Abstract
O6-Methylguanine-DNA methyltransferase (MGMT), a ubiquitous DNA repair protein, reverses mutagenic and cytotoxic effects of O6-alkylguanine in DNA induced by chemotherapeutic N-alkyl N-nitrosourea and procarbazine type drugs by dealkylating the adduct. MGMT expression is down-regulated by wild-type p53 (WTp53) in human tumor cells. Here we report that p53 sequesters the Sp1 transcription factor to prevent its binding to the cognate cis elements in the MGMT promoter and thus inhibits MGMT expression. Sp1 overexpression abrogated the inhibitory effect of p53 on the MGMT promoter activity in a dose-dependent manner. Stable interaction of Sp1 with WTp53 was observed in HCT116 cells. Moreover, WTp53 overexpression reduced the binding of the nuclear extract to the Sp1 consensus sequence, even though recombinant p53 alone did not bind to the same sequence. Taken together, these results suggest that sequestration of Sp1 could be one of the mechanisms by which p53 negatively regulates MGMT expression, thus enhancing sensitivity of tumor cells to O6-alkylguanine generating drugs.
Keywords: MGMT, p53, DNA repair, Sp1 transcription factor, drug resistance, alkylating agent
Alkylation of the O6-position of guanine in DNA, induced by methylating (temozolomide, procarbazine) and chloroethylating [carmustine (BCNU); nimustine, (ACNU)], is considered to be the most mutagenic and carcinogenic lesion (1, 2). Alkylating agents such as nitrogen mustards, procarbazine and nitrosoureas are among the most widely used chemotherapeutic agents. Mutagenic and cytotoxic adducts are removed from the O6 position of guanine by O6 methylguanine-DNA methyltransferase (MGMT) (3–6). MGMT is a ubiquitous DNA repair protein that acts in a stochiometric reaction in which an alkyl group attached to the O6 position of guanine is transferred to a specific cysteine residue in the protein’s active site [reviewed in (7, 8)]. This in situ dealkylation restores the original guanine in DNA while inactivating the protein MGMT (9). The MGMT level varies widely in both normal and tumor tissues and in tumor cell lines. Approximately 20% of in vitro transformed tumor cell lines have no detectable MGMT and are highly sensitive to methylating and chloroethylating agents; these are called Mer−/Mex− (10–14). Several studies showed that human MGMT down-regulation in human Mer−/Mex− cells was linked to the presence of methylated CpGs in the promoter region, while other studies indicated that regulation of MGMT expression was complex (15–18). Increased MGMT expression was observed in rat hepatoma cells after treatment with alkylating agents or UV light (19). One recent study showed that treatment with cysteine prodrugs and herbal antioxidant such as curcumin also increased the MGMT level in many cancer cells (20). Tumor cells with high levels of MGMT are known to have high resistance to BCNU, and depletion of MGMT activity by pseudosubstrates (such as O6-benzylguanine, O6-BG) reverses this resistance (4, 21, 22). Hence the level of MGMT expression could be a key determinant in tumor cell resistance to alkylating agents.
The MGMT promoter, like that of many housekeeping genes, does not have any TATA or CAAT boxes, but contain six putative Sp1-binding sequences, two glucocorticoid response-elements (GRE-1), and two activator protein-1 (AP-1) binding sequences (23–27). The involvement of Sp1, AP-1, GRE-1 in MGMT expression has been previously documented (26, 27). Interestingly, in spite of the lack of p53-binding sites in the MGMT promoter, several studies have explored the impact of tumor suppressor p53 on MGMT expression. P53 serves as a transcription factor, and has a wide range of functions in cell cycle regulation, cell cycle arrest, DNA repair and apoptosis through transactivation of specific genes in response to DNA damage (ionizing radiation, IR and UV light, chemotherapeutic agents) and other cellular stress signals (28, 29). P53 not only activates transcription of target genes (e.g. p21, GADD45) but also mediates repression of a wide range of viral and cellular gene promoters that do not contain consensus p53-binding sites (30–33). For example, p53 down-regulated basal expression of human MGMT and on the other hand, p53 was also shown to be required for MGMT up-regulation after ionizing radiation in rodent cells (34–36). Moreover, inverse correlation between MGMT expression and p53 in breast cancer and other tumor samples has been documented (37–39).
P53 is one of the most commonly mutated genes in human cancer and 50% or more of all human carcinomas have an inactive p53 allele and these tumors respond poorly to chemotherapy (40, 41). Interestingly, some mutant p53 proteins, which do not interact with p53 consensus binding sites, have been shown to up-regulate expression of certain genes (31). Therefore, altered gene expression resulting from p53 inactivation has significant consequences on cell cycle regulation and drug resistance. In spite of several studies showing the involvement of p53 as a negative regulator of MGMT in human tumor cells, the mechanism by which it regulates MGMT expression has not been elucidated. A clear understanding of this mechanism is essential for more effective use of alkylating agents in the treatment of tumors with wild-type (WT) or mutant p53.
Materials and Methods
Plasmids
The p53 expression vectors containing WT and mutant forms of p53 were kindly provided by Dr. A. J. Levine (University of Medicine and Dentistry of New Jersey, New Brunswick) and are described elsewhere (42). The Sp1 mammalian expression vector was a generous gift from Dr. R. Tijian (University of California, Berkeley). Generation of MGMT promoter-Luciferase reporter plasmids are described elsewhere (26).
Cell culture
The human colorectal adenocarcinoma line p53 null HCT116 P53−/− (a gift from Dr. B. Vogelstein, Johns Hopkins University School of Medicine) and human lung carcinoma Calu6 line (ATCC 2023CL) were grown in McCoy’s 5A and DMEM (Gibco Life Technologies, Carlsbad, CA, USA) medium supplemented with 10% fetal bovine serum (Sigma-Aldrich, St. Louis, Mo, USA), 100 U/ml penicillin, and 100 µg/ml streptomycin (Gibco Life Technologies) in a 5% CO2 incubator at 37°C.
Transfection and luciferase assay
HCT116 p53−/− cells were cotransfected with 500 ng of each MGMT-reporter plasmid and expression plasmid for WT or mutant (V143A and L22G, T23S) p53 or Sp-1 expression plasmids or equivalent amounts of empty vector using LipofectAMINE 2000 (GIBCO Life Technologies), according to the manufacturer’s instructions. At 48 h after transfection, cells were lysed with reporter lysis buffer (Promega, Madison, WI, USA) and the luciferase activity in the cell lysates was measured in a luminometer (AutoLumant LB593; Berthold, Oak Ridge, TN, USA) with a luciferase assay kit (Promega) according to the manufacturer’s protocol. We observed that CMV promoter-dependent β-galactosidase and thymidine kinase-promoter-dependent renilla-luciferase (pRL-TK; Promega) levels were affected due to overexpression of WT p53 for unknown reasons. We therefore could not use these reporter plasmids for normalizing transfection efficiency. The luciferase activity was normalized using the amount of protein in the lysates.
