Abstract
The micropylar endosperm cap covering the radicle in the mature seeds of most angiosperms acts as a constraint that regulates seed germination. Here, we report on a comparative seed biology study with the close Brassicaceae relatives Lepidium sativum and Arabidopsis thaliana showing that ethylene biosynthesis and signaling regulate seed germination by a mechanism that requires the coordinated action of the radicle and the endosperm cap. The larger seed size of Lepidium allows direct tissue-specific biomechanical, biochemical, and transcriptome analyses. We show that ethylene promotes endosperm cap weakening of Lepidium and endosperm rupture of both species and that it counteracts the inhibitory action of abscisic acid (ABA) on these two processes. Cross-species microarrays of the Lepidium micropylar endosperm cap and the radicle show that the ethylene-ABA antagonism involves both tissues and has the micropylar endosperm cap as a major target. Ethylene counteracts the ABA-induced inhibition without affecting seed ABA levels. The Arabidopsis loss-of-function mutants ACC oxidase2 (aco2; ethylene biosynthesis) and constitutive triple response1 (ethylene signaling) are impaired in the 1-aminocyclopropane-1-carboxylic acid (ACC)-mediated reversion of the ABA-induced inhibition of seed germination. Ethylene production by the ACC oxidase orthologs Lepidium ACO2 and Arabidopsis ACO2 appears to be a key regulatory step. Endosperm cap weakening and rupture are promoted by ethylene and inhibited by ABA to regulate germination in a process conserved across the Brassicaceae.
INTRODUCTION
The first Arabidopsis thaliana hormone mutants were identified using seed germination and dormancy phenotypes (Koornneef et al., 1982). These key seed traits are antagonistically regulated by abscisic acid (ABA) and gibberellins (GA). Subsequent work with the ABA- and GA-related mutants showed that, in general, while ABA induces dormancy and inhibits germination, GA releases dormancy and promotes germination (reviewed in Bewley, 1997; Kucera et al., 2005; Finch-Savage and Leubner-Metzger, 2006; Holdsworth et al., 2008). It also became evident that interactions between seed tissues are important for germination and dormancy and that these interactions are, at least in part, regulated by GA and ABA. The GA-ABA antagonism has become a hallmark of seed germination and dormancy, and mechanisms underlying the GA-ABA-related seed tissue interactions have been investigated in many publications (e.g., Ogawa et al., 2003; Liu et al., 2005; Müller et al., 2006; Penfield et al., 2006; Bethke et al., 2007; Piskurewicz et al., 2008).
Another major regulator of seed germination and plant development is ethylene (C2H4). In general, seedling growth is inhibited by ethylene as well as by ABA, and this growth inhibition by both hormones is well characterized at the molecular level (Etheridge et al., 2006; Nemhauser et al., 2006). In contrast with the situation in seedlings, ethylene is known to promote seed germination, and an ABA-ethylene antagonism is evident for seeds of Arabidopsis (Beaudoin et al., 2000; Ghassemian et al., 2000) and other species (Kucera et al., 2005; Matilla and Matilla-Vazquez, 2008). Interestingly, Arabidopsis mutants in ethylene signal transduction components have been recovered as enhancer and suppressor mutants of ABA-insensitive mutants with seed germination phenotypes (Beaudoin et al., 2000; Ghassemian et al., 2000). Among them is the constitutive triple response1 (ctr1) mutant, which has reduced ABA sensitivity of seed germination (Beaudoin et al., 2000). The first cloned plant hormone receptor was the Arabidopsis ethylene receptor ETR1 (Bleecker et al., 1988; Chang et al., 1993a). Ethylene-insensitive ethylene response1 (etr1) mutant seeds germinate poorly, their dormancy is enhanced, and their germination is ABA hypersensitive. This is, at least in part, due to a higher ABA content in the etr1 mutant seeds (Chiwocha et al., 2005). The ethylene action inhibitor 2,5-norbornadiene (NBD) binds specifically to ethylene receptors in direct competition with ethylene for the ethylene binding site (Sisler and Serek, 2003). Experiments with this inhibitor in many species, including Arabidopsis (Siriwitayawan et al., 2003), tobacco (Nicotiana tabacum; Leubner-Metzger et al., 1998), pea (Pisum sativum; Petruzzelli et al., 2000), and others (Kucera et al., 2005) show that the influence of ethylene receptors on the regulation of seed germination is a phylogenetically widespread phenomenon. Several hypotheses have been put forward to explain the mechanisms of ethylene action in germinating seeds and the ethylene-ABA antagonism (see Discussion), but unlike the GA–ABA interaction, the evidence for these hypotheses is circumstantial and the mechanisms are unknown.
The process of seed germination starts with imbibition of the dry seed and ends when the radicle has emerged through all the coats enveloping the embryo (Finch-Savage and Leubner-Metzger, 2006). In a mature Arabidopsis seed, the embryo is covered by two coats: a single layer of living endosperm cells (aleurone layer) and a dead testa (seed coat). Testa rupture and endosperm rupture are two sequential events during the germination of many species, including Arabidopsis, and endosperm rupture was found to be promoted by GA and inhibited by ABA. This seems to involve exchange of GA- and ABA-related signals between the embryo and endosperm (e.g., Liu et al., 2005; Müller et al., 2006; Okamoto et al., 2006; Penfield et al., 2006; Piskurewicz et al., 2008). This ABA-GA antagonism is clearly evident at the Arabidopsis whole-seed transcriptome level (Ogawa et al., 2003; Cadman et al., 2006; Finch-Savage et al., 2007). Here, we present a transcriptome analysis of the interaction between individual seed tissues during germination to further investigate hormone interactions during germination.
In seeds, where the endosperm acts as a mechanical barrier, endosperm weakening seems to be essential for endosperm rupture and radicle protrusion; these changes can be measured biomechanically as a decline in mechanical resistance of the micropylar endosperm (the endosperm layer covering the radicle tip, the cap) of seeds from a variety of families (e.g., Bewley, 1997; Toorop et al., 2000; da Silva et al., 2004; Finch-Savage and Leubner-Metzger, 2006). Seeds of the model species Arabidopsis are too small for such measurements, and this has limited progress in linking biomechanical and molecular studies. To help overcome this obstacle, we have demonstrated in a comparative seed biology study of Arabidopsis and its close relative Lepidium sativum (which has larger seeds than Arabidopsis) that the latter can be used as model system for studying both the molecular and biomechanical mechanisms of endosperm cap weakening (Müller et al., 2006, 2009). In this work, we showed by direct biomechanical measurement of Lepidium endosperm cap weakening that the weakening is promoted by GA and inhibited by ABA. An early embryo signal is required and sufficient to induce endosperm weakening, which afterwards appears to be an organ-autonomous process. Experiments with isolated endosperm caps showed that GA can replace the embryo signal, that de novo GA biosynthesis occurs in the endosperm, and that the weakening is regulated, at least in part, by the GA/ABA ratio. These biomechanical and physiological findings in Lepidium are in agreement with what is known for the spatial, temporal, and GA-mediated regulation of genes during Arabidopsis seed germination (Yamaguchi et al., 2001; Ogawa et al., 2003; Yamauchi et al., 2004). The genera Lepidium and Arabidopsis are closely related; they both belong to the lineage I clade of the Brassicaceae family (Franzke et al., 2009). Comparative Brassicaceae seed biology with Lepidium/Arabidopsis is therefore a powerful approach to investigate evolutionarily conserved mechanisms. We have adopted this approach in this work to study the ABA–ethylene interaction that influences endosperm cap weakening and rupture in both species.
Key genes involved in ABA biosynthesis and degradation have been investigated during Arabidopsis seed germination (e.g., Müller et al., 2006; Okamoto et al., 2006; Penfield et al., 2006; Holdsworth et al., 2008; Piskurewicz et al., 2008). ABA produced during seed maturation is degraded during the imbibition of nondormant Arabidopsis seeds and barley (Hordeum vulgare) grains by ABA 8'-hydroxylases (CYP707A; Okamoto et al., 2006; Barrero et al., 2009). Transcript expression of these hydroxylases in the barley grain coleorhiza, which covers the radicle, is associated with the transition from the dormant to the nondormant state (Barrero et al., 2009). The temporal and spatial transcript expression patterns of these hydroxylases in Arabidopsis seeds indicate that ABA degradation in the embryo and the endosperm is involved in the regulation of endosperm rupture (Okamoto et al., 2006). Penfield et al. (2006) investigated differential gene expression in the embryo and the endosperm of Arabidopsis seeds by transcriptome analysis, but this comparison was made only after radicle emergence (i.e., after the completion of germination). Nevertheless, a major conclusion of this study was that the observed transcript changes in the Arabidopsis endosperm layer is of importance for the ABA-inhibited transition from seed germination to seedling growth. This is in agreement with roles for the endosperm in Arabidopsis coat dormancy (Bethke et al., 2007) and in endosperm weakening/rupture of Lepidium/Arabidopsis (Liu et al., 2005; Müller et al., 2006, 2009). These findings show that the living endosperm layer, which is retained in the mature seeds of these Brassicaceae species, has an important developmental role.
Here, we report on a cross-species transcriptome analysis that shows that Arabidopsis microarrays can be used to investigate transcript expression patterns of separate Lepidium seed tissues during germination. As part of this analysis, we studied the role of hormone-related genes in the interaction between the endosperm cap and radicle. This analysis revealed a significant involvement of ethylene-related transcripts during Lepidium endosperm weakening. We therefore went on to investigate this further using biomechanical, physiological, enzyme, and transcript expression analyses to provide direct evidence for a novel mechanism of ethylene action during Lepidium seed germination. We found that ethylene promotes endosperm rupture by promoting endosperm weakening and by counteracting the ABA inhibition of this process. Parallel experiments with Arabidopsis mutants that are defective in ethylene biosynthesis and signaling provided further evidence to support our conclusion that endosperm cap weakening is a major target of ethylene.
RESULTS
Endosperm Cap Weakening, Endosperm Cap Hole Formation, and Endosperm Rupture Are Developmentally and Hormonally Regulated during Lepidium Seed Germination
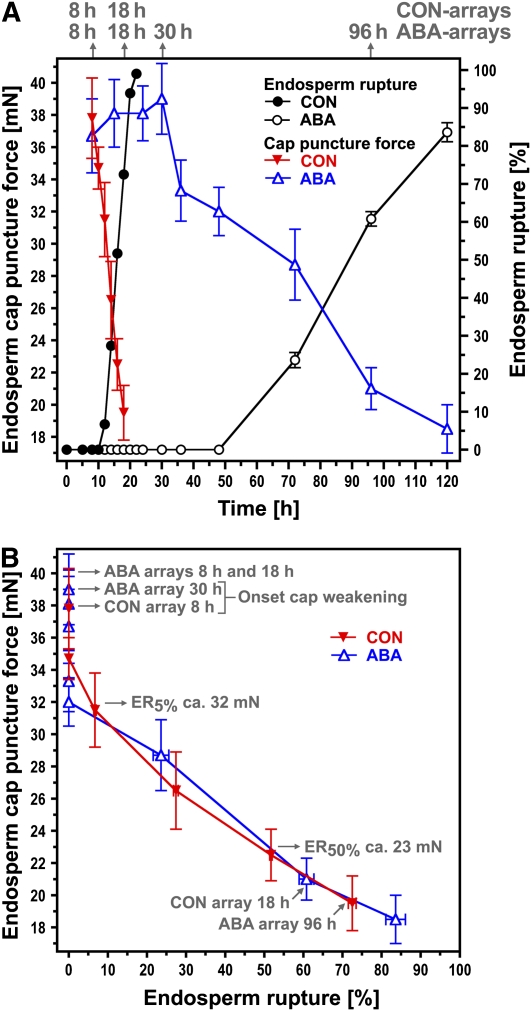
The time-course analysis in Figure 1 shows that endosperm cap weakening precedes endosperm rupture of Lepidium seeds when imbibed both without (CON) or with ABA added to the medium. The mechanical resistance of the micropylar endosperm (endosperm cap) was used as a measure of weakening and quantified by the puncture-force method (Müller et al., 2006). ABA treatment delayed the onset and decreased the rate of decline in endosperm cap weakening (Figure 1A). However, when the endosperm cap puncture force of the CON and ABA series was plotted against the percentage of endosperm rupture in the seed population, the same relationship was evident in both treatments (Figure 1B). The lowest endosperm cap puncture force measured in both treatments was ∼19 mN. By this point, the radicles of all seeds were able to generate sufficient force to overcome the resistance of the cap and emerge. The results presented in Figure 1 support our hypothesis that a defined degree of endosperm cap weakening is a prerequisite of endosperm rupture and that endosperm weakening is a major target of ABA-induced inhibition of germination.
Figure 1.
Time Course of Endosperm Cap Weakening and Endosperm Rupture of L. sativum FR1 and the Effect of ABA.
(A) Endosperm cap weakening and rupture of seeds incubated without (CON) or with 10 μM ABA added to the medium in continuous light at 18°C. Endosperm cap weakening was determined by measuring tissue resistance by the puncture force method at the times indicated. The time points (after the start of imbibition) at which tissues were dissected for the microarrays are indicated in gray. All seeds selected for the puncture force measurements and for the microarrays had completed testa rupture but intact endosperm caps. ABA did not affect testa rupture.
(B) The CON and ABA seed populations showed a very similar relationship between decreasing endosperm cap puncture force and increasing percentage of seeds showing endosperm rupture. Mean values ± se of 3 × 50 seeds for the endosperm rupture [%] and at least 50 endosperm caps for the puncture force (mN) measurements are presented.
