinant formin proteins. Lane 1, GST-AFH3 FH1FH2 protein; lane 2, GST-AFH3 FH2 protein.
(C) Time course of actin polymerization in the presence of AFH3 FH1FH2 monitored by pyrene fluorescence. Different concentrations of AFH3 FH1FH2 were added to 2.0 μM of 10% pyrene-labeled actin before initiation of actin polymerization. The concentration of AFH3 FH1FH2 from bottom to top is 0, 70, 90, 180, 250, 350, and 450 nM.
(D) Nucleation efficiency of AFH3 FH1FH2. The efficiency of nucleation for AFH3 FH1FH2 was determined at half-maximal actin polymerization according to Blanchoin et al. (2000).
(E) Time course of actin polymerization with 2.0 μM Arabidopsis profilin 4 in the presence or absence of AFH3 FH1FH2 monitored by pyrene fluorescence. Closed squares, 2.0 μM actin plus 2.0 μM Arabidopsis profilin 4; closed circles, 2.0 μM actin plus 2.0 μM Arabidopsis profilin 4 plus 500 nM AFH3 FH1FH2; closed triangles, 2.0 μM actin plus 2.0 μM Arabidopsis profilin 4 plus 1000 nM AFH3 FH1FH2.
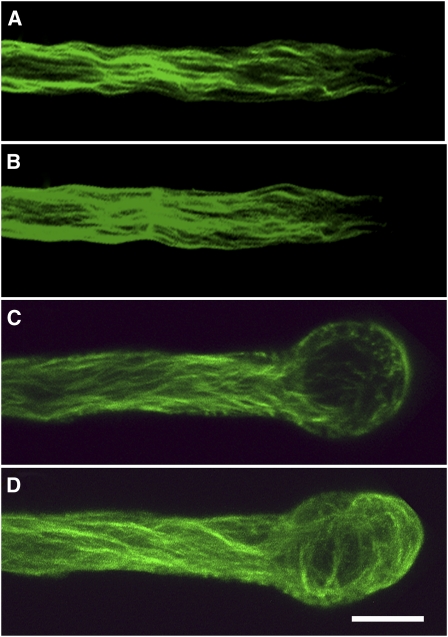
(F) and (G) Micrographs of actin filaments in the presence and absence of FH1FH2. (F) Actin alone; (G) actin plus 400 nM AFH3 FH1FH2. Bar in (F) = 10 μm.
(H) Actin filaments mean length (±se). Actin alone, white bars; Actin plus 200 nM AFH1 FH1FH2, black bars; Actin plus 200 nM AFH3 FH1FH2, gray bars; Actin plus 400 nM AFH3 FH1FH2, hatched bars.
The FH1FH2 of AFH3 Nucleated Actin Assembly
Based on the predicted structure of AFH3, we hypothesized that it is a functional formin that nucleates actin microfilaments. To test this hypothesis, we generated recombinant proteins containing the FH1FH2 domain (amino acids 253 to 736, AFH3 FH1FH2) and the FH2 domain (amino acids 307 to 736, AFH3 FH2) of FH3, respectively (Figure 1A). We were interested in testing the effect of the FH1FH2 domain of AFH3 on actin. To test this, N-terminal glutathione S-transferase (GST) fusion constructs were made and expressed in Escherichia coli (Figure 1B). Purified fusion proteins were used to test whether they nucleated actin assembly in vitro. Pyrene-labeled actin monomers were incubated with various concentrations of AFH3 FH1FH2 and AFH3 FH2 fusion proteins, and actin polymerization was monitored by pyrene fluorescence. As shown in Figure 1C, the AFH3 FH1FH2 fusion protein decreased the initial lag phase of actin polymerization in a dose-dependent manner, corresponding to active nucleation. However, the AFH3 FH2 fusion protein nucleated actin assembly very weakly (see Supplemental Figure 3A online). The behavior of the FH2 domain of AFH3 was similar to that of At FH5, which was also shown to be unable to nucleate actin from a pool of free actin monomers (Ingouff et al., 2005). The concentration of new barbed ends was strongly dependent on the presence of the AFH3 FH1FH2 fusion protein (Figure 1D).
It has been reported that the majority of actin monomer is buffered by profilin in some plant cells (Gibbon et al., 1999; Snowman et al., 2002; Wang et al., 2005; Chaudhry et al., 2007). It was imperative to test whether AFH3 is able to use the pool of actin bound to profilin for efficient nucleation of actin filaments. We performed the nucleation experiment in the presence of an equivalent molar concentration of Arabidopsis profilin 4. As shown in Figure 1E, the initial lag phase of actin polymerization was decreased in the presence of the AFH3 FH1FH2 fusion protein in a dose-dependent manner, confirming the active nucleation activity of the AFH3 FH1FH2 with actin-profilin. Elongation of actin filaments nucleated by the AFH3 FH1FH2 fusion protein with actin monomers bound to profilin confirms that these filaments were growing at their barbed ends. However, the FH2 domain of AFH3 did not nucleate actin polymerization in the presence of profilin. Filament length is inversely correlated with the concentration of nuclei. Accordingly, actin filaments were significantly shorter in the presence of the AFH3 FH1FH2 fusion protein (Figure 1G) than actin alone (Figure 1F), but the presence of AFH3 FH2 did not reduce the length of actin filaments (see Supplemental Figure 3Bb online), which is consistent with the pyrene-actin polymerization curve (see Supplemental Figure 3A online). The actin filaments' mean length (±se) in the absence and presence of 200 nM AFH1 FH1FH2, 200 nM AFH3 FH1FH2, and 400 nM AFH3 FH1FH2 is 14.4 ± 1.2 μm (n = 122), 3.4 ± 0.3 μm (n = 124), 4.8 ± 0.5 μm (n = 152), and 2.0 ± 0.2 μm (n = 123), respectively (Figure 1H).
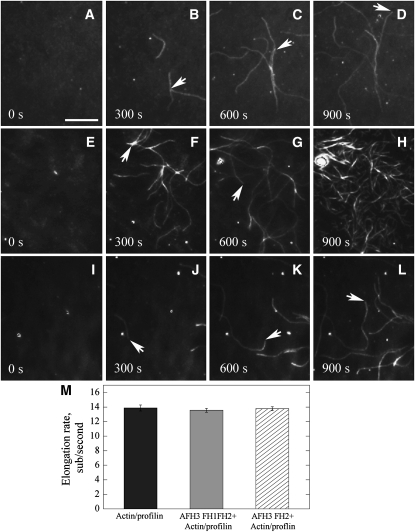
To further confirm the effect of AFH3 FH2 or AFH3 FH1FH2 on actin assembly, we employed total internal reflection fluorescence microscopy (TIRFM) to visualize actin polymerization directly. Actin polymerization was observed near the surface of the cover glass, which was coated with NEM-myosin alone or NEM-myosin plus either AFH3 FH1FH2 or AFH3 FH2. As shown in Figure 2, actin filaments on the cover glass were able to elongate rapidly during the 15-min observation period. The rates of actin elongation were determined. The rates of elongation were 13.9 ± 0.4 (n = 17) subunits per second for the actin/profilin complex (Figure 2M). In the presence of AFH3 FH2, the rates of actin elongation were determined to be 13.6 ± 0.4 (n = 15) subunits per second (Figure 2M), which is roughly the same as actin/profilin alone. At the end of 15-min window, the number of actin filaments did not increase significantly (Figure 2L; see Supplemental Movie 3 online) compared with that of actin/profilin alone (Figure 2D; see Supplemental Movie 1 online), confirming that AFH3 FH2 does not have nucleation activity, which is consistent with pyrene-actin polymerization curves (see Supplemental Figure 3A online). To determine the rates of actin elongation, only filaments attached by one end were selected to ensure that these filaments have a formin attached. The rates of actin elongation were determined to be 13.6 ± 0.4 (n = 12) in the presence of AFH3 FH1FH2 (Figure 2M). Noticeably, in the presence of AFH3 FH1FH2, the number of actin filaments in the field increases significantly, confirming its active nucleation activity (Figures 2E to 2H; see Supplemental Movie 2 online). However, we did not detect any buckle events in the presence of AFH3 FH1FH2, as reported previously for other formins (Kovar and Pollard, 2004; Kovar et al., 2006), which indicates that AFH3 is very likely a nonprocessive formin. Furthermore, according to these data, we were able to determine the nucleation efficiency of AFH3 FH1FH2 using the slope at half-maximal polymerization from the pyrene fluorescence curves (Figure 1C) based on a rate constant of association of 11.6 μM−1 s−1 (Figure 1D), generating a maximum of 0.0012 barbed ends per formin molecule (Figure 1D). AFH3 FH1FH2 is less efficient than that of AFH1 (Michelot et al., 2005), but it is roughly the same efficiency as that of At FH5 (Ingouff et al., 2005).
Figure 2.
Direct Visualization of the Effect of AFH3 FH2 or AFH3 FH1FH2 on Actin Nucleation and Actin Filament Elongation by TIRFM.
TIRFM was conducted with 1.5 μM ATP-Oregon-green-actin (33.3% labeled) incubated with 3 μM human profilin in the absence or presence of GST-AFH3 FH1FH2 or GST-AFH3 FH2. The time point of the captured image is indicated in seconds at the bottom, and white arrows indicate the elongating barbed end of the actin filament. Conditions: 10 mM imidazole, pH 7.0, 50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 50 mM DTT, 0.2 mM ATP, 50 μM CaCl2, 15 mM glucose, 20 μg/mL catalase, 100 μg/mL glucose oxidase, 1.0% methylcellulose. Bar = 10 μm.
(A) to (D) Time-lapse micrographs of profilin/Oregon-green-actin polymerization. NEM-myosin was added into a flow cell before the addition of 1.5 μM ATP-Oregon-green-actin (33.3% labeled) plus 3 μM human profilin.
(E) to (H) Time-lapse micrographs of the effect of AFH3 FH1FH2 on actin polymerization. NEM-myosin and AFH3 FH1FH2 (200 nM) were attached before addition of 1.5 μM ATP-Oregon-green-actin (33.3% labeled) plus 3 μM human profilin.
(I) to (L) Time-lapse micrographs of the effect of AFH3 FH1FH2 on actin polymerization. NEM-myosin and AFH3 FH2 (300 nM) were attached before addition of 1.5 μM ATP-Oregon-green-actin (33.3% labeled) plus 3 μM human profilin.