Preparation of cell extracts and Western blot analysis
Whole cell extracts were prepared as described elsewhere (43). Nuclear and cytosolic extracts were obtained as follows: Cells washed with cold phosphate-buffered saline were resuspended in buffer A, consisting of 10 mM [4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) (HEPES)], 10 mM KCl, 0.1 mM ethylenediaminetetraacetic acid (EDTA), 0.1 mM ethyleneglycol tetraacetic acid (EGTA), 1 mM dithiotheritol DTT, 0.5 mM phenylmethylsulphonyl fluoride (PMSF), and protease cocktail inhibitor tablet (Complete Mini; Roche, Nutely, New Jersey, USA) and allowed to swell on ice for 15 min. A volume of 2.5 µL of a 10% solution of Nonidet P40 was added to 400 µL cell suspension and the samples were vortexed vigorously for 10S. The homogenate was centrifuged for 30S. (20,800 x g at 4°C), and the supernatant (cytosol fraction) was separated. The pellets (nuclear fraction) were resuspended on 50 µL ice-cold buffer C (20 mM HEPES, 0.4 M NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, 1 mM PMSF, and Complete Mini-protease inhibitor tablets, and shaken vigorously at 4°C for 15 min and the supernatant (nuclear fraction) was collected. The samples were then centrifuged for 5 min (20,800 x g at 4°C). Extracts (nuclear and cytosolic fractions) were aliquoted and stored at −80°C. Cell extracts were subjected to SDS-PAGE (12.5% acrylamide) and transferred to a nitrocellulose membrane (Trans-Blot 0.2µm; Bio-Rad, Hercules, CA, USA). Western immunoblot analysis with mouse monoclonal anti-p53 antibody (Sc-DO-1 and PAb1801, (dilution 1:200) Santa Cruz Biotechnology, Santa Cruz, CA, USA); Pab421 (Biomol), Phospho Ser15-specific P53 antibody (Santa Cruz Biotechnology); rabbit polyclonal anti-Sp1 (dilution 1:2,000), and rabbit monoclonal anti-β-actin (Sigma; 1:1000 dilution) antibodies was carried out using enhanced chemiluminescence assay (ECL kit, Amersham Pharmacia, Piscataway, NJ, CA, USA). Lung carcinoma Calu6 cells were transfected with WT p53 expression plasmids or empty vector and at 48 h after transfection, cells were lysed and Western blot analysis was performed with polyclonal MGMT antibody as described elsewhere (25).
Co-immunoprecipitation
The nuclear extracts were immunoprecipitated with anti-Sp1 antibodies (Santa Cruz Biotechnology) or anti-FLAG M2 mouse antibody-conjugated agarose beads (Sigma) for 3 h. Immunoprecipitates of control and Sp1-FLAG or p53-FlAG-transfected cells were washed four times with TBS (50 mM Tris-HCl, pH 7.5; 150 mM NaCl) and boiled with Laemmli buffer (2% SDS and 100 µM β-mercaptoethanol). The supernatants were subjected to SDS-PAGE followed by Western blot analysis with mouse monoclonal anti-p53 (PAb1801) and anti-FLAG M2 antibodies.
Electrophoretic shift mobility analysis (EMSA)
EMSA was performed as described elsewhere (44) with some modifications.
The 5′ 32P-labeled’ oligo 5´-GCCCCGGCCCCGCCCCCGCGCG- 3´ (containing an Sp1-binding site present in the MGMT promoter) and 5´-GCTATGCGTTATTGAGCACGCG- 3´containing mutated (underlined) Sp-1 site were annealed with appropriate complementary stands to generate duplex oligo. The DNA (50 fmol) was then incubated with 10 ng p53 or nuclear extracts (5 µg) isolated from HCT116 p53−/− cells for 20 min at 25°C in a buffer containing 40 mM HEPES-KOH, pH 7.5, 50 mM KCl, 1 mM MgCl2, 0.5 mM EDTA, 0.5 mM DTT, 10% glycerol and 1 µg of poly (dI-dC) (Sigma). After electrophoresis in nondenaturing 5% polyacrylamide gels in Tris-borate buffer at 4°C, the gel was dried and the radioactivity quantitated by PhosphorImager (Molecular Dynamics, Sunnyvale, CA, USA) and analyzed using IMAGEQUANT software (Piscataway, NJ, USA).
Clonogenic survival assay
Colony-forming assays were performed as previously described (45). Briefly, exponentially growing glioblastoma p53-null LNZ308 and its derivative 2024 cells (46) seeded at a density of 250 to 300 cells/60-mm dish were allowed to attach. After 24 h, the cells were treated with 1 µg/ ml doxicycline for another 24 h and then exposed to increasing concentrations of temozolomide (TMZ) for 30 min, then transferred back to the normal medium. After 2 weeks, the colonies were fixed and stained with a crystal violet/formalin solution and counted for plotting the survival curve.
Results
Effect of overexpression of WTp53 on MGMT promoter activity
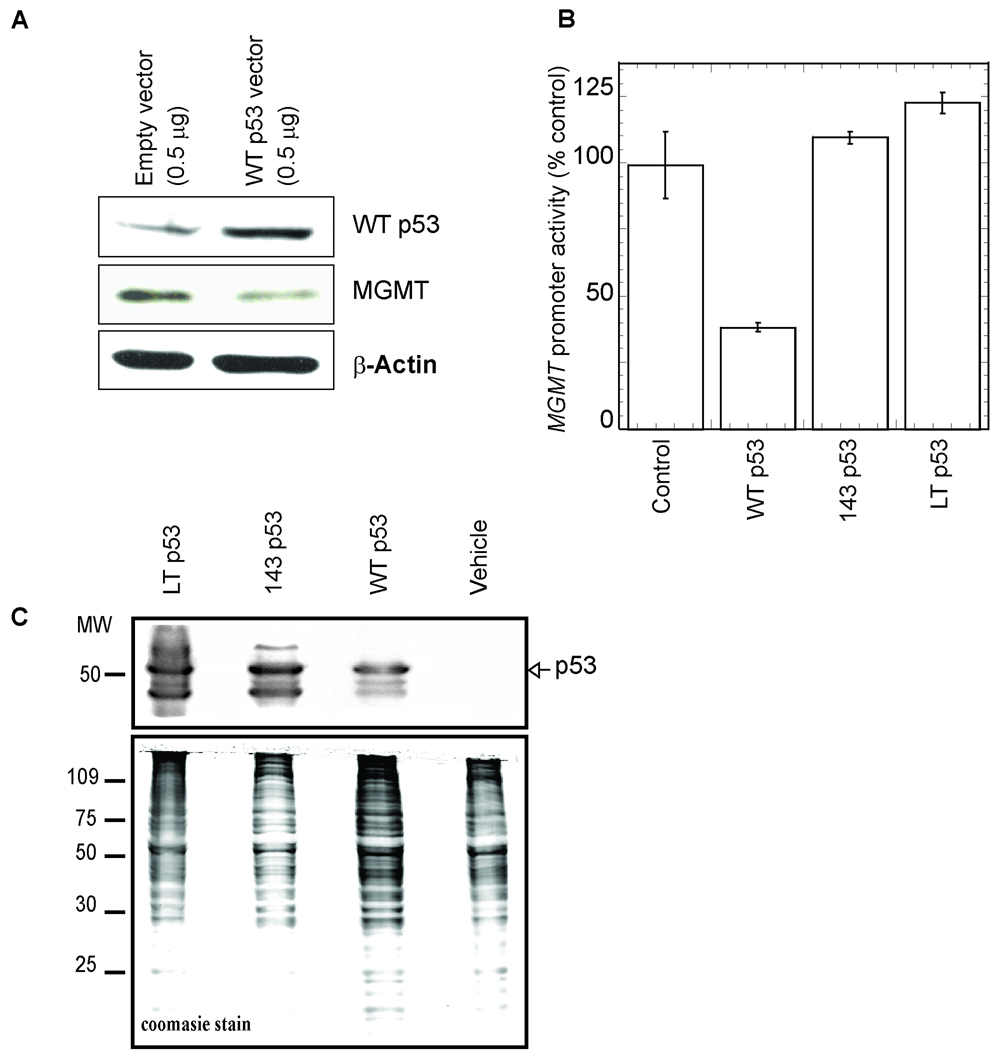
An earlier study showed that WT p53 induction down-regulated endogenous MGMT expression in several human tumor cell lines, including two isogenic p53-null cell lines (36). Consistent with this, we observed that overexpression of WT p53 down-regulated endogenous MGMT expression in the human pulmonary lung carcinoma Calu6 line (Figure 1A). To further investigate the effect of p53 on the MGMT promoter, we used a p53-null colon carcinoma cell line, HCT116 p53−/−. We transiently co-transfected HCT116 p53−/− cells with MGMT promoter-reporter (containing the 954 bp region upstream of the transcription initiation site) and WT p53 expression plasmids. Ectopic expression of WT p53 significantly reduced MGMT promoter activity (70% as compared to vector transfected control; Figure 1B). We also examined the effects of two p53 point mutants on MGMT promoter activity namely, Val143Ala (V143A), a missense mutation which inactivates p53 sequence-specific DNA-binding, and a Leu22Gln Trp23Ser (p53LT) double mutant which retains the ability to bind p53-cis sequences but lacks transcriptional activation ability (42). While WT p53 overexpression significantly reduced MGMT promoter-dependent luciferase expression, ectopic expression of the p53 mutants had no significant inhibitory effect (Figure 1B). Western analysis of p53−/− cell lysates, transiently transfected with p53 expression plasmids, revealed that the levels of mutant p53 were higher (2-fold; Figure 1C) than that of WT p53. These results indicate that in spite of the higher level, mutant-p53 could not inhibit the MGMT promoter.
