Abstract
IL-15 is an immunostimulatory cytokine trans-presented with the IL-15 receptor α chain to the shared IL-2/IL-15Rβ and common γ chains displayed on the surface of T cells and NK cells. To further define the functionally important regions of this cytokine, activity and binding studies were conducted on human IL-15 muteins generated by site-directed mutagenesis. Amino acid substitutions of the asparagine residue at position 72, which is located at the end of helix C, were found to provide both partial agonist and superagonist activity, with various non-conservative substitutions providing enhanced activity. Particularly, the N72D substitution provided a 4–5 fold increased in biological activity of the IL-15 mutein compared to the native molecule based on proliferations assays with cells bearing human IL-15Rβ and common γ chains. The IL-15N72D mutein exhibited superagonist activity through improved binding ability to the human IL-15Rβ chain. However, the enhanced potency of IL-15N72D was not observed with cells expressing the mouse IL-15Rα-IL-15Rβ-γc complex suggesting that this effect is specific to human IL-15 receptor. The enhanced biological activity of IL-15N72D was associated with more intense phosphorylation of Jak1 and Stat5 and better anti-apoptotic activity compared to the wild-type IL-15. IL-15N72D superagonist activity was also preserved when linked to a single-chain T cell receptor domain to generate a tumor-specific fusion protein. Thus, the human IL-15 superagonist muteins and fusions may create opportunities to construct more efficacious immunotherapeutic agents with clinical utility.
Keywords: IL-15, Cytokines, Human, Cytokine receptors, T cell receptors, Cell proliferation, Signal transduction, Apoptosis
Introduction
We have previously reported the construction of a soluble single-chain T cell receptor (scTCR)2-cytokine fusion protein (c264scTCR/IL-2) comprising IL-2 genetically linked to a soluble HLA-A*0201-restricted TCR recognizing a peptide of human p53 protein (1, 2). This c264scTCR/IL-2 fusion protein exhibits potent targeted anti-tumor activity in nude mice bearing human tumor xenografts that display cognate peptide/HLA complexes (1, 2). Our studies further support a model where c264scTCR/IL-2 activates immune cells expressing IL-2 receptors. Following stable interaction with cell-surface IL-2 receptors, c264scTCR/IL-2 fusion molecules increase accumulation of immune cells at the site of tumors displaying target peptide/HLA complexes where the immune cells mediate anti-tumor effects (2).
To further explore the anti-tumor activity of scTCR-cytokine fusions, we substituted the IL-2 portion of the molecule with the human IL-15 protein (hIL-15). IL-15 and IL-2 are structurally related and belong to the family of four α-helix bundle cytokines (3). The cell surface receptors for IL-15 and IL-2 comprise three subunits: the cytokine-specific receptor α chains (IL-15Rα and IL-2Rα, respectively) and the shared IL-15Rβ (also known as IL-2Rβ or CD122) and common γ(γc) chains. Despite sharing critical receptor and signaling components and overlapping activities supporting development, survival and function of T cells, B cells and NK cells, IL-15 and IL-2 exert fundamentally distinct functions (3, 4). IL-2 has critical roles in activation-induced cell death and in the development and maintenance of CD4+ and CD25+ regulatory T cells to prevent autoimmune diseases. In contrast, IL-15 plays an essential role in supporting the survival of CD44high CD8+ memory T cells for long-lasting, high-avidity T cell response to infections (4). The functional difference between IL-15 and IL-2, therefore, suggests that IL-15 could be more effective than IL-2 in augmentation of T-cell responses against cancer and infectious agents. The potent anti-tumor activity of IL-15 has recently been shown in various experimental cancer models in animals (5–7). Additionally, IL-15 is unique among cytokines due to its participation in a trans signaling mechanism in which IL-15Rα on monocytes or dendritic cells present IL-15 to neighboring NK or CD8+ T cells expressing IL-15Rβ-γc chains (3).
The use of recombinant hIL-15 (rhIL-15) as a therapeutic is precluded by its poor expression level in standard mammalian cell systems and the need for high doses to achieve biological responses in vivo (8, 9). To overcome this, we used targeted mutagenesis to improve the biological activity of hIL-15. Here, we report the characterization of a superagonist form of hIL-15 created by introducing an aspartic acid substitution of asparagine at the amino acid residue 72 of the mature protein.
Materials and Methods
Cell culture
CTLL-2, 32D, and TF-1 cells were purchased from American Type Culture Collection (ATCC). Phoenix Eco and 293GP cell lines were kindly provided by Dr. Richard Morgan (National Cancer Institute). CHO, Phoenix Eco and 293GP cells were cultured in IMDM complete medium [IMDM plus 10% FBS (Hyclone)]. 32D cells were cultured in IMDM complete medium plus 10% mouse IL-3 culture supplement (BD Bioscience). CTLL-2 and 32Dβ cells were cultured in IMDM complete medium plus 50 ng/ml of rhIL-2 or 10 ng/ml of rhIL-15. TF-1 and TF-1β cells were cultured in IMDM complete medium plus 2 ng/ml of human GM-CSF (R&D Systems). Mouse anti-human TCR Cβ mAb was produced from hybridoma BF1 8A3.31 (ATCC), purified with protein A-coupled Sepharose 4 Fast Flow column (GE Healthcare Life Sciences), and biotinylated with EZ-Link Sulfo-NHS-LC-Biotin (Pierce) according to manufacture’s instructions.
RT-PCR and mutagenesis
hIL-15, human IL-15Rα (hIL-15Rα), and human IL-15Rβ(hIL-15Rβ) genes were amplified by reverse transcriptase PCR (Invitrogen) from total RNA obtained from normal human donor PBMC using the RNeasy kit (Qiagen). The following primers were
used: hIL-15: forward-CACCTTGCCATAGCCAGCTCTTC
reverse-GTCTAAGCAGCAGAGTGATGTTTG;
hIL-15Rα: forward-AGTCCAGCGGTGTCCTGTGG
reverse-TGACGCGTTTAAGTGGTGTCGCTGTGCCCTG;
hIL-15Rβ: forward-TCCTTCCTCGGCTCCACCCTG
reverse-AGCTGCCTGCCTCCCACCCTG.
Site-specific mutants of hIL-15 were generated in a two step process where forward and reverse primers encoding specific codon mutation were used to amplify the hIL-15 gene segments by PCR using PfuUltra polymerase (Promega). The resulting PCR products were gel purified and used as templates in a second PCR reaction to amplify the entire hIL-15 mutein gene with forward and reverse primers containing restriction sites to facilitate cloning. Random mutations were also introduced into the hIL-15 gene at amino acid position 72 by PCR using the KOD Hot Start DNA Polymerase Kit (Calbiochem), plasmid DNA containing the hIL-15 gene isolated from a Dam+ E. coli strain as a template and 21 nucleotide mutagenesis primer with NNN at the position corresponding to codon 72. The resulting PCR products were digested with DpnI to remove the Dam-methylated template DNA.
Gene expression and protein production
The rhIL-15 used as a control in these studies was produced by cloning DNA sequences for hIL-15 into the pET28b plasmid and expressed as inclusion bodies in the E. coli host BL21-AI. The inclusion bodies were solubilized in 8 M guanidine hydrochloride and dilution-refolded in a redox buffer containing oxidized and reduced glutathiones. Refolded rhIL-15 was orthogonally purified using hydrophobic interaction (Butyl-650M), ion-exchange (Source 15Q, QXL) and size exclusion (Superdex 75) chromatographies. The final product was formulated in 25 mM Na2HPO4, 500 mM NaCl, pH 7.4. This refolded and purified rIL-15 was used in the IL-15-specific ELISA methods as a standard to quantitate and normalize rhIL-15 and rhIL-15 mutein concentrations in different preparations. The rhIL-15 and rhIL-15 muteins genes containing a 5’ ATG codon and 3’ TGA TAA stop codons were also cloned into the pDG-160 expression vector and transformed into E. coli DG-116 competent cells (10). The expression of rhIL-15 and rhIL-15 muteins under the control of the PL promoter was induced at 42°C. After cell lysis with sonication, inclusion bodies were separated, washed, and dissolved in 8 M urea, 60 mM MES buffer pH 6.5, 0.1 mM EDTA and 0.1 mM dithiothreitol. For renaturation, the solubilized proteins were dialyzed overnight against 10 mM phosphate buffer pH 7.0 containing 40 mM NaCl and 2.5 % sucrose. The refolded rhIL-15 and rhIL-15 mutein was further purified using Q sepharose chromatography.