(M) Plot of the elongation rates (±se) of actin filaments in the absence (black bar) or presence of AFH3 FH1FH2 (gray bar) or AFH3 FH2 (hatched bar).
AFH3 FH1FH2 Binds to the Barbed End of Actin Filaments and Prevents Actin Polymerization and Depolymerization from the Barbed End
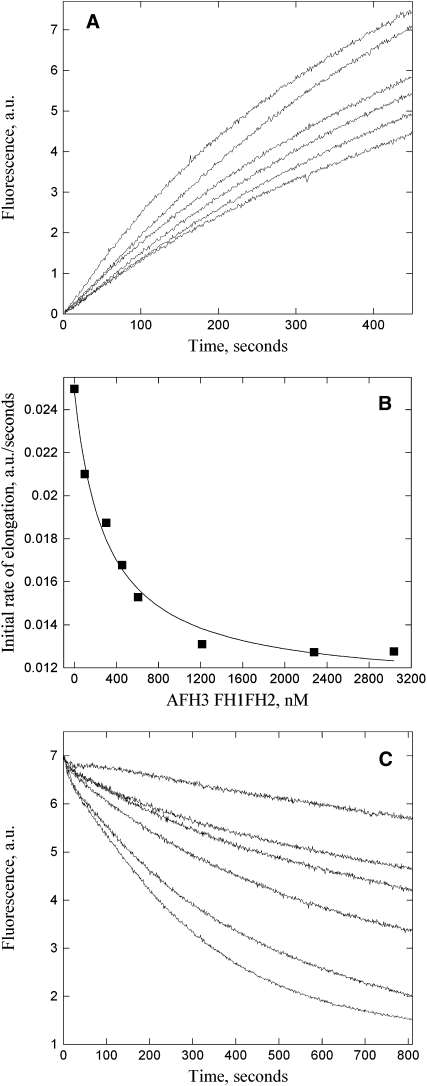
A key property of formins is their binding to the barbed end of actin filaments. We investigated AFH3 binding to the barbed end and the affinity for this binding using an actin filament elongation assay (Blanchoin et al., 2000). Freshly prepared seeds consisting of unlabeled actin filaments were incubated with various concentrations of AFH3 FH1FH2 (Figure 3A) before the addition of pyrene-labeled actin monomers. The decrease in the initial rate of elongation as a function of the concentration of AFH3 FH1FH2 suggests that the actin filaments' barbed ends are blocked by AFH3 FH1FH2 (Figure 3B). However, AFH3 FH2 did not block the actin filaments' barbed ends (see Supplemental Figure 3C online), consistent with the TIRFM data (Figures 2I to 2L). The apparent Kd value (±se) was determined to be 320.4 ± 49.1 nM (n = 4) for AFH3 FH1FH2.
Figure 3.
AFH3 FH1FH2 Binds to Barbed Ends and Blocks Actin Polymerization and Depolymerization from Barbed Ends.
(A) Kinetics of actin filament barbed-end elongation in the presence of AFH3 FH1FH2. Preformed actin filaments (0.8 μM) were incubated with various concentrations of AFH3 FH1FH2 before the addition of 1 μM pyrene-actin monomers. The concentration of AFH3 FH1FH2 from top to the bottom: 0, 300, 450, 600, 1200, and 2000 nM.
(B) Variation in the initial rate of elongation as a function of AFH3 FH1FH2 concentration. The data were fit with Equation 1 (see Methods) to determine the equilibrium dissociation constant value of 307 nM for AFH3 FH1FH2.
(C) Kinetics of actin depolymerization in the presence of various concentrations of AFH3 FH1FH2. AFH3 FH1FH2 was incubated with 5 μM F-actin for 5 min before dilution of the solution 25-fold into Buffer G. The concentration of AFH3 FH1FH2 from bottom to top: 0, 50, 70, 150, 200, and 400 nM.
The binding of AFH3 FH1FH2 to the barbed ends is expected to prevent actin depolymerization from actin filaments' barbed ends under certain conditions, such as dilution. To test this, a dilution-mediated depolymerization assay was performed. Indeed, AFH3 FH1FH2 decreases the initial rate of actin depolymerization in a dose-dependent manner (Figure 3C), and this result further supports its barbed-end capping activity. To rule out the possibility that bundling activity partially contributed to this decrease in depolymerization rate, a low-speed cosedimentation assay was employed to test whether AFH3 FH1FH2 has bundling activity, which has been shown for some formins (Harris et al., 2004; Michelot et al., 2005). Most of the actin remained in the supernatant (see Supplemental Figure 4, lane 1, online) and only very little actin sedimented at 13,500g in the actin alone sample (see Supplemental Figure 4, lane 2, online). In the presence of AFH1 FH1FH2, more polymerized actin sedimented (see Supplemental Figure 4, lane 8, online). However, in the presence of AFH3 FH1FH2, the amount of sedimented actin (see Supplemental Figure 4, lanes 4 and 6, online) was not significantly different from that of actin alone (see Supplemental Figure 4, lane 2, online). It suggests that AFH3 FH1FH2 does not have bundling activity. Therefore, the inhibition of AFH3 FH1FH2 on actin depolymerization is mainly due to its barbed-end capping activity.
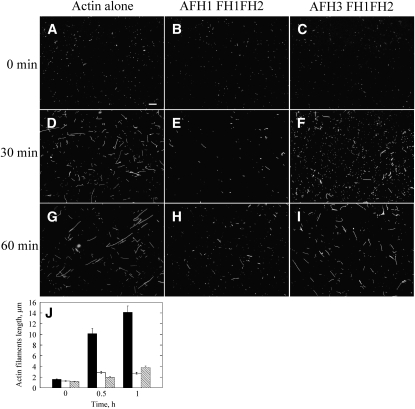
To explore the effect of AFH3 FH1FH2 on the behavior of actin barbed end further, an actin filament annealing assay was employed, which requires free actin ends. Phalloidin-stabilized actin filaments were physically sheared by a needle and then allowed to reanneal in the absence or presence of AFH1 FH1FH2 or AFH3 FH1FH2. As shown in Figure 4, AFH3 FH1FH2 slows actin filament reannealing (Figures 4C, 4F, and 4I), and AFH1 FH1FH2 has a stronger inhibitory effect on actin filament reannealing (Figures 4B, 4E, and 4H). The mean actin filament length (±se) at 0, 0.5, and 1 h is 1.6 ± 0.1 μm (n = 136), 10.1 ± 1.0 μm (n = 142), and 14.1 ± 1.2 μm (n = 123) for actin alone, 1.3 ± 0.1 μm (n = 133), 2.9 ± 0.2 μm (n = 119), and 2.7 ± 0.2 μm (n = 120) for AFH1 FH1FH2, and 1.2 ± 0.1 μm (n = 122), 2.0 ± 0.1 μm (n = 123), and 3.8 ± 0.3 μm (n = 136) for AFH3 FH1FH2, respectively (Figure 4J).
Figure 4.
AFH3 FH1FH2 Inhibits Reannealing of Actin Filaments.
(A) to (I) Four micromolar polymerized actin was stabilized with 4 μM rhodamine-phalloidin, sheared by a needle, and then allowed to reanneal in the presence of buffer, 300 nM AFH1 FH1FH2, or 300 nM AFH3 FH1FH2. Samples were diluted at the indicated time points and visualized by fluorescence microscopy. Bar in (A) = 10 μm and applies to (A) to (I).
(J) Mean length of actin filaments (±se) at different time points. Actin alone, black bars; Actin plus AFH1 FH1FH2, white bars; Actin plus AFH3 FH1FH2, crosshatched bars.
AFH3 Overexpression Enhanced the Formation of Actin Cables in Pollen Tubes
We next sought to determine the effect of AFH3 overexpression on actin organization in pollen tubes. The TAIR-annotated AFH3 was initially amplified, which lacks the C-terminal I and J subdomains of FH2. We were then interested in testing whether AFH3 deletions lacking these subdomains were able to induce polymerization in vivo. We generated several constructs from the 1848-bp cDNA clone that lacks these two subdomains (Figure 1A). These constructs were driven by the pollen-specific Lat52 promoter (Twell et al., 1991) and transiently expressed in tobacco (Nicotiana tabacum) pollen tubes using biolistics-mediated transformation (Higashida et al., 2004). Overexpression of both NΔFH1FH2-GFP (green fluorescent protein) and ΔFH1FH2-GFP induced widening of tubes and inhibited their elongation (see Supplemental Figure 5 online). The severity of the phenotype was correlated with the intensity of fluorescence in the pollen tube. The stronger the fluorescence in the pollen tube, the shorter and broader the pollen tube was (cf. Supplemental Figures 5B to 5A, and 5D to 5C online). These effects were similar to those induced by the overexpression of similar formin domains from At FH1 (Cheung and Wu, 2004). It was also reported that overexpression of FH1FH2 domains of yeast formins caused polarity defects and growth arrest (Evangelista et al., 2002b; Kovar et al., 2003). These observations suggest that the FH1FH2 domains of AFH3 may be functionally analogous to their counterparts in yeast and to those of AFH1.
To determine the effect of the overexpression of NΔFH1FH2 of AFH3 on actin organization in the pollen tube, GFP-fABD2 was used as a probe to image the actin cytoskeleton in living plant cells (Sheahan et al., 2004). GFP-fABD2 revealed thick longitudinal actin cables parallel to the growth axis in the shank of the control pollen tubes, but the actin cables never extended into the tip of the pollen tubes (Figure 5A, a median section; Figure 5B, a projection of confocal laser scanning sections). When Lat52:NFH1FH2 plasmids were cotransformed with Lat52:GFP-fABD2 into tobacco pollen tubes, the longitudinal actin cables appeared finer but more numerous than those of control pollen tubes (Figures 5A and 5B), and they extended into the apical and subapical regions of the pollen tube (Figure 5C, a median section; Figure 5D, a projection). The number of actin cables seen in the single optical section of a pollen tube expressing Lat52:NFH1FH2 (Figure 5C) was two to three times more than that of control pollen tube (Figure 5A). These changes were distinct from those caused by the overexpression of NFH1FH2 domains of AFH1, which induced supernumerary short actin cables that were not normally present in control pollen tubes (Cheung and Wu, 2004). These observations suggest that AFH3 and AFH1 may organize different populations of actin filaments, although they may both function to nucleate the formation of bundled actin filaments.
Figure 5.
Overexpression of AFH3-NΔFH1FH2 Induces the Formation of Excessive Actin Cables in Tobacco Pollen Tubes and Causes Swollen Pollen Tubes to Swell.