Figure 1.
A, Calu6 cells were transfected with expression plasmids for WT p53 or empty vector. At 48 h after transfection, the MGMT level in the cell extracts was analyzed by Western blotting with MGMT antibody. B, Inhibition of MGMT promoter activity by WT but not mutant p53. HCT116 p53−/− cells were transiently transfected with (0.5 µg) of MGMT promoter-luciferase plasmid (−954/+24) and 0.01 µg of WT p53 or mutant p53, or equivalent amount of empty vector. Luciferase activity was measured at 48 h after transfection and normalized for the amount of protein. The mean ± S.D. of five independent experiments performed in duplicate is shown. C, Western analysis of the p53 level in the transfected cell extracts used in (B).
Effect of p53 on the minimal MGMT promoter
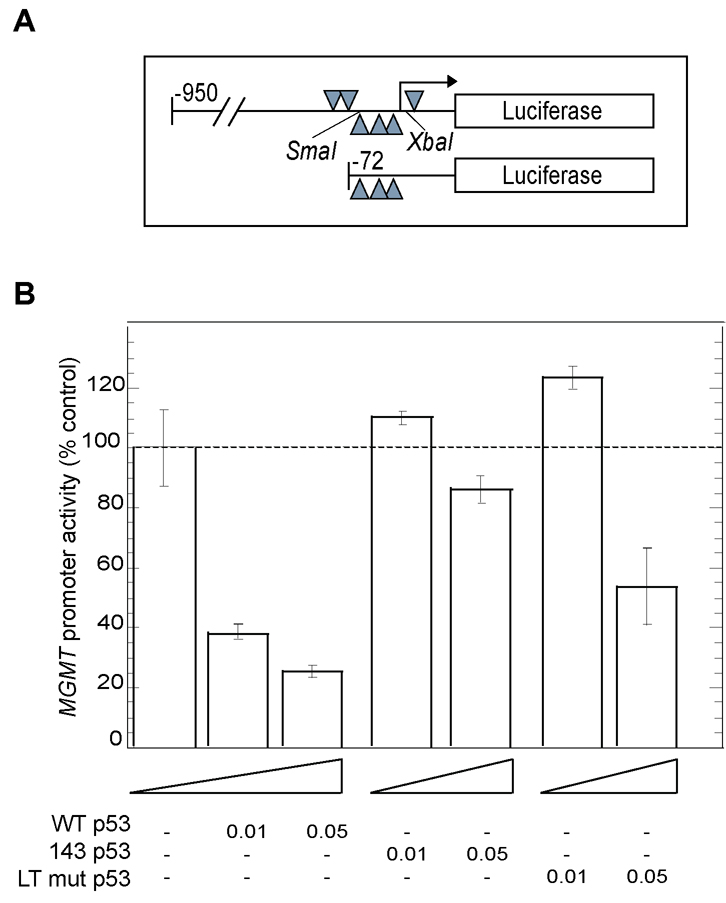
A 72 bp region upstream of the MGMT gene transcription initiation site, containing three Sp1-binding sites, was previously shown to be the minimal sequence required for reporter expression (26)(Figure 2A). To test whether WT p53 is also able to inhibit the minimal MGMT promoter, we transiently co-transfected HCT116 p53−/− cells with minimal MGMT promoter-luciferase reporter and WT- or mutant-p53 expression plasmids. The minimal MGMT promoter was down-regulated by WT p53 in a dose-dependent manner (up to 80% compared to vector transfected controls). While overexpression of the p53LT mutant did not significantly down-regulate MGMT promoter activity compared to WT p53 (although moderate down-regulation was observed at a higher dose), V143A mutant had no inhibitory effect on MGMT promoter activity, but rather slight activation of the MGMT promoter was observed (Figure 2B). These suggest that WT p53-mediated inhibition of MGMT occurs via the Sp1 transcription factor.
Figure 2. Effect of p53 on the minimal MGMT promoter.
A, Schematic diagram of the MGMT promoter 5´ regulatory region showing the location of Sp1-binding sites cloned upstream of the luciferase coding region. B, HCT116 p53−/− cells were transiently transfected with (0.5 µg) of minimal MGMT promoter-luciferase plasmid (−72/+24) and various amounts of WT p53 expression plasmid or mutant p53, or equivalent amount of empty vector. Luciferase activity was measured at 48 h after transfection and normalized for the amount of protein. The mean ± S.D. of five independent experiments performed in duplicate is shown.
