Abstract
The phytohormone auxin plays a major role in embryonic and postembryonic plant development. The temporal and spatial distribution of auxin largely depends on the subcellular polar localization of members of the PIN-FORMED (PIN) auxin efflux carrier family. The Ser/Thr protein kinase PINOID (PID) catalyzes PIN phosphorylation and crucially contributes to the regulation of apical-basal PIN polarity. The GTP exchange factor on ADP-ribosylation factors (ARF-GEF), GNOM preferentially mediates PIN recycling at the basal side of the cell. Interference with GNOM activity leads to dynamic PIN transcytosis between different sides of the cell. Our genetic, pharmacological, and cell biological approaches illustrate that PID and GNOM influence PIN polarity and plant development in an antagonistic manner and that the PID-dependent PIN phosphorylation results in GNOM-independent polar PIN targeting. The data suggest that PID and the protein phosphatase 2A not only regulate the static PIN polarity, but also act antagonistically on the rate of GNOM-dependent polar PIN transcytosis. We propose a model that includes PID-dependent PIN phosphorylation at the plasma membrane and the subsequent sorting of PIN proteins to a GNOM-independent pathway for polarity alterations during developmental processes, such as lateral root formation and leaf vasculature development.
INTRODUCTION
Postembryonic plant growth results in shapes that are not predictable by their previous embryonic architecture. Plants have evolved the outstanding ability to redefine the polarity of an already specified tissue, eventually leading to de novo postembryonic organ formation. The flexible nature of plant development most probably compensates for the plant's sessile lifestyle. A decisive role in establishing and redefining the polarity of plant tissues is played by the phytohormone auxin (Sauer et al., 2006; Scarpella et al., 2006). Spatial and temporal auxin accumulation (auxin gradients) determines positional cues for the presumptive sites of embryonic and postembryonic primordia development (Benková et al., 2003; Friml et al., 2003; Heisler et al., 2005). Hence, insights into the regulation of the auxin distribution and, subsequently, signaling are key to understanding this type of plant growth regulation.
The PIN-FORMED (PIN) auxin efflux carriers catalyze the cell-to-cell transport of auxin and largely mediate its spatial and temporal auxin distribution (Petrášek et al., 2006; reviewed in Tanaka et al., 2006). The coordinated polar localization of PIN proteins at different sides of the cell determines the direction of the auxin flux within a tissue (Wiśniewska et al., 2006). Thus, directional PIN activity has the capacity to translate cellular polarizing signals into polarity of the whole tissue. Moreover, the dynamic nature of the polar PIN localization regulates plant development by rearranging the auxin fluxes that initiate embryonic and postembryonic developmental programs (reviewed in Kleine-Vehn and Friml, 2008).
A valuable tool for unraveling polar PIN-targeting mechanisms is the fungal toxin brefeldin A (BFA), which is known to interfere with various vesicle trafficking processes in cells. The molecular targets of BFA are GDP-to-GTP exchange factors (GEFs) for small G proteins of the ADP-ribosylation factor (ARF) class that activates the ARF proteins and, thus, direct the formation of vesicle coats (reviewed in Donaldson and Jackson, 2000).
In Arabidopsis thaliana roots, because PIN1 exocytosis is sensitive to BFA, PIN1 accumulates rapidly and reversibly into so-called BFA compartments, hinting at a constitutive PIN1 cycling mechanism (Geldner et al., 2001). A green-to-red photoconvertible fluorescent reporter (EosFP) was used to capture the internalization of PIN proteins and their subsequent recycling to the plasma membrane, thus confirming that PIN proteins cycle constitutively between the plasma membrane and some endosomal compartments (Dhonukshe et al., 2007). This constitutive cycling mechanism might have several functions, such as PIN polarization after originally nonpolar secretion (Dhonukshe et al., 2008) or dynamic intracellular resorting of PIN proteins for transcytosis-like polarity alterations during plant development (Kleine-Vehn et al., 2008a).
In Arabidopsis, the BFA-sensitive endosomal ARF-GEF GNOM is required for the polar localization and recycling of PIN1 (Steinmann et al., 1999; Geldner et al., 2001). The inhibitory effect of BFA on PIN1 cycling in the root stele cells is due to its effect specifically on GNOM (Geldner et al., 2003). Hence, the vesicle transport regulator GNOM seemingly defines the recycling rate of the PIN1 protein to the basal (root apex-facing) side of the cell. Moreover, GNOM activity is also involved in dynamic transcytosis of PIN proteins from one side of the cell to the other, eventually regulating PIN-dependent tissue repolarization (Kleine-Vehn et al., 2008a).
Remarkably, while PIN1 localizes preferentially to the basal side of the cell, PIN2 is targeted to the apical (shoot apex-facing) side in the same cell, suggesting polarity determinants in the protein sequence itself (Wiśniewska et al., 2006). The polarity signals are probably related to the phosphorylation sites within the PIN proteins (F. Huang, M. Kemel-Zago, A. van Marion, C.G. Ampudia, and R. Offringa, unpublished data; Zhang et al., 2009) because the Ser/Thr protein kinase PINOID (PID) and protein phosphatase 2A (PP2A) act on PIN phosphorylation, thus determining the apical or basal PIN targeting, respectively (Friml et al., 2004; Michniewicz et al., 2007). The pid mutants display apical-to-basal PIN polarity shifts that cause defects during embryo and shoot organogenesis (Christensen et al., 2000; Benjamins et al., 2001; Friml et al., 2004), whereas PID gain-of-function specifically mistargets the PIN proteins to the apical sides of cells in the primary root, with auxin depletion from the root meristem and its collapse as a consequence (Benjamins et al., 2001; Friml et al., 2004). Similarly, basal-to-apical PIN polarity shifts in the primary root meristem can be observed in the loss-of-function mutants of the A regulatory subunits of PP2A (Michniewicz et al., 2007).
Recently, several Ser- and Thr-containing phosphorylation sites in conserved motifs have been identified in the PIN proteins. Using engineered dephosphorylated or phosphorylation-mimicking mutant PIN proteins, phosphorylation of these specific residues in the PIN proteins have been found to determine the apical-basal polar PIN localization (F. Huang, M. Kemel-Zago, A. van Marion, C.G. Ampudia, and R. Offringa, unpublished data; Zhang et al., 2009). However, the underlying mechanism of how the phosphorylation-based sequence modulation of PIN proteins affects their polar localization is unknown. In particular, the question of how the PID-dependent pathway relates to the GNOM-dependent recycling of PIN proteins remains to be addressed.
Here, we specifically investigated the interaction between the PID and the GNOM polar-targeting mechanisms. Based on our findings, we propose a model in which the PIN proteins are phosphorylated by PID, decreasing their affinity for the GNOM-dependent basal recycling pathway. The increased PIN affinity for the distinct and presumably ARF-GEF–dependent apical targeting pathway eventually initiates PIN transcytosis from the basal to the apical side of the cell. This mechanism might be functionally important for various dynamic PIN polarity alterations during plant development, such as during lateral root primordia formation or vascular development.