(A) and (B) A control pollen tube transformed with Lat52:GFP-fABD2 (1 μg), showing the actin cytoskeleton at the medial section of the pollen tube (A) and a projection of the whole pollen tube (B).
(C) and (D) A typical pollen tube cotransformed with Lat52:GFP-fABD2 (1 μg) and Lat52:NΔFH1FH2 (3 μg). A medial section of the pollen tube (C) and a projection of the whole pollen tube (D).
Bar = 5 μm.
Actin Cable Formation in the Shank of Arabidopsis Pollen Tubes Is Solely Dependent on AFH3
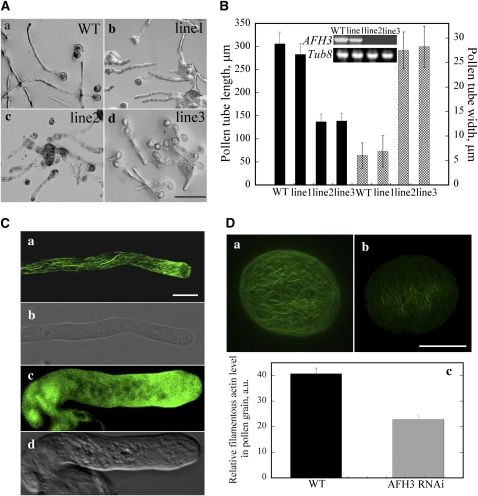
The increased number of longitudinal actin cables induced by AFH3 NFH1FH2 overexpression supports a role for AFH3 in the nucleation of actin filaments to form these actin cables. We next tested this hypothesis using RNA interference (RNAi)-induced suppression of AFH3 gene expression. An AFH3-specific 5′-end cDNA sequence was chosen for the generation of a Lat52:AFH3-RNAi construct. The construct was introduced into Arabidopsis plants by Agrobacterium tumefaciens–mediated transformation. RT-PCR analysis indicated that this RNAi construct specifically downregulated the expression of the AFH3 gene (Figure 6B, inset) and did not affect the expression of At FH5 in the flowers, which shares the highest sequence similarity with AFH3 (see Supplemental Figure 6 online). It did not affect the expression of another well-characterized formin AFH1 and a group II formin AFH13 in the flowers either (see Supplemental Figure 6 online). Pollen grains from the T2 homozygous transgenic plants were germinated in vitro, and their phenotypes were observed after culturing for 3 h. As shown in Figure 6A, RNAi-mediated suppression of AFH3 gene expression induced short and broadened pollen tubes. Occasionally, AFH3 RNAi tubes were swollen in the tips, but the majority of AFH3 RNAi tubes were of even width from the germination pore to the tip. As shown in Figure 6B, the severity of the RNAi-induced phenotype was correlated with the level of AFH3 suppression. RT-PCR analysis showed that RNAi lines 2 and 3 had stronger suppression of AFH3 expression than line 1 (Figure 6B, inset). The mean length (±sd) of pollen tubes was 305.7 ± 25.6 μm, 283.0 ± 23.2 μm, 136.9 ± 16.8 μm, and 138.7 ± 17.2 μm for the wild type and RNAi lines 1, 2, and 3, respectively. The mean tube width (±sd) was 6.0 ± 2.6 μm, 6.9 ± 3.2 μm, 27.5 ± 3.8 μm, and 28.3 ± 4.2 μm for the wild type, line 1, line 2, and line 3, respectively.
Figure 6.
Downregulation of AFH3 Expression Induces the Loss of Actin Cables in the Shank of the Pollen Tube and Consequently Inhibits Its Growth and Increases Its Width.
(A) Pollen tube phenotype typical of those from wild-type plants and different AFH3-RNAi lines after culturing in germination medium for 3 h; (a) wild type; (b) RNAi line 1; (c) RNAi line 2; (d) RNAi line 3. Bar = 100 μm for (a) to (d).
(B) Quantitative analysis of the effect of AFH3 knockdown on pollen tube length and width. Inset: RT-PCR analysis confirms the knockdown of AFH3 transcript in different AFH3 RNAi lines.
(C) Visualization of the actin cytoskeleton in pollen tubes stained with by Alexa-488 phalloindin. The actin cytoskeleton of a wild-type pollen tube (a) and AFH3 RNAi pollen tube (c) was visualized by confocal laser scanning microscopy after staining with Alexa-488-phalloidin. (b) and (d) show the corresponding bright-field images for (a) and (c) pollen tubes, respectively. Bar = 10 μm in (a) to (d).
(D) The filamentous actin level was substantially decreased in the pollen grains of the AFH3 RNAi transgenic plant. The actin cytoskeleton of a pollen grain from wild-type (a) and AFH3 RNAi (b) plants was visualized by confocal laser scanning microscopy after staining with Alexa-488-phalloidin. Confocal settings and image collection and display parameters were identical between pollen grains. Images shown are z-series stacks of all optical sections. Bar = 10 μm. In (c), the average pixel intensity was measured for the pollen grains by confocal laser scanning microscopy. About 100 pollen grains were measured for each genotype, and the mean values (±se) are plotted.
To examine whether AFH3 RNAi expression altered the organization of the actin cytoskeleton in pollen tubes, AFH3 RNAi pollen tubes (line 2) were stained with Alexa-488-phalloidin using the protocol of Lovy-Wheeler et al. (2005), which was shown to produce reproducible actin staining of pollen tubes. As shown in Figure 6Ca, the actin cables were arranged longitudinally in the shank of the wild-type pollen tube, and some fine actin filaments were present in the tip. The wild-type pollen tube was a thin tube structure with a clear zone at the tip (Figure 6Cb). However, the most prominent actin cables were almost completely absent in the shanks of AFH3 RNAi pollen tubes and were replaced with some actin filaments distributed irregularly in the shank. More fluorescence was detected at the tip of the AFH3 RNAi pollen tube (Figure 6Cc) than of the wild type, suggesting the accumulation of filamentous actin in this region. Taken together, our data suggest that AFH3 is responsible for the generation and/or maintenance of longitudinal actin cables in the shank of pollen tubes.
The Filamentous Actin Level Was Decreased in the Pollen Grains of AFH3-RNAi Transgenic Plants
The above data suggest that AFH3 is a functional formin responsible for the nucleation of bundled actin filaments in pollen tubes. If so, we would anticipate a reduction in the F-actin content in AFH3 RNAi pollen. To test this, we visualized F-actin with Alexa-488 phalloidin staining in individual pollen grains. Typically, actin filaments appeared brighter and denser in wild-type pollen grains (Figure 6Da) when compared with those in AFH3 RNAi transgenic pollen grains (Figure 6Db). The average fluorescence pixel intensity (±se) of individual grains was 40.7 ± 2.2 (n = 20) and 22.7 ± 1.7 (n = 20) for the wild type and AFH3 RNAi pollen grains, respectively (Figure 6Dc; P < 0.001). This result suggested that AFH3 indeed promotes actin polymerization to maintain the content of filamentous actin in pollen grains.
The Direction and Velocity of Cytoplasmic Streaming Was Altered in AFH3-RNAi Pollen Tubes
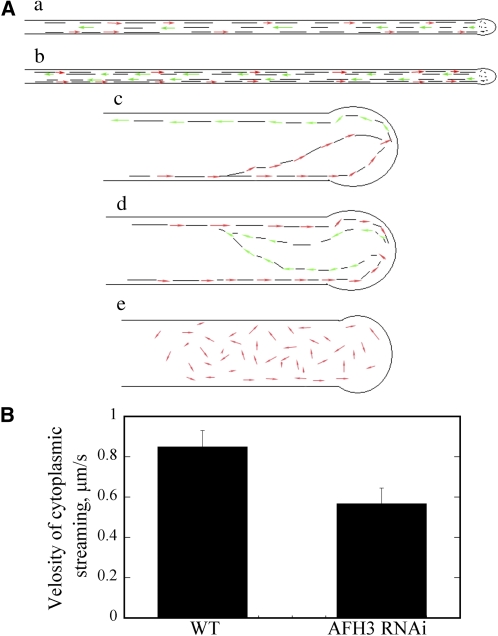
It has been proposed that cytoplasmic actin cables serve as a molecular track for cytoplasmic streaming in plant cells; however, the previous experiments that led to this conclusion all involved the use of actin-disrupting drugs, which generally destroy different F-actin populations in the cell. The AFH3 RNAi lines provided us with a tool to more specifically investigate a potential role for actin cables in the regulation of cytoplasmic streaming. As shown in the supplemental movies (see Supplemental Movies 4 and 5 online), there was rapid and reverse fountain cytoplasmic streaming in wild-type pollen tubes; particles moved to the apex along the tube cortex (red arrows) and moved back in the center of the cell (green arrows) once reaching the subapical region (Figure 7Aa; see Supplemental Movie 4 online), a pattern known as reverse fountain streaming (de Win et al., 1999). In some wild-type pollen tubes, particles move back right next to the previous track (green arrows) along the tube cortex once reaching the subapical region (Figure 7Ab; see Supplemental Movie 5 online). In AFH3 RNAi pollen tubes, the reserve fountain cytoplasmic streaming was disrupted; cytoplasmic streaming appeared random and irregular. Many particles traveled directly into the extreme apical region (see Supplemental Movies 6 to 8 online). We identified three distinct types of cytoplasmic streaming in AFH3 RNAi pollen tubes. The first one is shown in Figure 7Ac; particles moved forward to the pollen tube tip (red arrows) but were not restricted to the tube cortex region. Some particles moved to the pollen tube tip in the tube center and then moved backward at the cortical region of the opposite side of the pollen tube (green arrows) (see Supplemental Movie 6 online). The second is shown as in Figure 7Ad; particles moved toward the pollen tube tip along the cortical region (red arrows) and moved backward in the center of the tube (green arrows) but not parallel to the axis of growth (see Supplemental Movie 7 online). The third one is shown in Figure 7Ae; the reverse fountain pattern was disrupted completely, and particles moved sporadically in short distances (red arrows) (see Supplemental Movie 8 online). We also measured the velocity of cytoplasmic streaming from several independent pollen tubes and found that the average velocity of cytoplasmic streaming was 0.85 ± 0.08 μm/s (n = 20) and 0.57 ± 0.08 μm/s (n = 20) for wild-type and AFH3 RNAi pollen tubes, respectively (Figure 7B) (P = 0.018). Taken together, our data suggest that the disruption of the longitudinal actin cables altered the direction of cytoplasmic streaming and decreased the rate of cytoplasmic streaming.