To express hIL-15, hIL-15 muteins or the sushi domain of human IL-15Rα (hIL-15RαSu) as a TCR fusion protein, the respective coding sequences were use to replace the IL-2 gene in the c264scTCR/IL2 mammalian expression vector described previously (1). Sequence encoding a mutated human IgG1 hinge region (EPKSSDKTHTSPPSP) was used as a linker between the c264scTCR gene and the hIL-15, hIL-15 mutein or hIL-15RαSu coding regions. The resulting expression vectors were transfected into CHO cells followed by selection in medium containing 2 mg/ml G418 (Invitrogen). The soluble fusion proteins were purified from culture medium with an anti-human TCR Cβ mAb (BF1) coupled to a Sepharose 4 Fast Flow column as described previously (1).
To express hIL-15Rβ in 32D or TF-1 cells, the hIL-15Rβ coding region was cloned into the retrovirus expression vector pMSGV-1 (11) that was modified by introducing the puromycin resistant gene after the IRES region. The Phoenix Eco packaging cell line was used to produce pseudo virus to transduce 32D cells. The 293GP cell line co-transfected with pMD2-G to produce VSV-G env (12) was used to produce pseudo virus to transduce TF-1 cells. Cells transduced with pseudo virus were selected with puromycin and the hIL-15Rβ expression was verified by staining with anti-CD122 Ab (BD Bioscience). 32D cells stably expressing hIL-15Rβ were referred to as 32Dβ cells and TF-1 cells stably expressing hIL-15Rβ were referred to as TF-1β cells.
ELISA
The 96-well microtiter plates were coated with 3 µg/ml mouse anti-hIL-15 Ab (MAB647, R&D Systems) or 1 µg/ml scTCR/hIL-15RαSu in coating buffer (10 mM Tris, pH 8.5, 0.3 mM NaCl) for 2 h at 4°C. The plates were washed thoroughly, blocked with 1% BSA/PBS and incubated with increasing concentrations of hIL-15 or hIL-15 muteins for 30 min at room temperature. The plates were washed and incubated with 50 ng/ml of biotinylated mouse anti-hIL-15 antibody (BAM247, R&D Systems) for 30 min at room temperature. The plates were washed and incubated with HRP-conjugated streptavidin (Jackson ImmunoResearch) for 10 min at room temperature. The plates were washed and the reaction developed with addition of ABTS (Pierce) for 2–5 min. The plates were read with a microtiter reader (Molecular Devices) at 405 nm.
Cell proliferation assays
To measure cell proliferation, CTLL-2, 32Dβ, and TF-1β cells (2×104 cells/well) were incubated with increasing concentrations of rhIL-15, rhIL-15 muteins, scTCR/hIL-15, or scTCR/hIL-15 muteins for 48 h at 37°C. In some assays, the recombinant cytokines were mixed with an equal molar concentration of soluble human or mouse IL-15Rα-Fc chimera fusion protein (R&D Systems) and incubated for 10 min at 4°C prior to addition to the cells. Cell proliferation reagent WST-1 (Roche Applied Science) was added during the last 4 h of cell growth according to the manufacturer’s procedures. Conversion of WST-1 to the colored formazan dye by metabolically active cells was determined through absorbance measurements at 440 nm. The EC50 was determined with the dose-response curve generated from the experimental data by nonlinear regression variable slope curve-fitting with GraphPad Prizm4 software.
Flow cytometry, apotosis, and competitive binding
To measure hIL-15Rβ expression on the cell surface, 32D, 32Dβ, TF-1, and TF-1β cells were stained with biotinylated mouse anti-human CD122 mAb (0.5 µg/ml) for 20 min at 4°C. Following a wash step, the cells were then stained with PE-conjugated streptavidin (0.5 µg/ml) (Jackson ImmunoResearch) for 20 min. Cells were washed, resuspended and analyzed on a FACScan flow cytometry using CellQuest software (BD Bioscience).
To measure cell apoptosis, 32Dβ cells were seeded in 24-well plates at 1 × 105 cells/well and cultured in IMDM complete medium with increasing concentrations of rhIL-15 or rhIL-15N72D mutein for 48 h at 37°C. Cells were stained using the Annexin V-FITC Apoptosis Detection kit (BD Bioscience) according to manufacture’s protocol and analyzed by flow cytometry as described above.
To compare the binding ability of rhIL-15 and rhIL-15N72D to hIL-15 receptor complexes on cell surface, TF-1 or TF-1β cells were incubated with increasing concentrations of rhIL-15 or rhIL-15N72D for 15 min at 4°C and then stained with c264scTCR/hIL-15 for 30 min. The binding of c264scTCR/hIL-15 was in turn detected with the biotinylated anti-human TCR Cβ Ab for 15 min and PE-conjugated streptavidin (0.5 µg/ml) for 15 min. Fluorescence intensity was analyzed by flow cytometry as described above and the geometric mean was determined. The percent inhibition was calculated as the following formula: 100%×[(Pos − Exp)/(Pos-Neg)], where Exp is the binding of scTCR/hIL-15 in the presence of rhIL-15 or rhIL-15N72D; Neg is the binding of the secondary reagent only in the absence of scTCR/hIL-15 or rhIL-15, Pos is the binding of scTCR/hIL15 in the absence of rhIL-15 or rhIL-15N72D. The IC50 was determined with the dose-response curve generated from the experimental data by nonlinear regression variable slope curve-fitting with GraphPad Prizm4 software.
Immunoblotting of phosphorylated proteins
To reduce basal phosphorylation levels, TF-1β cells were first cultured in IMDM complete medium plus 0.1 ng/ml of GM-CSF overnight and then incubated in serum-free IMDM for an additional 4 h. The starved cells (4×106 cells/ml) were untreated or treated with 10 pM of rhIL-15 or rhIL-15N72D for 15 min. Cells were extracted in the cell lysis buffer [50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% NP-40, 2 mM EDTA, 2 mM Na3OV4, 2 mM NaF, 1x protease inhibitor mixture (Sigma-Aldrich)]. Protein concentration of cell extracts was determined with Lowery assays. Equal amounts of cell extracts (100 µg) were separated under reducing conditions by 8% Tris-glycine polyacrylamide gel electrophoresis and transferred onto a 0.2-µm nitrocellulose membrane (GE Healthcare). The membranes were blocked with Odyssey blocking buffer (LI-COR) for 1 hr and probed with the following rabbit anti-phosphoprotein antibodies: anti-pJak1 (Tyr 1022/1023) (Santa Cruz Biotechnology) or anti-pStat5 (Tyr 694) (Cell Signaling) in Odyssey blocking buffer overnight at 4°C. The same membranes were then re-probed using anti-native protein mouse monoclonal antibodies against Jak1 or Stat5 (BD Biosciences). Primary antibody binding to the membrane was detected by incubation with either IRDye 700DX-conjugated anti-rabbit IgG or IRDye 800-conjugated anti-mouse IgG (LI-COR), followed by visualization with the Odyssey scanner using Odyssey analysis software v1.1 (LI-COR). To correct for possible variation in the amount of protein loaded, the values are expressed as phosphoprotein/native protein ratios (e.g. pJak1/Jak1). Results are expressed as the fold increase with respect to the levels obtained from untreated cells.