RESULTS
The Localization of PID and PIN Suggests a Preferential Interaction at the Plasma Membrane
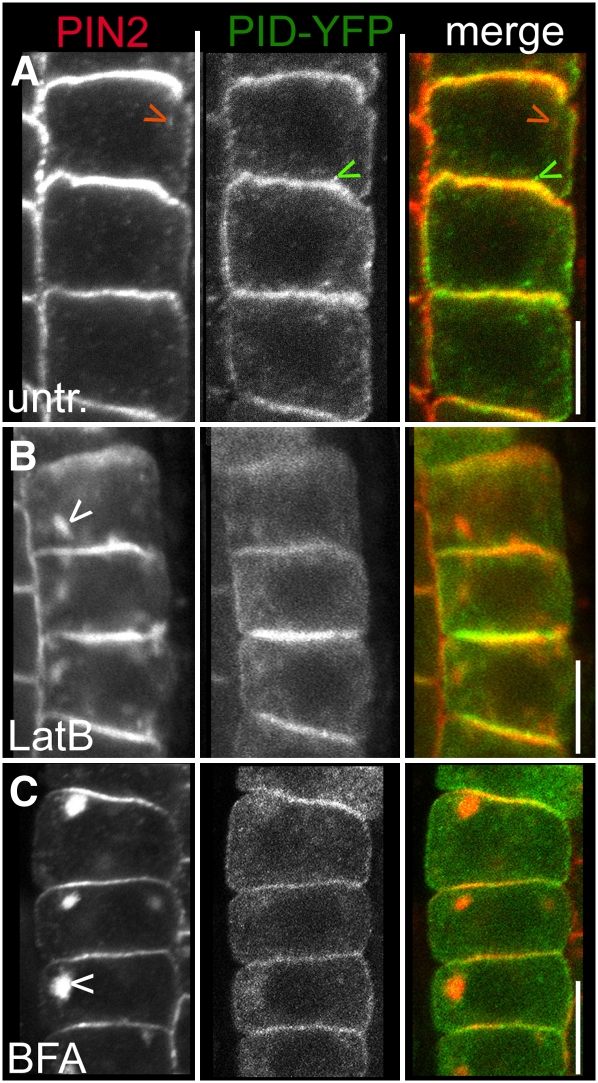
To address the question of how PID regulates the polar PIN targeting in planta, we initially investigated PID localization and targeting. PIN2 and PID display substantial colocalization at the plasma membrane (Michniewicz et al., 2007; Figure 1A). Because a strong internal PID-yellow fluorescent protein (YFP) signal was observed, we analyzed the endosomal PIN and PID distributions, but the colocalization of endogenous PIN2 and functional PID-YFP was not detectable in the endosomes (Michniewicz et al., 2007; Figure 1A). To elaborate on this observation, we induced PIN2 internalization in root epidermal cells. Whereas PIN2 strongly accumulated in intracellular compartments after latrunculin B–dependent actin depolymerization (Figure 1B) or BFA treatment (Figure 1C), PID localization was less sensitive, and accumulating endosomes were more dispersed (Figures 1B and 1C), resulting in only faint endoplasmic colocalization with PIN2.
Figure 1.
Subcellular Localization of PIN2 and PID in Root Epidermal Cells.
(A) to (C) Confocal images of anti-PIN2 and anti-GFP immunolocalizations. Bars = 10 μm.
(A) Pronounced colocalization of PIN2 (left panel/red in the merged image) and PID-YFP (middle panel/green in the merged image) at the plasma membrane, whereas no pronounced colocalization in the endosomes could be detected (indicated by red and green arrowheads for endosomal PIN2 and PID signal, respectively).
(B) and (C) Strong intracellular PIN2 localization (depicted by white arrowheads) induced by latrunculin B (B) and BFA (C) treatments but weaker and more dispersed intracellular PID-YFP accumulations.
Because no pronounced endosomal colocalization or cotrafficking of PID and PIN proteins could be induced, a largely PIN-independent endosomal trafficking or membrane recruitment of PID was assumed. Besides the weak endoplasmic colocalization, both PIN and PID proteins reside and colocalize mainly in/at the plasma membrane; therefore, we hypothesized that the plasma membrane was the most likely site of the PID-PIN interaction for the regulation of the PIN polarity. This finding suggests that PID-dependent PIN phosphorylation at the plasma membrane might affect PIN residence or recycling and, eventually, impose intracellular resorting of PIN proteins.
PID and GNOM Have Opposite Effects on PIN Polarity and Auxin Accumulation in the Primary Root
In animals and plants, polar targeting to the plasma membrane ultimately requires polar sorting processes at endosomes (Farr et al., 2009; reviewed in Kleine-Vehn and Friml, 2008). The predominant colocalization of PID and PIN at the plasma membrane suggests that the polar PIN distribution is regulated by PID upstream of the actual endosomal sorting event. Hence, we investigated how PID-dependent PIN phosphorylation relates to the constitutive PIN recycling that is presumably related to intracellular polar sorting.
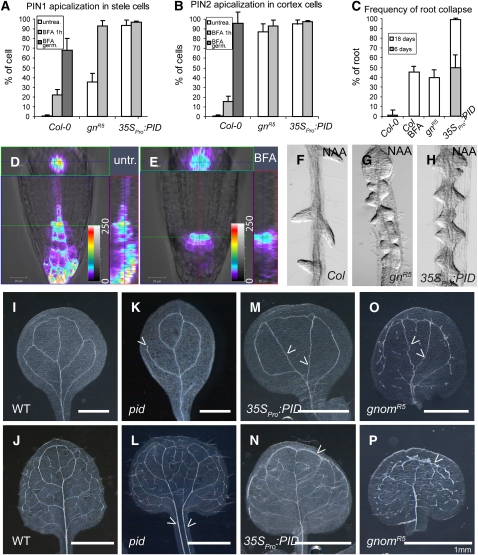
The endosomal ARF-GEF GNOM is known to mediate PIN recycling preferentially to the basal side of the cell, and pharmacologically or genetically reduced GNOM activity leads to a dynamic basal-to-apical PIN transcytosis (Kleine-Vehn et al., 2008a). Intriguingly, a similar apical PIN polarity shift is triggered by pp2aa loss-of-function (Michniewicz et al., 2007) and PID gain-of-function (in 35SPro:PID) (Friml et al., 2004; Figures 2A and 2B). In agreement with these similar cellular phenotypes, PID gain-of-function and reduced GNOM function (in gnomR5) also result in root meristem collapse (Benjamins et al., 2001; Geldner et al., 2004) (Figure 2C). Notably, the severity of the PIN apicalization in 35SPro:PID and gnomR5 correlated well with the frequency of root meristem abortion (Figures 2A to 2C). Moreover, auxin seemed to be depleted from the root tip after the increased rate of PIN phosphorylation in 35SPro:PID and pp2aa mutants (Michniewicz et al., 2007). Similarly, pharmacological inhibition of GNOM decreased auxin response in the root tip as monitored by DR5Pro:GFP (green fluorescent protein) activity (Figures 2D and 2E; see Supplemental Figures 1E and 1F online), consistent with the auxin depletion from the root tip by the upward-directed auxin flux. In line with the requirement of PIN-mediated auxin accumulation in the root tip for root meristem activity (Friml et al., 2002a), the expression pattern of quiescent center and columella markers was altered after BFA treatment (see Supplemental Figures 1A to 1D online). These and previous observations reveal that PID gain-of-function and gnom loss-of-function alleles have similar effects on PIN polarity, auxin distribution, and development of the primary root.
Figure 2.
Opposite Actions of GNOM and PID on PIN Polarity and Plant Development.
(A) and (B) Apicalization of PIN1 (A) in stele and PIN2 (B) in cortex cells of wild-type, partial loss of GNOM function [gnomR5, labeled gn(R5)], and PID gain-of-function (35SPro:PID) lines. White bars represent the untreated condition, and gray bars illustrate BFA treatment for 1 h (light gray) or germination on medium containing BFA (dark gray). At least 1000 stele and 400 cortex cells for each treatment or genotype (roots, n > 12) were counted. Error bars indicate sd.
(C) Frequency of primary root collapse in 35SPro:PID, BFA-treated wild-type plants and the weak gnomR5 allele after 18 d (and after 6 d for 35SPro:PID). Error bars indicate sd (n = 30 seedlings).