Figure 7.
Cytoplasmic Streaming Direction Was Altered and the Velocity of Cytoplasmic Streaming Was Reduced in AFH3 RNAi Pollen Tubes.
(A) Schematic representation of the cytoplasmic streaming pattern in wild-type ([a] and [b]) and AFH3 RNAi ([c] to [e]) pollen tubes. Red arrows represent the direction of cytoplasmic streaming that moves particles toward the tip of the pollen tube; green arrows represent the direction of cytoplasmic streaming that moves particles backward in the pollen tube; the black lines in the pollen tube indicate the cytoplasmic streaming track.
(B) The velocity of cytoplasmic streaming was analyzed by Image J software (http://rsbweb.nih.gov/ij/). Twenty cytoplasmic streaming particles from several independent pollen tubes were measured for each genotype. and the mean values (±se) are plotted.
DISCUSSION
Our study provides biochemical and genetic evidence that AFH3 nucleates actin for the formation of the longitudinal actin cables in pollen tubes. Our data suggest that the actin cables regulate cytoplasmic streaming and participate in the modulation of polarized pollen tube growth. This loss-of-function study demonstrates a direct linkage of formins with the organization of the cytoplasmic actin cables in plants. Furthermore, our findings provide genetic evidence that demonstrates the long-speculated function of cytoplasmic actin cables in the regulation of cytoplasmic streaming.
AFH3 Is a Specific Nucleator of Actin Filaments to Form the Axial Actin Cables
A most striking aspect of plant formins is the existence of a large number of isoforms (e.g., 21 formins are encoded in the Arabidopsis genome). Functional redundancy among some isoforms is likely but has not been demonstrated genetically. An interesting question about the function of these formin isoforms is whether a formin or a subset of formins specifically regulates the formation of one particular population of actin filaments, which could provide a mechanism for coordinately regulating multiple forms of the actin cytoskeleton in a specific cell type. At least three formins, AFH3, At FH5, and a group II formin AFH13, are expressed abundantly in Arabidopsis mature pollen (Honys and Twell, 2003; Pina et al., 2005). Given the existence of multiple populations of actin filaments in pollen tubes, it is possible that each of these formin isoforms may participate in the formation of a distinct actin structures in the pollen tube. The three formins present in fission yeast are also thought to be responsible for the construction of distinct actin arrays (Petersen et al., 1998; Feierbach and Chang, 2001; Kovar et al., 2003). Our data provide evidence that AFH3 specially activates the formation of the axial actin cables.
We found that AFH3 overexpression induced an excessive number of longitudinal actin cables in the pollen tube (Figures 5C and 5D). By contrast, AFH1 overexpression induced excessive actin polymerization near the PM and caused PM deformation consistent with its PM localization, and the AFH1-induced actin bundles were short (Cheung and Wu, 2004). Thus, AFH3 appears to be functionally distinct from AFH1. Loss of AFH1 function studies will be necessary to determine what specific forms of pollen actin are regulated by this formin. The study of the function of AFH3 is further extended by an RNAi approach. Our finding that actin cables almost completely disappeared in AFH3 RNAi pollen tubes and that the number of actin cables increased upon overexpression of AFH3 suggests that AFH3 is necessary and sufficient for the formation of longitudinal actin cables. An important question that needs to be answered about the AFH3-mediated formation of actin cables is where the site of AFH3-dependent actin nucleation is and whether there is a mechanism that spatially regulated AFH3-dependent actin nucleation. Subcellular localization of AFH3 may provide some insights into this question. Our attempt to confirm the intracellular localization of AFH3 using immunostaining failed because we were unable to produce a specific antibody that only detected AFH3 in Arabidopsis. Therefore, AFH3 localization needs to be determined by other methods in the future. Other actin binding proteins, such as fimbrin, may coordinate with AFH3 to form the longitudinal actin cables in the reverse fountain pattern. Nonetheless, future studies, such as the isolation of mutants that cause disoriented actin cables, will need to be conducted to fully understand the mechanism for the AFH3-dependent formation of actin cables in pollen tubes.
AFH3 May Have a Distinct Action on Actin Dynamics
The overall effect of formins on actin polymerization in vitro could vary greatly, but there exist some common features of in vitro activities for various formins, such as nucleating actin assembly from monomers and slowing the on and/or off rate of actin monomers from the barbed ends. Indeed, AFH3 FH1FH2 nucleates actin assembly efficiently from monomers (Figure 1C). However, the FH2 domain alone does not have such activity, in contrast with the FH2 domain of AFH1 (Michelot et al., 2005), but is similar to FH2 of At FH5 (Ingouff et al., 2005). This suggests that the FH1 domain may contribute to the ability of FH2 to bind actin monomer. The FH1 domain of AFH3 may be important for its dimer formation, which was demonstrated to be critical for its nucleation (Xu et al., 2004). AFH3 also nucleates actin assembly from monomeric G-actin bound to profilin, negating the inhibitory effect of profilin on spontaneous actin nucleation (Figure 1E). This result further indicates that AFH3 is a functional actin nucleator in vivo, since most monomeric G-actin is buffered by profilin in pollen tubes (Vidali and Hepler, 1997; Gibbon et al., 1999; Snowman et al., 2002).
AFH3 was also shown to have a capping activity, as it blocked actin elongation at its barbed end in a dose-dependent manner (Figure 3A). However, actin elongation occurred in the presence of AFH3 FH1FH2, which is similar to FH1FH2 of AFH1 and At FH5 (Ingouff et al., 2005; Michelot et al., 2005) and is in agreement with the leaky capping property previously reported for Bni1p (Zigmond et al., 2003). AFH3 FH1FH2 acts differently from the FH1FH2 domains of Cdc12, since Cdc12 FH1FH2 requires the binding of profilin to the FH1 domain to allow the addition of actin monomer into the barbed end (Kovar et al., 2003). Interestingly, the Kd value for AFH3 FH1FH2 is almost 10-fold higher than the Kd of AFH1 FH1FH2 under the same assay conditions (Michelot et al., 2005). This implies that it may be easier for actin monomer to access the AFH3 FH1FH2 capped barbed end than the AFH1 FH1FH2 capped barbed end. Indeed, the visualization of single actin filament growth in the presence of AFH3 FH1FH2 supports this notion, since the presence of AFH3 FH1FH2 on the barbed end did not change the rate of addition of actin/profilin into barbed ends (Figure 2M). To slow down barbed-end elongation, formins need to move with the elongated barbed end. This behavior leads to the establishment of a processive capping model proposed for formins (Kovar and Pollard, 2004; Romero et al., 2004; Kovar et al., 2006). However, none of the plant formins have been shown experimentally to bind processively to the barbed end of actin filaments. On the contrary, AFH1 was reported to be a nonprocessive formin, as it moves to the side of the growing filament following the initiation of actin nucleation at the barbed end (Michelot et al., 2006). As is the case for AFH3, it is very likely a nonprocessive formin, since we did not detect any buckle events of growing actin filaments in the presence of AFH3 FH1FH2. In the depolymerization assay, AFH3 blocks actin depolymerization in a dose-dependent manner (Figure 3C), and AFH3 FH1FH2 does not have bundling activity (see Supplemental Figure 4 online). Thus, the stabilization of actin filaments by AFH3 through its side binding is very unlikely. We therefore concluded that the barbed-end capping activity of AFH3 plays an important role in the stabilization of actin filaments. In summary, the effect of AH3 on actin dynamics is different from other Arabidopsis formins and formins from other species. Further structural characterization of AFH3 and visualization of the effect of AFH3 on single actin filaments will provide insights into the action mechanism for AFH3 on actin dynamics.
The AFH3-Dependent Actin Cables Regulate Cytoplasmic Streaming
Pharmacological experiments suggest that the cytoplasmic streaming in pollen tubes is actin dependent. Furthermore, the reverse fountain pattern of the pollen cytoplasmic streaming is consistent with the organization of the longitudinal actin cables. In this study, we demonstrated that the RNAi-mediated downregulation of AFH3 expression altered the direction and velocity of cytoplasmic streaming (Figures 7Ac to 7Ae), providing direct genetic evidence that the axial actin cables function as a molecular track for cytoplasmic streaming. Cytoplasmic streaming possesses a distinct polarity, occurring in a characteristic reverse fountain pattern in pollen tubes and root hairs (Pierson et al., 1996; de Win et al., 1999). In this scheme, transport is thought to be driven by myosin on the vesicle surface that moves along the actin filaments (Miller et al., 1995). In plants, cytoplasmic streaming is assumed to be performed by myosin XI (Shimmen and Yokota, 2004; Wang and Pesacreta, 2004). Because myosin XI is a plus-end (i.e., barbed-end) directed motor (Shimmen and Yokota, 2004), the tracks for the myosin would have to be bidirectional to facilitate reverse-fountain cytoplasmic streaming. In support of this notion, myosin subfragment 1 decoration experiments showed that actin plus ends face the tip along the cortical regions and reverse polarity in the middle of root hairs and pollen tubes (Tominaga et al., 2000; Lenartowska and Michalska, 2008). Both the velocity and the direction of cytoplasmic streaming were significantly compromised in AFH3 RNAi pollen tubes, in which the actin cables were reduced and disorganized. These observations are consistent with a role for the actin cables as tracks for actomyosin-mediated movement. However, a role for the dynamics of the actin cables in regulating cytoplasmic streaming cannot be ruled out.