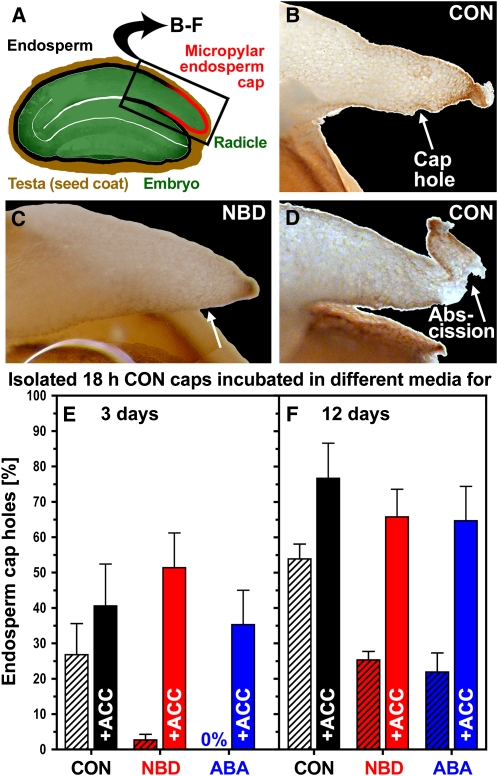
Once endosperm cap weakening has been induced by an early embryo signal, the isolated cap will continue to weaken further, and in the absence of the radicle (isolated caps; Figure 7 in Müller et al., 2006), puncture force values can be measured below the minimal value of 19 mN that allows rupture by the emerging radicle in whole seeds (Figure 1). In these isolated caps, this continued weakening finally results in a hole in the place where the radicle, if present, would have emerged (Figures 2A and 2B). This hole always occurs in the same place in direct proximity to the very tip of the endosperm cap and can be readily recorded without specialist biomechanical equipment to show the completion of weakening. The results presented in Figure 2 show that cap hole formation is developmentally and hormonally regulated and, as it occurs in isolated caps, is a cap-autonomous process that does not require the presence of a pushing radicle. The addition of hormones to the incubation medium affected the speed and completion of cap hole formation. For example, hole formation was promoted when the isolated caps were treated with GA (10 μM GA4+7 60.0 ± 9.4% compared with CON 26.9 ± 8.8% cap holes on day 3) or with 1-aminocyclopropane-1-carboxylic acid (ACC), the direct precursor of ethylene (Figure 2). In keeping with this, treatment with NBD, a well-characterized ethylene action inhibitor that binds to the ethylene receptors (Sisler and Serek, 2003), inhibited cap hole formation (Figure 2). The reversion of this inhibition by simultaneous treatment with NBD+ACC demonstrates the specificity of this effect for the ethylene action (i.e., ACC-derived ethylene displaces the bound NBD). This also demonstrates that ACC acts via its conversion to ethylene by ACC oxidase (ACO) enzyme activity. ABA treatment inhibited cap hole formation, but ACC reversed this inhibition as shown by simultaneous treatment with ABA+ACC. In general, cap hole formation was promoted by GA and ethylene and inhibited by ABA (Figure 2). In both Lepidium accessions that we analyzed (FR1 and FR14), endosperm cap hole formation was regulated by an ethylene-ABA antagonism. This suggests that ethylene-related transcripts/proteins are of importance for counteracting the ABA-induced inhibition of endosperm cap weakening, hole formation, and rupture.
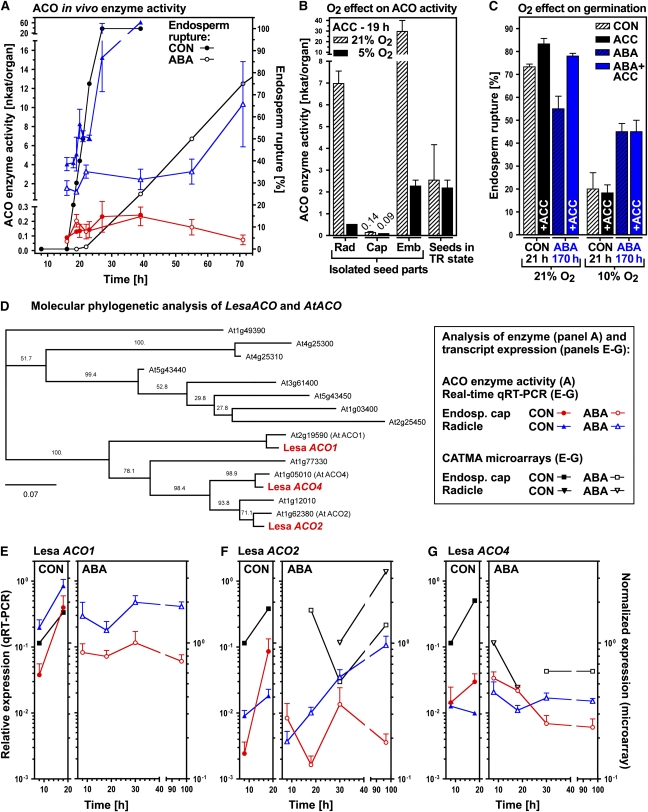
Figure 7.
Analysis of ACO Enzyme Activity and Transcript Expression in Specific Seed Tissues of L. sativum during Germination.
The framed box shows symbols used in (A) and (E) to (G). Mean values ± se ([A] to [C]) or +se (qRT-PCR; [E] to [G]) are presented for three ([A] to [C]) or four ([E] to [G]) biologically independent samples each with 50 seeds (C), 100 to 200 seed parts ([A] and [B]) or 1000 endosperm caps or 100 radicles ([E] to [G]) from seeds with ruptured testa but intact endosperm ([A], [B], and [E] to [G]). L. sativum FR14 ([A] to [D]) or FR1 ([E] to [G]) seeds were incubated at 24°C in continuous light. Note that the qRT-PCR and the microarrays were performed as independent experiments.
(A) Time course of in vivo ACO enzyme activities in endosperm caps and radicles of seeds incubated without (CON) and with 10 μM ABA; for comparison, the kinetics of endosperm rupture are presented. In vivo ACO enzyme activities of endosperm caps and radicles dissected from seeds at different time points were measured by subsequent organ incubation in medium with ACC (plus ABA for the ABA series).
(B) The effect of ambient and reduced oxygen atmospheres on the in vivo ACO enzyme activities of isolated seed parts compared with intact seeds. Note that only seeds in the testa rupture (TR) state were selected for the measurements (i.e., seeds with intact endosperm). No wound-induced ethylene production by the radicle/embryo was evident.
(C) The effect of oxygen on the promotion of endosperm rupture by ACC and the reversion of the ABA inhibition of endosperm rupture by ACC.
(D) Molecular phylogenetic analysis of Lepidium ACO and Arabidopsis ACO cDNA sequences. The bar (0.07) defines the number of substitutions per 100 amino acids. Lepidium ACO cDNA sequences and comparisons to the Arabidopsis orthologs are presented in Supplemental Figure 3 online.
(E) to (G) LesaACO transcript expression pattern determined by qRT-PCR in endosperm cap and radicle during incubation on medium without (CON) or with 10 μM ABA added. Relative ΔΔCt expression values based on the comparison with validated constitutive transcripts are presented.
Figure 2.
Endosperm Cap Hole Formation of L. sativum Is a Developmentally and Hormonally Regulated Process.
(A) to (D) Endosperm cap hole formation was investigated by dissecting micropylar endosperm caps ([A]; isolated caps) from L. sativum FR1 seeds imbibed for 18 h in CON medium.
(B) and (C) Subsequent incubation of isolated caps (radicle removed) for several days on CON medium (B) or on medium with 1 mM ACC resulted in endosperm cap hole formation (arrow), which was inhibited when the isolated caps were incubated in the presence of the ethylene action inhibitor NBD ([C]; 100 μL/L applied via the gas phase).
(D) Prolonged incubation of caps with holes on CON medium leads in many cases to abscission of the cap tip (arrow).
(E) and (F) Percentage of isolated caps that formed holes by 3 and 12 d, respectively. ACC promoted cap hole formation and completely reverted the inhibiting effects of NBD and 3 μM ABA when added in combination to the incubation medium of the isolated caps. Mean values ± se from two independent experiments with five and 12 isolated caps, respectively, are presented. All incubation conditions were continuous light at 18°C. Similar results were obtained for L. sativum FR14.
Spatial, Temporal, and ABA-Related Transcriptome Analysis in the Endosperm Cap and the Radicle Reveal Tissue-Related Differences in Transcript Expression Patterns and a Role for Ethylene during Endosperm Weakening and Endosperm Rupture
To investigate how Lepidium gene transcripts in specific seed tissues are temporally, spatially, and hormonally regulated, we hybridized Lepidium RNA samples to Arabidopsis CATMA 25K microarrays (Complete Arabidopsis Transcriptome Microarray, www.catma.org) (Hilson et al., 2004; Allemeersch et al., 2005). The RNA was extracted from specific Lepidium seed tissues at defined times during germination: Radicle (Rad), micropylar endosperm (Cap), and nonmicropylar endosperm (NonE). These tissues were collected after testa rupture, before and during endosperm weakening, but prior to endosperm rupture (i.e., only seeds with intact endosperms were used) (Figure 1A).
In the CON arrays, we compared Rad, Cap, and NonE at 8 h after the start of imbibition and therefore just prior to the onset of endosperm weakening, and at 18 h, just prior to endosperm rupture (Figure 1A). It is not possible to dissect the cap from seeds before 8 h, and by this time significant transcriptome changes may have already occurred to facilitate weakening in imbibed CON seeds. Preparing additional samples from seeds imbibed on ABA allows the dissection at earlier developmental stages. Therefore, in the ABA arrays, we compared Rad and Cap from seeds incubated in medium containing 10 μM ABA at 8, 18, and 30 h after the start of imbibition, in the period leading up to the onset of the ABA-inhibited endosperm weakening, and later at 96 h, just prior to endosperm rupture (Figure 1A). Each transcriptome approach (CON and ABA) may therefore identify different genes that are temporally and spatially regulated, and using both approaches enhances the likelihood of identifying genes important for endosperm weakening during seed germination. Normalized expression values for Lepidium were obtained in the CON arrays for 22,025 transcripts (see Supplemental Data Set 1 online) and in the ABA arrays for 19,794 transcripts (see Supplemental Data Set 2 online), and Lepidium gene transcripts refer to the putative Arabidopsis orthologs defined by having an Arabidopsis Genome Initiative (AGI) identifier such as At1g62380 and a gene ontology (GO) annotation associated with this AGI (www.Arabidopsis.org). Supplemental Data Sets 1 and 2 online provide the normalized values for the mean expression differences between time points and seed tissues. The microarray data, including the normalized intensity values for each microarray of our Lepidium work, were deposited in ArrayExpress.
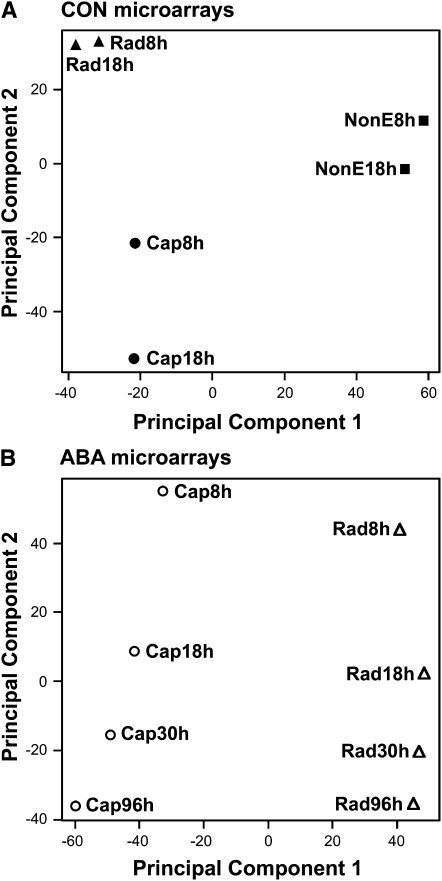
Principal component analysis (PCA) was used to search for global patterns in the Lepidium CON and ABA array expression data across all the gene transcripts (Figure 3). In both cases, the two components PC1 and PC2 accounted for >60% of the variance in gene expression. The CON arrays compared gene expression at the start and end of endosperm weakening only, whereas the ABA arrays also compared expression leading up to the start of weakening. In the CON arrays, PC1 separates the tissues that are arguably actively involved in the germination mechanism (Rad and Cap) from the storage component of the endosperm (NonE) (Figure 3A). In PC2, clear differences were observed between Rad and Cap, tissues whose interaction regulates the completion of germination. NonE was not considered in the ABA array experiment; therefore, PC1 of the ABA arrays clearly separated Rad and Cap. In the ABA arrays, PC2 then separated the times in the correct temporal order: 8 h → 18 h → 30 h → 96 h (Figure 3B). These clear patterns indicate that the data behave in an expected fashion with greatest differences in gene expression occurring between the tissues. The ABA array comparison indicates that the majority of changes in transcript numbers occur before endosperm weakening (i.e., 8 to 30 h). The very similar ordering of the time course between Rad and Cap tissues suggests that much of this change in the earlier stages of germination is common to the two tissues. Together, the ABA and CON array data suggest a greater temporal transcript expression difference in the cap than the radicle during the period of endosperm weakening leading to its rupture (i.e., CON, 8 to 18 h; ABA, 30 to 96 h).
Figure 3.
The Results of PCA Applied to CON and ABA Microarray Data of L. sativum FR1.
(A) PCA was applied to the expression of all informative genes on the CON microarrays (22,025 genes; see Supplemental Data Set 1 online) in various tissues and at various times after the start of imbibition, which is before and near the end of endosperm weakening, respectively (see Figure 1A), to look for global patterns of similarities and differences between the samples. PC1 and 2 accounted for 46 and 26% of the variance in gene expression, respectively.
(B) The results of PCA for the ABA microarrays (19,794 genes; see Supplemental Data Set 2 online) in various tissues and at various times, as indicated. Samples at 8, 18, and 30 h were taken before the start of endosperm weakening, and the sample at 96 h was taken near the end of endosperm weakening and before radicle emergence (see Figure 1A). PC1 and 2 accounted for 42 and 20% of the variance in gene expression, respectively.
The transcript abundance of individual genes in the CON and ABA array data (see Supplemental Data Sets 1 and 2 online) were compared for statistical significance of the mean expression differences between times and tissues using individual t tests and across treatments using F-tests (see Supplemental Data Sets 3 [CON arrays] and 4 [ABA arrays] online; for details, see Methods). The overall F-tests were used to identify those genes whose transcript numbers had significantly changed over time and/or between tissues. P values were adjusted for false discovery rate (Benjamini and Hochberg, 1995), and the resulting transcript lists (P values ≤ 0.1; F≤0.1 gene lists) contained 1350 transcripts for the CON array (see Supplemental Data Set 5 online) and 3530 transcripts for the ABA array (see Supplemental Data Set 6 online). These transcripts were considered to be either upregulated or downregulated between time points in the same tissue and/or differentially regulated between tissues. Application of the established seed-specific TAGGIT workflow, which allows annotation of transcriptome data sets by ascribing functional categories (as tags) that have previously been associated with seed maturation, dormancy, and germination (Carrera et al., 2007; Holdsworth et al., 2008) to these four gene lists (see Supplemental Data Sets 1, 2, 5, and 6 online), provided marked differences in their proportional representations of the functional categories (see Supplemental Figure 1 online). For reasons of brevity and in line with the topic of this article, only the “germination” and “hormone” categories will be considered.