(D) and (E) Confocal z-stack analysis and subsequent fluorescence intensity profiling (red/yellow denotes high fluorescence intensities and blue/purple denotes low) of the auxin-responsive promoter element DR5revPro:GFP in untreated (D) and BFA-treated (E) seedlings. The central image shows a single medium confocal section, while the top (green) and right (red) insets represent the radial (green line) and longitudinal (red line) distributions of the signal, respectively, giving three-dimensional information of the signal intensity. Under untreated conditions, DR5 signal is the highest in the quiescent center and outermost tier of columella cells (D). By contrast, BFA-dependent inhibition of GNOM function leads to depletion of the response maximum in the root tip and radial expansion of the signal (E).
(F) to (H) Synthetic auxin 1-naphthyl acetic acid (NAA) treatment induces enhanced, but spaced, lateral root development in wild-type seedlings (F). By contrast, both gnomR5 (G) and 35SPro:PID (H) mutants display defective primordium spacing and development after NAA treatment.
(I) to (P) Vascular development of cotyledons ([I], [K], [M], and [O]) and leaves ([J], [L], [N], and [P]) in wild-type ([I] and [J]), pid ([K] and [L]), 35SPro:PID ([M] and [N]), and gnomR5 ([O] and [P]) backgrounds. Arrows point out vein discontinuity (K), vein polarity defects ([L], [M], and [O]), and enhanced cortical vascularization ([N] and [P]).
PID and GNOM Have Opposite Effects on the Formation of Lateral Roots and Vascular Tissues
Dynamic PIN polarity alterations during lateral root development have been shown to depend on GNOM; accordingly, gnom partial loss-of-function alleles display severely reduced numbers of lateral roots (Benková et al., 2003; Geldner et al., 2004; Kleine-Vehn et al., 2008a). This is also seen in 35SPro:PID lines (Benjamins et al., 2001), but the primary root collapse precedes lateral root induction in the 35SPro:PID background (Benjamins et al., 2001; Friml et al., 2004), making the interpretation of this observation difficult. Therefore, we studied PID function in lateral root development prior to the root collapse by inducing lateral root organogenesis with auxin. We observed an aberrant development of lateral root primordia and strongly reduced primordium spacing after auxin treatment in PID gain-of-function lines (Figures 2F to 2H), undistinguishable from those in weak gnom alleles (Geldner et al., 2004).
Dynamic PIN polarity rearrangements also accompany and regulate leaf vascular development (Scarpella et al., 2006). In accordance with the anticipated function of GNOM in dynamic PIN polarity alterations (Kleine-Vehn et al., 2008a), weak gnom mutant alleles display severe defects in leaf vasculature development (Koizumi et al., 2000; Geldner et al., 2004) (Figures 2O and 2P). Therefore, we analyzed PID involvement in leaf vasculature development. Both PID gain-of-function and pid loss-of-function seedlings showed defects in leaf vasculature development (Figures 2I to 2N), suggesting that PID might also regulate dynamic PIN polarity alterations during vascular development. Intriguingly, 35SPro:PID seedlings had lost the main vein polarity in cotyledons and ectopic vasculature development at the leaf margin, largely phenocopying the gnomR5 partial loss-of-function mutant (Figures 2M to 2P, arrowheads). These and previous observations show that PID gain-of-function and gnom loss-of-function alleles similarly affect lateral root organogenesis and vascular tissue development.
PID and GNOM Show an Antagonistic Genetic Interaction
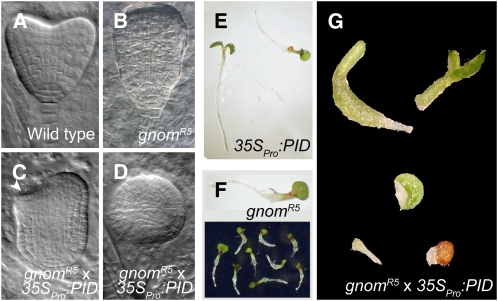
The similar effects of PID gain-of-function and gnom loss-of-function on polar PIN deposition, auxin distribution, and plant development suggest an antagonistic action of PID and GNOM. To examine this, we crossed the PID gain-of-function line (Figure 3E) with the gnomR5 mutant line (Figure 3F). The gnomR5 mutant embryos were defective in apical and basal embryo patterning (Figures 3A and 3B). However, none of the mutants displayed rootless development, and only one-third of the gnom mutant seedlings had fused cotyledons (Geldner et al., 2004; Figure 3F). The 35SPro:PID gnomR5 double mutants showed notably stronger embryonic apical-basal patterning phenotypes, being defective in root and shoot formation (Figures 3C and 3D). In the most affected embryos, apical-basal polarity had disappeared completely (Figure 3D), strongly resembling the gnom complete loss-of-function mutant (Shevell et al., 1994; Steinmann et al., 1999). Consistently with the defects observed during embryo development, the 35SPro:PID gnomR5 double mutants showed pronounced patterning defects during seedling development, resulting in an early developmental arrest (Figure 3G). Remarkably, all observed aspects of the 35SPro:PID gnomR5 phenotypes were markedly stronger than those of each of the single mutants and resembled those of the gnom complete loss-of-function mutant (Shevell et al., 1994; Steinmann et al., 1999). This synergistic genetic interaction between PID gain-of-function and gnom partial loss-of-function together with the similar phenotypes of the single alleles in a range of developmental and cellular processes suggest an antagonistic action of PID and GNOM in the same process.
Figure 3.
Genetic Interaction of PID and GNOM.
(A) to (D) Patterning defects in mutant embryos compared with wild-type embryos (A). Apical and basal patterning defects in gnomR5 mutant embryos (B). More severe apical-to-basal embryonic patterning defects in 35SPro:PID gnomR5 double mutants (C), sometimes leading to the complete loss of apical basal patterning (D).
(E) and (F) Viable 35SPro:PID (E) and gnomR5 (F) seedlings.
(G) Strongly affected seedling morphology and growth arrest in 35SPro:PID gnomR5 double mutants, resembling full gnom knockout mutants.
GNOM Localization Is Independent of PID Activity
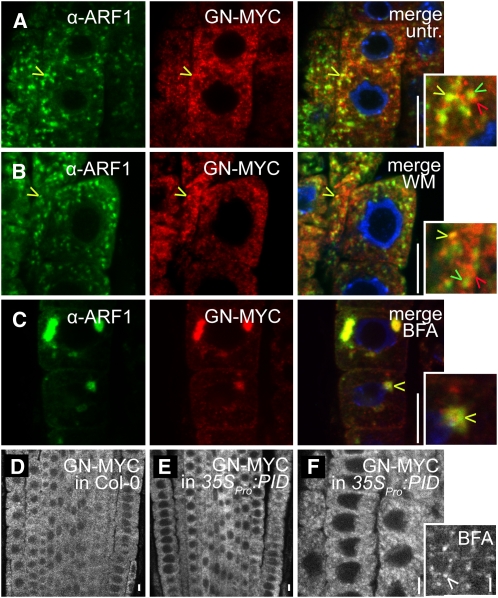
Next, we addressed the mechanism underlying the antagonistic action of PID and GNOM. One possibility is that PID might regulate subcellular GNOM localization and thus influence its activity. We initially analyzed the localization of GNOM that resides in an endosomal compartment, functionally defined as recycling endosome (Geldner et al., 2003). Myc-tagged GNOM proteins (GNOM-MYC) were found close to, and regularly colocalized with, anti-ARF1–labeled endomembranes (Figure 4A; see Supplemental Figures 2A to 2D online). This partial colocalization of ARF1 and GNOM might indicate a potential function for the polar PIN targeting at ARF1-positive endosomes. In agreement with this assumption, a GTP-locked, constitutively active arf1QL mutant is defective in polar PIN targeting (Xu and Scheres, 2005; Kleine-Vehn et al., 2008b).
Figure 4.