Potential Roles for AFH3-Dependent Actin Cables in Polarized Pollen Tube Growth
Pollen tubes elongate rapidly, requiring a tightly regulated intracellular polar transport system, which delivers cellular materials to the tip of pollen tubes for membrane expansion and cell wall synthesis. Pollen tubes were wider and shorter than wild-type pollen tubes and swelled at the tip in AFH3 RNAi expressing pollen tubes, suggesting that disruption of the actin cables may impair the intracellular polar transport system. The alteration in the direction and velocity of cytoplasmic streaming in AFH3 RNAi pollen tubes may contribute to a defect in the polar transport system. Early cytological studies revealed a dramatic cytoplasmic compartmentalization in pollen tubes (e.g., there is a relatively smooth cytoplasm restricted to an inverted cone region at the apex of the pollen tubes, called the clear zone) (Hepler et al., 2001). The clear zone essentially disappeared in AFH3 RNAi pollen tubes, causing organelles to move into the extreme tip region of pollen tubes (Figures 6Ac, 6Ad, and 6Cd). Thus, the AFH3-dependent actin cables seem to be important for organizing cytoplasmic compartmentalization, and the disruption of this cytoplasmic zonation could also explain the reduced growth polarization in AFH3 RNAi pollen tubes. Finally, the reduction in actin cable formation in AFH3 RNAi pollen tubes may also affect the dynamics of other actin populations in pollen tubes. The disruption of the actin cables may elevate the concentration of actin monomers in the pollen tube, which could subsequently be used for an increased polymerization of dynamic tip F-actin in pollen tubes. Interestingly, the phenotype of AFH3 RNAi resembles that of Arabidopsis RIC4 overexpression, which causes an increase in the accumulation of dynamic tip F-actin (Gu et al., 2005). To test whether the accumulation of the dynamic tip F-actin contributes to the growth depolarization induced by AFH3 RNAi expression, pollen tubes were subjected to LatB treatment. Indeed, with an increasing concentration of LatB (up to 0.8 nM), the length of pollen tube increases and the width of pollen tube decreases (see Supplemental Figure 7 online), indicating that addition of LatB partially suppress the growth depolarization induced by AFH3 RNAi expression.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana ecotype Columbia plants were grown in an air-conditioned growth room at 22°C under 16-h-light/8-h-dark cycles. Seed germination and selection were performed as described previously by Sundaresan et al. (1995). Briefly, surface-sterilized seeds were plated onto Murashige and Skoog agar medium supplemented or not with antibiotics. For hygromycin selection, 20 mg/L of hygromycin (Roche) was added. Plant transformation was performed according to Bechtold and Pelletier (1998).
cDNA Isolation and Recombinant Plasmid Construction
Arabidopsis total RNA was purified from inflorescence tissues using a Total RNA Isolation Kit (Promega). The cDNA was prepared by reverse transcription with MMLV reverse transcriptase (Promega) according to the manufacturer's recommendations. The cDNA sequence of the AFH3 gene was referenced to design oligonucleotide primers. The primers for amplification of AFH3 NFH1FH2 were 5′-CGGGATCCATGGGGAGATTGAGATTAGC-3′ (the forward oligonucleotide), containing a BamHI site (underlined), and 5′-CGGGATCCACCCCGTAACCACTTGAAG-3′ (the reverse oligonucleotide), containing a BamHI site (underlined). The primers for amplification of AFH3 FH1FH2 were 5′-CGGGATCCGAGTTTTCAACGGCTGAAT-3′ (the forward oligonucleotide), containing a BamHI site (underlined), and the same reverse oligonucleotide as the reverse oligonucleotide of AFH3 NFH1FH2. The amplified products were A-tailed and cloned into the pGEM-T vector. The sequence was verified by sequencing. The cDNA fragments were subcloned into a binary vector under the control of a pollen-specific promoter LAT52 (Twell et al., 1991) by excision with BamHI/BamHI to produce LAT52:NFH1FH2-GFP and LAT52:FH1FH2-GFP constructs.
To generate the AFH3 RNAi construct, the sense and antisense fragments were amplified with the following primers: 5′-CGGGATCCATGGGGAGATTGAGATTAGC-3′ (the sense forward oligonucleotide), containing the BamHI site (underlined), 5′-CACTTCCTCTGGAGATGCATCTG-3′ (the sense reverse oligonucleotide), containing the NsiI site (underlined), 5′-ATTTAAATGAGGAGGTGCTGATCTTC-3′ (the antisense reverse oligonucleotide), containing the SwaI site (underlined), and 5′-CGCCATGGATGGGGAGATTGAGATTAGC-3′ (the antisense forward oligonucleotide), containing the NcoI site (underlined). The sense fragment was excised with BamHI and NsiI and cloned into pFGC5941 digested with the same restriction enzyme pair. Subsequently, the antisense fragment was excised with NcoI and SwaI and cloned into pFGC5941 containing the sense fragment. For pollen-specific expression, the cauliflower mosaic virus 35S promoter was replaced with the LAT52 promoter excised with EcoRI and NcoI. The aforementioned constructs were introduced into Agrobacterium tumefaciens GV3101 by electroporation and transformed into Arabidopsis ecotype Columbia. T2 homozygous plants were selected for analysis.
Pollen Microprojectile Bombardment
Particle bombardment-mediated transient expression in tobacco (Nicotiana tabacum) pollen was performed as described previously (Fu et al., 2001). Tobacco pollen grains were freshly harvested and suspended in pollen suspension medium [0.01 mM CaCl2, 0.01 mM Ca(NO3)2, 1 mM MgSO4, 0.02% mM boric acid, and 18% (w/v) sucrose, pH 7.0]. Approximately 6 mg of pollen was bombarded with different amounts of plasmid DNA coated on 1-nm (diameter) gold particles using the BIOLISTIC-PDS-1000/He particle delivery system (Bio-Rad) at a pressure of 1100 p.s.i. The bombarded pollen grains were immediately transferred to pollen germination medium and incubated at 26°C in the dark until observation.
Analysis of Overexpression Phenotypes of Various AFH3 Domains and F-Actin Visualization in Tobacco Pollen Tubes
Approximately 5 h after bombardment, tubes expressing LAT52:GFP, LAT52: NFH1FH2-GFP, and LAT52:FH1FH2-GFP were identified using epifluorescence microscopy, imaged, and analyzed as described previously (Fu et al., 2001). The degree of depolarized growth was determined by measuring the diameter of the widest region of the tube, and the degree of polar growth was determined by measuring the length of pollen tubes. Transient expression of GFP-fABD2 (modified from construct used by Sheahan et al. [2004] by replacing 35S with Lat52) was used to visualize F-actin in tobacco pollen tubes. A Zeiss LSM510 META system was used for confocal imaging. GFP fluorescence was visualized using the argon-ion laser at 4% of the maximum power, combined with the main dichroic beam splitter HFT 488 nm, and a band-pass 505- to 530-nm emission filter. All confocal images were analyzed using Metamorph v4.5 and processed using Adobe Photoshop CS3.
RT-PCR Analysis
Total RNA was isolated from inflorescence tissues of wild-type and AFH3-RNAi transgenic plants with TRIzol reagent (Invitrogen). RNA quality and quantity were analyzed by electrophoresis and spectrophotometry. For cDNA synthesis, 3 μg of total RNA from different tissue was used for reverse transcription with AMV reverse transcriptase (Takara) according to the manufacturer's recommendations. One microliter of the reaction product was then used as template for PCR amplification with primers 152BaA 5′-CGGGATCCACCCCGTAACCACTTGAAG -3′ and R152SacS 5′-GAAGGTAATGAGCTCCCAGTGGAG-3′ to amplify a 581-bp cDNA fragment of the 3′ terminal of AFH3 to confirm the expression level of AFH3. Tub8 was also used as an internal control with primers 5′-CTTCGTATTTGGTCAATCCGGTGC-3′ and 5′-GAACATGGCTGAGGCTGTCAAGTA-3′ to amplify about a 1-kb cDNA fragment. PCR products were evaluated by 1% agarose gel electrophoresis and stained with ethidium bromide.
Quantifying Cytoplasmic Streaming in Pollen Tubes
Pollen grains were germinated on a thin layer of germination medium [1 mM CaCl2, 1 mM Ca(NO3)2, 1 mM H3BO3, 1 mM MgSO4, 18% sucrose, and 0.5% agarose, pH 7.0] for 4 h at 28°C in the dark. The cytoplasmic streaming of pollen tubes was observed using a Leica DMI6000CS microscope equipped with a ×63 1.2–numerical aperture water objective, and digital images were collected with a Photometrics cascade II 512 CCD camera (Major Instruments) using LAS AF software. Images were captured at 0.25-s intervals over a total of 6 min. To calculate the velocity of particles in the pollen tubes, images of cytoplasmic streaming from wild-type and AFH3 RNAi pollen tubes were processed for analysis of individual particle movement with Image J software (http://rsbweb.nih.gov/ij/, version 1.38). To determine the velocity of particles, only the particles exhibiting continuous movement were selected at random. Particles of AFH3 RNAi pollen tubes exhibiting Brownian movement were not selected for analysis.
Arabidopsis Pollen Tube Growth Measurement
Flowers collected from Arabidopsis plants 2 weeks after bolting were used for the examination of pollen tube phenotypes. Pollen was germinated on standard agar medium containing 2 mM Ca2+ [with an equal molar ratio of CaCl2 and Ca(NO3)2] or various concentrations of Ca2+ as described previously (Li et al., 1999). To determine the effect of LatB on pollen tube growth, various concentrations of LatB were added to the germination medium. Approximately 5 h after germination, images of pollen tubes were recorded using a cooled CCD camera (model C4742-95; Hamamatsu) attached to an Eclipse inverted microscope (model TE300; Nikon). The images were analyzed using the MetaMorphv4.5 measurement function. For each experiment, ∼100 pollen tubes were chosen randomly for length measurement.
Protein Production
To express the fusion protein for AFH3 FH1FH2 and AFH3 FH2, the coding sequences for these two constructs were amplified by RT-PCR. Arabidopsis total RNA was purified from the flower using a Total RNA Isolation Kit (Promega). Primers were designed based on the cDNA sequence. The primers used for amplification of AFH3 FH1FH2 were as follows: AFH3 FH1FH2 forward, 5′-GGGGATCCGAGTTTTCAACGGCTGAATCGTCA-3′ (with the BamHI site underlined), AFH3 FH1FH2 reverse, 5′-GAGAATTCTTAATGGTGATGGTGATGGTGGTTCGTCGTCTT-3′ (with the EcoRI site underlined), AFH3 FH2 forward, 5′-CGGGATCCCCTAAAGGTGCGGCTCCA-3′ (with the BamHI site underlined), and AFH3 FH2 reverse, 5′-GAGAATTCTTAATGGTGATGGTGATGGTGGTTCGTCGTCTT-3′ (with the EcoRI site underlined). Amplified sequences were A-tailed and cloned into the pMD19-T vector at the corresponding sites designed in the primers (TaKaRa Biotechnology). The cDNA sequences were verified by sequencing. For expression of AFH3 FH1FH2 and AFH3 FH2 proteins in Escherichia coli, the pGEX-KG-AFH3 FH1FH2 and pGEX-KG-AFH3 FH2 constructs were created by cloning the coding sequences into the pGEX-KG vector; both the constructs and the vector were prepared by digestion with BamHI and EcoRI. Fusion proteins were expressed in the Rosseta Strain of E. coli (Invitrogen) by overnight induction with 0.4 mM isopropyl β-d-thiogalactopyranoside at 25°C. The GST fusion protein was affinity purified with the glutathione-Sepharose resin according to the manufacturer's protocol (Amersham Biosciences). They were further purified with a nickel resin (Novagen) according to the manufacturer's instructions. The purified proteins were dialyzed against buffer containing 2 mM Tris-HCl, pH 8.0, 100 mM KCl, 0.01% NaN3, 0.2 mM CaCl2, 0.2 mM ATP, and 0.2 mM DTT, separated into aliquots, frozen in liquid nitrogen, and stored at −80°C. The protein was clarified further by centrifugation at 200,000g for 1 h before use. Protein samples were analyzed by SDS-PAGE and visualized by staining the gels with Coomassie Brilliant Blue R 250 (Sigma-Aldrich).