When all transcripts (see Supplemental Data Sets 1 and 2 online) were compared with the regulated transcripts (see Supplemental Data Sets 5 and 6 online), the functional categories “germination” and “hormones” are overrepresented in the latter (CON array; see Supplemental Figure 1A online), and ABA affected this upward shift (ABA array; see Supplemental Figure 1C online). Among the hormone-related transcripts, the functional category “ethylene” was of particular interest. The relative number of ethylene-related transcripts was increased in the regulated transcripts compared with the list of all transcripts in both arrays: from 2.2 to 3.3% for CON and from 2.2 to 2.6% for ABA (see Supplemental Figure 1 online). Interestingly, when only transcripts that are regulated differently in the radicle and the endosperm are considered, the percentages of ethylene-related transcripts almost double in the CON array between 8 and 18 h after the start of imbibition from 1.1 to 2.1% (see Supplemental Figure 1B online). These results suggest that ethylene-related transcripts may play important spatial and temporal roles during endosperm rupture and in counteracting the ABA-induced inhibition.
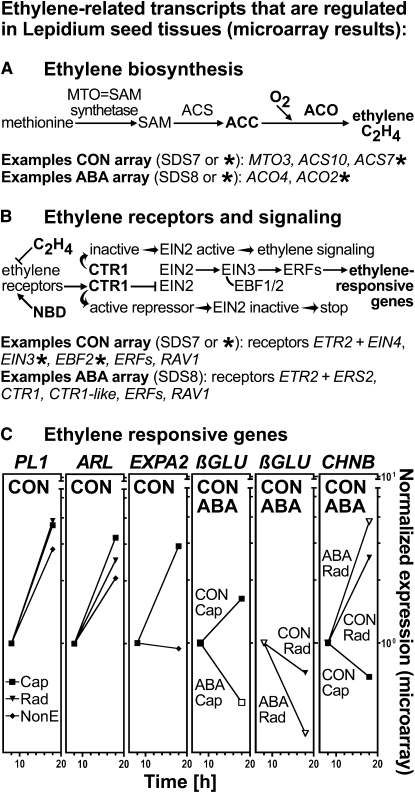
The role of ethylene was further analyzed by comparing regulated transcripts identified in the Lepidium seed tissues to gene lists known to be regulated by ethylene and ABA in Arabidopsis seedlings (Nemhauser et al., 2006). Of the 1350 regulated genes of the CON array, subsets of 24, 274, and 20 genes were also regulated in Arabidopsis seedlings by ethylene only, ABA only, or both hormones, respectively (see Supplemental Data Set 7 online). Similar subset proportions, 34, 578, and 54 genes, respectively, were shown for the ABA array regulated genes (3530) (see Supplemental Data Set 8 online). These subsets contain ethylene biosynthesis, signaling, and responsive genes that may be putative targets for investigating the ethylene–ABA interactions during Lepidium germination (Figure 4; see Supplemental Data Sets 7 and 8 online). These include (1) ACO2 and ACO4 orthologs of ACO, a key enzyme of ethylene biosynthesis. The ACO2 ortholog is not regulated in Arabidopsis seedlings by ethylene or ABA (Figure 4A) and thus may play an important and specific role in counteracting the ABA-induced inhibition of endosperm rupture. (2) CTR1, a negative regulator of the ethylene signal transduction pathway (Figure 4B), which is regulated in both seeds and seedlings by ABA. Therefore, CTR1 may be important for the ABA interactions in seeds and seedlings. (3) Putative ethylene-responsive downstream genes, including Pectate Lyase1 (PL1), Argos-like (ARL), Expansin A2 (EXPA2), β-1,3-glucanase (βGLU), and chitinase B (CHNB) (Figure 4C), for which roles in endosperm weakening and/or radicle growth during seed germination or in organ growth of seedlings have been proposed (e.g., Leubner-Metzger et al., 1998; Leubner-Metzger, 2003; Ogawa et al., 2003; Hu et al., 2006; Penfield et al., 2006; Carrera et al., 2008).
Figure 4.
Ethylene-Related Regulated Transcripts in L. sativum FR1 Seed Tissues.
The regulated transcript data sets of the Lepidium seed arrays (see Supplemental Data Sets 5 [CON, 1350 transcripts] and 6 [ABA, 3530 transcripts] online) were analyzed in two ways. First, TAGGIT analysis (see Supplemental Figure 1 online) was performed, and transcripts of the functional category “ethylene” were indentified in this analysis. Second, comparison to Arabidopsis transcriptome data sets known to be regulated in seedlings by ethylene and/or ABA (Nemhauser et al., 2006) provided transcript subsets that are regulated in seeds as well as by ethylene and/or ABA in seedlings (see Supplemental Data Sets 7 [CON array] and 8 [ABA array] online; SDS7 and SDS8 in Figure 4). These ethylene-related transcripts were considered further in (A) to (C). Examples of these ethylene-related transcripts shown in (A) and (B) are from the CON array and ABA array, as indicated. Those that are not marked are regulated in seedlings by ethylene and/or ABA and therefore appear on the subset lists SDS7 and SDS8, whereas those that are marked with an asterisk are not and therefore do not appear in subset lists (see Venn diagrams in SDS7 and SDS8).
(A) Key steps in ethylene biosynthesis include the oxygen-requiring conversion of ACC to ethylene by ACO.
(B) Key steps in ethylene signaling include ethylene binding to the receptors, which can be blocked by the ethylene action inhibitor NBD. In the absence of ethylene or with NBD bound to the ethylene receptors activating CTR1, a negative regulator of the downstream signaling pathway and the ethylene responses are blocked. Upon ethylene binding, the receptors and consequently CTR1 are inactive and the downstream signaling pathway factors (EIN2, EIN3, and ERFs) become active and mediate the expression of genes that facilitate the ethylene responses.
(C) Normalized expression values for transcripts of selected ethylene responsive genes that are regulated in seeds (CON and ABA arrays; see Supplemental Data Sets 3 and 4 online, respectively). ACS, ACC synthase; ERE, ethylene-responsive element.
Ethylene Biosynthesis and Signaling Are Both Involved in Counteracting the ABA-Mediated Inhibition of Endosperm Rupture
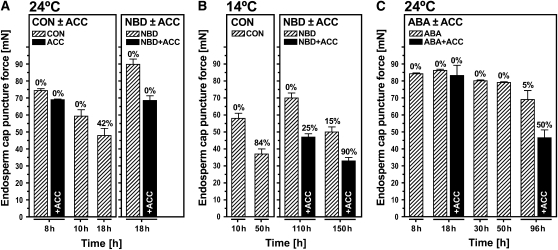
To test if ethylene biosynthesis and/or signaling are involved in endosperm rupture and in counteracting the ABA-mediated inhibition of endosperm rupture, we analyzed the time courses of Lepidium (Figure 5) and Arabidopsis (Figure 6) seed germination. Different concentrations of ABA were used as the two species differ in their dose responses (Müller et al., 2006), and ACC was used at a saturating concentration to enhance its enzymatic conversion to ethylene by endogenous ACO. Neither the addition of ACC, the ethylene action inhibitor NBD, nor ABA affected testa rupture of Lepidium and Arabidopsis (Figure 6; see Supplemental Figure 2 online). However, ACC promoted endosperm rupture, and both NBD and ABA markedly delayed it in both species. The inhibition was reversed by simultaneous addition of ACC or C2H4 (Figures 5 and 6). Thus, ethylene or ACC counteract the ABA-induced inhibition. In addition, as the ethylene action inhibitor NBD can only be displaced from the receptor binding site by ethylene itself (Sisler and Serek, 2003), this result demonstrates that ACC acts via its conversion to ethylene by ACO and suggests that ethylene signaling is required for the response.
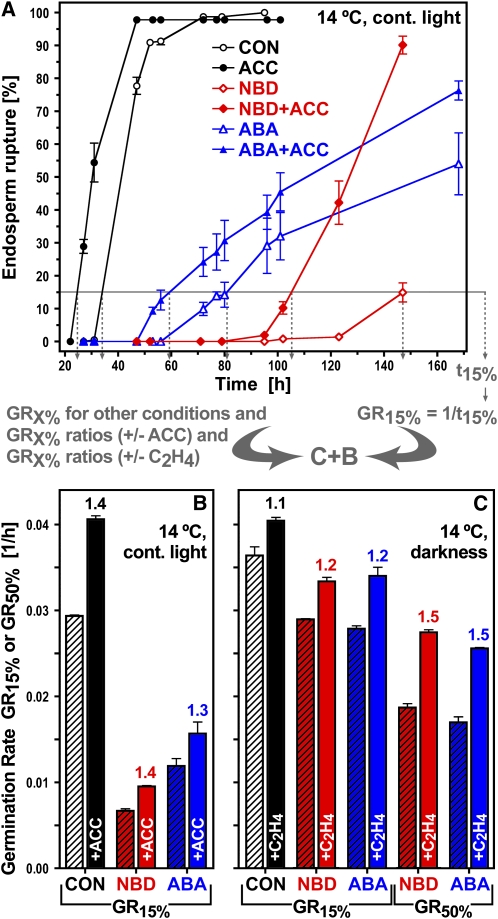
Figure 5.
The Effect of Ethylene, ACC, and ABA on the Time Course of Endosperm Rupture and on the Germination Rates GRX% of L. sativum FR14 Seeds.
(A) and (B) The times to reach 15 or 50% endosperm rupture (t15% or t50%) of the seed population were determined from the time courses of endosperm rupture in medium without (CON) and with additions (NBD, ABA, ACC, C2H4, or combinations). Germination rates GR15% or GR50% were then calculated (GRX% = 1/tX%) and used in subsequent analyses (gray arrows). The effect of ACC on time course (A) and GR15% (B) of seeds treated without (CON) or with NBD or ABA added.
(C) The effect of C2H4 on GR15% and GR50% of seeds imbibed without (CON) or with NBD or ABA. Numbers above the columns are fold increases in GRX% from the addition of ACC or C2H4 (GRX% ratios ±ACC or ±C2H4). Medium additions, as indicated: 5 μM ABA, 1 mM ACC, 100 μL/L NBD, and 70 μL/L ethylene. Mean values ± se of 3 × 50 seeds are presented.
Figure 6.
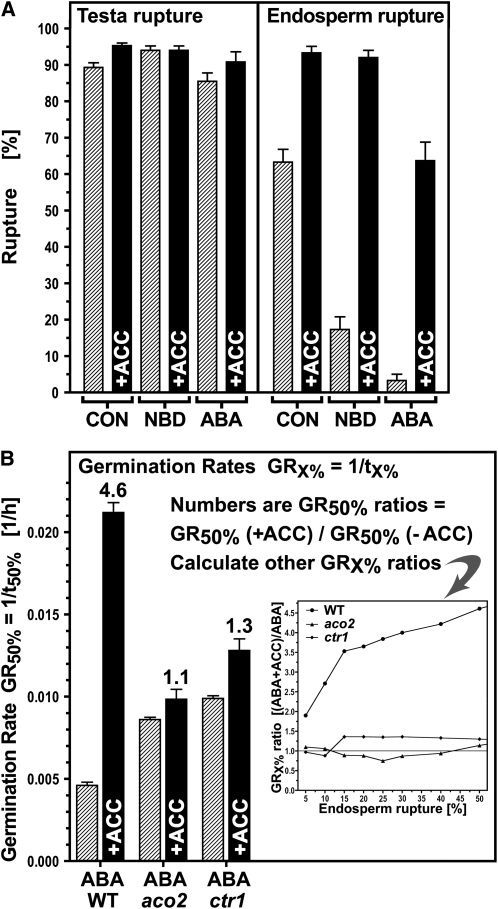
The Effect of Ethylene and ABA on the Germination of Arabidopsis: Wild type (Col) and Ethylene-Related Mutants (aco2 and ctr1).
(A) The effect of the ethylene precursor ACC on testa and endosperm rupture of wild-type seeds incubated on medium without (CON) or with NBD or ABA for 38 h.
(B) The effect of ACC on the germination rates GR50% (= 1/t50%) of wild-type and ethylene-related mutants incubated on medium with ABA. Numbers above the columns are GR50% ratios (ABA±ACC). Inset: GRX% ratios at other percentages of endosperm rupture. Incubation conditions: continuous light, 24°C, no cold stratification. Medium additions, as indicated: 1 μM ABA, 1 mM ACC, and 100 μL/L NBD. Mean values ± se of 3 × 50 seeds are presented. For detailed time courses, see Supplemental Figure 2 online.
We tested whether ethylene and the ethylene-ABA antagonism are important for endosperm rupture in different environmental conditions and whether early and late germinating seeds within the Lepidium populations have different ethylene responsiveness (Figure 5). This was achieved by quantifying the ethylene-mediated increase in germination rates GRx% at defined percentages (x% = 15 and 50%; Figure 5) of endosperm rupture. The GR15% (=1/t15%) values in continuous light show that ACC increased the CON, NBD, and ABA values between 1.3- and 1.4-fold (Figure 5B). When seeds were imbibed in darkness, the ACC- and C2H4-mediated increases in GR15% were between 1.1- and 1.2-fold for CON, NBD, and ABA (Figure 5C). Interestingly, the GR50% ratios of the NBD and ABA series were higher at 1.5-fold (Figure 5C). This demonstrates that ethylene binding to the receptors and signaling are important for optimal endosperm rupture and that seeds germinating later within the population appear to have an increase in ethylene responsiveness.
These findings were confirmed by experiments with Arabidopsis ethylene mutant seeds (Figure 6B; see Supplemental Figure 2 online). The seeds of Arabidopsis aco2 knockout (ethylene biosynthesis) and the ctr1 loss-of-function (ethylene signaling) mutants were compared with those of the wild type in time-course experiments. There was no appreciable difference on CON medium (see Supplemental Figure 2 online), but there were marked differences in the effects of ACC on ABA-containing medium (Figure 6B). In the wild type, ACC increased the GR50% 4.6-fold compared with the aco2 and ctr1 mutants, where ratios were only 1.1 and 1.3, respectively. In wild-type seeds, the GRx% ratios increased steadily from 5 to 50% (Figure 6B, inset), whereas for the aco2 mutant, these GRx% ratios remained around 1, demonstrating that there was no ACC effect. Therefore, the expression of ACO2 and ACO2-mediated ethylene biosynthesis are required for counteracting the ABA-induced inhibition of endosperm rupture. For the ctr1 mutant, in which ethylene signaling via CTR1 is always on, the GRx% ratios also remained roughly constant at ∼1.3, indicating that signaling via CTR1 is the major pathway for counteracting ABA-induced inhibition. However, the retention of a low level of ethylene responsiveness suggests that an additional minor pathway may be involved. One possible candidate is the putative CTR1-like Ser/Thr protein kinase whose transcript accumulation is enhanced by ABA in Lepidium endosperm caps (see Supplemental Figure 4E online). Such interactions have been observed elsewhere; for example, CTR1-like genes interact with the ethylene receptors and play roles in ethylene signaling and development in tomato (Solanum lycopersicum) seedlings (Lin et al., 2008).