PID-Independent GNOM Localization.
(A) to (F) Confocal images of anti-ARF1 and anti-MYC immunolocalizations. Bars = 10 μm.
(A) to (C) Partial ARF1 (green) and GNOM (GN-MYC) (red) colocalization in untreated seedlings (A) and seedlings treated with wortmannin (B) or BFA (C). Insets display enlarged regions highlighting colocalizing (yellow arrowheads) and noncolocalizing (red and green arrowheads) endosomes.
(D) and (E) Similar expression pattern of GNOM (GN-MYC under endogenous promoter) in the wild type (D) and 35SPro:PID (E).
(F) Normal subcellular distribution and response to BFA (BFA treatment is shown in the inset) of GNOM-MYC in 35SPro:PID-expressing seedlings. White arrowhead indicates GNOM accumulation in the BFA compartment.
By means of the endocytic tracer FM4-64 that labels GNOM-positive endosomes only within 10 to 15 min (Geldner et al., 2003), GNOM had been found not to reside at early endosomes (Chow et al., 2008). In support of this, the early endosomal ARF-GEF BEN1/MIN7 does not colocalize with GNOM (Tanaka et al., 2009).
Whereas both ARF1 and GNOM-MYC showed a pronounced sensitivity to BFA (Figure 4C), they were not sensitive to treatment with the phosphatidylinositol-3-kinase inhibitor wortmannin that targets late endocytic trafficking to the vacuole by affecting the prevacuolar compartment (PVC) maturation (Matsuoka et al., 1995; daSilva et al., 2005; Kleine-Vehn et al., 2008b) (Figure 4B). These findings imply that GNOM and its potential substrates of the ARF1 subclass do not function at the PVC and substantiate previous assumptions that GNOM acts at a late endosome required for PIN recycling (Geldner et al., 2003; Chow et al., 2008) but is not directly involved in PVC-dependent PIN trafficking to the lytic vacuole for degradation (Kleine-Vehn et al., 2008c).
Notably, the PID overexpression did not visibly alter the expression (Figures 4D and 4E) nor the subcellular distribution of GNOM (Figure 4F). We used the stabilizing effect of BFA on ARF/ARF-GEF intermediates to study the GNOM functionality in the PID gain-of-function line. GNOM proteins accumulated normally after BFA treatments in 35SPro:PID, indicating that the overall interaction of GNOM and ARF proteins was largely unaffected (Figure 4F, inset). These findings suggest that PID does not directly regulate the subcellular GNOM localization or its BFA-sensitive ARF binding activity.
PID and PP2A Antagonistically Affect PIN Sorting into a GNOM-Independent Pathway
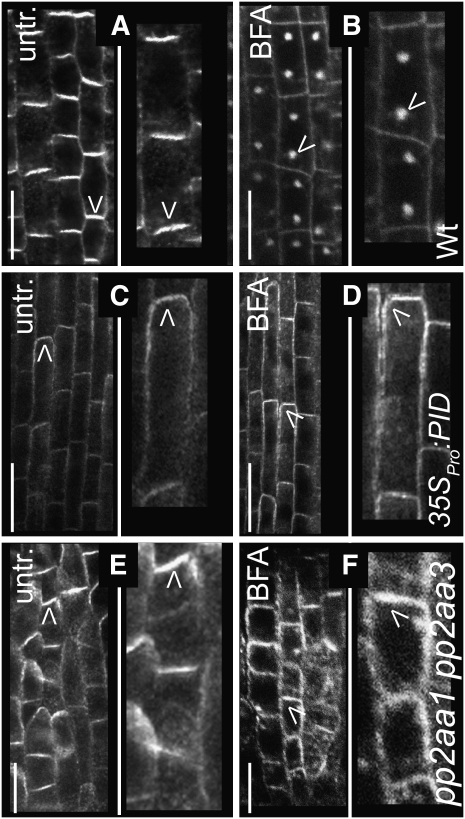
To investigate further which mechanism connected PID and GNOM functions, we assessed the GNOM-dependent recycling of PIN1 protein in PID gain-of-function mutants. We used BFA, which in wild-type inhibits GNOM and consequently causes rapid intracellular PIN1 accumulation (Geldner et al., 2003) (Figures 5A and 5B). While GNOM remained sensitive to BFA (Figure 4F), the effect of BFA on PIN1 localization was severely compromised in 35SPro:PID lines, as manifested by the strongly reduced BFA-induced PIN1 internalization and PIN1 retention at the plasma membrane (Figures 5C and 5D; see Supplemental Figure 3A online).
Figure 5.
BFA-Insensitive PIN Trafficking by Antagonistic PID and PP2A Action.
(A) to (F) Confocal images of anti-PIN1 immunolocalizations. Arrowheads indicate the most pronounced localization of endogenous PIN1 at the apical/basal cell side or in BFA compartments. Bars = 10 μm.
(A) Basal PIN1 localization in stele cells of untreated wild-type seedlings.
(B) Rapid PIN internalization after BFA (50 μM, 1 h) treatment in wild-type seedlings.
(C) Apical PIN1 localization in untreated stele cells as a consequence of PID gain of function.
(D) Largely BFA-insensitive PIN1 localization in 35SPro:PID, displaying a reduced accumulation in BFA compartments and persistent labeling of the plasma membrane.
(E) Preferentially apical PIN1 localization due to partial loss of PP2A function in untreated pp2aa1 pp2aa3 double mutants.
(F) Reduced sensitivity to BFA of PIN1 trafficking in pp2aa1 pp2aa3 double mutants.
The partial loss of the phosphatase activity in pp2aa1 pp2aa3 double mutants similarly reduced the BFA-induced PIN1 accumulation in BFA compartments (Figures 5E and 5F; see Supplemental Figure 3A online). This finding suggests that gain of PID kinase and loss of PP2A phosphatase activity both result in PIN recruitment to a BFA-insensitive and, hence, GNOM-independent apical targeting pathway.
PID and PP2A Antagonistically Determine the Affinity of PIN Proteins for Distinct Apical and Basal Targeting Machineries
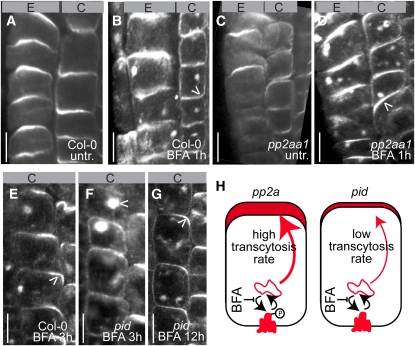
BFA preferentially interferes with basal PIN recycling and, hence, leads in time to artificial recruitment to the apical targeting pathway and basal-to-apical translocation of basal cargos, reminiscent of transcytosis in animal epithelial cells (Kleine-Vehn et al., 2008a). The reduced sensitivity of PIN protein trafficking to BFA treatments in 35SPro:PID and pp2aa hints at a favored recruitment of phosphorylated PIN proteins to a BFA-insensitive, GNOM-independent recycling pathway. In an alternative scenario, BFA could also preferentially inhibit PIN internalization at the apical side of the cell, reducing PIN accumulation in BFA compartments. To distinguish between these two scenarios, and the GNOM function was partially inhibited by BFA, and the PIN basal-to-apical shift was analyzed. A mild 1-h BFA treatment induced only weak or no basal-to-apical PIN2 transcytosis in the lower cortex cells of wild-type seedlings (Figures 6A and 6B; see Supplemental Figure 3B online). By contrast, PP2A phosphatase-deficient pp2aa1 mutants showed an enhanced BFA-induced basal-to-apical PIN transcytosis in cortex cells (Figures 6C and 6D; see Supplemental Figure 3B online). In pid loss-of-function mutants, the apical PIN incidence after prolonged (3 h) GNOM inhibition was lower than that of the wild type (Figures 6E and 6F; see Supplemental Figure 3B online). However, PID was not absolutely required for the BFA-induced apical shift of PIN proteins because a complete apical PIN polarity shift was induced after 12 h of BFA treatment in pid mutants as well (Figure 6G), indicating a functional redundancy within the PID pathway or a partial PID kinase-independent BFA-induced PIN transcytosis. These data suggest that a higher PIN2 phosphorylation state in a pp2aa1 mutant background (Michniewicz et al., 2007) promotes the PIN affinity for the apical targeting machinery, whereas a reduced PIN phosphorylation level in a pid mutant background results in decreased PIN affinity for the apical targeting pathway (Figure 6H). Hence, we conclude that the PID and PP2A activities determine the affinity of the PIN proteins for the basal GNOM-dependent or the apical GNOM-independent polar targeting pathways.