P53-mediated inhibition of MGMT is relieved by overexpression of Sp1
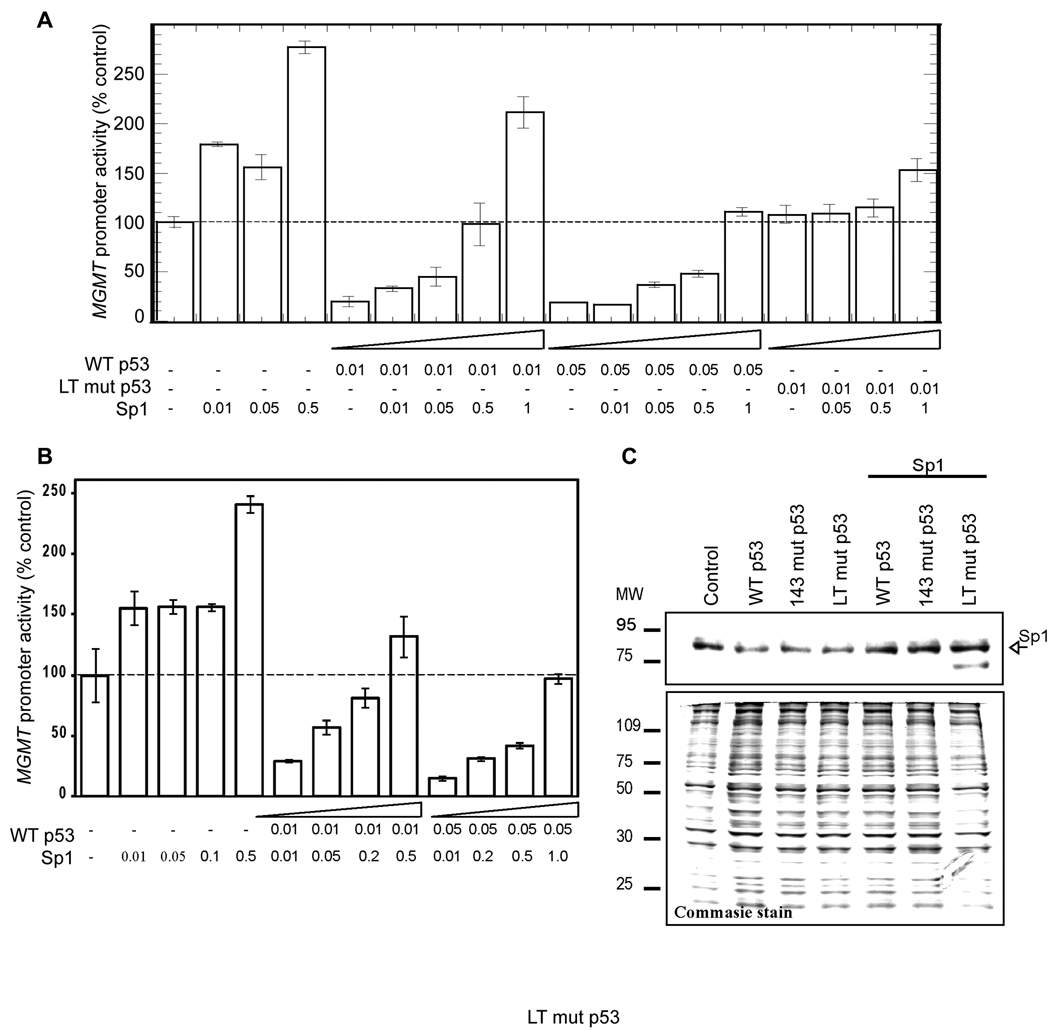
To investigate whether p53-induced down-regulation of MGMT could be overcome by ectopic overexpression of Sp1, we co-transfected HCT116 p53−/− cells with WT p53 and Sp1 expression plasmids together with MGMT promoter-luciferase plasmid. We observed that Sp1 overexpression relieved WT p53-induced down-regulation of MGMT promoter activity in a dose-dependent manner (Figure 3A). To further confirm that p53-induced down-regulation of MGMT was achieved by regulation of Sp1-binding to the MGMT promoter, we used the minimal MGMT promoter plasmid containing only three Sp1-binding sites proximal to the transcription initiation site. Sp1 overexpression abrogated p53-mediated inhibition of the minimal MGMT promoter suggesting possible interference by p53 of endogenous Sp1 binding to the MGMT promoter (Figure 3B). Western analysis using Sp1-specific antibody in HCT116 p53−/− cells, showed that overexpression of WT p53 or p53 mutants did not affect endogenous or ectopic Sp1 expression (Figure 3C).
Figure 3. Effect of overexpression of Sp1 on P53-mediated inhibition of MGMT promoter.
HCT116 p53−/− cells co-transfected with WT p53 (0.01 µg or 0.05 µg) and various amounts of Sp1 expression plasmids together with the longer (−954/+24; A) or the minimal (−72/+24; B) MGMT promoter-luciferase plasmid. Luciferase activity was measured at 48 h after transfection and normalized for the amount of protein. The mean ± S.D. of five independent experiments performed in duplicate is shown. C, Western analysis of the Sp1 level in the extracts of HCT116 p53−/− transfected with WT or mutant p53 expression plasmids.
Effect of p53 expression on Sp1 binding to its consensus sequence on the MGMT promoter
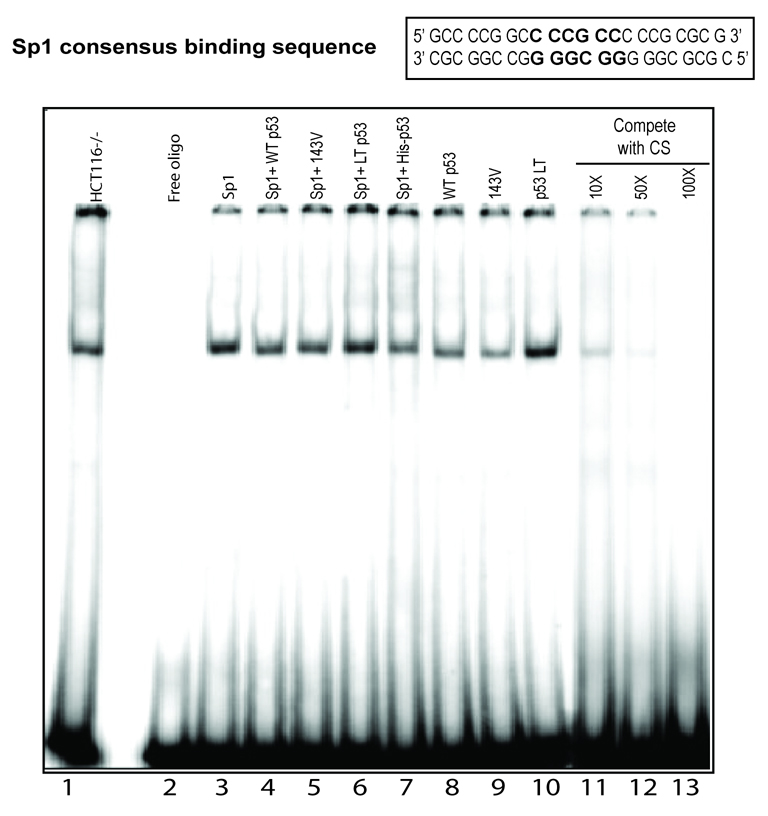
The observation that Sp1 overexpression overcame the p53-mediated down-regulation of MGMT raised two possibilities: namely, either p53 competes with Sp1 for binding to one or more of the Sp1 consensus binding sites in the MGMT promoter, or p53 sequesters Sp1 thereby reducing its availability for binding to the MGMT promoter. To test whether p53 interferes with Sp1 binding to its consensus site in the MGMT promoter, we performed EMSA using a consensus Sp1-binding sequence present in the MGMT promoter. Significant reduction in the binding of the nuclear extract of HCT116 p53−/− cells with ectopic overexpression of WT p53 was observed (Figure 4; compare lane 1 versus 8). In contrast, overexpression of the p53LT mutant had no effect on the Sp1 binding (compare lane 1 versus 10). Interestingly, overexpression of the V143A mutant also showed reduced Sp1 binding (lane 9). As expected, ectopic overexpression of Sp1 increased the binding of nuclear extract (compare lane 1 versus 3), however coexpression of Sp1 with WT p53 significantly reduced the binding (compare lane 3 versus 4) while the p53LT mutant had no significant effect (lane 6). It is worth noting that the recombinant p53 polypeptide alone did not bind to the Sp1 consensus oligo (data not shown). However, addition of recombinant His-tagged p53 protein to the nuclear extract inhibited the binding to the Sp1 oligo (compare lane 3 versus 7). The specificity of the banding was tested by competition with cold oligonucleotides containing the Sp1 consensus binding sequence (lanes 11–13).
Figure 4. Effect of p53 expression on Sp1 binding.
Representative EMSA with nuclear extracts of p53−/− cells transfected with empty vector or WT p53 expression plasmid using 32P-labeled duplex oligo containing a SP1 sequence in the MGMT promoter.