Results
Construction of single-chain TCR and human IL-15 fusion protein
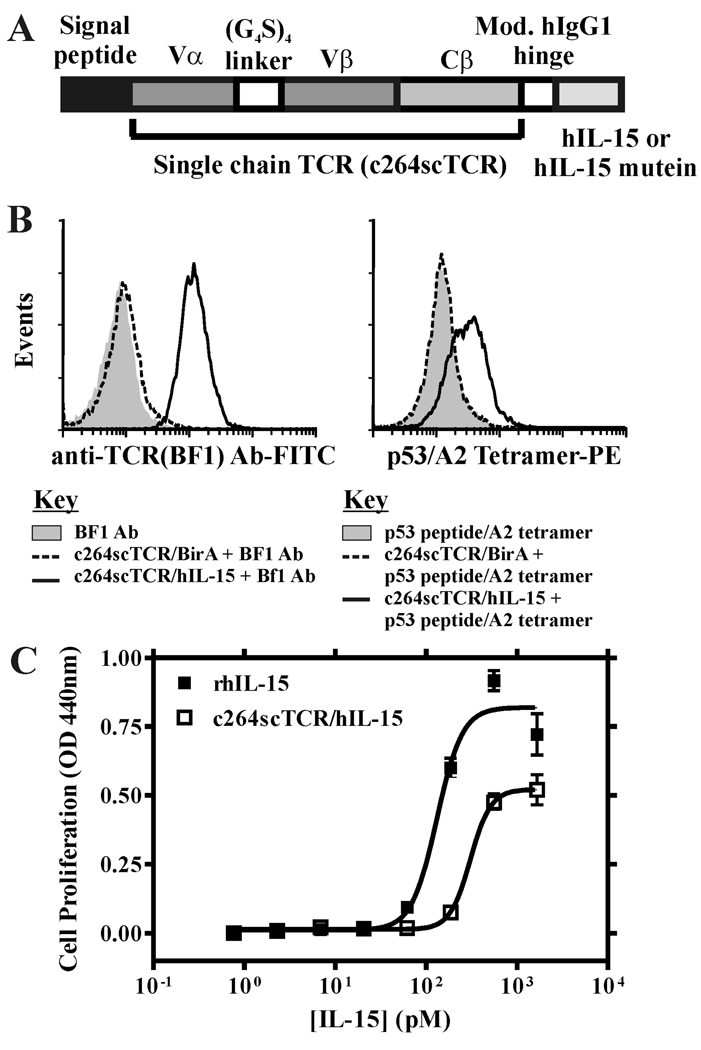
A fusion protein was constructed by fusing hIL-15 to a three-domain, HLA-A*0201 restricted chimeric TCR specific for a p53 peptide antigen. This fusion protein is similar to the c264scTCR/IL-2 molecule described previously (1) except that the coding regions of IL-2 and the linker between the scTCR and IL-2 have been replaced with that of hIL-15 and the modified human IgG1 hinge sequence. This fusion protein is designated as c264scTCR/hIL-15 (Fig. 1A). The gene for this fusion protein was stably transfected into CHO cells and the protein was purified from cell culture media by immunoaffinity chromatography as previously described (1).
FIGURE 1.
Characterization of c264scTCR/hIL-15 fusion proteins. A, Schematic representation of the fusion protein of c264scTCR/hIL-15 or c264scTCR/hIL-15N72D. B, CTLL-2 cells were incubated with or without c264scTCR/birA or c264scTCR/hIL-15 initially and then detected with either biotinylated anti-human TCR Cβ (BF1) and PE-conjugated streptavidin (left) or PE-conjugated p53 (aa264-272)/HLA-A2 tetramer (right). C, 32Dβ cells were incubated with increasing concentrations of c264scTCR/hIL-15 (□) or recombinant hIL-15 (■) for 48 h prior to addition of WST-1 for 4 h and cell proliferation was quantitated by absorbance reading at 440 nm to assess formazan levels. The data points shown are means (±SEM) of triplicate samples and the lines represent sigmoidal dose-response curve fit for EC50 determination. The results are representative of at least three experiments.
The hIL-15 receptor binding capability of the hIL-15 domain of the c264scTCR/hIL-15 fusion protein was studied using flow cytometry analysis. CTLL-2 cells expressing mouse IL-15 receptor complex (i.e. mIL-15Rα-mIL-15Rβ-mγc) were incubated either with c264scTCR/hIL-15 or c264scTCR/BirA (13) and stained with the antibody BF-1 that recognizes an epitope on the human TCR Cβ domain of the c264TCR (1). As shown in Fig. 1B (left panel), the CTLL-2 cells were stained positively with c264TCR/hIL-15, but not with c264scTCR/BirA, indicating that the hIL-15 portion of c264scTCR/hIL-15 is capable of binding the mouse IL-15 receptor complex. When the PE-labeled p53 (aa 264–272) peptide-loaded HLA-A*0201 tetramers were used to substitute the BF-1 antibody in the flow cytometry analysis, we also found that the tetramers positively stained the c264scTCR/hIL-15, but not the c264scTCR/BirA-loaded CTLL-2 cells (Fig. 1B; right panel). These data also indicate that the TCR portion of c264scTCR/hIL-15 fusion is capable of recognizing its cognate peptide/HLA molecules.
The hIL-15 biological activity of the fusion protein was examined with cell proliferation assays using 32Dβ cells. As shown in Fig. 1C, the ability of c264TCR/hIL-15 to support the growth of 32Dβ was concentration dependent exhibiting half maximal stimulation (EC50) at about 300 pM compared to 130 pM for rhIL-15. The observed two-fold difference in relative activity of the c264TCR/hIL-15 fusion protein and free cytokine is less than that reported for other IL-15 fusion proteins (14, 15) and may be due to a number of factors currently under investigation including differences in steric interactions and/or cytokine internalization and recycling. Overall, these data indicate that presence of the scTCR does not severely affect the biological functions of the hIL-15 in the c264scTCR/hIL-5 fusion molecule. Therefore, the scTCR domain of c264scTCR/hIL-15 was used as a detection tag in flow cytometry or cell-based activation analysis to characterize the biological functions of hIL-15.
The agonist effect of asparagine-to-aspartic acid substitution at amino acid residue 72 of IL-15
The molecular interactions between IL-15 and its receptor chain IL-15Rα have been determined by high-resolution, x-ray diffraction structural analysis (16, 17). However, due to the high-affinity interaction between IL-15 and IL-15Rα and the manner in which the IL-15-IL-15Rα complex is transpresented to the IL-15Rβ-γc complex on T lymphocytes and NK cells, we believe that further improvement in the affinity between IL-15 and IL-15Rα may not have a significant impact on the stimulation of the IL-15R signaling machinery in vivo. Therefore, we were particularly interested in improving the interaction between the IL-15 and IL-15Rβ since this receptor chain is known to be responsible for stimulating Jak-1-, Stat5- and AKT-dependent signaling pathways that support cellular survival and proliferation (18–21).
We used the known molecular structure of the interaction between human IL-2 (hIL-2) and IL-2Rβ and sequence alignment of hIL-2 and hIL-15 to identify the targeted sequences for site-specific mutagenesis. The amino acid residues Asp-20 on helix A, Asn-88 and Glu-95 on helix C of hIL-2 were known to be involved in interactions with hIL-2Rβ (22, 23). Amino acid sequence alignment between hIL-2 and hIL-15 identified the amino acid residues Asp-8 on helix A, Asn-65 and Asn-72 on helix C of hIL-15 as likely equivalents to that of hIL-2 mentioned above. Structure-function analysis of hIL-15 support the biological importance of Asp-8 and Asn-65, the amino acid residues we identified using this method (24–26).