Figure 6.
PP2A and PID Modulate the BFA-Induced Transcytosis of PIN Proteins.
(A) to (G) Confocal images of anti-PIN2 immunolocalizations. Arrowheads indicate the most pronounced localization of endogenous PIN2 at the apical/basal side of the cell or in BFA compartments. E, epidermal cell files; C, cortical cell files. Bars = 10 μm.
(A) to (D) Wild-type seedlings (untreated in [A]) display only weak basal-to-apical transcytosis of PIN2 in cortex cells after 1 h of BFA (50 μM) treatment (B). Enhanced basal-to-apical transcytosis of PIN2 in cortex cells of pp2a mutants (untreated in [C]) after 1 h of BFA (50 μM) treatment (D).
(E) to (G) Preferential apical PIN2 localization in lower cortex cells of wild-type seedlings after 3 h of 50 μM BFA incubation (E). Strong PIN2 accumulation in BFA compartments and reduced basal-to-apical transcytosis of PIN2 in cortex cells in pid mutants (F). PIN2 recruitment to the apical side of the cell in lower cortex cells after prolonged BFA treatments (12 h) in the pid mutant background (G).
(H) Scheme depicting altered affinity (depicted by thickness of the arrow) of PIN proteins for the apical targeting machinery and subsequent PIN transcytosis rate (depicted at the upper side of the cell) in pp2aa1 and pid mutants.
[See online article for color version of this figure.]
PIN's Phosphorylation Status Determines Its Differential Recruitment to GNOM-Dependent or GNOM-Independent Pathways
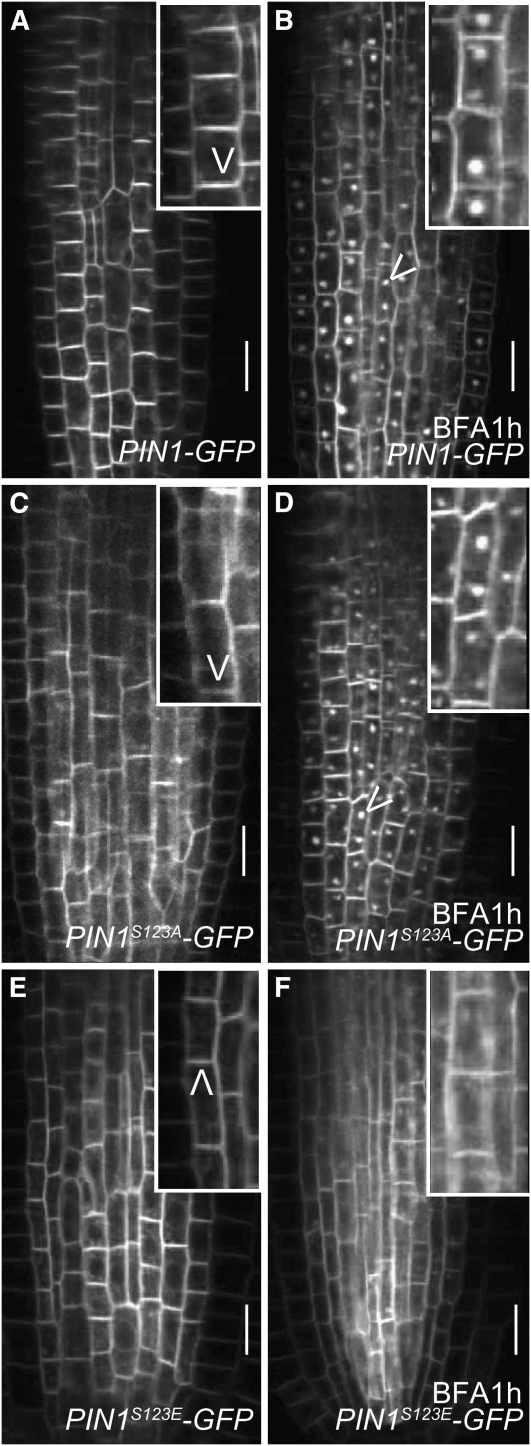
Next, we examined whether reduced PIN recruitment to the GNOM-dependent basal targeting pathway is directly determined by its PID-dependent phosphorylation state. Recently, the phosphorylation sites in PIN1 that are target of PID have been identified and verified with unphosphorylated PIN1S123A-GFP or phosphorylation-mimicking PIN1S123E-GFP constructs (F. Huang, M. Kemel-Zago, A. van Marion, C.G. Ampudia, and R. Offringa, unpublished data). Phosphomimicking PIN1S123E-GFP showed largely BFA-insensitive trafficking (Figures 7E and 7F) compared with PIN1-GFP (Figures 7A and 7B), suggesting that phosphorylated PIN proteins get recruited to a GNOM-independent pathway. By contrast, nonphosphorylated PIN1S123A-GFP localization remained sensitive to BFA (Figures 7C and 7D). In agreement with the differential recruitment to the GNOM-dependent basal or GNOM-independent apical pathways, PIN mutant proteins mimicking constitutive phosporylation (PIN1S123E-GFP) or dephosphorylation (PIN1S123A-GFP) showed a preferential apical or basal localization, respectively (Figures 7C and 7E) (F. Huang, M. Kemel-Zago, A. van Marion, C.G. Ampudia, and R. Offringa, unpublished data).
Figure 7.
BFA-Independent PIN Targeting by Phosphorylation-Based Sequence Modification.
(A) to (F) Confocal live cell imaging of PIN1-GFP variants. Bars = 10 μm.
(A) and (B) PIN1-GFP localization in the root (untreated in [A]) is sensitive to BFA treatment, leading to PIN1-GFP accumulation in BFA compartments (B).
(C) and (D) Nonphosphorylated PIN (PIN1S123A-GFP) targeting in untreated (C) and BFA-treated seedlings (D).
(E) and (F) Phosphorylation-mimicking PIN1S123E-GFP localization in untreated (E) and BFA-treated seedlings (F), indicating that the phosphorylated PIN proteins preferentially traffic in a BFA-resistant manner.
This finding indicates that the PID-dependent PIN phosphorylation state determines the differential PIN recruitment to ARF-GEF–dependent pathways. Nonphosphorylated PIN proteins are preferential cargos for the BFA-sensitive ARF-GEF GNOM-dependent basal polar targeting pathway. By contrast, PID-dependent PIN phosphorylation (Michniewicz et al., 2007; F. Huang, M. Kemel-Zago, A. van Marion, C.G. Ampudia, and R. Offringa, unpublished data) reduces the affinity of PIN proteins for the basal GNOM-dependent recycling pathway, leading to BFA-insensitive apical targeting.