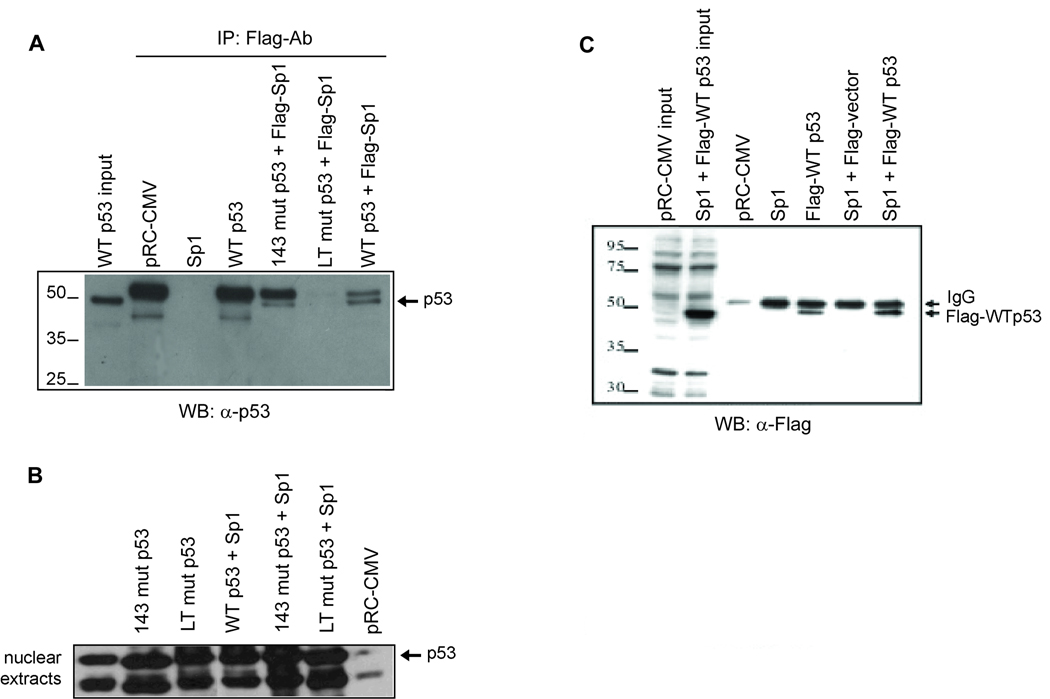
Stable interaction of WT p53, but not mutant with Sp1 in HCT 116 cells
WT p53 and some of its mutants are known to form heterocomplexes with Sp1 (47). Because direct interaction between p53 and Sp1 has been reported for the WT and some mutant forms of p53, we decided to investigate whether the differential effects of WT and mutant p53 on MGMT promoter activity were due to variable affinity for interactions with Sp1. To test this, we performed co-immunoprecipitation analysis of p53 in extracts of HCT116 p53−/− cells transfected with WT or mutant p53 and FLAG-Sp1 expression plasmids. Western analysis of the immunoprecipitate with p53 antibody showed the presence of WT p53 but not the p53LT mutant in the Sp1 immunoprecipitate (Figure 5A). Comparison of the p53 level in the starting nuclear extracts (Figure 5B) indicated that V143A p53 mutant also interacted with Sp1, although not with similar affinity to the WT p53. Together, these results indicate that Sp1 interacts strongly with WTp53 but not the p53LT mutant. To further confirm these results, we performed reciprocal co-immunoprecipitation with Sp1 antibody and showed the presence of FLAG-tagged WT p53 in the Sp1 immunoprecipitate (Figure 5C). Taken together, these results strongly suggest that WT p53 but not its p53LT mutant interacts with Sp1 in vivo.
Figure 5. Stable interaction of WT p53, but not mutant p53 with Sp1.
A, Extracts of HCT116 p53−/− cells co-transfected with Sp1-FLAG and WT or mutant p53 expression plasmids or empty vector were immunoprecipitated with FLAG antibody and the immunoprecipitates were analyzed for the presence of p53 by Western analysis with a p53 antibody. B, Western analysis of the p53 levels in the nuclear extracts used for immunoprecipitation. C, Extracts of HCT116 p53−/− cells co-transfected with Sp1 expression plasmid and FLAG-tagged WT p53 expression plasmids or empty vector were immunoprecipitated with Sp1 antibody and the immunoprecipitates were analyzed for the presence of p53 by Western analysis with a FLAG antibody.
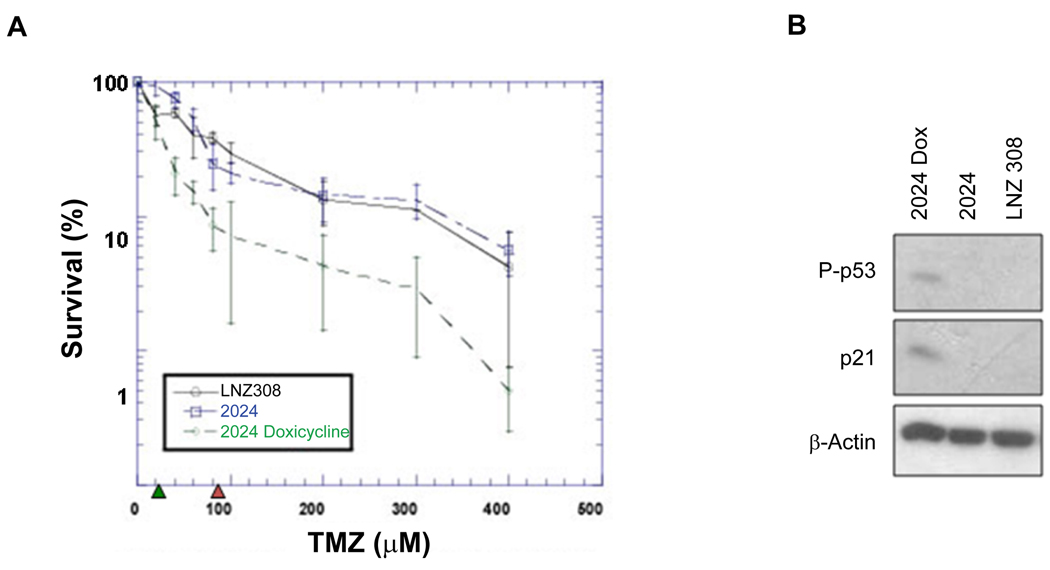
Induction of WT p53 enhances sensitivity of glioblastoma cells to the alkylating agent temozolomide
Because MGMT-mediated repair of O6-alkylguanine prevents the formation of cytotoxic lesion induced by many alkylating agents, we examined the effect of p53-mediated down-regulation of MGMT level on cell sensitivity to temozolomide (TMZ) which produces O6-methylguanine adduct. We have used human glioblastoma p53-null LNZ308 cell line and its derivatives isogenic 2024 cell line carrying a doxicycline (Dox)-inducible p53 expression system (46). Induction of p53 in 2024 cells with Dox enhances phosphorylated p53 level which concomitant with p21 expression (Figure 6B). We determined the cologenic survival of these cells after exposure of increasing doses of TMZ. A four-fold reduction of IC50 (20 µM) for TMZ was observed after induction of p53 with Dox in 2024 cells compared to noninduced 2024 (IC50, 80 µM) or parental p53-null LNZ308 cells (Figure 6A). These data indicate that changes in the p53 level alter the sensitivity to alkylating agent.
Figure 6. Induction of WT p53 sensitizes human tumor cells to the alkylating agent temozolomide.