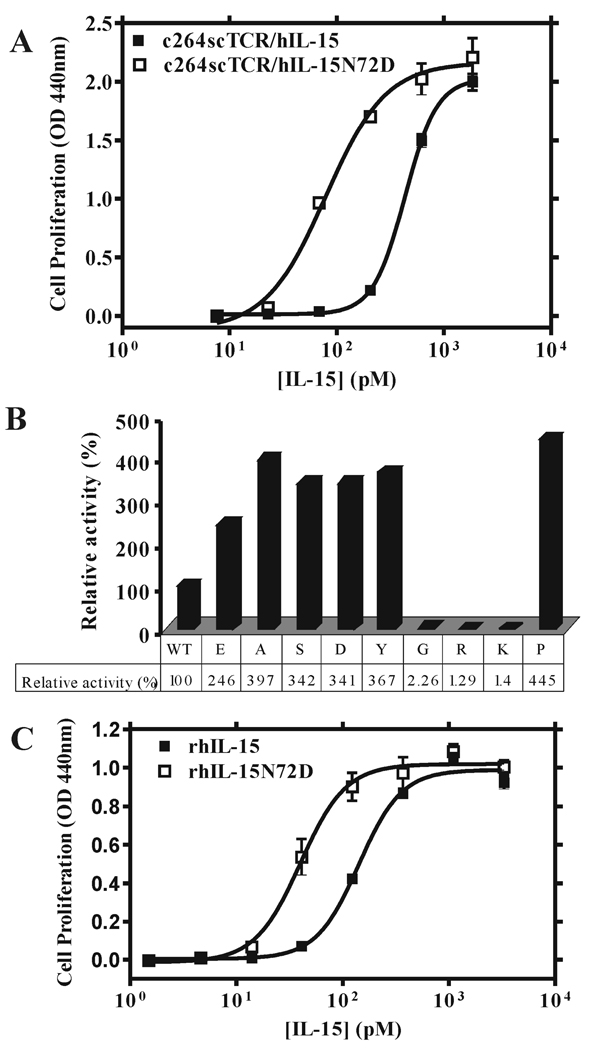
PCR-based mutagenesis was then employed to introduce multiple mutations into the coding region of hIL-15 at amino acid residues Asp-8, Asn-65 and Asn-72 on the c264scTCR/hIL-15 fusion molecule. The muteins were then produced in CHO cells, purified, and used in 32Dβ cell proliferation assays. As shown in Table I, fusion proteins with hIL-15 mutations at amino acid 8 or 65 completely lost their biological activity to support the proliferation of 32Dβ cells whereas the hIL-15N72D fusion mutein provided better cell stimulatory activity. Compared to the c264scTCR/hIL-15 fusion protein, c264scTCR/hIL-15N72D displayed greater than a five-fold increase in bioactivity for promoting 32Dβ cell growth (EC50: 79 pM for c264scTCR/hIL-15N72D vs. 430 pM for c264scTCR/hIL-15) (Fig. 2A). Additional substitutions at the amino acid residue 72 were further introduced and their biological activities were also assessed to support of 32Dβ cell proliferation (Fig. 2B). Among the nine muteins, the glycine, arginine and lysine substitutions were found to exhibit substantially weaker cell stimulation activity than that of the wild-type hIL-15. The other substitutions, however, including the acid aspartic acid and glutamic acid substitutions, enhanced the cell proliferation activity of their corresponding hIL-15 muteins. Interestingly, the alanine substitution did not significantly affect the cell proliferation activity of the hIL-15 mutein. Since alanine has no side chain, this finding suggests that the Asn-72 side chain may not contact the hIL-15Rβ as we initially thought.
Table I.
Bioactivity of hIL-15 muteins
| Analog | Relative activitya in 32Dβ cells (%) |
Relative activitya in CTLL-2 cells (%) |
|---|---|---|
| N72D | 618.5 | 110.2 |
| D8N | 0 | 0 |
| N65D | 0 | 0 |
| N65A | 0 | 0 |
Relative activity represent relative change in EC50 of mutein compared to wild type hIL-15 based on cytokine-dependent cell proliferation assays. The results are representative of at least three experiments.
FIGURE 2.
Effect of wild-type hIL-15 and muteins on 32Dβ cell proliferation. 32Dβ cells (hIL-15Rβ-mγ-positive) were incubated with increasing concentrations of hIL-15-containing proteins for 48 h and proliferation assays were performed as described in Figure 1. A, Dose-responses curves of 32Dβ cell proliferation assessed with different concentrations of c264scTCR/hIL-15 wild type (■) and c264scTCR/hIL-15N72D (□) fusions. B, Proliferation assays of 32Dβ cells using c264scTCR/hIL-15 and nine hIL-15 mutein fusion proteins were carried out. The activity based on EC50 of c264scTCR/hIL15 wild-type fusion was normalized to 100% and the relative activities of mutein fusions were determined. C, Dose-responses curves of 32Dβ cell proliferation assessed with different concentrations of rhIL-15 wild type (■) and rhIL-15N72D (□) proteins. The results are representative of at least three independent experiments.
To assess whether the agonist effect of hIL-15N72D is dependent on the fusion protein format, we directly expressed rhIL-15 and rhIL-15N72D mature proteins in E. coli as insoluble inclusion bodies using a either T7 promoter-driven or temperature-controlled λ PL expression vectors (10, 27). The inclusion bodies were purified and refolded as described in the Materials and Methods. The biological activity of refolded rhIL-15 produced by either of the expression methods was equivalent (data not shown). The activities of rhIL-15 and rhIL-15N72D proteins were compared in the 32Dβ cell proliferation assays. As shown in Fig. 2C, rhIL-15N72D exhibits three to four-fold stronger activity than that of rhIL-15 in 32Dβ cell proliferation assays (EC50: 39.9 pM for rhIL-15N72D vs. 142 pM for rhIL-15). These data demonstrate that the effect of the N72D mutation on hIL-15 is not dependent of the c264TCR fusion domain on c264scTCR/hIL-15 molecule.
N72D effect is restricted to human IL-15Rβ
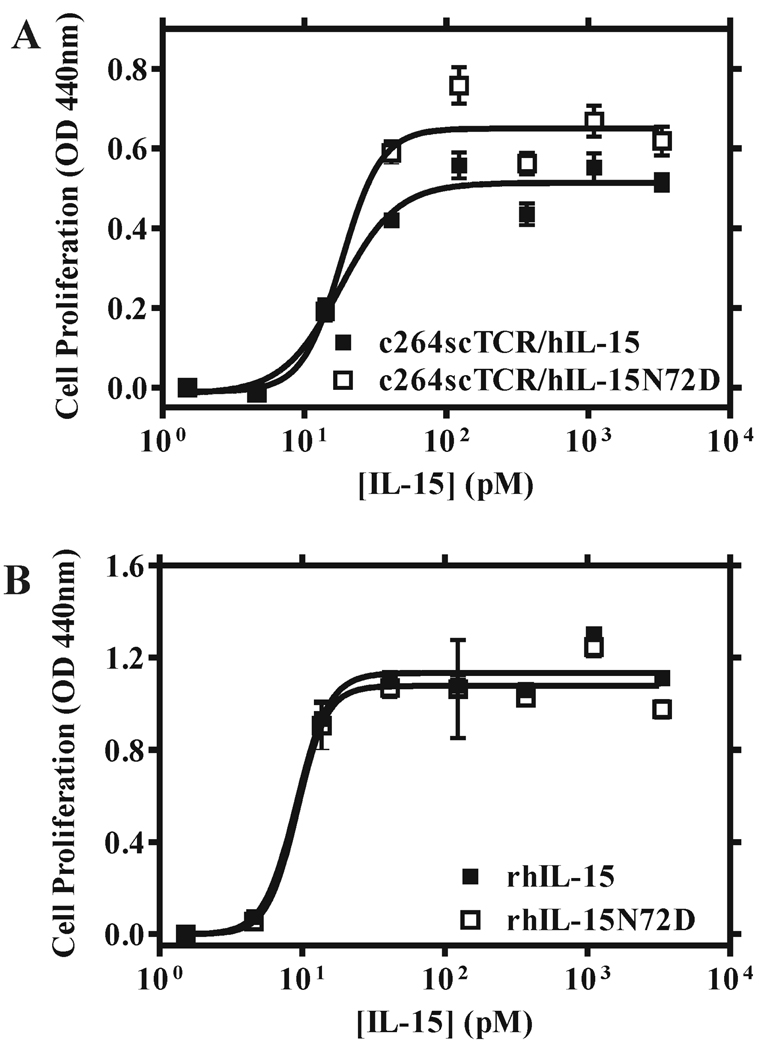
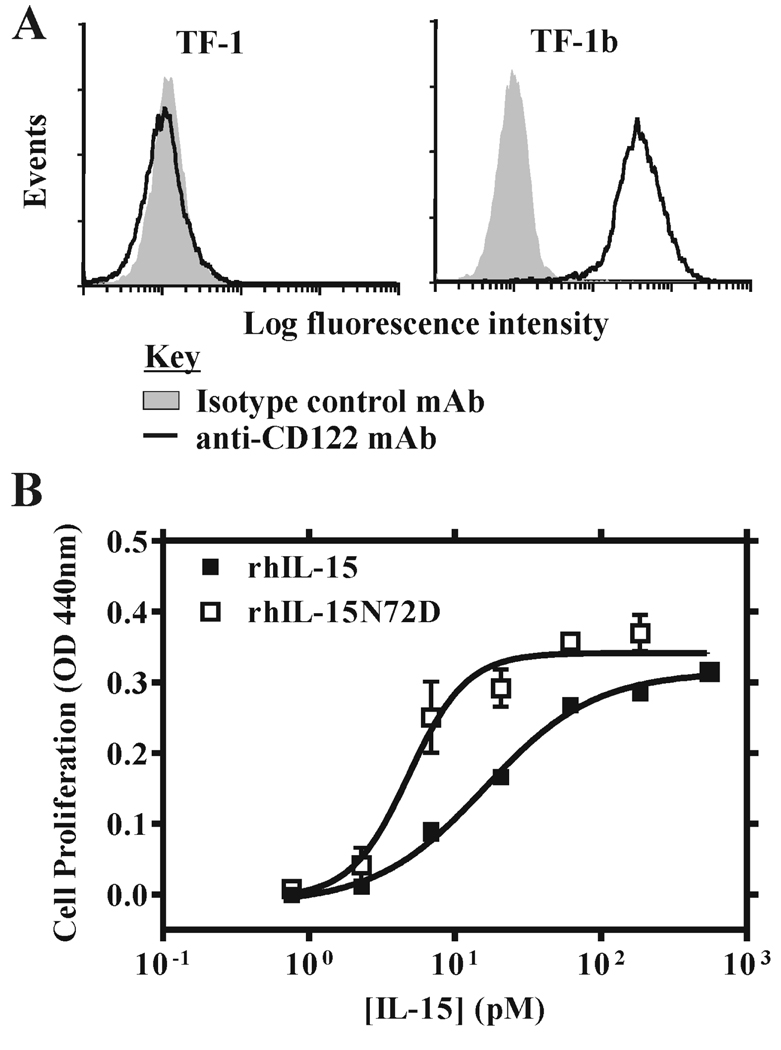