DISCUSSION
PID and GNOM Antagonistically Regulate PIN Polarity and Plant Development
The Ser/Thr kinase PID and the ARF-GEF GNOM are the most prominent regulators of polar PIN targeting identified to date. PID plays a decisive role in apical-basal polar PIN targeting and phosphorylates PIN proteins at specific sites (Friml et al., 2004; Michniewicz et al., 2007; F. Huang, M. Kemel-Zago, A. van Marion, C.G. Ampudia, and R. Offringa, unpublished data). The mechanism by which the PID-dependent PIN phosphorylation regulates polar PIN delivery and its relation to the GNOM-dependent PIN subcellular trafficking has been elusive so far.
In a remarkable analogy, PID gain-of-function and gnom loss-of-function lead to ectopic apical PIN localization in Arabidopsis root cells (Friml et al., 2004; Kleine-Vehn et al., 2008a), resulting in auxin depletion from the root tip and root meristem collapse. By contrast, pid loss-of-function and GNOM activity favor basal PIN targeting (Friml et al., 2004; Kleine-Vehn et al., 2008a). In agreement with the subcellular phenotype, the developmental defects in gnom partial loss-of-function mutants and PID gain-of-function lines were similar. Lateral root primordia spacing and leaf vascular development were altered in both, and both showed collapse of the primary root meristem. Furthermore, PID overexpression in gnom partial loss-of-function mutants induced phenotypes reminiscent of the gnom complete loss-of-function phenotypes, further suggesting that PID might affect GNOM-dependent processes. Our data indicate that GNOM and PID are part of the same mechanism for the polar PIN delivery because, intriguingly, they interact synergistically in gain-of-function and loss-of-function mutants, implying that they regulate the PIN polarity in an antagonistic manner.
PID Induces PIN Sorting into a GNOM-Independent Pathway
Morphological and cellular phenotypes as well as genetic interaction revealed that PID and GNOM are part of the same mechanism that regulates polar PIN delivery. However, PID does not appear to alter either GNOM localization or its activity. PID kinase gain of function or PP2A phosphatase partial loss of function reduces the BFA sensitivity of PIN1 localization at the apical plasma membrane, suggesting that PIN phosphorylation promotes its BFA-insensitive, GNOM-independent targeting. This assumption was confirmed by site-directed mutagenesis of PID-targeted phosphorylation sites in PIN1 (F. Huang, M. Kemel-Zago, A. van Marion, C.G. Ampudia, and R. Offringa, unpublished data), where phosphomimicking versions of PIN1-GFP show a largely BFA-insensitive trafficking. These observations correlate with previous findings that increased PID kinase or decreased PP2A phosphatase activities (Friml et al., 2004; Michniewicz et al., 2007) or phosphorylation-mimicking mutations (F. Huang, M. Kemel-Zago, A. van Marion, C.G. Ampudia, and R. Offringa, unpublished data) promote the presence of PIN proteins at the apical polar domain. Consistent with this, the PID-dependent PIN phosphorylation leads to PIN protein recruitment into a GNOM-independent trafficking pathway and finally to apical PIN delivery.
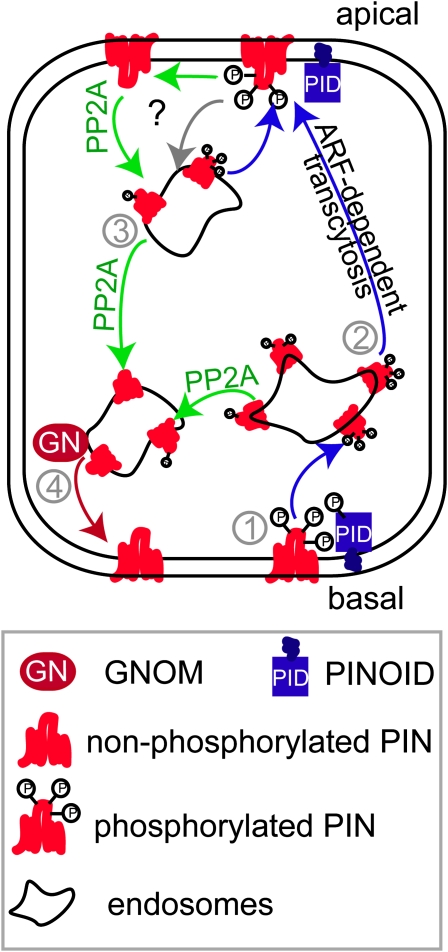
Proposed Model for PID/GNOM-Dependent Polar PIN Transcytosis
Polar sorting of cargos as we learned from animal models typically takes place at the endosomal level in so-called sorting endosomes (Farr et al., 2009). However, the PID localization is most pronounced at the plasma membrane, implying that PID does not directly function at the place of polar PIN sorting. Consequently, PID might not affect the polar targeting of de novo synthesized and, presumably, apolarly secreted PIN proteins (Dhonukshe et al., 2008). We propose that PID regulates the PIN localization upstream of its polar sorting by phosphorylating PIN proteins at the plasma membrane. Following their internalization into endosomes, the phosphorylated PIN proteins have a reduced affinity for the GNOM-dependent basal recycling pathway (Figure 8). As a result, the phosphorylated PIN proteins are recognized and, to a large extent, recruited to the apical GNOM-independent trafficking pathway that still might require ARF1-type proteins (Kleine-Vehn et al., 2008b). This alternative recruitment leads to basal-to-apical PIN transcytosis (Figure 8). The close PID homologs PID2, WAG1, and WAG2 kinases might similarly contribute to polar PIN resorting during plant development, but the redundancy or specificity of these components still needs to be investigated.
Figure 8.
Model of PID and GNOM-Dependent Intracellular PIN Sorting.
PID might phosporylate PIN proteins preferentially at the plasma membrane (1). Phosporylated PIN proteins get internalized into endosomes but fail to get sorted to the ARF-GEF GNOM-dependent basal recycling pathway (4). Phosphorylated PIN proteins display an enhanced affinity for a GNOM-independent, but ARF-dependent, apical targeting pathway, eventually leading to basal-to-apical PIN transcytosis (2). PP2AA function can counteract (3) the PID-dependent PIN phosphorylation, leading to GNOM-dependent (GN) basal recycling of PIN proteins (4).
[See online article for color version of this figure.]
PID activity could be counteracted by the PP2A phosphatase activity that eventually restores the PIN recruitment to the GNOM-dependent basal recycling pathway. Since PP2AA components can act on multiple, most likely also PIN-unrelated, substrates (Zhou et al., 2004) and is broadly distributed within the cell (Michniewicz et al., 2007), its place and mode of action in PIN polar targeting is still unclear.
The proposed mechanism for PID- and GNOM-dependent polar PIN targeting seems to be functionally important for plant development because PIN proteins undergo dynamic polarity alterations during various developmental processes, such as gravitropism, lateral root primordium formation, and leaf vascular development (Friml et al., 2002b; Benková et al., 2003; Heisler et al., 2005; Sauer et al., 2006; Scarpella et al., 2006). These processes require both GNOM and PID activity, suggesting that they are both involved in dynamic PIN polarity alterations. We hypothesize that PID/PP2A-dependent PIN phosphorylation or dephosphorylation might induce rapid PIN polarity changes during plant development via a GNOM-dependent and GNOM-independent transcytosis-like mechanism (Figure 8).
Polar targeting and transcytosis of polar plasma membrane cargos have been extensively studied in animal epithelial cells. A prominent example is albumin that transcytoses from the luminal (apical) to the abluminal (basolateral) sides of the cell (Hu and Minshall, 2009). Intriguingly, GP60, a plasma membrane–located albumin binding protein, becomes phosphorylated at the plasma membrane after its binding to albumin (Tiruppathi et al., 1997). Subsequently, a SRC kinase–dependent signaling cascade initiates the internalization of the GP60/albumin at the apical side of the cell, leading to its transcytosis and exocytosis at the basolateral side (Hu and Minshall, 2009).