A, Clonogenic survival assay of the human glioblastoma LNZ308 p53-null cells and its derivates 2024 cells in the p53-induced (+DOX) and p53-uninduced (−DOX) states after exposure to different concentrations of temozolomide. The data points show the average ± SD values of three independent experiments performed in triplicates dishes. B, Western analysis of the phosphorylated (Ser15) p53, p21 levels in the extracts of cells used in A.
Discussion
MGMT expression ranges from a high level in some tumor cells to an undetectable level in Mex− tumor cells (10, 13, 14). The level of MGMT expression plays a decisive role in protection of cells from toxicity to alkylating agents (4). Elucidating the molecular basis for this variation of expression is extremely important from both basic and clinical perspectives. In addition to its function in cell cycle regulation, p53 plays an important role in the induction of apoptosis after genotoxic damage. Because p53 plays a central role in sensing DNA damage and promoting DNA repair, the possibility that MGMT may be regulated by p53 had received much attention. Several previous studies showed negative regulation of MGMT expression by WT p53. This study confirms and further expands the initial observation by Harris et al. and others that WT p53 down-regulates human MGMT expression (34–36). Consistent with this, Srivenugopal et al. (36) conclusively showed that tetracycline regulated overexpression of p53 protein in a p53-null MGMT-proficient human lung tumor cells (H1299) curtails the transcription of the endogenous MGMT gene and alters their sensitivity to alkylating agents (36). Down-regulation of endogenous MGMT expression by WT p53 was further demonstrated in isogeneic p53 models. These included the H460 human lung cancer cell line, in which p53 function had been disrupted by the E6 papillomavirus oncoprotein, and GM47.23 glioblastoma cell line in which the stably integrated WT p53 gene could be activated by dexamethasone. Consistent with this, we have shown that overexpression of WT p53 also down-regulated endogenous MGMT level in lung cancer Calu6 cells. However, the mechanism by which p53 controls MGMT expression is not known. In this report we provide the first evidence that WT p53 sequesters Sp1 and abrogates MGMT expression and thus provides a molecular basis for WT p53-mediated MGMT regulation.
We have shown that ectopic expression of WT p53 but not mutant p53 in p53-null (HCT116 p53−/−) colon carcinoma cells suppresses basal MGMT promoter-dependent luciferase activity. This indicated that p53-mediated repression of promoter activity occurs at the transcription level. The V143A p53 mutant lacking DNA binding activity did not down-regulate the basal MGMT promoter which strongly suggests that the DNA-binding of p53 is a prerequisite for its repressor activity. However, the L22G T23S (p53LT) double mutant with normal DNA-binding activity, but lacking trans-acting activity, is also unable to repress the MGMT promoter as much as the WT p53 which suggests that both DNA-binding and transactivation abilities of p53 are required for the repression (42). It was shown earlier that some mutant p53 polypeptide do not degrade as fast as WT p53. Consistent with this, Western analysis showing that p53 mutants (48) are more stable than the WT p53 protein eliminated a possible reason that the former’s inability to repress the MGMT promoter was due to reduced stability.
We showed that p53 represses the minimal MGMT promoter that does not contain any consensus p53-binding site. Therefore, p53-mediated repression of promoter activity appears to be executed without direct binding of p53 to the MGMT promoter. The observation that WT p53 can repress minimal MGMT promoter containing only three Sp1 transcription factor-binding sites raised the possibility that p53-mediated repression could be mediated via Sp1. Consistent with this Sp1 overexpression abrogated p53-mediated repression of MGMT promoter in a dose-dependent manner. The Sp1 sites are often found in the promoters of housekeeping genes such as MGMT, and needed for its basal promoter activity. The MGMT gene promoter lacking TATA and CAAT boxes has 10 Sp1-binding sites (24).
The observation that Sp1 overexpression relieved p53-mediated down-regulation of MGMT raises two possibilities: p53 may compete with Sp1 for binding to one or more of the Sp1 consensus binding sites in the MGMT promoter or it may sequester Sp1 and thus reduce its availability to bind the MGMT promoter. The fact that addition of recombinant p53 to the nuclear extracts reduced the binding while recombinant p53 alone could not bind to that sequence ruled out the possibility that p53 competes with Sp1 for binding to the same sequences present on the MGMT promoter. It thus appears that repression may be achieved through the physical interaction of WT p53 with Sp1 that prevents Sp1 from binding to the MGMT promoter. This is supported by the observation that WT p53 forms a stable complex with Sp1 in vivo and overexpression of Sp1 overcame the inhibitory effect of p53. However, we have also shown that V143A p53 interacted with and inhibited Sp1 binding, although it was unable to down-regulate MGMT promoter activity suggesting that p53 may down-regulate the MGMT promoter through multiple mechanisms including sequestration of Sp1. P53 has been shown to repress many promoters that lack p53 specific cis elements indicating that the mechanisms of p53-mediated repression are varied and complex (31, 49–51). Such mechanisms could include interference with transcriptional activators, interference with the basal transcription machinery, compaction of the chromatin structure at the promoter site by recruitment of histone deacetylases and other histone-modifying enzymes (31). We have shown earlier that the basal MGMT promoter activity is strongly dependent on p300/CBP, a transcriptional co-activator with histone acetyltransferase activity and treatment with tricostatin A, a histone deacetylase inhibitor, activates MGMT expression through alteration of chromatin structure. Because p53 was shown to interact with p300/CBP and p53-mediated recruitment of mSin3A/HDAC complex was shown to be responsible for repression of many promoters (52), it is likely that p53 sequesters p300/CBP or recruits deacetylase complex to down-regulate the MGMT promoter. In any case, our study suggests that sequestration of Sp1 could be one of the mechanisms by which p53 negatively regulates MGMT expression and thus enhances the sensitivity of tumor cells to alkylating drugs.
Although reports on p53 gene defects and MGMT expression have been inconsistent, the inverse correlation between the expression of MGMT and p53 gene in breast cancer and other tumors has been documented (37–39). It is interesting to speculate that induction of WT p53 in tumors would cause drastic reduction of the MGMT level, which should increased cytotoxicity of the alkylating drugs. Consistent with this, one recent study by Roos et al showed that TMZ-induced glioma cell death and apoptosis is greatly dependent on p53 (53). It was shown that WT p53 expressing glioma cells were more sensitive to TMZ than their p53 mutant-containing counterpart. It is important to note that radiation and O6-methylating drugs administered concomitantly are being used in glioma therapy. Indeed one study indicated that the effect was additive (54). Therefore, combination of radiation and alkylating drugs should augment the apoptotic function of WT p53, resulting in significant potentiation of the cytotoxicity of alkylating drugs due to modulation of MGMT level.
Acknowledgements
We thank Robert Tijian, University of California, Berkeley, for providing the Sp1 expression plasmid. This work was supported by R01 ES07572, R01 CA53791, P50 ES66076 (to SM), American Heart Association grant 0565008Y (to KB).