ACO Gene Expression, in Vivo ACO Activity, and Ethylene Production Are Regulated in Germinating Lepidium Seeds in a Tissue-Specific and ABA-Related Manner
The above results prompted us to analyze Lepidium ACOs during germination. Radicles and endosperm caps were dissected from seeds at various time points during incubation both without and with ABA and were then further incubated on ACC or ABA+ACC, respectively (Figure 7A). In vivo ACO enzyme activity accumulated in the radicle in line with the progress of endosperm rupture on both control and ABA medium. On a per organ basis, the in vivo ACO enzyme activity of the endosperm cap was much lower compared with that of the radicle (Figure 7A), but it was similar on a per dry weight basis. In contrast with the radicle, in vivo ACO enzyme activity in the cap increased from 16 to 40 h after the start of imbibition and thereafter decreased in the presence of ABA.
Ethylene production by ACO requires oxygen and Figure 7B shows that the in vivo ACO enzyme activities of isolated radicles, endosperm caps, and embryos are higher at ambient oxygen concentration (21%) compared with low oxygen concentration (5%). In agreement with this, the promotion of endosperm rupture by ACC and the reversion of the ABA-induced inhibition of endosperm rupture by ACC was only evident at ambient oxygen concentration (21%) but not at low oxygen concentration (10%; Figure 7C). These results support the hypothesis that ethylene is produced by the seed ACOs and plays an important role in facilitating endosperm rupture.
Three functional ACOs, namely, ACO1, ACO2, and ACO4/EFE (ethylene-forming enzyme; Alonso and Ecker, 2001), and 13 sequences with ACO similarity are known in Arabidopsis (Figure 7D; see Supplemental Data Set 9 online). Array results for putative Lepidium ACO orthologs (see Supplemental Data Sets 1 to 4 online) are presented in Figures 7E to 7G. The CON arrays showed that transcripts of Lepidium orthologs of Arabidopsis ACO1, ACO2, and ACO4 accumulated in the endosperm cap between 8 and 18 h. In the CON arrays, no other transcripts with ACO sequence similarity showed a significant regulation in the cap, and no significant upregulation was evident in the radicle (see Supplemental Data Sets 1 to 4 online). The cDNAs of the Lepidium orthologs of these genes were cloned and, based on the highest BLAST hits and the molecular phylogenetic analysis, named Lepidium ACO1, ACO2, and ACO4, respectively (Figure 7D; see Supplemental Figure 3 online). The putative proteins contain the characteristic domain of 2OG-Fe(II)-oxygenases with conserved Fe2+ binding site motifs for the substrates ACC and O2 as well as the cosubstrate binding site motif (see Supplemental Figure 3 online; Seo et al., 2004). We therefore conclude that Lepidium ACO1, ACO2, and ACO4 are functional ACOs and that they represent true Lepidium orthologs of the corresponding Arabidopsis genes.
To analyze ACO transcript expression patterns in Lepidium seeds and to verify the microarray results, we performed quantitative RT-PCR (qRT-PCR). In agreement with the CON array results, transcripts of Lepidium ACO2, ACO1, and ACO4 genes accumulated 35-, 10-, and 2-fold, respectively, in the endosperm cap between 8 and 18 h after the start of imbibition (Figures 7E to 7G). In the CON series, Lepidium ACO2 and ACO1 transcripts accumulated 2- and 4-fold in the radicle, whereas Lepidium ACO4 was not regulated. The qRT-PCR showed that upregulation of Lepidium ACO1 was inhibited by ABA in both organs, while Lepidium ACO2 upregulation was not inhibited in the radicle, causing ∼6-fold higher Lepidium ACO2 transcript contents at 96 h of culture in medium containing ABA compared with 18 h in CON medium. In the endosperm cap, ABA inhibited Lepidium ACO2 accumulation. ABA also inhibited Lepidium ACO4 upregulation in the endosperm cap but had no effect on the low and roughly constant Lepidium ACO4 transcript levels in the radicle. Based on this ABA-insensitive expression pattern of ACO2 in the Lepidium radicle and the impaired reversion of the ABA inhibition of endosperm rupture by ACC in the Arabidopsis aco2 knockout mutant, we conclude that ethylene production by the ACO2 enzymes plays an important role in the ethylene-ABA antagonism in both species.
To further test the validity of the array data, we investigated the expression of key genes for ABA degradation and GA biosynthesis, two processes that are a hallmark of germinating seeds (Kucera et al., 2005). ABA 8'-hydroxylases, encoded by the CYP707A gene family, have four members in Arabidopsis and are the key catabolic enzymes for ABA degradation during seed germination (Kushiro et al., 2004; Okamoto et al., 2006). These publications show that CYP707A2 and CYP707A3 are very important in Arabidopsis seeds. We therefore cloned the cDNAs of their putative Lepidium orthologs and analyzed their expression by qRT-PCR (see Supplemental Figure 4A online). The expression of the putative orthologs Arabidopsis CYPA707A2/Lepidium CYP707A2 was higher in seed tissues of both species compared with Arabidopsis CYPA707A3/Lepidium CYP707A3, and these levels declined during seed imbibition of both species. ABA induced CYP707A2 accumulation in Arabidopsis seeds (Kushiro et al., 2004) and in Lepidium radicles (see Supplemental Figure 4A online). Interestingly, just prior to endosperm rupture of ABA-imbibed Lepidium seeds, CYP707A2 and CYP707A3 transcripts accumulated in the endosperm cap. This is in keeping with previous work that showed that ABA treatment of Arabidopsis mutants impaired in ABA degradation, including the cyp707a2 mutant, specifically delayed endosperm rupture (Müller et al., 2006).
GA3 oxidases catalyze the final key step of GA biosynthesis, and the transcripts of the GA3OX2 putative orthologs accumulate prior to endosperm rupture in the radicles of Arabidopsis (Yamaguchi et al., 2001; Ogawa et al., 2003; Cadman et al., 2006) and Lepidium (CON arrays, t test, P = 0.02; see Supplemental Data Sets 1 and 3 online). The finding that the transcript expression patterns of the CYP707A2, CYP707A3, and GA3OX2 putative orthologs are similar in the two species suggests that they serve evolutionarily conserved roles.
Transcript Expression Patterns of Ethylene Signaling and Responsive Genes Support the Importance of the Ethylene-ABA Antagonism during Seed Germination
Our findings above in both species suggest that ethylene sensitivity increases over time in response to ABA. This is further supported by the observation that transcript levels of the ethylene receptor ETR2 and of the negative regulator of ethylene responses Lepidium CTR1 (the Lepidium cDNA was cloned and qRT-PCR performed) decreased upon ABA treatment of Lepidium endosperm caps and radicles (see Supplemental Figures 4B and 4D online). This decrease in CTR1 transcript levels suggests enhanced ethylene signaling via downstream positive factors (Ethylene Insensitive2 [EIN2]⇒ EIN3 ⇒ethylene-responsive element binding transcription factors [ERFs]; Figure 4B). Accumulation of EIN3 Binding F-Box2 (EBF2) transcripts (CON) and the inhibition of this accumulation by ABA (see Supplemental Figure 4C online) is further evidence for enhanced ethylene signaling. In the absence of ethylene, EBF2 targets EIN3 for degradation, and this EBF2–EIN3 interaction is inhibited in the presence of ethylene. EBF2 is known to accumulate during ethylene signaling to prevent excess accumulation of EIN3 (Potuschak et al., 2003; Konishi and Yanagisawa, 2008). The ABA-induced inhibition of the EBF2 accumulation (see Supplemental Figure 4C online) therefore suggests that there is no excess of EIN3 to trigger this feedback loop. Downstream of EIN3, transcripts of six (CON arrays) and 18 (ABA arrays) ERF (see Supplemental Data Sets 3 and 4 online) as well as Related to ABI3/VP1 (RAV1; an AP2/EREBP-type transcription factor with ABI3/VP1-like domain; see Supplemental Figure 4G online) transcription factors of the AP2/EREBP family (Etheridge et al., 2006) are significantly regulated and may therefore mediate the expression of ethylene-responsive downstream genes.
The GPCR-type G protein genes GTG1 and GTG2 encode ABA receptors, and double mutants lacking these ABA receptors exhibit ABA-hyposensitive germination (Pandey et al., 2009). Transcripts of the putative Lepidium GTG2 ortholog accumulated in the endosperm cap early during ABA-inhibited germination (see Supplemental Figure 4F online), which suggests that the ABA sensitivities also increased over time in response to ABA. ABA inhibits the transcript expression of endosperm cap-specific genes like the ethylene-inducible β-1,3-glucanase βGLU in tobacco (Leubner-Metzger et al., 1998; Leubner-Metzger, 2003) or the expansin EXPA2 in Arabidopsis (Ogawa et al., 2003; Penfield et al., 2006; Carrera et al., 2008). ABA-inhibitable transcript accumulation of Lepidium βGLU and EXPA2 was evident in the endosperm cap during germination (Figure 4C), and, like in tobacco, transcripts of chitinase CHNB accumulated in an ABA-insensitive manner only in the radicle. ABA affected pectate lyase PL1 transcript accumulation in a tissue-specific manner (i.e., it inhibited accumulation in the endosperm cap and promoted it in the radicle) (see Supplemental Figure 4H online). In ripening fruits, pectin-related cell wall weakening is enhanced by ethylene, and in seeds it is proposed to be involved in endosperm weakening (Kucera et al., 2005). The ARL gene regulates cell expansion and organ growth in response to brassinosteroids (Hu et al., 2006) and is ethylene induced in Arabidopsis seedlings (Nemhauser et al., 2006). ARL transcripts accumulate in the Lepidium endosperm cap, and this accumulation is inhibited by ABA (see Supplemental Figure 4I online).
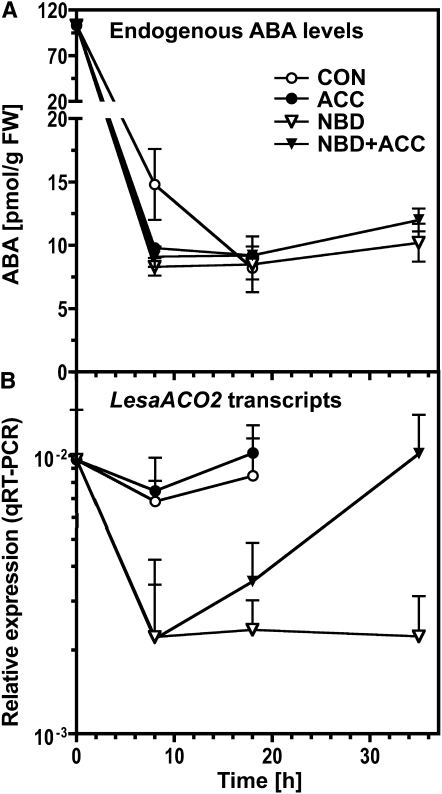
Seeds of ethylene-related mutants differ in the ABA sensitivities of their germination responses (Beaudoin et al., 2000; Ghassemian et al., 2000), and etr1 mutant seeds differ in ABA contents to wild-type seeds (Chiwocha et al., 2005). To test if ethylene affects ABA levels in Lepidium seeds, we measured the effects of treatments with ACC (i.e., generation of higher ethylene contents) and NBD (i.e., inhibition of ethylene signaling) on ABA content (Figure 8A). These measurements show that in the control and all three treatments (ACC, NBD, and NBD+ACC), the ABA concentrations declined rapidly during imbibition to a consistent level by 18 h. The rate of decline was similar in the treatments, but slightly less rapid in the control. The lack of difference between the contrasting treatments indicates that ethylene has little or no impact of physiological importance on endogenous ABA concentrations. In agreement with this, we did not find any ethylene-mediated regulation of transcript levels of key regulatory genes for ABA biosynthesis (Nine-cis-epoxycarotenoid dioxygenase9, Abscisic aldehyde oxidase3, Short-chain dehydrogenase reductase1; Nambara and Marion-Poll, 2005; Toh et al., 2008) or ABA degradation (Lepidium CYP707A2 and Lepidium CYP707A3; see previous section) (see Supplemental Figure 5 online). Taken together, these results demonstrate that ethylene did not affect the ABA metabolism of Lepidium seeds during germination. It must therefore act on the reversion by interfering with ABA signaling.
Figure 8.
The Effect of ACC, NBD, or NBD+ACC on Endogenous ABA Contents and ACO2 Transcript Expression during Germination of L. sativum FR14.
(A) Endogenous ABA concentrations of whole seeds incubated without (CON) or with 1 mM ACC or 100 μL/L NBD or ACC+NBD added to the medium in continuous light at 18°C. FW, fresh weight.
(B) Lepidium ACO2 transcript expression pattern determined by qRT-PCR in whole seeds. Relative ΔΔCt expression values based on the comparison with validated constitutive transcripts are presented.
Mean values ± se (A) or +se (B) are presented for three biologically independent samples each with 50 seeds.
Treatment of Lepidium seeds with ACC did not affect the ACO2 transcript levels of whole seeds (Figure 8B), but NBD caused a decrease in the ACO2 transcript levels and the reversion by NBD+ACC demonstrates that this is an ethylene-specific effect. The maintenance of ACO2 transcript expression therefore requires a basal level of ethylene signaling.
Endosperm Cap Weakening Precedes Lepidium Endosperm Rupture and Is Regulated by an ABA-Ethylene Antagonism
Figure 9 shows, by direct biomechanical measurements, that ethylene signaling is important for Lepidium endosperm cap weakening. Binding of the ethylene action inhibitor NBD to the receptors inhibited this weakening, an effect that was to a large extent prevented by simultaneous treatment of NBD with ACC at 24°C (Figure 9A) and 14°C (Figure 9B).
Figure 9.
The Effect of ACC, NBD, ABA, or Their Combinations on Endosperm Cap Weakening and Endosperm Rupture of L. sativum FR14.
Endosperm cap weakening (columns) and rupture (percentages above columns) of seeds incubated without (CON) or with 1 mM ACC or 10 μM ABA added to the medium in continuous light at 24°C ([A] and [C]) or 14°C (B). Endosperm cap weakening was determined by measuring tissue resistance by the puncture force method at the times indicated. All the seeds selected for the puncture force measurements had completed testa rupture but still had intact endosperm caps; ACC, NBD, and ABA did not appreciably affect testa rupture. Mean values ± se of 3 × 50 seeds for the endosperm rupture and at least 50 endosperm caps for the puncture force measurements are presented. Note that the puncture force values measured for the endosperm cap weakening of L. sativum FR14 were approximately twofold compared with L. sativum FR1 (Figure 1). These higher values are accession-specific absolute differences, but the relative decreases of the two accessions are similar.