When CTLL-2 cells expressing the intact mouse IL-15 receptor complex (i.e. mIL-15Rα-mIL-15Rβ-mγc) were used in proliferation assays, the relative activities of c264TCR/hIL-15 and rhIL-15 proteins were similar to those observed with 32Dβ cells; though, as expected, the proteins were 15–20 fold more potent due to the presence of the high affinity mIL-15Rα chain (Table II). Consistent with results of previous reports (24–26), the hIL-15 Asp-8 and Asn-65 muteins were incapable of supporting growth of CTLL-2 cells (Table I). However, in contrast to 32Dβ cell assays, the c264scTCR/hIL-15N72D fusion protein did not exhibit significantly better biological activity compared with the c264scTCR/hIL-15 for CTLL-2 cell proliferation (Table I). The EC50s for both hIL-15N72D and wild-type hIL-15 fusion proteins were approximately 18 pM in assays with CTLL-2 cells (Figure 3A). Similarly, no difference in biological activities for the free rhIL-15 and rhIL-15N72D cytokines was observed with this cell line (EC50s were approximately 9 pM for both rhIL-15N72D and wild-type rhIL-15) (Fig. 3B). As indicated above, the CTLL-2 cells express the mIL-15Rα-mIL-15Rβ-mγc complex whereas 32Dβ cells express mγc and transfected hIL-15Rβ (28). Therefore, we speculate that the increased agonist effect of hIL-15N72D was either masked by the presence of the high affinity mIL-15Rα subunit or required the hIL-15Rβ. To distinguish these possibilities, we introduced the hIL-15Rβ gene using a recombinant retroviral vector into the human erythroleukemia cell TF-1 (29). TF-1 cells express endogenous hIL-15Rα and hγc. The transfectants carrying hIL-15Rβ were identified using the anti-CD122 mAb in flow cytometry analysis (Fig. 4A) and the resulting cell line is designated TF-1β. When TF-1β cells were used in cell proliferation assays for rhIL15 and rhIL15N72D, we observed that the rhIL-15N72D exhibited three to four-fold better activity than that of wild-type rhIL-15 (Fig. 4B; EC50 = 4.8 pM for rhIL-15N72D vs. EC50 = 16.0 pM for rhIL-15). When the parental TF-1 cells were used in the cell proliferation assay, no activity was observed for either rhIL15N72D or rhIL-15. Although the relative levels of the IL-15Rα and IL-15Rβ-γc subunits may influence the responses observed, the finding that the rhIL-15N72D mutein showed enhanced activity on 32D-β(hIL-15Rβ-mγc positive) and TF-1β(hIL-15Rα-hIL-15Rβ-hγc positive) cells but not CTLL-2 cells (mIL-15Rα-mIL-15Rβ-mγc positive) suggests that hIL-15Rβ is the target of the superagonist activity of the hIL-15N72D mutein.
FIGURE 3.
Effect of hIL-15 wild type and N72D mutein proteins on CTLL-2 cell proliferation. CTLL-2 cells (mIL-15Rα-mIL-15Rβ-mγc-positive) were incubated with increasing concentrations of hIL-15-containing proteins for 48 h and proliferation assays were performed as described in Figure 1. A, Dose-responses curves of CTLL-2 cell proliferation assessed with different concentrations of c264scTCR/hIL-15 wild type (■) and c264scTCR/hIL-15N72D (□) fusions. B, Dose-responses curves of CTLL-2 cell proliferation assessed with different concentrations of rhIL-15 wild type (■) and rhIL-15N72D (□) proteins. The results are representative of at least three experiments.
FIGURE 4.
Effect of hIL-15 wild type and N72D mutein proteins on TF-1β cell proliferation. A, TF-1 cells and TF-1β cells were stained with anti-CD122 (black thick line) or an isotype control (gray shade) to verify expression of the hIL-15Rβ gene. B, TF-1β cells (hIL-15Rα-hIL-15Rβ-hγc-positive) were incubated with increasing concentrations of rhIL-15 wild type (■) and rhIL-15N72D (□) for 48 h and proliferation assays were performed as described in Figure 1. The results are representative of at least three experiments.
When comparing the effect of rhIL-15 on 32Dβ cell growth with rhIL-15N72D mutein on TF-1β cell growth, it appears that the combined effects of the hIL-15/hIL-15Rα complex formation and rhIL-15N72D superagonist activity resulted in a ~30-fold increase in biological activity over rhIL-15 alone (Table II). To investigate this further, we tested the activity of rhIL-15 and rhIL-15 N72D on 32Dβ cell proliferation in the presence and absence of soluble IL-15Rα-IgG Fc protein. Preassociation of IL-15 with IL-15Rα-Fc fusions or other soluble IL-15Rα molecules has previously been shown to enhance the biological activity of IL-15 in vitro and in vivo (5–7, 30, 31). Consistent with these reports, we found that addition of either mouse or human IL-15Rα-Fc fusion protein to equal molar concentrations of rhIL-15 provided about a 3-fold shift in the IL-15-dependent proliferation response of 32Dβ cells (Table III). In this study, the rhIL-15N72D mutein displayed ~7 fold lower EC50 than wild-type rhIL-15 and the superagonist activity was enhanced further by preassociation with either mouse or human IL-15Rα-Fc molecules. Overall, the rhIL-15N72D/hIL-15Rα-Fc complex exhibited a 10-fold lower EC50 than rhIL-15 alone for supporting 32Dβ cell growth. The fact that similar effects were seen with soluble mouse and human IL-15Rα-Fc suggesting that the differences in hIL-15N72D activity observed on CTLL-2 and TF-1β proliferation were not due to the species-specific differences in the IL-15Rα subunit.