Actin was purified from rabbit skeletal muscle acetone powder (Spudich and Watt, 1971), and monomeric Ca-ATP-actin was purified by Sephacryl S-300 chromatography at 4°C in G buffer (5 mM Tris-HCl, pH 8.0, 0.2 mM ATP, 0.1 mM CaCl2, 0.5 mM DTT, and 0.1 mM imidazole) (Pollard, 1984). Actin was labeled on Cys-374 with pyrene iodoacetamide (Pollard, 1984) and Oregon-green 488 iodoacetamide (Amann and Pollard, 2001). AFH1 FH1FH2 was purified according to Michelot et al. (2005), and Arabidopsis profilin 4 was purified roughly according to the purification procedure of maize (Zea mays) recombinant profilin (Gibbon et al., 1997).
Actin Nucleation Assay
Actin nucleation was performed essentially as described by Higgs et al. (1999). Mg-ATP-monomeric actin or actin-profilin complexes (10% pyrene labeled) were polymerized at room temperature in the presence or absence of AFH3 by the addition of one-tenth volume of 10× KMEI (500 mM KCl, 10 mM MgCl2, 10 mM EGTA, and 100 mM imidazole-HCl, pH 7.0). Then, changes in pyrene fluorescence were determined using a QuantaMaster Luminescence QM 3 PH Fluorometer (Photon Technology International).
Elongation Assay to Determine the Affinity of AFH3 FH1FH2 for the Barbed End of Filamentous Actin
Equivalent amounts of actin filaments were incubated for a few minutes at room temperature with various concentrations of AFH3 FH1FH2. Elongation was initiated by the addition of pyrene-labeled actin monomers to the actin filament mixture. The affinity of the fusion proteins for the barbed end of actin filaments was determined by the variation of the initial rate of elongation as a function of the concentration of AFH3 using Equation 1:
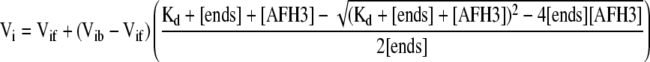 |
where Vi is the observed rate of elongation, Vif is the rate of elongation when all the barbed ends are free, Vib is the rate of elongation when all the barbed ends are capped, [ends] is the concentration of barbed ends, and [AFH3] is the concentration of AFH3. The data were modeled by Kaleidagraph version 3.6 software (Synergy Software).
Depolymerization Assay
The depolymerization assay was adopted according to Huang et al. (2003). F-actin at 5 μM (50% pyrene-labeled) was mixed with various concentrations of AFH3 FH1FH2, incubated at room temperature for 5 min, and diluted 25-fold into Buffer G at room temperature. The decrease in pyrene fluorescence accompanying actin depolymerization was monitored for 800 s after dilution.
Fluorescence Microscopy of Actin Filaments
Actin filaments labeled with fluorescent phalloidin were observed in vitro by epifluorescence illumination as previously reported (Blanchoin et al., 2000). Actin at 4 μM alone or together with AFH3 FH1FH2 or AFH1 FH1FH2 was polymerized in 50 mM KCl, 2 mM MgCl2, 1 mM EGTA, 0.2 mM ATP, 0.2 mM CaCl2, 0.5 mM DTT, 3 mM NaN3, and 10 mM imidazole, pH 7.0, at 25°C for 30 min and labeled with 4 μM rhodamine-phalloidin (Sigma-Aldrich) during polymerization. The polymerized F-actin was diluted to 10 nM in fluorescence buffer containing 10 mM imidazole-HCl, pH 7.0, 50 mM KCl, 1 mM MgCl2, 100 mM DTT, 100 mg/mL glucose oxidase, 15 mg/mL glucose, 20 mg/mL catalase, and 0.5% methylcellulose. A dilute sample of 3 μL was applied to a 22 × 22-mm cover slip coated with poly-l-Lys (0.01%). Actin filaments were observed by epifluorescence illumination with an IX71 microscope (Olympus) equipped with a ×60 1.42–numerical aperture oil objective, and digital images were collected with a Retiga EXi Fast 1394 CCD camera (QImaging) using Image-Pro Express 6.3 software. Filament length was measured using Image J (version 1.38) software.
An actin filament annealing experiment was performed roughly according to Huang et al. (2003). Actin (4 μM) was polymerized in the presence of equal molar amounts of rhodamine-phalloidin for 30 min at room temperature. The actin filaments were then sheared physically with a needle in the absence or presence of 300 nM AFH1 FH1FH2 or 300 nM AFH3 FH1FH2. Actin filaments were then visualized at various time points by fluorescence microscopy.
Direct Visualization of Actin Assembly by TIRFM
We performed the TIRFM assay as previously reported (Kovar and Pollard, 2004). Actin filaments were observed by epifluorescence illumination with a DMI6000CS microscope (Leica) equipped with a ×100 1.46–numerical aperture HC PLANs objective, and digital images were taken with a Photometrics cascade II 512 CCD camera (Major Instruments) using LAS AF software. The interval time between subsequent images was 15 s. Filament length was measured using Image J software (http://rsbweb.nih.gov/ij/, version 1.38).
The flow cell was made as described previously (Amann and Pollard, 2001). The slide was coated with 100 nM NEM-Myosin in the absence or presence of AFH3 FH1FH2 and AFH3 FH2 for 2 to 4 min and was subsequently equilibrated with 1% BSA. Then, the slide was buffered with 1× TIRFM buffer (10 mM imidazole, pH 7.0, 50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 50 mM DTT, 0.2 mM ATP, 50 μM CaCl2, 15 mM gluocose, 20 μg/mL catalase, 100 μg/mL glucose oxidase, and 1.0% methylcellulose). Finally, 1.5 μM ATP-Mg-actin (containing 33.3% Oregon-green-ATP-Mg-actin) was injected into the flow cell. The image was acquired as soon as the focal plane was found.
Imaging of Actin Filaments and F-Actin Quantification
Pollen grains were stained with Alexa-488-phalloidin (Invitrogen) as described by Snowman et al. (2002). Images were collected with a Leica TCS SP5 confocal laser scanning microscope equipped with a ×100 1.46–numerical aperture HC PLANs objective. The fluorescent phalloidin was excited with the 488-nm line of an argon laser, 0.11-μm optical sections were scanned and captured, and three Kalman-filtered scans were averaged for each optical section. Projections of the optical sections through an individual pollen grain were compiled using LAS AF software (version 1.8.0, build 1346). To quantify F-actin in pollen grains, images of pollen grains from wild-type and AFH3-RNAi transgenic plants were processed to determine the average pixel intensity with LAS AF software. For visualization of the actin cytoskeleton in pollen tubes, pollen was geminated for 5 h in pollen germination medium and then simultaneously fixed and permeabilized for 30 min with a buffer composed of 100 mM PIPES, 0.6 mM N-(maleimidobenzoyloxy)-succini-mide, 5 mM MgSO4, 0.5 mM CaCl2, 0.05% Triton X-100, 1.5% formaldehyde, and 0.05% glutaraldehyde at pH 9.0 for 0.5 to 1 h. The growth medium was completely removed before adding the fixative. The fixative was then washed out two times with buffer containing 100 mM PIPES, 5 mM MgSO4, 10 mM EGTA, and 0.05% Triton X-100 at pH 7.0 (wash buffer). Pollen tubes were stained in wash buffer containing 500 nM Alexa 488–phalloidin (Molecular Probes) for 1 h, washed twice with wash buffer, and mounted in wash buffer. Fluorescence of F-actin was observed with a confocal laser scanning microscope (Zeiss) using the 488-nm excitation line of an argon laser.
Accession Numbers
Sequence data from this article can be found in GenBank/EMBL databases under the following accession numbers: AFH3 (ACQ91096), Bni1p (NP_014128), AFH1 (AAF14548), and At FH5 (AAK68741).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. The Coding Region Nucleotide Sequence of AFH3 and Its Encoded Amino Acid Sequence.
Supplemental Figure 2. The FH2 Domain of AFH3 Is Highly Conserved Compared with FH2 Domains of Other Formins.
Supplemental Figure 3. AFH3 FH2 Has Hardly Detectable Actin Nucleation Activity and Does Not Cap Barbed Ends of Actin Filaments.
Supplemental Figure 4. AFH3 FH1FH2 Does Not Bundle Actin Filaments.
Supplemental Figure 5. Overexpression of Both NΔFH1FH2-GFP and ΔFH1FH2-GFP Arrest Pollen Tube Growth and Cause Swollen Pollen Tubes.
Supplemental Figure 6. The Transcript Level of AFH1, At FH5, and AFH13 Does Not Decrease in AFH3 RNAi Flowers.
Supplemental Figure 7. LatB Partially Suppresses the Phenotype of AFH3 RNAi Pollen Tubes.
Supplemental Movie 1. Time Lapse of Profilin/Oregon-Green-Actin Polymerization.
Supplemental Movie 2. Time Lapse of the Effect of AFH3 FH1FH2 on Profilin/Oregon-Green-Actin Polymerization.
Supplemental Movie 3. Time Lapse of the Effect of AFH3 FH2 on Profilin/Oregon-Green-Actin Polymerization.
Supplemental Movie 4. This Movie Corresponds to the Pattern of Cytoplasmic Streaming of the Wild-Type Pollen Tube Shown in Figure 7Aa.
Supplemental Movie 5. This Movie Corresponds to the Pattern of Cytoplasmic Streaming of the Wild-Type Pollen Tube Shown in Figure 7Ab.