Treatment with ACC also reversed the ABA-induced inhibition of Lepidium endosperm weakening (Figure 9C). Although ACO is present in ABA-treated seeds at 18 h (prior to the onset of endosperm weakening; Figure 7A), ACC did not reverse the ABA-induced inhibition of endosperm cap weakening during the early germination phase (18 h), but it reversed the ABA-induced inhibition of endosperm cap weakening during the late germination phase (Figure 9C). This temporal difference may be explained by the finding that ABA not only inhibited the slope of endosperm cap weakening, but also its onset (Figures 1A and 9C). Prior to the onset of endosperm cap weakening (i.e., in the early phase of ABA-inhibited germination), the ABA-mediated mechanism that inhibits endosperm cap weakening is pronounced and the counteraction by ethylene may be ineffective. Ethylene becomes effective after the onset of ABA-inhibited endosperm cap weakening, and this could explain the increased ethylene responsiveness for counteracting ABA in seeds of the population that germinate later.
DISCUSSION
Cross-Species Transcriptome Analysis Is a Useful Tool to Identify Regulated Genes and to Generate Hypotheses in Comparative Brassicaceae Biology
Comparative transcriptomics by cross-species microarray hybridization has emerged as a useful tool to compare closely related species for which no specific arrays are available (reviewed in Van de Mortel and Aarts, 2006; Bar-Or et al., 2007; Broadley et al., 2008). Arabidopsis- and tomato-spotted cDNA microarrays have been successfully used in cross-species analyses with Brassica napus seeds and several Solanaceae fruits, respectively. The authors concluded that longer probes facilitate cross-species hybridization but also that cDNA-based probes have disadvantages (Van de Mortel and Aarts, 2006; Bar-Or et al., 2007). Spotted PCR-amplified gene-specific tag (GST)–based microarrays like the CATMA 25K microarrays (containing 150- to 500-bp Arabidopsis GSTs) provide a good alternative to cDNA- and oligomer-based microarrays and provide a near-full coverage of the Arabidopsis genome (Allemeersch et al., 2005; Van de Mortel and Aarts, 2006; Bar-Or et al., 2007). The use of these CATMA microarrays for cross-species microarray hybridization was shown to be effective in recent work by Slotte et al. (2007), in which Capsella bursa-pastoris accessions differing in flowering time were compared at the transcriptome level. Cross-species microarray hybridization has also been successful for Brassica and Thlaspi using oligomer-based Affymetrix ATH1 GeneChips (Broadley et al., 2008). It is thought that the transcriptome divergence correlates positively with evolutionary distance between taxa, and both of these genera are more distantly related to Arabidopsis than Lepidium, which like Capsella is from the lineage-I clade of Brassicaceae (Franzke et al., 2009). Both Capsella and Lepidium gave high present rates with genomic DNA (see Methods) using the CATMA array: 60.5% for Capsella (Slotte et al., 2007) and 70.9% for Lepidium (this work; see Methods for details).
When GO-based seed-specific TAGGIT annotation (Carrera et al., 2007; Holdsworth et al., 2008) was applied to our Lepidium arrays, we found that ethylene-related transcripts were overrepresented in the lists of regulated genes (see Supplemental Figure 1 online). Several of these ethylene-related transcripts were also regulated by ethylene and ABA in Arabidopsis seedlings (Nemhauser et al., 2006), but others (e.g., ACO2) were not and therefore may constitute genes that are specifically important during seed germination (Figure 4). To verify the transcript expression pattern of the arrays, we compared them to the corresponding qRT-PCR results obtained with independent biological RNA samples from a separate experiment. We investigated 10 Lepidium putative orthologs by qRT-PCR (Lepidium ACO1, ACO2, ACO4, CYP707A2, CYP707A3, CTR1, CTR1-like, PL1, ARL, and EF1a). For these, there were 33 significant expression patterns between two time points (up, down, and unchanged) in the microarrays, and 82% of these patterns were confirmed by qRT-PCR (Figure 7; see Supplemental Figure 4 online). We conclude that cross-species microarray hybridization with the CATMA platform is a useful tool for heterologous transcriptomics with Lepidium, as it was with Capsella (Slotte et al., 2007).
A comparison of the Lepidium cDNAs of ACO1, ACO2, ACO4, CTR1, CYP707A2, and CYP707A3 (see Supplemental Figure 3 online and GenBank entries) with the corresponding Arabidopsis putative orthologs showed 80 to 90% pairwise identity at the mRNA and amino acid level. Several of the Lepidium putative orthologs analyzed in this work are known in Arabidopsis to be key genes for germination and exhibited similar spatial and temporal expression in both species, for example, genes involved in ABA degradation (ABA 8'-hydroxylases CYP707A2 and CYP707A3) and GA biosynthesis (GA3 oxidase GA3OX2) and the GA-inducible expansin EXPA2 in the micropylar endosperm cap (this work; Yamaguchi et al., 2001; Ogawa et al., 2003; Kushiro et al., 2004; Kucera et al., 2005; Cadman et al., 2006; Okamoto et al., 2006; Penfield et al., 2006; Carrera et al., 2008). Taken together, the high sequence similarities and the similar transcript expression pattern of GA- and ABA-related genes suggest that the mechanisms of the GA-ABA antagonism of endosperm rupture may be evolutionarily conserved and that true orthologs of key genes mediate these processes in related Brassicaceae species. The work presented indicates that the involvement of ACO2 orthologs in the ethylene-ABA antagonism of endosperm rupture is also evolutionarily conserved.
Ethylene Biosynthesis during Seed Germination Is Regulated by ACO and Involved in Counteracting the Inhibiting Effects of ABA on Endosperm Rupture
Increased ethylene evolution and/or ACO accumulation is associated with the germination of many seeds (e.g., Leubner-Metzger et al., 1998; Petruzzelli et al., 2000; Chiwocha et al., 2005; Matilla et al., 2005; Hermann et al., 2007; Iglesias-Fernandez and Matilla, 2009). Ethylene evolution during seed germination is regulated by ACO, which catalyzes the final rate-limiting step in ethylene biosynthesis (Kucera et al., 2005; Matilla and Matilla-Vazquez, 2008). When the transcript levels of Lepidium and Arabidopsis ACOs are compared in germinating seeds, their abundance was ACO1 > ACO2 > ACO4 in both species (Figures 7E to 7G; and Arabidopsis eFP Browser Seed at http://bbc.botany.utoronto.ca). Not only was ACO4 less abundant, but it also did not exhibit germination-associated regulation; therefore, the ACO1 and ACO2 orthologs appear to be the major ACOs in seeds. Arabidopsis ACO1 transcripts accumulate in the embryo and endosperm of germinated Arabidopsis wild-type seeds, this accumulation is inhibited by ABA, and seeds of ABA-insensitive mutants exhibit high levels of ACO1 transcript accumulation (Arabidopsis eFP Browser results based on Ogawa et al., 2003; Nakabayashi et al., 2005; Penfield et al., 2006; Carrera et al., 2008). Lepidium ACO1 transcripts (Figure 7E) are regulated in a similar manner, with accumulation in the endosperm cap and the radicle prior to endosperm rupture and inhibition by ABA in both tissues. In contrast with ACO1, ACO2 transcript expression in Arabidopsis seeds differed considerably between microarrays from different labs (Arabidopsis eFP Browser results): Yamauchi et al. (2004) describes upregulation in whole seeds, and Penfield et al. (2006) found that ACO2 accumulated mainly in the embryo of germinated seeds, and this upregulation was inhibited by ABA. Lepidium ACO2 transcripts (Figure 7F) accumulate in the endosperm cap and the radicle prior to endosperm rupture, but their accumulation is inhibited by ABA only in the endosperm cap.
Compared with in vivo Lepidium ACO enzyme activities, the corresponding Lepidium ACO transcript expression pattern is complex. In the radicle, Lepidium ACO1 and ACO2 transcripts accumulate in parallel with the in vivo ACO enzyme activity accumulation (CON). ACO1 transcript accumulation is inhibited by ABA, as is the ACO activity accumulation in the radicle, but ACO2 transcript accumulation is not inhibited by ABA. The late accumulation of in vivo ACO enzyme activity in the radicle of ABA-imbibed seeds could therefore be due to translation of ACO2 mRNA. In the endosperm cap, ACO1 and ACO2 transcripts accumulate in the CON series, and this accumulation is inhibited by ABA. Translation of ACO1 and ACO2 transcripts combined with mechanisms that regulate ACO protein stability could therefore explain the ACO activities in the radicle and endosperm cap of Lepidium seeds. In contrast with pea seeds, where a positive feedback loop involving ACO promotes ethylene biosynthesis (Petruzzelli et al., 2000), no such autocatalytic enhancement was evident for the ACO in germinating sugar beet (Beta vulgaris; Hermann et al., 2007) or the ACO2 orthologs of the Brassicaceae species Sisymbrium officinale (Iglesias-Fernandez and Matilla, 2009) and Lepidium (Figures 7 and 8). We found, however, that a basal level of ethylene signaling is required to maintain ACO2 transcript levels during germination.
These findings for Arabidopsis and Lepidium suggest that the importance of the ACO2 orthologs in seed germination could be a widespread phenomenon within the Brassicaceae. The sequences of ACO2 orthologs are known for four Brassicaceae species (see Supplemental Figure 3D online), and the transcript accumulation pattern of both Brassica rapa (Br ACO1) and S. officinale (So ACO2) are associated with seed germination responses (Matilla et al., 2005). S. officinale seeds, like those of Arabidopsis and Lepidium, also have a single layer of endosperm, and So ACO2 transcripts accumulate prior to the onset of endosperm rupture (Iglesias-Fernandez and Matilla, 2009). In Arabidopsis, we found that ACC cannot reverse the ABA-induced inhibition of endosperm rupture in the Arabidopsis aco2 mutant. We also found that in Lepidium, sustained ABA-independent ACO2 transcript accumulation in the radicle leads to increased in vivo ACO activity late during germination. In addition, ACC can only counteract the ABA-induced inhibition of endosperm weakening late during germination. S. officinale, Arabidopsis, and Lepidium (Liu et al., 2005; Müller et al., 2006; Bethke et al., 2007; Iglesias-Fernandez and Matilla, 2009) are all endospermic Brassicaceae seeds. The collective evidence from these three species suggests an evolutionarily conserved role for ethylene and the ACO2 orthologs in endosperm cap weakening and rupture in these endospermic Brassicaceae species.
Ethylene Signaling via CTR1 Regulates Downstream Gene Expression That Is Important for Optimal Seed Germination and for Mediating the ABA-Ethylene Antagonism
Ethylene perception by the endosperm cap was identified as a requirement for optimal weakening and rupture by direct biomechanical measurement of Lepidium endosperm weakening in experiments where ethylene binding to its receptors was blocked by the ethylene action inhibitor NBD. Further support for this conclusion came from decreased ETR2 transcript levels upon ABA treatment in Lepidium radicles and endosperm caps (see Supplemental Figure 4B online) and Arabidopsis embryos and endosperms (Arabidopsis eFP Browser results based on Penfield et al., 2006). Collectively, these results suggest that the ethylene sensitivities of the two tissues increased over time in response to ABA, which is in agreement with the effects of NBD on the endosperm rupture of tobacco (Solanaceae; Leubner-Metzger et al., 1998) and lettuce (Lactuca sativa, Asteraceae; Abeles, 1986). Furthermore, Arabidopsis seeds of ethylene-insensitive receptor mutants (e.g., etr1) have drastically increased ABA contents and exhibit ABA-hypersensitive germination (Beaudoin et al., 2000; Chiwocha et al., 2005). It is also known that ethylene can act by regulating ABA metabolism and signaling in seedlings of various plants (e.g., Yang and Choi, 2006; Cheng et al., 2009). In Lepidium seeds, we found no appreciable effect of ethylene on the ABA contents itself or the transcript expression pattern of key regulatory genes of ABA biosynthesis and degradation; therefore, ethylene must act during germination by interference with ABA signaling.
In Arabidopsis seedlings, the ethylene receptors interact with the single downstream Ser/Thr protein kinase CTR1. We confirmed the result of Beaudoin et al. (2000) that seed germination of the Arabidopsis loss-of-function ctr1 mutant exhibits reduced ABA sensitivity. We extended this knowledge by finding that not only the promotion of endosperm rupture, but also the reversion of the ABA-induced inhibition are mediated, at least in part, by the CTR1-associated ethylene signaling pathway. Lepidium CTR1 transcript levels in endosperm caps (this work) and Arabidopsis CTR1 in endosperms (Arabidopsis eFP Browser results based on Penfield et al., 2006) decrease upon ABA treatment, which suggests enhanced ethylene signaling and downstream responsive gene expression. Ethylene signaling and responses are mediated by the positive transcription factor EIN3, which is regulated in its turnover by the F-box proteins EBF1 and EBF2 (Guo and Ecker, 2003; Potuschak et al., 2003) and by downstream AP2/EREBP-type transcription factors (Etheridge et al., 2006). Their transcript expression patterns in Lepidium seeds are described in Results together with downstream ethylene-responsive genes, including the cell wall–modifying proteins β-1,4-mannanase, β-1,3-glucanase, and expansin and pectin-hydrolyzing enzymes that have putative endosperm cap weakening functions (Bewley, 1997; Finch-Savage and Leubner-Metzger, 2006; Matilla and Matilla-Vazquez, 2008). These transcript expression patterns in the endosperm cap of germinating seeds of Lepidium (Figure 4; see Supplemental Figures 4C and 4H online) are in agreement with a role for ethylene and/or ABA in regulating their expression and thereby affecting cap weakening.
Endosperm Cap Weakening Is Phylogenetically Widespread and Regulated by Conserved Mechanisms That Are Inhibited by ABA and Promoted by GA and Ethylene
In all species that have been investigated, endosperm weakening preceded radicle protrusion and was promoted by GA (Bewley, 1997; Finch-Savage and Leubner-Metzger, 2006). By contrast, the ABA response may differ between angiosperm clades. Biomechanical measurements of the ABA response in tomato (Toorop et al., 2000) and coffee (Coffea arabica; da Silva et al., 2004) suggest that endosperm weakening is biphasic in these seeds of the asterid clade: the first phase is ABA insensitive, and the second phase is inhibited by ABA. By contrast, a one-phase ABA-inhibited endosperm weakening is observed in Lepidium seeds (rosid clade; this work; Müller et al., 2006, 2009). Based on our comparative seed biology approach, we speculate that the endospermic Brassicaceae seeds have evolved to retain the ABA-inhibitable mechanism(s) found in both clades but not the ABA-insensitive phase of endosperm weakening. Our work on Lepidium endosperm cap weakening analyzed the Brassicaceae transcriptome of seed tissues during germination (prior to the completion of endosperm rupture) and of the radicle and cap tissues. A major conclusion from these results is that ABA-inhibited endosperm cap weakening and hole formation are promoted by ethylene.