Table II.
hIL-15 and hIL-15N72D bioactivity and protein binding properties
| Cell line | IL-15R | EC50/IC50 of Purified Protein (pM) | |||
|---|---|---|---|---|---|
| rhIL-15 | c264scTCR/ IL-15 |
rhIL-15N72D | c264scTCR/ IL-15N72D |
||
| 32Dβ | hβ/mγc | 132a, 142a | 308a, 434a | 39.9a | 79.0a |
| CTLL-2 | mα/mβ/mγc | 9.1a | 17.9a | 9.2a | 18.4a |
| TF-1β | hα/hβ/hγc | 16.0a, 3.8b | ND | 4.8a, 0.5b | ND |
| TF-1 | hα/hγc | 18.0b | ND | 17.5b | ND |
EC50 of cytokine-dependent cell proliferation assay.
IC50 of cell surface binding assay.
ND: not determined
Table III.
Effect of soluble IL-15Rα-Fc on the activity of rhIL-15 and rhIL-15N72D
| Added Proteins | EC50 on 32Dβ cell proliferation (pM) |
Relative activity a compared to rhIL-15 (%) |
|---|---|---|
| rhIL-15 | 117.8 | 100 |
| rhIL-15 + hIL-15Rα-Fc | 39.5 | 298 |
| rhIL-15 + mIL-15Rα-Fc | 40.7 | 289 |
| rhIL-15N72D | 16.2 | 727 |
| rhIL-15N72D + hIL-15Rα-Fc | 11.5 | 1024 |
| rhIL-15N72D + mIL-15Rα-Fc | 11.7 | 1007 |
Relative activity represent relative percent change in EC50 compared to wild type rhIL-15 based on cytokine-dependent 32Dβ cell proliferation assays. The results are representative of three independent experiments.
N72D substitution increases human IL-15 binding affinity to human IL-15Rβ
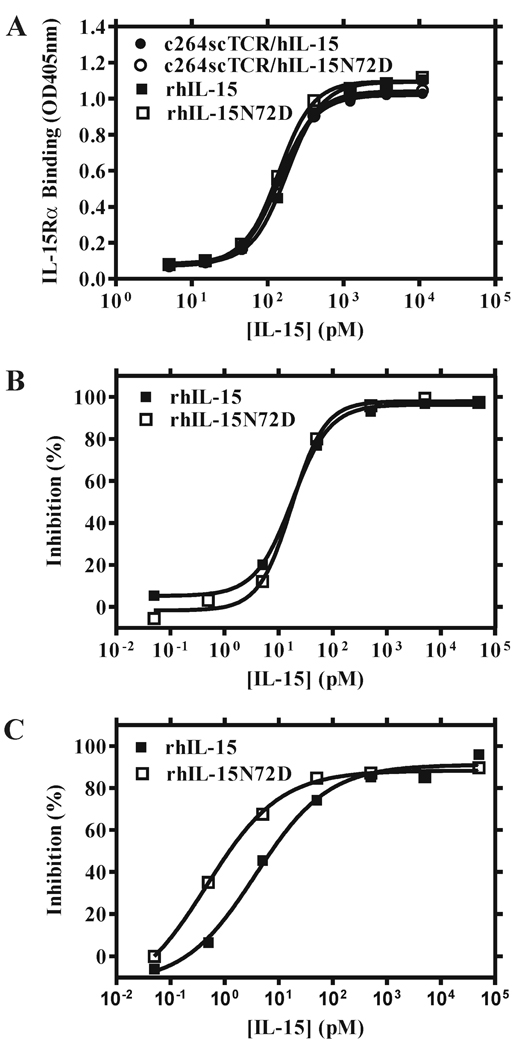
To verify that the N72D substitution does not change the binding affinity of hIL-15 to hIL-15Rα, we constructed a fusion protein by substituting the hIL-15 coding region of c264scTCR/hIL-15 with the sushi-binding domain of hIL-15Rα. This fusion molecule is designated as c264scTCR/hIL-15RαSu and was used in an ELISA-based method to examine the hIL-15Rα-binding capability of the hIL-15 and hIL-15N72D proteins. When c264scTCR/hIL-15, c264scTCR/hIL-15N72D, rhIL-15 or rhIL-15N72D were added to c264scTCR/hIL-15RαSu-coated microtiter plates in a final concentration ranging from 10 to 104 pM and probed by biotinylated anti-hIL-15 mAb and HRP-conjugated streptavidin, we found that all of the proteins bound equally well to hIL-15RαSu (Fig. 5A). Thus, the N72D mutation did not enhance the binding of hIL-15 to hIL-15Rα. This was further confirmed in flow cytometry-based competition analysis using cell lines expressing hIL-15Rα. As shown in Fig 5B, rhIL-15 and rhIL-15N72D compete equally well with c264scTCR/hIL-15 for cell surface binding to TF-1 cells expressing hIL-15Rα and hγ chains. Each protein competed with binding at an IC50 of approximately 18 pM. In contrast, when TF-1β cells carrying the intact hIL-15Rα-hIL-15Rβ-hγ complex were used, rhIL-15N72D exhibits almost 8-fold greater competitive binding activity than rhIL-15 (Fig. 5C; IC50 = 0.48 pM for rhIL-15N72D vs. IC50 = 3.8 pM for rhIL-15). A similar increase in competitive binding activity for rhIL-15N72D compared to rhIL-15 wild-type protein was observed with the 32Dβ cell line (data not shown). Table II provides a summary of the activities of hIL-15 wild type and the N72D mutein on cell proliferation and binding in the context of cells bearing different IL-15R subunits. Taken together, these results indicate that the N72D mutation does not affect interactions with hIL-15Rα but substantially increases the binding affinity of hIL-15 to hIL-15Rβ.
FIGURE 5.
hIL-15N72D exhibits higher binding affinity than wild type hIL-15 to hIL-15Rβ-hγc complexes. A, The binding activity of IL-15 and IL-15 muteins to c264scTCR/IL-15RαSu-coated wells was determined in an ELISA-based assay. No differences in the binding of rhIL-15 (■), rhIL-15N72D (□), c264scTCR/hIL-15 fusion protein (●) or c264scTCR/hIL-15N72D fusion protein (○) were detected. B & C, Cell-based competitive binding assays were conducted using TF-1 (hIL-15Rα-hγc-positive) (B) or TF-1β(hIL-15Rα-hIL-15Rβ-hγc-positive) (C) cells. Binding of a fixed concentration (500 pM) of c264scTCR/hIL-15 was competed with increasing concentrations of rhIL-15 wild type (■) or rhIL-15N72D (□) proteins. Cell-bound c264scTCR/hIL-15 was then detected by staining with biotinylated anti-human TCR Cβ Ab-PE-conjugated streptavidin followed by flow cytometry analysis. Geo-mean of fluorescence intensity (MFI) was used to calculate the percent binding inhibition at different concentrations of rhIL-15N72D or rhIL-15 protein. The results are representative of at least three experiments.