Supplemental Movie 6. This Movie Corresponds to the Pattern of Cytoplasmic Streaming of the AFH3 RNAi Pollen Tube Shown in Figure 7Ac.
Supplemental Movie 7. This Movie Corresponds to the Pattern of Cytoplasmic Streaming of the AFH3 RNAi Pollen Tube Shown in Figure 7Ad.
Supplemental Movie 8. This Movie Corresponds to the Pattern of Cytoplasmic Streaming of the AFH3 RNAi Pollen Tube Shown in Figure 7Ae.
Supplemental Movie Legend.
Supplemental Methods.
Supplementary Material
Acknowledgments
This work was supported by grants from the Ministry of Science and Technology (2007CB947600), the National Natural Science Foundation of China (30771088, 30500044, and 30821007), and China Agriculture University (a seed research grant 2005022). Z.Y. was supported by grants from the U.S. National Institute of General Medical Sciences and Department of Energy, and S.H. was supported by the Chinese Academy of Sciences (hundred talents program). We thank Laurent Blanchoin (The Centre National de la Recherche Scientifique, Grenoble, France) for AFH1 plasmids and Dave Kovar (University of Chicago) for invaluable advice on setting up the TIRFM assay. We also thank Yan Zhao (China Agricultural University) for the initial cloning of AFH3 cDNA and Jin Yan (Institute of Botany, Chinese Academy of Sciences) for the help on actin staining of pollen tubes.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) are: Shanjin Huang (sjhuang@ibcas.ac.cn) or Zhenbiao Yang (zhenbiao.yang@ucr.edu).
Online version contains Web-only data.
References
- Amann, K.J., and Pollard, T.D. (2001). Direct real-time observation of actin filament branching mediated by Arp2/3 complex using total internal reflection fluorescence microscopy. Proc. Natl. Acad. Sci. USA 98 15009–15013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banno, H., and Chua, N.H. (2000). Characterization of the arabidopsis formin-like protein AFH1 and its interacting protein. Plant Cell Physiol. 41 617–626. [DOI] [PubMed] [Google Scholar]
- Bechtold, N., and Pelletier, G. (1998). In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 82 259–266. [DOI] [PubMed] [Google Scholar]
- Blanchoin, L., Amann, K.J., Higgs, H.N., Marchand, J.B., Kaiser, D.A., and Pollard, T.D. (2000). Direct observation of dendritic actin filament networks nucleated by Arp2/3 complex and WASP/Scar proteins. Nature 404 1007–1011. [DOI] [PubMed] [Google Scholar]
- Blanchoin, L., and Staiger, C.J. (2008). Plant formins: Diverse isoforms and unique molecular mechanism. Biochim. Biophys. Acta doi:10.1016/j.bbamcr.2008.09.015. [DOI] [PubMed]
- Cardenas, L., Lovy-Wheeler, A., Kunkel, J.G., and Hepler, P.K. (2008). Pollen tube growth oscillations and intracellular calcium levels are reversibly modulated by actin polymerization. Plant Physiol. 146 1611–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casella, J.F., Flanagan, M.D., and Lin, S. (1981). Cytochalasin D inhibits actin polymerization and induces depolymerization of actin filaments formed during platelet shape change. Nature 293 302–305. [DOI] [PubMed] [Google Scholar]
- Chaudhry, F., Guerin, C., von Witsch, M., Blanchoin, L., and Staiger, C.J. (2007). Identification of Arabidopsis cyclase-associated protein 1 as the first nucleotide exchange factor for plant actin. Mol. Biol. Cell 18 3002–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chereau, D., Boczkowska, M., Skwarek-Maruszewska, A., Fujiwara, I., Hayes, D.B., Rebowski, G., Lappalainen, P., Pollard, T.D., and Dominguez, R. (2008). Leiomodin is an actin filament nucleator in muscle cells. Science 320 239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung, A.Y., and Wu, H.M. (2004). Overexpression of an Arabidopsis formin stimulates supernumerary actin cable formation from pollen tube cell membrane. Plant Cell 16 257–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole, R.A., and Fowler, J.E. (2006). Polarized growth: Maintaining focus on the tip. Curr. Opin. Plant Biol. 9 579–588. [DOI] [PubMed] [Google Scholar]
- Cvrckova, F. (2000). Are plant formins integral membrane proteins? Genome Biol. 1 research001.1–research001.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvrckova, F., Novotny, M., Pickova, D., and Zarsky, V. (2004). Formin homology 2 domains occur in multiple contexts in angiosperms. BMC Genomics 5 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeks, M.J., Hussey, P.J., and Davies, B. (2002). Formins: Intermediates in signal-transduction cascades that affect cytoskeletal reorganization. Trends Plant Sci. 7 492–498. [DOI] [PubMed] [Google Scholar]
- Deeks, M.J., Cvrckova, F., Machesky, L.M., Mikitova, V., Ketelaar, T., Zarsky, V., Davies, B., and Hussey, P.J. (2005). Arabidopsis group Ie formins localize to specific cell membrane domains, interact with actin-binding proteins and cause defects in cell expansion upon aberrant expression. New Phytol. 168 529–540. [DOI] [PubMed] [Google Scholar]
- de Win, A.H., Pierson, E.S., and Derksen, J. (1999). Rational analyses of organelle trajectories in tobacco pollen tubes reveal characteristics of the actomyosin cytoskeleton. Biophys. J. 76 1648–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelista, M., Blundell, K., Longtine, M.S., Chow, C.J., Adames, N., Pringle, J.R., Peter, M., and Boone, C. (1997). Bni1p, a yeast formin linking cdc42p and the actin cytoskeleton during polarized morphogenesis. Science 276 118–122. [DOI] [PubMed] [Google Scholar]
- Evangelista, M., Pruyne, D., Amberg, D.C., Boone, C., and Bretscher, A. (2002. a). Formins direct Arp2/3-independent actin filament assembly to polarize cell growth in yeast. Nat. Cell Biol. 4 260–269. [DOI] [PubMed] [Google Scholar]
- Evangelista, M., Pruyne, D., Amberg, D.C., Boone, C., and Bretscher, A. (2002. b). Formins direct Arp2/3-independent actin filament assembly to polarize cell growth in yeast. Nat. Cell Biol. 4 32–41. [DOI] [PubMed] [Google Scholar]
- Favery, B., Chelysheva, L.A., Lebris, M., Jammes, F., Marmagne, A., De Almeida-Engler, J., Lecomte, P., Vaury, C., Arkowitz, R.A., and Abad, P. (2004). Arabidopsis formin AtFH6 is a plasma membrane-associated protein upregulated in giant cells induced by parasitic nematodes. Plant Cell 16 2529–2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feierbach, B., and Chang, F. (2001). Roles of the fission yeast formin for3p in cell polarity, actin cable formation and symmetric cell division. Curr. Biol. 11 1656–1665. [DOI] [PubMed] [Google Scholar]
- Fox, J.E., and Phillips, D.R. (1981). Inhibition of actin polymerization in blood platelets by cytochalasins. Nature 292 650–652. [DOI] [PubMed] [Google Scholar]
- Fu, Y., Wu, G., and Yang, Z. (2001). Rop GTPase-dependent dynamics of tip-localized F-actin controls tip growth in pollen tubes. J. Cell Biol. 152 1019–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbon, B.C., Kovar, D.R., and Staiger, C.J. (1999). Latrunculin B has different effects on pollen germination and tube growth. Plant Cell 11 2349–2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbon, B.C., Ren, H., and Staiger, C.J. (1997). Characterization of maize (Zea mays) pollen profilin function in vitro and in live cells. Biochem. J. 327 909–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, Y., Fu, Y., Dowd, P., Li, S., Vernoud, V., Gilroy, S., and Yang, Z. (2005). A Rho family GTPase controls actin dynamics and tip growth via two counteracting downstream pathways in pollen tubes. J. Cell Biol. 169 127–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, E.S., Li, F., and Higgs, H.N. (2004). The mouse formin, FRLalpha, slows actin filament barbed end elongation, competes with capping protein, accelerates polymerization from monomers, and severs filaments. J. Biol. Chem. 279 20076–20087. [DOI] [PubMed] [Google Scholar]
- Hepler, P.K., Vidali, L., and Cheung, A.Y. (2001). Polarized cell growth in higher plants. Annu. Rev. Cell Dev. Biol. 17 159–187. [DOI] [PubMed] [Google Scholar]
- Higaki, T., Sano, T., and Hasezawa, S. (2007). Actin microfilament dynamics and actin side-binding proteins in plants. Curr. Opin. Plant Biol. 10 549–556. [DOI] [PubMed] [Google Scholar]
- Higashida, C., Miyoshi, T., Fujita, A., Oceguera-Yanez, F., Monypenny, J., Andou, Y., Narumiya, S., and Watanabe, N. (2004). Actin polymerization-driven molecular movement of mDia1 in living cells. Science 303 2007–2010. [DOI] [PubMed] [Google Scholar]
- Higgs, H.N., Blanchoin, L., and Pollard, T.D. (1999). Influence of the C terminus of Wiskott-Aldrich syndrome protein (WASp) and the Arp2/3 complex on actin polymerization. Biochemistry 38 15212–15222. [DOI] [PubMed] [Google Scholar]
- Honys, D., and Twell, D. (2003). Comparative analysis of the Arabidopsis pollen transcriptome. Plant Physiol. 132 640–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, S., Blanchoin, L., Kovar, D.R., and Staiger, C.J. (2003). Arabidopsis capping protein (AtCP) is a heterodimer that regulates assembly at the barbed ends of actin filaments. J. Biol. Chem. 278 44832–44842. [DOI] [PubMed] [Google Scholar]
- Ingouff, M., Fitz Gerald, J.N., Guerin, C., Robert, H., Sorensen, M.B., Van Damme, D., Geelen, D., Blanchoin, L., and Berger, F. (2005). Plant formin AtFH5 is an evolutionarily conserved actin nucleator involved in cytokinesis. Nat. Cell Biol. 7 374–380. [DOI] [PubMed] [Google Scholar]
- Kleinebrecht, J., Selow, J., and Winkler, W. (1982). The mouse mutant limb-deformity (ld). Anat. Anz. 152 313–324. [PubMed] [Google Scholar]
- Kovar, D.R., Harris, E.S., Mahaffy, R., Higgs, H.N., and Pollard, T.D. (2006). Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell 124 423–435. [DOI] [PubMed] [Google Scholar]