Several hypotheses have been proposed to explain the mechanism(s) of ethylene action in germinating seeds (reviewed in Kucera et al., 2005; Matilla and Matilla-Vazquez, 2008). The major effect of ethylene could be the promotion of radial cell expansion in the embryonic hypocotyl, increased seed respiration, decreased seed base water potential, or enhanced expression of cell wall hydrolases in the endosperm cap. Ethylene promotes the rupture of the thin lettuce endosperm and, although there is no experimental evidence provided for this, Abeles (1986) speculated that ethylene acts by promoting radial cell expansion in the embryonic hypocotyl. Pavlista and Haber (1970) showed that chemical inhibition of lettuce endosperm weakening causes embryo buckling (i.e., the embryos expand without protrusion and remain encased in the unruptured endosperm envelope). Dutta and Bradford (1994) showed that ACC treatment decreased the seed base water potential of lettuce with both an intact and a slit endosperm. Interestingly, slitting of the endosperm improves lettuce germination and abolished the promotion of radicle protrusion by ACC. These findings (Pavlista and Haber, 1970; Dutta and Bradford, 1994) point to a role of ethylene in promoting endosperm rupture with the thin lettuce endosperm as a major target. In agreement with this, ethylene enhances the expression of cell wall–modifying proteins, including β-1,4-mannanase in lettuce and endospermic S. officinale seeds (Bewley, 1997; Matilla and Matilla-Vazquez, 2008; Iglesias-Fernandez and Matilla, 2009) and β-1,3-glucanase in the endosperm cap of tobacco (Leubner-Metzger et al., 1998; Leubner-Metzger, 2003; Finch-Savage and Leubner-Metzger, 2006).
The work of Bethke et al. (2007) shows that the endosperm is sufficient and necessary to confer Arabidopsis coat dormancy. Ultrastructural cellular changes in the micropylar endosperm, including vacuolation and changes that suggest thinning and weakening of cell walls or even cell separation, occur only when coat dormancy has been released. These processes are confined to the micropylar endosperm region (cap), and the vacuolation was inhibited by ABA (Bethke et al., 2007). In agreement with this, endosperm cap weakening in Lepidium seeds is a highly localized event at the location of radicle protrusion. In the absence of the radicle (detached endosperm cap), a hole still forms in the same position very close to the extreme tip of the cap (this work; Müller et al., 2009). Lepidium endosperm cap weakening, endosperm rupture, and endosperm cap hole formation are constituents of a developmental process associated with the regulation of germination. We have shown that these three subprocesses are inhibited by ABA, and this inhibition is counteracted by ethylene. The results are presented as a schematic model (Figure 10) that explains (1) the ethylene action in endospermic Brassicaceae seeds, (2) the ethylene-ABA antagonism regulating endosperm rupture, and (3) part of the interaction between the radicle and the endosperm cap. Endosperm cap weakening is a major target for ethylene action.
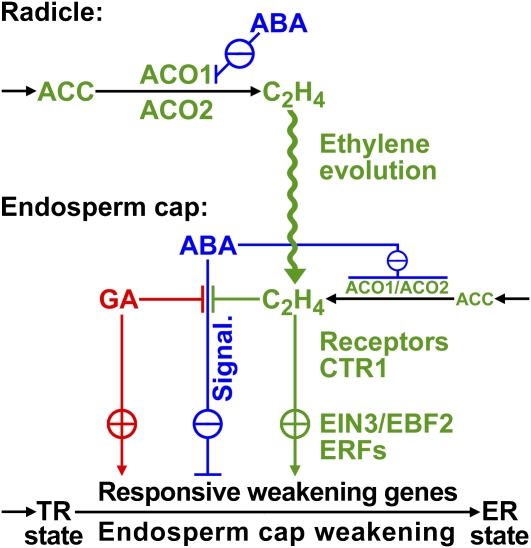
Figure 10.
Proposed Model for the Hormonal Regulation of Endosperm Cap Weakening and Rupture.
According to our working model, endosperm cap weakening is required during the transition from testa rupture (TR) to endosperm rupture (ER), and endosperm cap weakening in turn requires ethylene biosynthesis and signaling. Endosperm cap weakening is a developmental process that is regulated by interactions between the cap and the radicle. GA, as an embryo signal, releases coat dormancy (if present) and induces the cap weakening process. Thereafter, weakening is a cap-autonomous process, and the rate of this process is regulated by the GA-ABA and ethylene-ABA antagonisms. The involvement of GA as antagonist of ABA in seed germination is well established in different Brassicaceae (e.g., Yamaguchi et al., 2001; Ogawa et al., 2003; Chiwocha et al., 2005; Müller et al., 2006; Iglesias-Fernandez and Matilla, 2009). GA biosynthesis may be more important in the early phase of germination, and ethylene biosynthesis may be more important for the late phase of the process. Ethylene biosynthesis in germinating seeds is regulated differently in the radicle and the endosperm cap, and this effect is mediated by ACO2 gene expression in the following way: although both ACO2 and ACO1 are active in the radicle and endosperm cap, the total activity is greater in the former. The radicle produces ethylene in excess, and this is targeted at the endosperm cap. ABA delays the ACO activity in the radicle and inhibits ACO1 transcript accumulation, but ABA does not inhibit ACO2 transcript accumulation. The later increase in ACO activity in the radicle of ABA-treated seeds is therefore due to ACO2, and the ethylene produced promotes endosperm cap weakening by antagonizing the ABA inhibition. In the endosperm cap, ABA inhibits ACO2 and ACO1 transcript accumulation. A basal level of ethylene signaling is required to maintain ACO2 transcript levels in seeds. Ethylene does not affect the seed ABA levels and therefore must counteract the ABA-induced inhibition of endosperm rupture by interfering with ABA signaling. High degrees of GA and ethylene sensitivity of the endosperm cap are associated with the after-ripened (post coat dormancy) seed state and are prerequisites for ethylene-enhanced expression of ABA-inhibitable downstream weakening genes in the cap. The products of the weakening genes cause endosperm cap weakening by cell wall loosening and cell separation that finally leads to endosperm rupture and radicle emergence.
METHODS
Plant Material, Germination, and Puncture Force Measurements
After-ripened garden cress Lepidium sativum FR1 (Gartenkresse, einfache) and FR14 (Keimsprossen) seeds (Juliwa) were incubated in Petri dishes on two layers of filter paper with 6 mL 1/10 Murashige and Skoog salts in continuous white light (∼100 μmol·s−1·m−2) as described by Müller et al. (2006) at the temperatures indicated. The two Lepidium cultivars differ in that FR1 is self-incompatible, while self-fertilization is possible in FR14; however, both cultivars exhibit equal physiological responses to the hormonal treatments. As we will use FR14 in future experiments that include genetic transformation, we have cloned the cDNAs from this cultivar. After-ripened Arabidopsis thaliana seeds were incubated without cold stratification in continuous light on the same medium solidified with 1% (w/v) agar-agar at 24°C. Arabidopsis ecotype Columbia (Col) and mutant seeds in the Col background were used. Homozygous ctr1-1 (N8057) seeds were obtained from the Nottingham Arabidopsis Stock Centre (NASC) (www.Arabidopsis.org). Homozygous seeds of the aco2 line (SALK_027311, NASC stock center; Alonso and Ecker, 2001) were also used. Where indicated, cis-S(+)-ABA (Duchefa), gibberellin A4+7 (GA4+7; Duchefa), ACC (Sigma-Aldrich), NBD (Sigma-Aldrich), or ethylene (Air Liquide) was added at the concentrations indicated; NBD or ethylene was added via the gas phase in air-tight vessels (Hermann et al., 2007). Testa rupture and endosperm rupture were scored using a binocular microscope. Puncture force measurements were performed as described by Müller et al. (2006).
In Vivo ACO Enzyme Activity Assay and Ethylene Measurements
Lepidium radicles, endosperm caps, or embryos (Figure 2A) were dissected from imbibed seeds at the times indicated. For the in vivo ACO enzyme activity assays, the organs (100 radicles or 200 endosperm caps) were imbibed at 24°C in 10-mL air-tight vessels with rubber seals containing 100 μL of water supplemented with 1 mM ACC ± 10 μM ABA, as indicated. Ethylene production or the conversion of ACC to ethylene was measured as described by Salmen Espindola et al. (1994). A 1-mL gas sample was taken from each flask and injected into a gas chromatograph (Hewlett Packard 5890, series II) equipped with a flame ionization detector and an activated alumina column (6-mm internal diameter, 50-cm long, 50 to 80 mesh) for ethylene determination. The carrier gas was nitrogen, the column temperature was 90°C, and the detector temperature was 110°C. The ACC was omitted in the medium when in vivo ethylene evolution of seeds was measured.
Analysis of Endogenous ABA Contents
L. sativum FR14 seeds were ground using 3-mm tungsten carbide beads (Retsch) with a MM 301 vibration mill (Retsch) at a frequency of 27.5 Hz for 3 min. An internal standard [50 pmoL of (+)-3′,5′,5′,7′,7′,7′-2H6-ABA] and 1 mL of cold methanol/water/acetic acid (80/19/1, v/v) extraction solvent were added to the samples. After 24 h of shaking in the dark at 4°C, the homogenates were centrifuged (20,000 rpm, 5 min, 4°C), and the pellets were then reextracted in 1 mL of the same extraction solvent for 60 min. The supernatants were transferred to a fresh glass tube and dried under vacuum. Extracts were dissolved in 100 μL 99% methanol:1% acetic acid (v/v) topped up to 1 mL with 99% water:1% acetic acid (v/v), purified by solid-phase extraction on an Oasis HLB cartridges (60 mg, 3 mL; Waters), and evaporated to dryness in a Speed-Vac (UniEquip). Subsequently, the evaporated samples were methylated, purified by ABA-specific immunoaffinity extraction (Hradecká et al., 2007), and analyzed by UPLC-ESI(+)-MS/MS (Turečková et al., 2009).
RNA Isolation from Lepidium Seed Tissues
For each sample, ∼1000 Lepidium endosperm caps (Cap), ∼1000 nonmicropylar endosperms (NonE), or ∼100 radicles (Rad) were collected at the times indicated, frozen in liquid nitrogen, and stored at −80°C. Total RNA extraction was performed and followed by quantity and quality control analyses as described (Chang et al., 1993b; Cadman et al., 2006). Four biological replicate RNA samples were used for downstream applications.
Microarray Experimental Design
We performed two separate microarray experiments. The first compared Cap, NonE, and Rad at 8 and 18 h of imbibition on germination medium, and the arrays used in this experiment were termed CON arrays. The second compared Cap and Rad at 8, 18, 30, and 96 h of imbibition on 10 μM ABA and were termed ABA arrays. Each experiment used four biological replicates. Hybridizations were performed according to the description below. For the CON experiment, the two time points for each tissue were directly compared on four microarrays, balanced for color (Cy3 and Cy5; see Supplemental Methods online). For each tissue in the ABA experiment, all time points were directly compared with each other on one microarray each, and for each time point the two tissues were compared on one microarray. Each treatment was balanced for color. This design can be thought of as four interlinked loops.
Cross-Species CATMA Microarrays and Lepidium RNA Hybridization
RNA was prepared in the following way for microarray hybridization. The Ambion MessageAmp II aRNA amplification kit (AM1751; Applied Biosystems) was used according to the manual with 2 μg of L. sativum FR1 total RNA as template to generate antisense amplified RNA (aRNA) (Van Gelder et al., 1990). The quality and quantity of the aRNA was checked by running an aliquot on a 2100 Bioanalyzer (Agilent). The microarrays used carried GST fragments generated using gene-specific primers identified by the CATMA Consortium (www.catma.org; Hilson et al., 2004; Allemeersch et al., 2005). CATMA version 2 arrays with 24,576 GSTs were used for the CON experiment, while CATMA version 3 arrays with 30,343 GSTs were used for the ABA experiment. The aRNA was labeled and the CATMA microarrays were hybridized according to the method described by Lim et al. (2007) and detailed in the Supplemental Methods online. The microarrays were scanned using an Affymetrix 428 array scanner at 532 nm (Cy3) and 635 nm (Cy5). Scanned data were quantified using Imagene version 4.2 software (BioDiscovery; www.biodiscovery.com/). Microarray data were deposited in ArrayExpress (www.ebi.ac.uk/microarray/) under accession numbers E-TABM-745 (CON) and E-TABM-743 (ABA).
Lepidium and Arabidopsis Genomic DNA Hybridization to CATMA Microarrays
The hybridization of genomic DNA and subsequent determination of Lepidium genes present on the microarray were performed as described in the Supplemental Methods online. A total of 21,527 probes out of the 30,343 spotted on the microarray (70.9%) were identified as having significant hybridization for L. sativum FR1 and therefore as being present. The corresponding number for Arabidopsis Cvi was 28,146 (93.0%). Microarray data were deposited in ArrayExpress (www.ebi.ac.uk/microarray/) under accession number E-TABM-744.
RNA Microarray Data Handling and Analysis
Data from the two experiments (CON and ABA) were analyzed separately using a similar approach, but differing according to the different experimental designs used and the availability of genomic DNA hybridization data for CATMA v3 arrays used in the ABA experiment. In both cases, spot intensity data from Imagene were analyzed using the limma package in Bioconductor (Smyth, 2005). The data were initially screened so that only probes that had a CATMA identifier and could be assigned to an Arabidopsis gene, defined by having an AGI identifier and a TAIR7 GO description (www.Arabidopsis.org), were included. Background correction was performed using the normexp method, which is analogous to RMA. Within-array normalization (Smyth and Speed, 2003) was performed using print tip loess. In the CON experiment, between-array normalization was performed using quantile normalization on the A values. For the ABA experiment, probes that had shown no significant response in the genomic DNA microarrays were weighted out of the normalization and analysis. The two filtering steps resulted in lists (see Supplemental Data Sets 1 and 2 online, respectively) containing 22,025 and 19,794 genes for the CON and ABA experiments, respectively. The presented data are log (base 2) transformed. These gene lists were then used in all downstream analyses. The data are available at ArrayExpress (www.ebi.ac.uk/microarray/) under accession numbers E-TABM-745 (CON) and E-TABM-743 (ABA).