References
- 1.Ludlum DB. DNA alkylation by the haloethylnitrosoureas: nature of modifications produced and their enzymatic repair or removal. Mutation Res. 1990;233:117–126. doi: 10.1016/0027-5107(90)90156-x. [DOI] [PubMed] [Google Scholar]
- 2.Erickson LC, Laurent G, Sharkey NA, Kohn KW. DNA cross-linking and monoadduct repair in nitrosourea-treated human tumour cells. Nature. 1980;288:727–729. doi: 10.1038/288727a0. [DOI] [PubMed] [Google Scholar]
- 3.Mitra S, Kaina B. Regulation of repair of alkylation damage in mammalian genomes. Prog Nucleic Acid Res Molec Biol. 1993;44:109–142. doi: 10.1016/s0079-6603(08)60218-4. [DOI] [PubMed] [Google Scholar]
- 4.Kaina B, Christmann M, Naumann S, Roos WP. MGMT: key node in the battle against genotoxicity, carcinogenicity and apoptosis induced by alkylating agents. DNA repair. 2007;6:1079–1099. doi: 10.1016/j.dnarep.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 5.Pegg AE. Repair of O(6)-alkylguanine by alkyltransferases. Mutation Res. 2000;462:83–100. doi: 10.1016/s1383-5742(00)00017-x. [DOI] [PubMed] [Google Scholar]
- 6.Dunn WC, Tano K, Horesovsky GJ, Preston RJ, Mitra S. The role of O6- alkylguanine in cell killing and mutagenesis in Chinese hamster ovary cells. Carcinogenesis. 1991;12:83–89. doi: 10.1093/carcin/12.1.83. [DOI] [PubMed] [Google Scholar]
- 7.Mitra S. MGMT: a personal perspective. DNA repair. 2007;6:1064–1070. doi: 10.1016/j.dnarep.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olsson M, Lindahl T. Repair of alkylated DNA in Escherichia coli. Methyl group transfer from O6-methylguanine to a protein cysteine residue. J Biol Chem. 1980;255:10569–10571. [PubMed] [Google Scholar]
- 9.Foote RS, Mitra S, Pal BC. Demethylation of O6-methylguanine in a synthetic DNA polymer by an inducible activity in Escherichia coli. Biochem Biophys Res Comm. 1980;97:654–659. doi: 10.1016/0006-291x(80)90314-9. [DOI] [PubMed] [Google Scholar]
- 10.Yarosh DB, Foote RS, Mitra S, Day RS., 3rd Repair of O6-methylguanine in DNA by demethylation is lacking in Mer− human tumor cell strains. Carcinogenesis. 1983;4:199–205. doi: 10.1093/carcin/4.2.199. [DOI] [PubMed] [Google Scholar]
- 11.Day RS, 3rd, Ziolkowski CH, Scudiero DA, Meyer SA, Lubiniecki AS, Girardi AJ, Galloway SM, Bynum GD. Defective repair of alkylated DNA by human tumour and SV40-transformed human cell strains. Nature. 1980;288:724–727. doi: 10.1038/288724a0. [DOI] [PubMed] [Google Scholar]
- 12.Schwartz JL, Turkula T, Sagher D, Strauss B. The relationship between O6- alkylguanine alkyltransferase activity and sensitivity to alkylation-induced sister chromatid exchanges in human lymphoblastoid cell lines. Carcinogenesis. 1989;10:681–685. doi: 10.1093/carcin/10.4.681. [DOI] [PubMed] [Google Scholar]
- 13.Citron M, Decker R, Chen S, Schneider S, Graver M, Kleynerman L, Kahn LB, White A, Schoenhaus M, Yarosh D. O6-Methylguanine-DNA methyltransferase in human normal and tumor tissue from brain, lung, and ovary. Cancer Res. 1991;51:4131–4134. [PubMed] [Google Scholar]
- 14.Sagher D, Karrison T, Schwartz JL, Larson RA, Strauss B. Heterogeneity of O6- alkylguanine-DNA alkyltransferase activity in peripheral blood lymphocytes: relationship between this activity in lymphocytes and in lymphoblastoid lines from normal controls and from patients with Hodgkin's disease or non-Hodgkin's lymphoma. Cancer Res. 1989;49:5339–5344. [PubMed] [Google Scholar]
- 15.Wang Y, Kato T, Ayaki H, Ishizaki K, Tano K, Mitra S, Ikenaga M. Correlation between DNA methylation and expression of O6-Methylguanine-DNA methyltransferase gene in cultured human tumor cells. Mutation Res. 1992;273:221–230. doi: 10.1016/0921-8777(92)90083-f. [DOI] [PubMed] [Google Scholar]
- 16.Qian X, von Wronski MA, Brent TP. Localization of methylation sites in the human O6-methylguanine-DNA methyltransferase promoter: correlation with gene suppression. Carcinogenesis. 1995;16:1385–1390. doi: 10.1093/carcin/16.6.1385. [DOI] [PubMed] [Google Scholar]
- 17.Bhakat KK, Mitra S. CpG methylation-dependent repression of the human O6- Methylguanine-DNA methyltransferase gene linked to chromatin structure alteration. Carcinogenesis. 2003;24:1337–1345. doi: 10.1093/carcin/bgg086. [DOI] [PubMed] [Google Scholar]
- 18.Costello JF, Futscher BW, Kroes RA, Pieper RO. Methylation-related chromatin structure is associated with exclusion of transcription factors from and suppressed expression of the O6-Methylguanine DNA methyltransferase gene in human glioma cell lines. Mol Cell Biol. 1994;14:6515–6521. doi: 10.1128/mcb.14.10.6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fritz G, Tano K, Mitra S, Kaina B. Inducibility of the DNA repair gene encoding O6- methylguanine-DNA methyltransferase in mammalian cells by DNA-damaging treatments. Molecular and cellular biology. 1991;11:4660–4668. doi: 10.1128/mcb.11.9.4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niture SK, Velu CS, Smith QR, Bhat GJ, Srivenugopal KS. Increased expression of the MGMT repair protein mediated by cysteine prodrugs and chemopreventative natural products in human lymphocytes and tumor cell lines. Carcinogenesis. 2007;28:378–389. doi: 10.1093/carcin/bgl155. [DOI] [PubMed] [Google Scholar]
- 21.Kaina B, Ochs K, Grosch S, Fritz G, Lips J, Tomicic M, Dunkern T, Christmann M. BER, MGMT, and MMR in defense against alkylation-induced genotoxicity and apoptosis. Prog Nucleic Acid Res Mol Biol. 2001;68:41–54. doi: 10.1016/s0079-6603(01)68088-7. [DOI] [PubMed] [Google Scholar]
- 22.Dolan ME, Pegg AE. O6-Benzylguanine and its role in chemotherapy. Clin Cancer Res. 1997;3:837–847. [PubMed] [Google Scholar]
- 23.Tano K, Shiota S, Collier J, Foote RS, Mitra S. Isolation and structural characterization of a cDNA clone encoding the human DNA repair protein for O6- alkylguanine. Proc Natl Acad Sci USA. 1990;87:686–690. doi: 10.1073/pnas.87.2.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harris LC, Potter PM, Tano K, Shiota S, Mitra S, Brent TP. Characterization of the promoter region of the human O6-Methylguanine-DNA methyltransferase gene. Nucleic acids research. 1991;19:6163–6167. doi: 10.1093/nar/19.22.6163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bhakat KK, Mitra S. Regulation of the human O(6)-methylguanine-DNA methyltransferase gene by transcriptional coactivators cAMP response element-binding protein-binding protein and p300. J Biol Chem. 2000;275:34197–34204. doi: 10.1074/jbc.M005447200. [DOI] [PubMed] [Google Scholar]