The N72D mutation improves the anti-apoptotic and signal transduction activities of IL-15
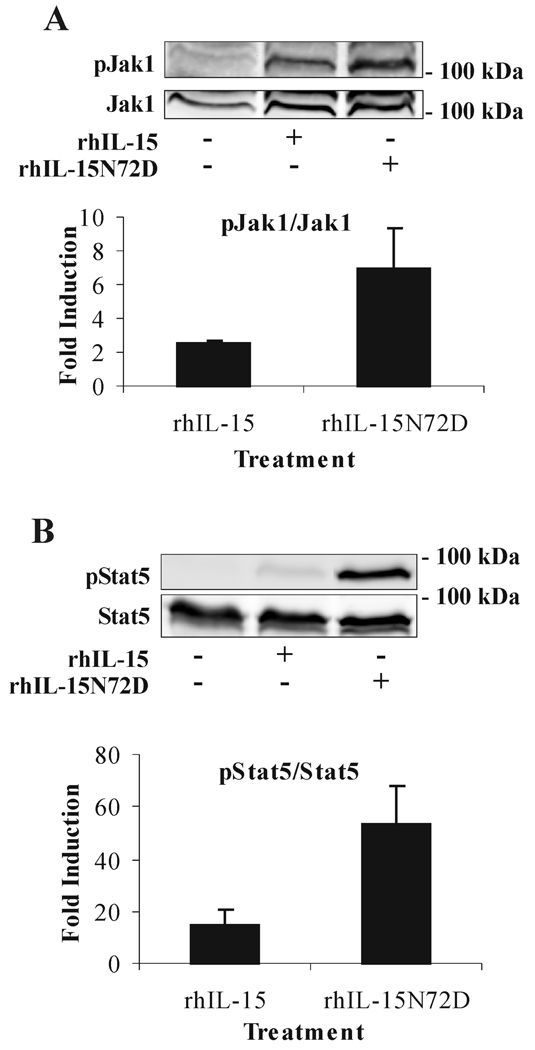
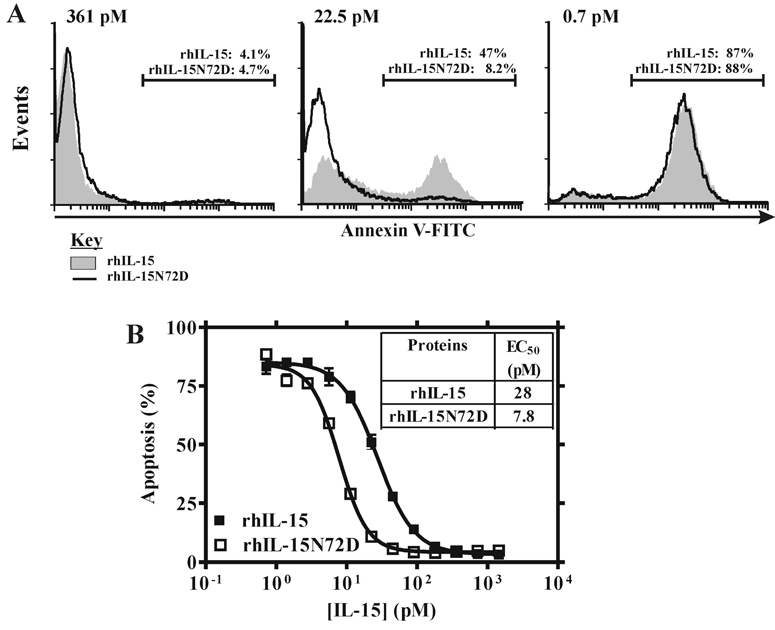
To examine whether the N72D mutation increases the cell signaling potency of hIL-15, TF-1β cells were activated with equal and rate-limited amounts of hIL-15N72D or hIL-15. The phosphorylation intensity was compared for Jak1 and Stat5, two signal transducers involved in the signaling pathway of the IL-15 receptor. As shown in Fig. 6, both Jak1 and Stat5 were phosphorylated at a higher level by the rhIL-15N72D than by rhIL-15 indicating the superiority of the mutein for inducing IL-15R-dependent cell signaling. Similarly, rhIL-15N72D provided a better concentration-dependent decrease in apoptosis of 32Dβ cells than was observed with rhIL-15 (Fig.7, EC50 = 7.8 pM for rhIL-15N72D and EC50 = 27.5 pM for rhIL-15). Overall, the hIL-15N72D mutein exhibited enhanced cell signaling and 3-fold greater anti-apoptotic activity than wild type hIL-15.
FIGURE 6.
Enhanced signal transduction via IL-15R by rhIL-15N72D. TF-1β cells were incubated with medium alone, 10 pM of rhIL-15 or rhIL-15N72D for 15 min at 37°C. Cell extracts were analyzed by Western blotting using anti-phospho-Jak1 (pJak1) antibody (A) or anti-phospho-Stat5 (pStat5) antibody (B). Membranes were then reprobed with antibodies recognizing the native proteins (Jak1 or Stat5). The results are representative of three independent experiments. To correct for possible variation in the amount of protein loaded, values were normalized and are expressed as pJak1/Jak1 or pStat5/Stat5 ratios. pJak1/Jak1 and pStat5/Stat5 levels were determined by densitometry including correction for background. Results are plotted as the mean fold induction (±SEM) in the normalize protein ratios observed in treated compared to untreated cells.
FIGURE 7.
rhIL-15N72D provides increased anti-apoptotic activity compared to rhIL-15. Apoptosis was evaluated by Annexin V cell-surface expression by flow cytometry. A, Histograms representing Annexin V staining of 32Dβ cells following growth for 48 h in the presence of 361 pM, 22.5 pM, or 0.7 pM of rhIL-15 (gray shade) or rhIL-15N72D (black thick line). B, The percentage of apoptotic cells observed at each concentration of rhIL-15N72D (□) or rhIL-15 wild type (■) protein was plotted and EC50 values determined from the fit curve. The data shown represent the means (±SEM) of three independent experiments.
Discussion
Structural analysis has revealed detailed similarities in the four helical domains of IL-15 and IL-2 and has provided information on specific interactions of these cytokines with corresponding unique receptor α subunits and shared receptor β-γc chains. Additionally, molecular model-driven mutagenesis has been further used to characterize these cytokines and has lead to the identification of IL-15 muteins that interact with IL-15Rα with higher affinity and more potent biological activity for supporting IL-15-responsive cell growth (25). However, the utility of such muteins is unclear. In vivo, IL-15 is transpresented by antigen-presenting cells expressing IL-15Rα to neighboring NK or CD8+ T-cells expressing IL-15Rβ-γc chains. This characteristic in combination with the already very high affinity of unmutated IL-15 for IL-15Rα (i.e. Kd ~10 pmolar) (3) and likelihood that in vivo responses are limited by free IL-15Rα levels (5) make it almost impossible to substantially increase the density of IL-15-IL-15Rα complexes on antigen-presenting cells by supplying stronger IL-15Rα-binding IL-15 muteins extracellularly. Therefore, we have focused on extending this mutagenesis approach to identify amino acid residues in the hIL-15 helix C domain that enhance binding to the hIL-15Rβ chain to increase IL-15 functional activity. In this study, we demonstrate that one of the resulting hIL-15 mutein with the N72D mutation is able to retain the binding to hIL-15Rα and exhibited enhanced cell binding and biological activity on cells expressing either IL-15Rβ-γc or IL-15Rα-IL-15Rβ-γc complex. Thus this mutein as a free cytokine and in the context of transpresentation by IL-15Rα-bearing cells is expected to provide increased immunostimulatory activity to IL-15Rβ-γc-positive immune effector cells.
Studies have shown that changes in conserved amino acid residues in the center of IL-15 helix A or C lead to an abrogation of functional activity presumably due to the loss of their binding ability to IL-15Rβ (24–26). Similar findings were observed with the TCR/hIL-15 muteins described here where amino acid substitutions at Asp-8 (helix A) or Asn-65 (helix C) resulted in molecules that completely lacked the ability to support cytokine-dependent cell growth (Table I) and inhibited the activity of unmutated hIL-15 on cells expressing IL-15Rα-IL-15Rβ-γc complex (data not shown). These effects are consistent with the involvement of the equivalent residues in hIL-2 that form the hydrogen bond network with side chains of hIL-15Rβ and bound water molecules (22, 23). A similar approach of using structural comparisons of hIL-2 and hIL-15 led us and others to suggest that the asparagine at position 72, located two helical turns from the Asn-65 residue, may also play a role in the binding hIL-15Rβ(16). However, our data indicate that substitutions at this position do not abrogate the hIL-15 biological activity. In fact, changes from Asn-72 to several different amino acids including Ala and Pro generate more active molecules. These findings suggest that the Asn-72 side chain is either not involved or may interfere with binding to hIL-15Rβ. In hIL-2, a glutamic acid residue at the equivalent position (aa 95) forms a salt bridge with the Arg side chain at position 41 of hIL-15Rβ (22, 23). The observation that the hIL-15N72D mutein exhibits high biological activity in our studies suggests that the newly introduced Asp side chain may enable the hIL-15 to form a similar salt bridge with the Arg side chain at position 41 of hIL-15Rβ. The substitution of Asn-72 for Arg or Lys resulted in less active molecules indicating that basic side chains are not favored at this position and support our suggestion that a negatively charged amino acid side chain is required to form such an additional salt bridge with hIL-15Rβ. Together these results demonstrate that changes at position 72 of hIL-15 can give rise to cytokine variants with tunable activity.