For both experiments, the data were analyzed as a linear model (Smyth, 2004), and for the CON experiment, the analysis was adjusted for the intraspot correlation. The variance estimates were adjusted using empirical Bayes estimates of the per-spot variability for use in differential expression analyses. For the CON experiment, four sets of contrasts between treatments were examined. Three of these were considered the differences between tissues, imbibed separately for 8 and 18 h and for the average of the values of these two time points. For each of these three sets of contrasts, the differences between each of the three pairs of tissues were calculated. The fourth set of contrasts consisted of comparisons between the 8 and 18 h imbibed treatments for each of the three tissues separately. For the ABA experiment, four sets of contrasts between treatments were examined. Three of these were considered the time course data, separately for each tissue and for the average of the two tissues. For each of these three sets of contrasts, the differences between the tissue(s) at 18 and 8 h, 30 and 18 h, and 96 and 30 h were calculated. The fourth set of contrasts consisted of comparisons between the tissues at each of the four time points. For each set of contrasts, the significance of the individual comparisons was assessed with t tests, and the comparisons were combined into an overall F-test, which assessed whether there were significant differences across the comparisons in each set of contrasts. Both the individual t tests and the F-tests were adjusted for multiple comparisons using the method of Benjamini and Hochberg (1995) (see Supplemental Data Sets 3 and 4 online). PCA was performed in R (http://www.r-project.org) for each experiment to compare the tissues and time points across the probes.
List Creation: Significantly Regulated Genes and Seed-Specific TAGGIT Workflow
We compared the transcript numbers of individual genes in our seed tissue array data (CON array: 22,025 genes; see Supplemental Data Sets 1 and 3 online; ABA array: 19,794 genes; see Supplemental Data Sets 2 and 4 online) across treatments using F-tests to identify those whose transcript numbers had significantly changed. P values were adjusted for false discovery rate (Benjamini and Hochberg, 1995), and the resulting transcript lists (P values ≤ 0.1) contained 1350 genes for the CON array (see Supplemental Data Set 5 online) and 3530 genes for the ABA array (see Supplemental Data Set 6 online). These genes were considered to be up- or downregulated either between time points in the same tissue and/or differentially regulated between tissues. Application of the GO-based established seed-specific TAGGIT workflow (Carrera et al., 2007; Holdsworth et al., 2008) provided proportional representations of genes in functional categories from the different Lepidium transcriptome data sets (see Supplemental Data Sets 3 to 6 and Supplemental Figure 1 online).
Microarray Verification by qRT-PCR
The transcript expression of selected genes was quantified by qRT-PCR, which was conducted according to the requirements described by Udvardi et al. (2008). Four biological replicates of L. sativum FR1 endosperm cap or radicle RNA were used for each time point and treatment. Five micrograms of RNA was reverse transcribed in a 50-μL reaction using both random hexamers (2.5 μM) and oligo(dT)16 (2.5 μM) according to the Superscript III Kit instructions (Invitrogen). Aliquots of 1 μL were then used for each quantitative PCR reaction. For quantification with the ABI PRISM 7300 sequence detection system (Applied Biosystems), the ABsolute QPCR SYBR Green ROX Mix (ABgene) was used according to the manufacturer's instructions. Single product amplification was validated by a melting curve according to the manufacturer's instructions. The qPCR efficiency (E) for every single well was determined by analysis of the raw data with the LinRegPCR program, and relative expression for each well was calculated as E−Ct. Expression data were normalized to the geometric mean (geomean) of two validated housekeeping genes, EF1a und ACT7. The relative expression result for every sample was then calculated as ΔΔCt = E−Ct sample/geomean E−Ct standards (Pfaffl, 2001). Mean values and se were calculated from the four biological replicates and are shown in the figures. For comparison, normalized microarray expression differences between time points are presented based on t tests in which a P value ≤ 0.1 was considered as significant up- or downregulation and a P value ≥ 0.9 was considered as no change. Primer sequences for the qRT-PCR are presented in Supplemental Table 1 online.
cDNA Cloning of Lepidium Sequences
First-strand cDNA was synthesized in 50-μL reactions with 2.5 μM oligo(dT)16, 2.5 μM random hexamers, and 5 μg total RNA from L. sativum FR14 seed tissues as a template, according to the instructions of the Superscript III reverse transcriptase kit (Invitrogen). Primers for subsequent PCR cloning of cress cDNAs were designed based on the corresponding CATMA-GST sequence (www.catma.org; Hilson et al., 2004; Allemeersch et al., 2005). Primer sequences (5′→3′) were LesaACO1 (ACO-F1, 5′-GATCAAAGAGAGAGAGATGGAG-3′; ACO-R1, 5′-TGGATACAAGAGCTTTGGAGC-3′), LesaACO2 (ACO2-F-whole, 5′-ATGGAGAAGAACATGAAGTTTCC-3′; ACO2-R-whole, 5′-TTAGAAAGTCTCTACGGCTGC-3′), LesaACO4 (aa-f1, RARWTAGAGAAGYTGKCRGAG; aa-r2, AGTCGSTKCYHGGRTTRTAG). To verify the Lepidium cDNA sequences, at least three independent cDNA clones were sequenced for each Lepidium ortholog (see Supplemental Figure 3 online).
Sequence Alignments and Molecular Phylogenetic Analysis
The bioinformatics software Geneious Pro 4.5.4 (www.geneious.com) was used as sequence analysis platform. The program Geneious Align was used for sequence alignments, with the Blosum62 matrix for the amino acid sequence alignment. The Geneious Tree Builder was used for construction of the phylogenetic tree, using the Jukes-Cantor model and the neighbor-joining method for tree construction and bootstrap resampling with 1000 trials for statistical support of the nodes, shown as consensus support (%) on the branches of the tree. The rooting was based on midpoint. Molecular phylogenetic analysis of the 13 Arabidopsis ACO and the three Lepidium ACO sequences was achieved by comparison of the amino acid sequence of the iron binding conserved domain, which is specific for the 2OG-Fe(II) oxygenase family (see Supplemental Figure 3 and Supplemental Data Set 9 online; Seo et al., 2004).
Accession Numbers
The L. sativum FR14 cDNAs isolated and described here have been deposited in GenBank/EMBL data libraries under the following accession numbers: LesaACO1 (GQ221031), LesaACO2 (GQ221032), LesaACO4 (GQ221033), LesaCTR1 (GQ221030), LesaCYP707A2 (GQ221028), and LesaCYP707A3 (GQ221029). Arabidopsis genes and Arabidopsis Genome Initiative numbers are as follows: ACTIN7 (At5g09810), ARL putative cell expansion gene (At2g44080), CTR1 (At5g03730), putative CTR1-like Ser/Thr protein kinase (At4g24480), CYP707A1 (At4g19230), CYP707A2 (At2g29090), CYP707A3 (At5g45340), CYP707A4 (At3g19270), EIN3 (At3g20770), EBF2 (At5g25350), EBF1 (At2g25490), transcription elongation factor 1-α (At5g60390), EIN2 (At5g03280), ETR1 (At1g66340), ETR2 (At3g23150), ERS1 (At2g40940), ERS2 (At1g04310), EIN4 (At3g04580), EXPA2 (At5g05290), GPCR-type G protein1 (At1g64990), GPCR-type G protein2 (At4g27630), G PROTEIN ALPHA SUBUNIT1 (At2g26300), GA3-OXIDASE2 (At1g80340), βGLU (At2g27500), CHNB (At3g12500), PL1 (At1g04680), RAV1 (At3g25730), NINE-cis-EPOXYCAROTENOID DIOXYGENASE9 (At1g78390), ABSCISIC ALDEHYDE OXIDASE3 (At2g27150), and SHORT-CHAIN DEHYDROGENASE REDUCTASE1 (also known as ABA2; At1g52340).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. TAGGIT Analysis of Functional Categories in the Lepidium sativum FR1 Transcriptome Data Sets.
Supplemental Figure 2. The Effect of Ethylene and ABA on the Germination of Arabidopsis thaliana: Wild-Type (Col) and Ethylene-Related Mutants (aco2 and ctr1).
Supplemental Figure 3. Brassicaceae ACO Sequence Comparisons.
Supplemental Figure 4. Analysis of Transcript Expression by qRT-PCR and Microarray Analysis in Specific Seed Tissues of Lepidium sativum FR1 during Germination.
Supplemental Figure 5. Analysis of the Transcript Expression of Key ABA Metabolism Genes, in Whole Lepidium sativum FR14 Seeds, by qRT-PCR Following Treatment with ACC or NBD.
Supplemental Table 1. Primer List for Transcript Expression Analysis by qRT-PCR.
Supplemental Data Set 1. Normalized Mean Expression Value Transcripts Used in the Analysis of the CON Arrays (22,025 Gene List).
Supplemental Data Set 2. Normalized Mean Expression Value Transcripts Used in the Analysis of the ABA Arrays (19,794 Gene List).
Supplemental Data Set 3. Statistical (t Tests, F-Tests) and TAGGIT Analysis of the CON Array Transcripts (22,025 Gene List).
Supplemental Data Set 4. Statistical (t Tests, F-Tests) and TAGGIT Analysis of the ABA Array Transcripts (19,794 Gene List).
Supplemental Data Set 5. Significantly Regulated Transcripts in the CON Arrays (1350 Gene List) Based on Adjusted P Values (≤0.1) from F-Tests.
Supplemental Data Set 6. Significantly Regulated Transcripts in the ABA Arrays (3530 Gene List) Based on Adjusted P Values (≤0.1) from F-Tests.
Supplemental Data Set 7. Venn Diagram Comparison of Transcripts Regulated in Arabidopsis Seedlings by Ethylene and/or ABA and in the Lepidium Seed CON Arrays.
Supplemental Data Set 8. Venn Diagram Comparison of Transcripts Regulated in Arabidopsis Seedlings by Ethylene and/or ABA and in the Lepidium Seed ABA Arrays.
Supplemental Data Set 9. Amino Acid Sequences Used to Generate the Phylogenetic Tree Presented in Figure 7D.
Supplemental Methods. Detailed Description of (1) aRNA Labeling and CATAM Microarray Hybridization and (2) Genomic DNA Hybridization to CATMA Microarrays.
Supplementary Material
Acknowledgments
We thank Anita Rott and Florian Wüst (University Freiburg, Germany) for expert technical help and Alex Tabrett and Jim Beynon for enabling access to CATMA arrays at Warwick Horticulture Research International. Our work is funded by grants from the Deutsche Forschungsgemeinschaft (Grant DFG LE720/6) and the Deutscher Akademischer Austauschdienst (Grant DAAD D/0628197) to G.L.-M., the Wissenschaftliche Gesellschaft Freiburg to G.L.-M. and A.L., the UK Department for the Environment, Food, and Rural Affairs and the Biotechnology and Biological Sciences Research Council (Grant BB/E006418/1) to W.E.F.-S., the Hubert Curien Program (Proscope Grant 14896 UL) to F.C., and the Czech Ministry of Education Grant MSM 6198959216 to M.S., which are gratefully acknowledged.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Gerhard Leubner-Metzger (gerhard.leubner@biologie.uni-freiburg.de).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Abeles, F.B. (1986). Role of ethylene in Lactuca sativa cv. 'Grand Rapids' seed germination. Plant Physiol. 81 780–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allemeersch, J., et al. (2005). Benchmarking the CATMA microarray. A novel tool for Arabidopsis transcriptome analysis. Plant Physiol. 137 588–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso, J.M., and Ecker, J.R. (2001). The ethylene pathway: A paradigm for plant hormone signaling and interaction. Sci. STKE 2001 RE1. [DOI] [PubMed] [Google Scholar]
- Bar-Or, C., Czosnek, H., and Koltai, H. (2007). Cross-species microarray hybridizations: A developing tool for studying species diversity. Trends Genet. 23 200–207. [DOI] [PubMed] [Google Scholar]
- Barrero, J.M., Talbot, M.J., White, R.G., Jacobsen, J.V., and Gubler, F. (2009). Anatomical and transcriptomic studies of the coleorhiza reveal the importance of this tissue in regulating dormancy in barley. Plant Physiol. 150 1006–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoin, N., Serizet, C., Gosti, F., and Giraudat, J. (2000). Interactions between abscisic acid and ethylene signaling cascades. Plant Cell 12 1103–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc., B 57 289–300. [Google Scholar]
- Bethke, P.C., Libourel, I.G.L., Aoyama, N., Chung, Y.-Y., Still, D.W., and Jones, R.L. (2007). The Arabidopsis aleurone layer responds to nitric oxide, gibberellin, and abscisic acid and is sufficient and necessary for seed dormancy. Plant Physiol. 143 1173–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley, J.D. (1997). Breaking down the walls - A role for endo-β-mannanase in release from seed dormancy? Trends Plant Sci. 2 464–469. [Google Scholar]
- Bleecker, A.B., Estelle, M.A., Somerville, C., and Kende, H. (1988). Insensitivity to ethylene conferred by a dominant mutation in Arabidopsis thaliana. Science 241 1086–1089. [DOI] [PubMed] [Google Scholar]
- Broadley, M.R., White, P.J., Hammond, J.P., Graham, N.S., Bowen, H.C., Emmerson, Z.F., Fray, R.G., Iannetta, P.P., McNicol, J.W., and May, S.T. (2008). Evidence of neutral transcriptome evolution in plants. New Phytol. 180 587–593. [DOI] [PubMed] [Google Scholar]
- Cadman, C.S.C., Toorop, P.E., Hilhorst, H.W.M., and Finch-Savage, W.E. (2006). Gene expression profiles of Arabidopsis Cvi seed during cycling through dormant and non-dormant states indicate a common underlying dormancy control mechanism. Plant J. 46 805–822. [DOI] [PubMed] [Google Scholar]
- Carrera, E., Holman, T., Medhurst, A., Dietrich, D., Footitt, S., Theodoulou, F.L., and Holdsworth, M.J. (2008). Seed after-ripening is a discrete developmental pathway associated with specific gene networks in Arabidopsis. Plant J. 53 214–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrera, E., Holman, T., Medhurst, A., Peer, W., Schmuths, H., Footitt, S., Theodoulou, F.L., and Holdsworth, M.J. (2007). Gene expression profiling reveals defined functions of the ABC transporter COMATOSE late in phase II of germination. Plant Physiol. 143 1669–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, C., Kwok, S.F., Bleecker, A.B., and Meyerowitz, E.M. (1993. a). Arabidopsis ethylene-response gene ETR1: Similarity of product to two-component regulators. Science 262 539–544. [DOI] [PubMed] [Google Scholar]