- Kovar, D.R., Kuhn, J.R., Tichy, A.L., and Pollard, T.D. (2003). The fission yeast cytokinesis formin Cdc12p is a barbed end actin filament capping protein gated by profilin. J. Cell Biol. 161 875–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar, D.R., and Pollard, T.D. (2004). Insertional assembly of actin filament barbed ends in association with formins produces piconewton forces. Proc. Natl. Acad. Sci. USA 101 14725–14730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y.J., Szumlanski, A., Nielsen, E., and Yang, Z. (2008). Rho-GTPase-dependent filamentous actin dynamics coordinate vesicle targeting and exocytosis during tip growth. J. Cell Biol. 181 1155–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y.J., and Yang, Z. (2008). Tip growth: Signaling in the apical dome. Curr. Opin. Plant Biol. 11 662–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenartowska, M., and Michalska, A. (2008). Actin filament organization and polarity in pollen tubes revealed by myosin II subfragment 1 decoration. Planta 228 891–896. [DOI] [PubMed] [Google Scholar]
- Li, H., Lin, Y., Heath, R.M., Zhu, M.X., and Yang, Z. (1999). Control of pollen tube tip growth by a Rop GTPase-dependent pathway that leads to tip-localized calcium influx. Plant Cell 11 1731–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovy-Wheeler, A., Wilsen, K.L., Baskin, T.I., and Hepler, P.K. (2005). Enhanced fixation reveals the apical cortical fringe of actin filaments as a consistent feature of the pollen tube. Planta 221 95–104. [DOI] [PubMed] [Google Scholar]
- Michelot, A., Derivery, E., Paterski-Boujemaa, R., Guerin, C., Huang, S., Parcy, F., Staiger, C.J., and Blanchoin, L. (2006). A novel mechanism for the formation of actin-filament bundles by a nonprocessive formin. Curr. Biol. 16 1924–1930. [DOI] [PubMed] [Google Scholar]
- Michelot, A., Guerin, C., Huang, S., Ingouff, M., Richard, S., Rodiuc, N., Staiger, C.J., and Blanchoin, L. (2005). The formin homology 1 domain modulates the actin nucleation and bundling activity of Arabidopsis FORMIN1. Plant Cell 17 2296–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, D.D., Scordilis, S.P., and Hepler, P.K. (1995). Identification and localization of three classes of myosins in pollen tubes of Lilium longiflorum and Nicotiana alata. J. Cell Sci. 108 2549–2563. [DOI] [PubMed] [Google Scholar]
- Mullins, R.D., Heuser, J.A., and Pollard, T.D. (1998). The interaction of Arp2/3 complex with actin: Nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc. Natl. Acad. Sci. USA 95 6181–6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul, A., and Pollard, T. (2008). The role of the FH1 domain and profilin in formin-mediated actin-filament elongation and nucleation. Curr. Biol. 18 9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen, J., Nielsen, O., Egel, R., and Hagan, I.M. (1998). FH3, a domain found in formins, targets the fission yeast formin Fus1 to the projection tip during conjugation. J. Cell Biol. 141 1217–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson, E.S., Miller, D.D., Callaham, D.A., van Aken, J., Hackett, G., and Hepler, P.K. (1996). Tip-localized calcium entry fluctuates during pollen tube growth. Dev. Biol. 174 160–173. [DOI] [PubMed] [Google Scholar]
- Pina, C., Pinto, F., Feijo, J.A., and Becker, J.D. (2005). Gene family analysis of the Arabidopsis pollen transcriptome reveals biological implications for cell growth, division control, and gene expression regulation. Plant Physiol. 138 744–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard, T.D. (1984). Polymerization of ADP-actin. J. Cell Biol. 99 769–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard, T.D., Blanchoin, L., and Mullins, R.D. (2001). Actin dynamics. J. Cell Sci. 114 3–4. [DOI] [PubMed] [Google Scholar]
- Pruyne, D., Evangelista, M., Yang, C., Bi, E., Zigmond, S., Bretscher, A., and Boone, C. (2002). Role of formins in actin assembly: nucleation and barbed-end association. Science 297 612–615. [DOI] [PubMed] [Google Scholar]
- Quinlan, M.E., Heuser, J.E., Kerkhoff, E., and Mullins, R.D. (2005). Drosophila Spire is an actin nucleation factor. Nature 433 382–388. [DOI] [PubMed] [Google Scholar]
- Redmond, T., Tardif, M., and Zigmond, S.H. (1994). Induction of actin polymerization in permeabilized neutrophils. Role of ATP. J. Biol. Chem. 269 21657–21663. [PubMed] [Google Scholar]
- Romero, S., Le Clainche, C., Didry, D., Egile, C., Pantaloni, D., and Carlier, M.F. (2004). Formin is a processive motor that requires profilin to accelerate actin assembly and associated ATP hydrolysis. Cell 119 419–429. [DOI] [PubMed] [Google Scholar]
- Sagot, I., Klee, S.K., and Pellman, D. (2002). Yeast formins regulate cell polarity by controlling the assembly of actin cables. Nat. Cell Biol. 4 42–50. [DOI] [PubMed] [Google Scholar]
- Sheahan, M.B., Staiger, C.J., Rose, R.J., and McCurdy, D.W. (2004). A green fluorescent protein fusion to actin-binding domain 2 of Arabidopsis fimbrin highlights new features of a dynamic actin cytoskeleton in live plant cells. Plant Physiol. 136 3968–3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimmen, T., and Yokota, E. (2004). Cytoplasmic streaming in plants. Curr. Opin. Cell Biol. 16 68–72. [DOI] [PubMed] [Google Scholar]
- Snowman, B.N., Kovar, D.R., Shevchenko, G., Franklin-Tong, V.E., and Staiger, C.J. (2002). Signal-mediated depolymerization of actin in pollen during the self-incompatibility response. Plant Cell 14 2613–2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich, J.A., and Watt, S. (1971). The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J. Biol. Chem. 246 4866–4871. [PubMed] [Google Scholar]
- Sundaresan, V., Springer, P., Volpe, T., Haward, S., Jones, J.D., Dean, C., Ma, H., and Martienssen, R. (1995). Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev. 9 1797–1810. [DOI] [PubMed] [Google Scholar]
- Symons, M.H., and Mitchison, T.J. (1991). Control of actin polymerization in live and permeabilized fibroblasts. J. Cell Biol. 114 503–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tellam, R., and Frieden, C. (1982). Cytochalasin D and platelet gelsolin accelerate actin polymer formation. A model for regulation of the extent of actin polymer formation in vivo. Biochemistry 21 3207–3214. [DOI] [PubMed] [Google Scholar]
- Tominaga, T., Sahai, E., Chardin, P., McCormick, F., Courtneidge, S.A., and Alberts, A.S. (2000). Diaphanous-related formins bridge Rho GTPase and Src tyrosine kinase signaling. Mol. Cell 5 13–25. [DOI] [PubMed] [Google Scholar]
- Twell, D., Yamaguchi, J., Wing, R.A., Ushiba, J., and McCormick, S. (1991). Promoter analysis of genes that are coordinately expressed during pollen development reveals pollen-specific enhancer sequences and shared regulatory elements. Genes Dev. 5 496–507. [DOI] [PubMed] [Google Scholar]
- Vidali, L., and Hepler, P.K. (1997). Characterization and localization of profilin in pollen grains and tubes of Lilium longiflorum. Cell Motil. Cytoskeleton 36 323–338. [DOI] [PubMed] [Google Scholar]
- Vidali, L., and Hepler, P.K. (2001). Actin and pollen tube growth. Protoplasma 215 64–76. [DOI] [PubMed] [Google Scholar]
- Wang, H.Y., Yu, Y., Chen, Z.L., and Xia, G.X. (2005). Functional characterization of Gossypium hirsutum profilin 1 gene (GhPFN1) in tobacco suspension cells. Characterization of in vivo functions of a cotton profilin gene. Planta 222 594–603. [DOI] [PubMed] [Google Scholar]
- Wang, Z., and Pesacreta, T.C. (2004). A subclass of myosin XI is associated with mitochondria, plastids, and the molecular chaperone subunit TCP-1alpha in maize. Cell Motil. Cytoskeleton 57 218–232. [DOI] [PubMed] [Google Scholar]
- Watanabe, N., Madaule, P., Reid, T., Ishizaki, T., Watanabe, G., Kakizuka, A., Saito, Y., Nakao, K., Jockusch, B.M., and Narumiya, S. (1997). p140mDia, a mammalian homolog of Drosophila diaphanous, is a target protein for Rho small GTPase and is a ligand for profilin. EMBO J. 16 3044–3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wear, M.A., Schafer, D.A., and Cooper, J.A. (2000). Actin dynamics: Assembly and disassembly of actin networks. Curr. Biol. 10 R891–R895. [DOI] [PubMed] [Google Scholar]
- Welch, M.D., Iwamatsu, A., and Mitchison, T.J. (1997). Actin polymerization is induced by Arp2/3 protein complex at the surface of Listeria monocytogenes. Nature 385 265–269. [DOI] [PubMed] [Google Scholar]
- Xu, Y., Moseley, J.B., Sagot, I., Poy, F., Pellman, D., Goode, B.L., and Eck, M.J. (2004). Crystal structures of a Formin Homology-2 domain reveal a tethered dimer architecture. Cell 116 711–723. [DOI] [PubMed] [Google Scholar]
- Yahara, I., Harada, F., Sekita, S., Yoshihira, K., and Natori, S. (1982). Correlation between effects of 24 different cytochalasins on cellular structures and cellular events and those on actin in vitro. J. Cell Biol. 92 69–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalovsky, S., Bloch, D., Sorek, N., and Kost, B. (2008). Regulation of membrane trafficking, cytoskeleton dynamics, and cell polarity by ROP/RAC GTPases. Plant Physiol. 147 1527–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z. (2008). Cell polarity signaling in Arabidopsis. Annu. Rev. Cell Dev. Biol. 24 551–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi, K., Guo, C., Chen, D., Zhao, B., Yang, B., and Ren, H. (2005). Cloning and functional characterization of a formin-like protein (AtFH8) from Arabidopsis. Plant Physiol. 138 1071–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zigmond, S.H., Evangelista, M., Boone, C., Yang, C., Dar, A.C., Sicheri, F., Forkey, J., and Pring, M. (2003). Formin leaky cap allows elongation in the presence of tight capping proteins. Curr. Biol. 13 1820–1823. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.