- 26.Biswas T, Ramana CV, Srinivasan G, Boldogh I, Hazra TK, Chen Z, Tano K, Thompson EB, Mitra S. Activation of human O6-Methylguanine-DNA methyltransferase gene by glucocorticoid hormone. Oncogene. 1999;18:525–532. doi: 10.1038/sj.onc.1202320. [DOI] [PubMed] [Google Scholar]
- 27.Boldogh I, Ramana CV, Chen Z, Biswas T, Hazra TK, Grösch S, Grombacher T, Mitra S, Kaina B. Regulation of expression of the DNA repair gene O6-Methylguanine-DNA methyltransferase via protein kinase C-mediated signaling. Cancer Res. 1998;58:3950–3956. [PubMed] [Google Scholar]
- 28.Jin S, Levine AJ. The p53 functional circuit. Journal of cell science. 2001;114:4139–4140. doi: 10.1242/jcs.114.23.4139. [DOI] [PubMed] [Google Scholar]
- 29.Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 30.el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 31.Ho J, Benchimol S. Transcriptional repression mediated by the p53 tumour suppressor. Cell Death Differentiation. 2003;10:404–408. doi: 10.1038/sj.cdd.4401191. [DOI] [PubMed] [Google Scholar]
- 32.Chan TA, Hwang PM, Hermeking H, Kinzler KW, Vogelstein B. Cooperative effects of genes controlling the G(2)/M checkpoint. Genes Dev. 2000;14:1584–1588. [PMC free article] [PubMed] [Google Scholar]
- 33.Kastan MB, Zhan Q, el-Deiry WS, Carrier F, Jacks T, Walsh WV, Plunkett BS, Vogelstein B, Fornace AJ., Jr A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell. 1992;71:587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- 34.Harris LC, Remack JS, Houghton PJ, Brent TP. Wild-type p53 suppresses transcription of the human O6-Methylguanine-DNA methyltransferase gene. Cancer Res. 1996;56:2029–2032. [PubMed] [Google Scholar]
- 35.Grombacher T, Eichhorn U, Kaina B. p53 is involved in regulation of the DNA repair gene O6-Methylguanine-DNA methyltransferase (MGMT) by DNA damaging agents. Oncogene. 1998;17:845–851. doi: 10.1038/sj.onc.1202000. [DOI] [PubMed] [Google Scholar]
- 36.Srivenugopal KS, Shou J, Mullapudi SR, Lang FF, Jr, Rao JS, Ali-Osman F. Enforced expression of wild-type p53 curtails the transcription of the O(6)- methylguanine-DNA methyltransferase gene in human tumor cells and enhances their sensitivity to alkylating agents. Clin Cancer Res. 2001;7:1398–1409. [PubMed] [Google Scholar]
- 37.Osanai T, Takagi Y, Toriya Y, Nakagawa T, Aruga T, Iida S, Uetake H, Sugihara K. Inverse correlation between the expression of O6-Methylguanine-DNA methyl transferase (MGMT) and p53 in breast cancer. Jp J Clin Oncol. 2005;35:121–125. doi: 10.1093/jjco/hyi036. [DOI] [PubMed] [Google Scholar]
- 38.Hengstler JG, Tanner B, Moller L, Meinert R, Kaina B. Activity of O(6)- methylguanine-DNA methyltransferase in relation to p53 status and therapeutic response in ovarian cancer. Int J Cancer. 1999;84:388–395. doi: 10.1002/(sici)1097-0215(19990820)84:4<388::aid-ijc10>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 39.Guo W, Liu X, Lee S, Park NH. High O6-Methylguanine methyl transferase activity is frequently found in human oral cancer cells with p53 inactivation. International journal of oncology. 1999;15:817–821. doi: 10.3892/ijo.15.4.817. [DOI] [PubMed] [Google Scholar]
- 40.Ko LJ, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 41.Steele RJ, Lane DP. P53 in cancer: a paradigm for modern management of cancer. Surgeon. 2005;3:197–205. doi: 10.1016/s1479-666x(05)80041-1. [DOI] [PubMed] [Google Scholar]
- 42.Lin J, Chen J, Elenbaas B, Levine AJ. Several hydrophobic amino acids in the p53 amino-terminal domain are required for transcriptional activation, binding to mdm-2 and the adenovirus 5 E1B 55-kDa protein. Genes Dev. 1994;8:1235–1246. doi: 10.1101/gad.8.10.1235. [DOI] [PubMed] [Google Scholar]
- 43.Horton JK, Roy G, Piper JT, Van Houten B, Awasthi YC, Mitra S, Alaoui-Jamali MA, Boldogh I, Singhal SS. Characterization of a chlorambucil-resistant human ovarian carcinoma cell line overexpressing glutathione S-transferase mu. Biochemical pharmacology. 1999;58:693–702. doi: 10.1016/s0006-2952(99)00142-2. [DOI] [PubMed] [Google Scholar]
- 44.Miyashita T, Reed JC. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell. 1995;80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 45.Bocangel DB, Finkelstein S, Schold SC, Bhakat KK, Mitra S, Kokkinakis DM. Multifaceted resistance of gliomas to temozolomide. Clin Cancer Res. 2002;8:2725–2734. [PubMed] [Google Scholar]
- 46.Kessler R, Hamou MF, Albertoni M, de Tribolet N, Arand M, Van Meir EG. Identification of the putative brain tumor antigen BF7/GE2 as the (de)toxifying enzyme microsomal epoxide hydrolase. Cancer Res. 2000;60:1403–1409. [PubMed] [Google Scholar]
- 47.Borellini F, Glazer RI. Induction of Sp1-p53 DNA-binding heterocomplexes during granulocyte/macrophage colony-stimulating factor-dependent proliferation in human erythroleukemia cell line TF-1. J Biol Chem. 1993;268:7923–7928. [PubMed] [Google Scholar]
- 48.Muller P, Hrstka R, Coomber D, Lane DP, Vojtesek B. Chaperone-dependent stabilization and degradation of p53 mutants. Oncogene. 2008;27:3371–3383. doi: 10.1038/sj.onc.1211010. [DOI] [PubMed] [Google Scholar]
- 49.Kanaya T, Kyo S, Hamada K, Takakura M, Kitagawa Y, Harada H, Inoue M. Adenoviral expression of p53 represses telomerase activity through down-regulation of human telomerase reverse transcriptase transcription. Clin Cancer Res. 2000;6:1239–1247. [PubMed] [Google Scholar]
- 50.Webster NJ, Resnik JL, Reichart DB, Strauss B, Haas M, Seely BL. Repression of the insulin receptor promoter by the tumor suppressor gene product p53: a possible mechanism for receptor overexpression in breast cancer. Cancer Res. 1996;56:2781–2788. [PubMed] [Google Scholar]
- 51.Zhang L, Yu D, Hu M, Xiong S, Lang A, Ellis LM, Pollock RE. Wild-type p53 suppresses angiogenesis in human leiomyosarcoma and synovial sarcoma by transcriptional suppression of vascular endothelial growth factor expression. Cancer Res. 2000;60:3655–3661. [PubMed] [Google Scholar]
- 52.Murphy M, Hinman A, Levine AJ. Wild-type p53 negatively regulates the expression of a microtubule-associated protein. Genes Dev. 1996;10:2971–2980. doi: 10.1101/gad.10.23.2971. [DOI] [PubMed] [Google Scholar]
- 53.Roos WP, Batista LF, Naumann SC, Wick W, Weller M, Menck CF, Kaina B. Apoptosis in malignant glioma cells triggered by the temozolomide-induced DNA lesion O6-Methylguanine. Oncogene. 2007;26:186–197. doi: 10.1038/sj.onc.1209785. [DOI] [PubMed] [Google Scholar]
- 54.Hermisson M, Klumpp A, Wick W, Wischhusen J, Nagel G, Roos W, Kaina B, Weller M. O6-Methylguanine DNA methyltransferase and p53 status predict temozolomide sensitivity in human malignant glioma cells. J Neurochem. 2006;96:766–776. doi: 10.1111/j.1471-4159.2005.03583.x. [DOI] [PubMed] [Google Scholar]