The question remains whether the phosphorylation of GP60 in animal cells, in analogy to the PIN phosphorylation in plants, negatively affects the recycling of albumin-bound GP60 back to the apical side of the cell, subsequently promoting its translocation to the basolateral side by endosomal resorting. Similarly, it is tempting to speculate that after PID-dependent PIN phosphorylation, an SRC-like signaling cascade could also regulate the rate of PIN internalization in plant cells. Hence, future research will unravel whether spatial and temporal PID kinase activity also contributes to other processes, such as the regulation of PIN endocytosis/exocytosis rates at the different sides of the cell.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana (ecotype Columbia or Landsberg erecta) plants and seedlings were grown in growth chambers under long-day conditions (16 h light/8 h darkness) at 23°C. Experiments were performed on 5-d-old seedlings grown on vertically oriented plates containing Arabidopsis medium (AM) consisting of half-strength Murashige and Skoog agar and 1% sucrose (pH 5.8). Seedlings were incubated in AM medium supplemented with 50 μM BFA in 24-well cell culture plates for 1 h (except that Figures 6E to 6G seedlings are incubated for 3 or 12 h as indicated in the figure legend). BFA-germinated seedlings were treated with 25 μM (used in Figures 2A to 2E). Latrunculin B at 20 μM (Figure 1) and wortmannin at 30 μM (Figure 4) was similarly applied for 3 h. Control treatments contained equal amounts of solvent (dimethylsulfoxide). For in vivo analysis of the development of individual lateral root primordia, 3- to 5-d-old seedlings were transferred to slides with a thin layer of half-strength Murashige and Skoog medium with 0.5% agarose, supplemented with NAA (1 μM; 48 h), and incubated 24 to 48 h in a humid chamber. Chloralhydrate was used for tissue clearing in Figures 2I to 2P. The Axio imager (Carl Zeiss Micoimaging) was used for microscopy. Primary root collapse was scored by root curling and growth arrest. For all comparisons, independent experiments were done at least in triplicate with the same significant results. Representative images are shown. The following mutants and transformants have been described previously: GNOM-MYC (Geldner et al., 2003; used in Figure 4); gnomR5 (Geldner et al., 2004; used in Figures 2 and 3); DR5revPro:GFP (Friml et al., 2003; used in Figure 2); PIN1-GFP (Benková et al., 2003; used in Figure 7); piden197 and 35SPro:PID-21 (Benjamins et al., 2001; used in Figures 2 and 3); pp2aa1 (used in Figure 6), pp2aa1 pp2aa3 (used in Figure 5), and PID-YFP (used in Figure 1) (Michniewicz et al., 2007); and PIN1S123A-GFP and PIN1S123E-GFP (F. Huang, M. Kemel-Zago, A. van Marion, C.G. Ampudia, and R. Offringa, unpublished data; used in Figure 7).
Expression and Localization Analyses
Whole-mount immunofluorescence preparations were done as described (Friml et al., 2003). Antibodies were diluted as follows: anti-PIN1 (1:2000) (Paciorek et al., 2005; used in Figure 5), anti-PIN2 (1:2000) (Abas et al., 2006; used in Figure 6), anti-GFP (1:300; Molecular Probes; used in Figure 1), anti-ARF1 (1:1000) (Pimpl et al., 2000; used in Figure 4), and anti-c-myc (1:500; Roche Diagnostics; used in Figure 4). FITC- and CY3-conjugated anti-rabbit secondary antibodies (Dianova) were diluted 1:500 and 1:600, respectively; GFP was visualized in 5% glycerol without fixation (live cell imagining) and analyzed with a confocal laser scanning microscopy (TCS SP2; Leica Microsystems; LSM 710; Carl Zeiss Micoimaging). Images were processed in Adobe Photoshop and Illustrator cs2. Statistical analyses were done with Office Excel 2003 (Microsoft).
Accession Numbers
Sequence data from this article can be found in The Arabidopsis Information Resource (http://www.arabidopsis.org/) or GenBank/EMBL databases under the following accession numbers: ARF1 (AT1G23490), GNOM (AT1G13980), PID (AT2G34650), PIN1 (AT1G73590), PIN2 (AT5G57090), PP2AA1 (AT1G25490), and PP2AA3 (AT1G13320).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. BFA-Induced Changes in the Primary Root Meristem.
Supplemental Figure 2. Evaluation of Anti-ARF1 and GNOM-MYC Colocalization.
Supplemental Figure 3. BFA-Dependent PIN Localization in Various Mutant Backgrounds.
Supplementary Material
Acknowledgments
We thank David Robinson for generously donating antibody-based subcellular markers, Gerd Jürgens and Ben Scheres for sharing published material, Martine De Cock for help in preparing the manuscript, and the European Arabidopsis Stock Centre for seed stock supply. This work was supported by grants from the EMBO Young Investigator Program (to J.F.) and the Odysseus Programme of the Research Foundation-Flanders (to J.F.), by a Chinese Science Council scholarship (to F.H.), and by the Japanese Society for Plant Sciences (to S.N.). J.K.-V. is indebted to the Friedrich Ebert Stiftung for a personal fellowship.
The author responsible for distribution of materials integral to the findings presented in the article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Jiří Friml (jiri.friml@psb.vib-ugent.be).
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Abas, L., Benjamins, R., Malenica, N., Paciorek, T., Wiśniewska, J., Moulinier-Anzola, J.C., Sieberer, T., Friml, J., and Luschnig, C. (2006). Intracellular trafficking and proteolysis of the Arabidopsis auxin-efflux facilitator PIN2 are involved in root gravitropism. Nat. Cell Biol. 8 249–256. [DOI] [PubMed] [Google Scholar]
- Benjamins, R., Quint, A., Weijers, D., Hooykaas, P., and Offringa, R. (2001). The PINOID protein kinase regulates organ development in Arabidopsis by enhancing polar auxin transport. Development 128 4057–4067. [DOI] [PubMed] [Google Scholar]
- Benková, E., Michniewicz, M., Sauer, M., Teichmann, T., Seifertová, D., Jürgens, G., and Friml, J. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115 591–602. [DOI] [PubMed] [Google Scholar]
- Chow, C.-M., Neto, H., Foucart, C., and Moore, I. (2008). Rab-A2 and Rab-A3 GTPases define a trans-Golgi endosomal membrane domain in Arabidopsis that contributes substantially to the cell plate. Plant Cell 20 101–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen, S.K., Dagenais, N., Chory, J., and Weigel, D. (2000). Regulation of auxin response by the protein kinase PINOID. Cell 100 469–478. [DOI] [PubMed] [Google Scholar]
- daSilva, L.L.P., Taylor, J.P., Hadlington, J.L., Hanton, S.L., Snowden, C.J., Fox, S.J., Foresti, O., Brandizzi, F., and Denecke, J. (2005). Receptor salvage from the prevacuolar compartment is essential for efficient vacuolar protein targeting. Plant Cell 17 132–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhonukshe, P., Aniento, F., Hwang, I., Robinson, D.G., Mravec, J., Stierhof, Y.-D., and Friml, J. (2007). Clathrin-mediated constitutive endocytosis of PIN auxin efflux carriers in Arabidopsis. Curr. Biol. 17 520–527. [DOI] [PubMed] [Google Scholar]
- Dhonukshe, P., et al. (2008). Generation of cell polarity in plants links endocytosis, auxin distribution and cell fate decisions. Nature 456 962–966. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Donaldson, J.G., and Jackson, C.L. (2000). Regulators and effectors of the ARF GTPases. Curr. Opin. Cell Biol. 12 475–482. [DOI] [PubMed] [Google Scholar]