- Chang, S., Puryear, J., and Cairney, J. (1993. b). A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Rep. 11 113–116. [Google Scholar]
- Cheng, W.H., Chiang, M.H., Hwang, S.G., and Lin, P.C. (2009). Antagonism between abscisic acid and ethylene in Arabidopsis acts in parallel with the reciprocal regulation of their metabolism and signaling pathways. Plant Mol. Biol. 71 61–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiwocha, S.D.S., Cutler, A.J., Abrams, S.R., Ambrose, S.J., Yang, J., Ross, A.R.S., and Kermode, A.R. (2005). The etr1-2 mutation in Arabidopsis thaliana affects the abscisic acid, auxin, cytokinin and gibberellin metabolic pathways during maintenance of seed dormancy, moist-chilling and germination. Plant J. 42 35–48. [DOI] [PubMed] [Google Scholar]
- da Silva, E.A.A., Toorop, P.E., van Aelst, A.C., and Hilhorst, H.W.M. (2004). Abscisic acid controls embryo growth potential and endosperm cap weakening during coffee (Coffea arabica cv. Rubi) seed germination. Planta 220 251–261. [DOI] [PubMed] [Google Scholar]
- Dutta, S., and Bradford, K.J. (1994). Water relations of lettuce seed thermoinhibition. II. Ethylene and endosperm effects on base water potential. Seed Sci. Res. 4 11–18. [Google Scholar]
- Etheridge, N., Hall, B.P., and Schaller, G.E. (2006). Progress report: Ethylene signaling and responses. Planta 223 387–391. [DOI] [PubMed] [Google Scholar]
- Finch-Savage, W.E., Cadman, C.S.C., Toorop, P.E., Lynn, J.R., and Hilhorst, H.W.M. (2007). Seed dormancy release in Arabidopsis Cvi by dry after-ripening, low temperature, nitrate and light shows common quantitative patterns of gene expression directed by environmentally specific sensing. Plant J. 51 60–78. [DOI] [PubMed] [Google Scholar]
- Finch-Savage, W.E., and Leubner-Metzger, G. (2006). Seed dormancy and the control of germination. New Phytol. 171 501–523. [DOI] [PubMed] [Google Scholar]
- Franzke, A., German, D., Al-Shehbaz, I.A., and Mummenhoff, K. (2009). Arabidopsis family ties: Molecular phylogeny and age estimates in Brassicaceae. Taxon 58 1–13. [Google Scholar]
- Ghassemian, M., Nambara, E., Cutler, S., Kawaide, H., Kamiya, Y., and McCourt, P. (2000). Regulation of abscisic acid signaling by the ethylene response pathway in Arabidopsis. Plant Cell 12 1117–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, H., and Ecker, J.R. (2003). Plant responses to ethylene gas are mediated by SCFEBF1/EBF2-dependent proteolysis of EIN3 transcription factor. Cell 115 667–677. [DOI] [PubMed] [Google Scholar]
- Hermann, K., Meinhard, J., Dobrev, P., Linkies, A., Pesek, B., Heß, B., Machackova, I., Fischer, U., and Leubner-Metzger, G. (2007). 1-Aminocyclopropane-1-carboxylic acid and abscisic acid during the germination of sugar beet (Beta vulgaris L.) - A comparative study of fruits and seeds. J. Exp. Bot. 58 3047–3060. [DOI] [PubMed] [Google Scholar]
- Hilson, P., et al. (2004). Versatile gene-specific sequence tags for Arabidopsis functional genomics: Transcript profiling and reverse genetics applications. Genome Res. 14 2176–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdsworth, M.J., Bentsink, L., and Soppe, W.J.J. (2008). Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol. 179 33–54. [DOI] [PubMed] [Google Scholar]
- Hradecká, V., Novák, O., Havlíček, L., and Strnad, M. (2007). Immunoaffinity chromatography of abscisic acid combined with electrospray liquid chromatography-mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 847 162–173. [DOI] [PubMed] [Google Scholar]
- Hu, Y., Poh, H.M., and Chua, N.H. (2006). The Arabidopsis ARGOS-LIKE gene regulates cell expansion during organ growth. Plant J. 47 1–9. [DOI] [PubMed] [Google Scholar]
- Iglesias-Fernandez, R., and Matilla, A. (2009). After-ripening alters the gene expression pattern of oxidases involved in the ethylene and gibberellin pathways during the early imbibition of Sisymbrium officinale L. seeds. J. Exp. Bot. 60 1645–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi, M., and Yanagisawa, S. (2008). Ethylene signaling in Arabidopsis involves feedback regulation via the elaborate control of EBF2 expression by EIN3. Plant J. 55 821–831. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., Jorna, M.L., Brinkhorst-van der Swan, D.L.C., and Karssen, C.M. (1982). The isolation of abscisic acid (ABA) deficient mutants by selection of induced revertants in non-germinating gibberellin sensitive lines of in Arabidopsis thaliana (L.). Heynh. Theor. Appl. Genet. 61 385–393. [DOI] [PubMed] [Google Scholar]
- Kucera, B., Cohn, M.A., and Leubner-Metzger, G. (2005). Plant hormone interactions during seed dormancy release and germination. Seed Sci. Res. 15 281–307. [Google Scholar]
- Kushiro, T., Okamoto, M., Nakabayashi, K., Yamagishi, K., Kitamura, S., Asami, T., Hirai, N., Koshiba, T., Kamiya, Y., and Nambara, E. (2004). The Arabidopsis cytochrome P450 CYP707A encodes ABA 8'-hydroxylases: Key enzymes in ABA catabolism. EMBO J. 23 1647–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leubner-Metzger, G. (2003). Functions and regulation of β-1,3-glucanase during seed germination, dormancy release and after-ripening. Seed Sci. Res. 13 17–34. [Google Scholar]
- Leubner-Metzger, G., Petruzzelli, L., Waldvogel, R., Vögeli-Lange, R., and Meins, F., Jr. (1998). Ethylene-responsive element binding protein (EREBP) expression and the transcriptional regulation of class I β-1,3-glucanase during tobacco seed germination. Plant Mol. Biol. 38 785–795. [DOI] [PubMed] [Google Scholar]
- Lim, P.O., Kim, Y., Breeze, E., Koo, J.C., Woo, H.R., Ryu, J.S., Park, D.H., Beynon, J., Tabrett, A., Buchanan-Wollaston, V., and Nam, H.G. (2007). Overexpression of a chromatin architecture-controlling AT-hook protein extends leaf longevity and increases the post-harvest storage life of plants. Plant J. 52 1140–1153. [DOI] [PubMed] [Google Scholar]
- Lin, Z., Alexander, L., Hackett, R., and Grierson, D. (2008). LeCTR2, a CTR1-like protein kinase from tomato, plays a role in ethylene signalling, development and defence. Plant J. 54 1083–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, P.-P., Koizuka, N., Homrichhausen, T.M., Hewitt, J.R., Martin, R.C., and Nonogaki, H. (2005). Large-scale screening of Arabidopsis enhancer-trap lines for seed germination-associated genes. Plant J. 41 936–944. [DOI] [PubMed] [Google Scholar]
- Matilla, A., Gallardo, M., and Puga-Hermida, M.I. (2005). Structural, physiological and molecular aspects of heterogeneity in seeds: a review. Seed Sci. Res. 15 63–76. [Google Scholar]
- Matilla, A.J., and Matilla-Vazquez, M.A. (2008). Involvement of ethylene in seed physiology. Plant Sci. 175 87–97. [Google Scholar]
- Müller, K., Linkies, A., Vreeburg, R.A.M., Fry, S.C., Krieger-Liszkay, A., and Leubner-Metzger, G. (2009). In vivo cell wall loosening by hydroxyl radicals during cress (Lepidium sativum L.) seed germination and elongation growth. Plant Physiol. 150 1855–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, K., Tintelnot, S., and Leubner-Metzger, G. (2006). Endosperm-limited Brassicaceae seed germination: Abscisic acid inhibits embryo-induced endosperm weakening of Lepidium sativum (cress) and endosperm rupture of cress and Arabidopsis thaliana. Plant Cell Physiol. 47 864–877. [DOI] [PubMed] [Google Scholar]
- Nakabayashi, K., Okamoto, M., Koshiba, T., Kamiya, Y., and Nambara, E. (2005). Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: Epigenetic and genetic regulation of transcription in seed. Plant J. 41 697–709. [DOI] [PubMed] [Google Scholar]
- Nambara, E., and Marion-Poll, A. (2005). Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 56 165–185. [DOI] [PubMed] [Google Scholar]
- Nemhauser, J.L., Hong, F., and Chory, J. (2006). Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126 467–475. [DOI] [PubMed] [Google Scholar]
- Ogawa, M., Hanada, A., Yamauchi, Y., Kuwahara, A., Kamiya, Y., and Yamaguchi, S. (2003). Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell 15 1591–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto, M., Kuwahara, A., Seo, M., Kushiro, T., Asami, T., Hirai, N., Kamiya, Y., Koshiba, T., and Nambara, E. (2006). CYP707A1 and CYP707A2, which encode ABA 8'-hydroxylases, are indispensable for a proper control of seed dormancy and germination in Arabidopsis. Plant Physiol. 141 97–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey, S., Nelson, D.C., and Assmann, S.M. (2009). Two novel GPCR-type G proteins are abscisic acid receptors in Arabidopsis. Cell 136 136–148. [DOI] [PubMed] [Google Scholar]
- Pavlista, A.D., and Haber, A.H. (1970). Embryo expansion without protrusion in lettuce seeds. Plant Physiol. 46 636–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield, S., Li, Y., Gilday, A.D., Graham, S., and Graham, I.A. (2006). Arabidopsis ABA INSENSITIVE4 regulates lipid mobilization in the embryo and reveals repression of seed germination by the endosperm. Plant Cell 18 1887–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petruzzelli, L., Coraggio, I., and Leubner-Metzger, G. (2000). Ethylene promotes ethylene biosynthesis during pea seed germination by positive feedback regulation of 1-aminocyclopropane-1-carboxylic acid oxidase. Planta 211 144–149. [DOI] [PubMed] [Google Scholar]
- Pfaffl, M.W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29 e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskurewicz, U., Jikumaru, Y., Kinoshita, N., Nambara, E., Kamiya, Y., and Lopez-Molina, L. (2008). The gibberellic acid signaling repressor RGL2 inhibits Arabidopsis seed germination by stimulating abscisic acid synthesis and ABI5 activity. Plant Cell 20 2729–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potuschak, T., Lechner, E., Parmentier, Y., Yanagisawa, S., Grava, S., Koncz, C., and Genschik, P. (2003). EIN3-dependent regulation of plant ethylene hormone signaling by two Arabidopsis F box proteins: EBF1 and EBF2. Cell 115 679–689. [DOI] [PubMed] [Google Scholar]
- Salmen Espindola, L., Noin, M., Corbineau, F., and Côme, D. (1994). Cellular and metabolic damage induced by desiccation in recalcitrant Araucaria angustifolia embryos. Seed Sci. Res. 4 193–201. [Google Scholar]
- Seo, Y.S., Yoo, A., Jung, J., Sung, S.K., Yang, D.R., Kim, W.T., and Lee, W. (2004). The active site and substrate-binding mode of 1-aminocyclopropane-1-carboxylate oxidase determined by site-directed mutagenesis and comparative modelling studies. Biochem. J. 380 339–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siriwitayawan, G., Geneve, R.L., and Downie, A.B. (2003). Seed germination of ethylene perception mutants of tomato and Arabidopsis. Seed Sci. Res. 13 303–314. [Google Scholar]
- Sisler, E.C., and Serek, M. (2003). Compounds interacting with the ethylene receptor in plants. Plant Biol. 5 473–480. [Google Scholar]
- Slotte, T., Holm, K., McIntyre, L.M., Lagercrantz, U., and Lascoux, M. (2007). Differential expression of genes important for adaptation in Capsella bursa-pastoris (Brassicaceae). Plant Physiol. 145 160–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth, G.K. (2004). Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3 Article 3. [DOI] [PubMed]
- Smyth, G.K. (2005). Limma: Linear models for microarray data. In Bioinformatics and Computational Biology Solutions Using R and Bioconductor, R. Gentleman, V. Carey, S. Dudoit, R. Irizarry, and W. Huber, eds (New York: Springer), pp. 397–420.
- Smyth, G.K., and Speed, T.P. (2003). Normalization of cDNA microarray data. Methods 31 265–273. [DOI] [PubMed] [Google Scholar]
- Toh, S., et al. (2008). High temperature-induced abscisic acid biosynthesis and its role in the inhibition of gibberellin action in Arabidopsis seeds. Plant Physiol. 146 1368–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toorop, P.E., van Aelst, A.C., and Hilhorst, H.W.M. (2000). The second step of the biphasic endosperm cap weakening that mediates tomato (Lycopersicon esculentum) seed germination is under control of ABA. J. Exp. Bot. 51 1371–1379. [PubMed] [Google Scholar]
- Turečková, V., Novák, O., and Strnad, M. (2009). Profiling ABA metabolites in Nicotiana tabacum L. leaves by ultra-performance liquid chromatography-electrospray tandem mass spectrometry. Talanta 80 390–399. [DOI] [PubMed] [Google Scholar]
- Udvardi, M.K., Czechowski, T., and Scheible, W.-R. (2008). Eleven golden rules of quantitative RT-PCR. Plant Cell 20 1736–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Mortel, J.E., and Aarts, M.G. (2006). Comparative transcriptomics - Model species lead the way. New Phytol. 170 199–201. [DOI] [PubMed] [Google Scholar]
- Van Gelder, R.N., von Zastrow, M.E., Yool, A., Dement, W.C., Barchas, J.D., and Eberwine, J.H. (1990). Amplified RNA synthesized from limited quantities of heterogeneous cDNA. Proc. Natl. Acad. Sci. USA 87 1663–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, S., Kamiya, Y., and Sun, T.P. (2001). Distinct cell-specific expression patterns of early and late gibberellin biosynthetic genes during Arabidopsis seed germination. Plant J. 28 443–453. [DOI] [PubMed] [Google Scholar]
- Yamauchi, Y., Ogawa, M., Kuwahara, A., Hanada, A., Kamiya, Y., and Yamaguchi, S. (2004). Activation of gibberellin biosynthesis and response pathways by low temperature during imbibition of Arabidopsis thaliana seeds. Plant Cell 16 367–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, S.H., and Choi, D. (2006). Characterization of genes encoding ABA 8'-hydroxylase in ethylene-induced stem growth of deepwater rice (Oryza sativa L.). Biochem. Biophys. Res. Commun. 350 685–690. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.