- Farr, G.A., Hull, M., Mellman, I., and Caplan, M.J. (2009). Membrane proteins follow multiple pathways to the basolateral cell surface in polarized epithelial cells. J. Cell Biol. 186 269–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml, J., Benková, E., Blilou, I., Wisniewska, J., Hamann, T., Ljung, K., Woody, S., Sandberg, G., Scheres, B., Jürgens, G., and Palme, K. (2002. a). AtPIN4 mediates sink driven auxin gradients and root patterning in Arabidopsis. Cell 108 661–673. [DOI] [PubMed] [Google Scholar]
- Friml, J., Vieten, A., Sauer, M., Weijers, D., Schwarz, H., Hamann, T., Offringa, R., and Jürgens, G. (2003). Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426 147–153. [DOI] [PubMed] [Google Scholar]
- Friml, J., Wiśniewska, J., Benková, E., Mendgen, K., and Palme, K. (2002. b). Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415 806–809. [DOI] [PubMed] [Google Scholar]
- Friml, J., et al. (2004). A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science 306 862–865. [DOI] [PubMed] [Google Scholar]
- Geldner, N., Anders, N., Wolters, H., Keicher, J., Kornberger, W., Muller, P., Delbarre, A., Ueda, T., Nakano, A., and Jürgens, G. (2003). The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 112 219–230. [DOI] [PubMed] [Google Scholar]
- Geldner, N., Friml, J., Stierhof, Y.-D., Jürgens, G., and Palme, K. (2001). Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature 413 425–428. [DOI] [PubMed] [Google Scholar]
- Geldner, N., Richter, S., Vieten, A., Marquardt, S., Torres-Ruiz, R.A., Mayer, U., and Jürgens, G. (2004). Partial loss-of-function alleles reveal a role for GNOM in auxin transport-related, post-embryonic development of Arabidopsis. Development 131 389–400. [DOI] [PubMed] [Google Scholar]
- Heisler, M.G., Ohno, C., Das, P., Sieber, P., Reddy, G.V., Long, J.A., and Meyerowitz, E.M. (2005). Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr. Biol. 15 1899–1911. [DOI] [PubMed] [Google Scholar]
- Hu, G., and Minshall, R.D. (2009). Regulation of transendothelial permeability by Src kinase. Microvasc. Res. 77 21–25. [DOI] [PubMed] [Google Scholar]
- Kleine-Vehn, J., Dhonukshe, P., Sauer, M., Brewer, P., Wiśniewka, J., Paciorek, T., Benková, E., and Friml, J. (2008. a). ARF GEF-dependent transcytosis and polar delivery of PIN auxin carriers in Arabidopsis. Curr. Biol. 18 526–531. [DOI] [PubMed] [Google Scholar]
- Kleine-Vehn, J., and Friml, J. (2008). Polar targeting and endocytic recycling in auxin-dependent plant development. Annu. Rev. Cell Dev. Biol. 24 447–473. [DOI] [PubMed] [Google Scholar]
- Kleine-Vehn, J., Langowski, L., Wiśniewska, J., Dhonukshe, P., Brewer, P.B., and Friml, J. (2008. b). Cellular and molecular requirements for polar PIN targeting and transcytosis in plants. Mol. Plant 1 1056–1066. [DOI] [PubMed] [Google Scholar]
- Kleine-Vehn, J., Leitner, J., Zwiewka, M., Sauer, M., Abas, L., Luschnig, C., and Friml, J. (2008. c). Differential degradation of PIN2 auxin efflux carrier by retromer-dependent vacuolar targeting. Proc. Natl. Acad. Sci. USA 105 17812–17817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi, K., Sugiyama, M., and Fukuda, H. (2000). A series of novel mutants of Arabidopsis thaliana that are defective in the formation of continuous vascular network: calling the auxin signal flow canalization hypothesis into question. Development 127 3197–3204. [DOI] [PubMed] [Google Scholar]
- Matsuoka, K., Bassham, D.C., Raikhel, N.V., and Nakamura, K. (1995). Different sensitivity to wortmannin of two vacuolar sorting signals indicates the presence of distinct sorting machineries in tobacco cells. J. Cell Biol. 130 1307–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michniewicz, M., et al. (2007). Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell 130 1044–1056. [DOI] [PubMed] [Google Scholar]
- Paciorek, T., Zažímalová, E., Ruthardt, N., Petrášek, J., Stierhof, Y.-D., Kleine-Vehn, J., Morris, D.A., Emans, N., Jürgens, G., Geldner, N., and Friml, J. (2005). Auxin inhibits endocytosis and promotes its own efflux from cells. Nature 435 1251–1256. [DOI] [PubMed] [Google Scholar]
- Petrášek, J., et al. (2006). PIN proteins perform a rate-limiting function in cellular auxin efflux. Science 312 914–918. [DOI] [PubMed] [Google Scholar]
- Pimpl, P., Movafeghi, A., Coughlan, S., Denecke, J., Hillmer, S., and Robinson, D.G. (2000). In situ localization and in vitro induction of plant COPI-coated vesicles. Plant Cell 12 2219–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer, M., Balla, J., Luschnig, C., Wišniewska, J., Reinöhl, V., Friml, J., and Benková, E. (2006). Canalization of auxin flow by Aux/IAA-ARF-dependent feed-back regulation of PIN polarity. Genes Dev. 20 2902–2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpella, E., Marcos, D., Friml, J., and Berleth, T. (2006). Control of leaf vascular patterning by polar auxin transport. Genes Dev. 20 1015–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevell, D.E., Leu, W.-M., Gillmor, C.S., Xia, G., Feldmann, K.A., and Chua, N.-H. (1994). EMB30 is essential for normal cell division, cell expansion, and cell adhesion in Arabidopsis and encodes a protein that has similarity to Sec7. Cell 77 1051–1062. [DOI] [PubMed] [Google Scholar]
- Steinmann, T., Geldner, N., Grebe, M., Mangold, S., Jackson, C.L., Paris, S., Gälweiler, L., Palme, K., and Jürgens, G. (1999). Coordinated polar localization of auxin efflux carrier PIN1 by GNOM ARF GEF. Science 286 316–318. [DOI] [PubMed] [Google Scholar]
- Tanaka, H., Dhonukshe, P., Brewer, P.B., and Friml, J. (2006). Spatiotemporal asymmetric auxin distribution: a means to coordinate plant development. Cell. Mol. Life Sci. 63 2738–2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, H., Kitakura, S., De Rycke, R., De Groodt, R., and Friml, J. (2009). Fluorescence imaging-based screen identifies ARF GEF component of early endosomal trafficking. Curr. Biol. 19 391–397. [DOI] [PubMed] [Google Scholar]
- Tiruppathi, C., Song, W., Bergenfeld, M., Sass, P., and Malik, A.B. (1997). Gp60 activation mediates albumin transcytosis in endothelial cells by tyrosine kinase-dependent pathway. J. Biol. Chem. 272 25968–25975. [DOI] [PubMed] [Google Scholar]
- Wiśniewska, J., Xu, J., Seifertová, D., Brewer, P.B., Růžička, K., Blilou, I., Rouquié, D., Benková, E., Scheres, B., and Friml, J. (2006). Polar PIN localization directs auxin flow in plants. Science 312 883. [DOI] [PubMed] [Google Scholar]
- Xu, J., and Scheres, B. (2005). Dissection of Arabidopsis ADP-RIBOSYLATION FACTOR1 function in epidermal cell polarity. Plant Cell 17 525–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., Nodzyński, T., Pěnčík, A., Rolčík, J., and Friml, J. (2009). PIN phosphorylation is sufficient to mediate pin polarity and direct auxin transport. Proc. Natl. Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed]
- Zhou, H.W., Nussbaumer, C., Chao, Y., and DeLong, A. (2004). Disparate roles for the regulatory A subunit isoforms in Arabidopsis protein phosphatase 2A. Plant Cell 16 709–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.