There is additional evidence that certain residues in the hIL-15 helix C can affect interactions with the hIL-15Rα chain (25, 26). Muteins carrying substitutions at positions 66 and 67 were found to have reduced binding to hIL-15Rα, hγc-positive TF-1 cells and no biological activity to support TF-1β cell growth. These residues are located in the hydrophobic core of helix C and contact the adjacent helix B involved in hIL-15Rα binding. In contrast, position 72 lies on the solvent-exposed side of helix C and is not predicted to influence the hIL-15Rα binding domain (16). This was verified by this study using cell and hIL-15Rα protein binding assay where the hIL-15N72D mutein showed equivalent hIL-15Rα binding activity as wild type hIL-15. It is also possible that binding of hIL-15Rα affects hIL-15 helix C-hIL-15Rβ interactions. This has been postulated to be the case for IL-2 since formation of the IL-2-IL-2Rα complex causes a conformational change in IL-2 helix C that may allow it to bind IL-2Rβ more effectively (22). Interestingly, IL-2Rα itself does not make any contact with IL-2Rβ or γc chain. Hence the induced conformational change in helix C may be the sole factor providing the increased biological activity of the high affinity IL-2-IL-2Rα-IL-2Rβ-γc complex. Interactions between hIL-15Rα and hIL-15 also result in higher binding affinity to hIL-15Rβ-hγc complex and increased biological potency (32). It remains to be determined if these effects are due to a similar change in hIL-15 conformation and/or interactions of the hIL-15Rα with other receptor chains. Regardless of whether this process occurs or not, the same four to five-fold increase in potency was observed when comparing hIL-15N72D with wild type hIL-15 in hIL-15Rβ -bearing cells with and without the IL-15Rα chain (i.e. 32Dβ and TF-1β cells, see Table II), suggesting that the enhancement of the N72D mutation in IL-15 is independent of hIL-15Rα-mediated effects. Additionally, based on the 132 pM EC50 of rhIL-15 in 32Dβ cell proliferation assays compared with the 4.8 pM EC50 of rhIL-15 N72D in TF-1β cell proliferation assays (Table II), the combined effects of the hIL-15-hIL-15Rα complex formation and hIL-15N72D superagonist activity appeared to result in a 30-fold increase in biological activity. As discussed below, the distinct nature of these effects may be advantageous in generating highly potent immunotherapeutic molecules.
Comparison of rIL-15N72D activity in cells expressing the human or murine IL-15Rα-IL-15Rβ-γc complex indicated that the superagonist activity is likely specific to hIL-15Rβ. While the critical Asp-8 and Asn-65 residues of IL-15 and their putative binding sites in IL-15Rβ [based on the IL-2-IL-2Rβ structure (22, 23)] are conserved between human and mouse, variations in the amino acid residues at position 72 of IL-15 (human: Asn, mouse: Ser) and position 41 of IL-15Rβ (human: Arg, mouse; Leu) suggest that our proposed stabilizing interactions between the N72D mutein and hIL-15Rβ is not formed with the mIL-15Rβ chain. Species-specific differences in the activities of human and mouse IL-15 with cells expressing IL-15R complexes have been described previously (33). In particular, hIL-15 was about 700-fold more active than mIL-15 when signaling through the hIL-15Rβ-γc complex whereas mIL-15 was 200-fold more active than hIL-15 when signaling through the mIL-15Rβ-mγc. It is likely that residues outside of the conserved central helical positions of IL-15 play a key role in the binding interactions and are responsible for the reported species-specific differences (17). To further assess the importance of the C-terminal domain of helix C, efforts are underway to determine whether mutations at position 72 and adjacent residues of mIL-15 helix C are capable of conferring the partial or superagonist activity observed with the hIL-15. Studies using such muteins in mouse models may provide correlative information on potential effects of hIL-15D72N mutein on human immune responses. Additionally, based on the similar binding relationships between four-helix bundle cytokines and γc chain receptors, modification of the C-terminal domain of helix C in other cytokines, including IL-2 and IL-4, may result in partial or super-agonists with enhanced clinical utility.
Function analysis of the binding of paired cytokines, IL-2 and IL-15 or IL-4 and IL-13, to shared receptor complexes suggest that different affinities of the cytokines for each receptor chain and varying concentrations of the cytokines and plasma membrane-bound receptors affect the receptor signaling processes and the resulting biological responses (34, 35). When the cell signaling potentials of rhIL-15 and rhIL-15N72D proteins were compared in TF-1β cells, the rhIL-15N72D mutein stimulated Jak1 and Stat5 phosphorylation in a faster and more intense manner than wild-type rhIL-15. Additionally, the mutein conferred better anti-apoptotic activity than the wild type IL-15 in 32Dβ cells. Since these responses are associated with both IL-15Rβ and γ chains (36, 37), these results suggest that improved binding of the hIL-15N72D mutein to hIL-15Rβ results in a more abundant and/or more stable formation of hIL-15-hIL-15Rβ-hγc complex that is capable of enhancing cell signaling via both receptor chains. When comparing the activity of IL-2 and IL-15 in antigen-activated T cells, Cornish et al. reported that both cytokines were equivalent in stimulating cell proliferation but IL-2 was much more potent at inducing amino acid uptake and protein synthesis than IL-15 (34). It was suggested that these differences were due the distinct ability of each cytokine to bind its receptor and modulate receptor levels resulting in more transient intracellular signaling by IL-15 compared to IL-2. Evaluation of the hIL-15 muteins, particularly the hIL-15N72D superagonist and hIL-15N72R partial agonist, in a similar human immune cell system should provide additional insight into the role of hIL-15-hIL-15Rβ interactions in IL-15-stimulated responses.
A recent review named twelve immunotherapy drugs that could potentially cure cancer (38). When ranked in order of importance, IL-15 was first on list due to its ability to maintain long-lasting T-cell responses to pathogens by supporting the survival of CD8+ cells. Several different approaches are being developed to exploit the therapeutic potential of IL-15. Administration of IL-15 in combination with immunostimulants, chemotherapy or adoptive cell therapies has been shown to provide anti-tumor activity in animal models (39–43). However, the short half-life and need for high doses to achieve biological responses is a major obstacle for therapeutic use of IL-15. A targeted approach designed to concentrate the biological activity of IL-15 at the tumor site may be able to overcome this limitation. For example, a single-chain antibody-IL15 fusion protein directed against the EDB domain of fibronectin was shown to have activity against subcutaneous and metastatic tumors in mice (15). In addition, we have previously demonstrated that hIL-2 fused to a soluble TCR domain that recognized a p53 peptide could provide more potent anti-tumor activity against p53+ subcutaneous tumor xenografts than rhIL-2 alone (1). Both tumor antigen-specific targeting and improved biological half-life contributed to the anti-tumor activity of the TCR/IL-2 fusion protein (2). This approach also shows efficacy at lower doses, thereby reducing the systemic side effects associated with cytokine treatment. We found that a similar fusion could be generated between the p53 peptide-specific TCR and hIL-15, wherein both TCR and hIL-15 domains (including hIL-15N72D superagonist) retained functional activity. Recently, it has been shown that various types of preformed complexes between IL-15 and soluble IL-15Rα could also exhibit enhanced biological activity and increased anti-tumor responses in animal models (5–7, 30, 31). Our cell-based assays indicate that the activity of the rhIL-15 N72D and TCR/hIL-15N72D muteins could be augmented further through interaction with soluble hIL-15Rα (Table III, data not shown), suggesting that these agonist effects act through different mechanism that can be combined in a single strategy. To this end, we have created various fusion molecules to exploit the opportunities of using the hIL-15N72D mutein with the p53-specific TCR molecule as a targeted immunotherapeutic. Successful evaluation of these molecules in tumor efficacy models may allow the advancement of this improved targeted therapeutic approach into clinical testing in patients with p53+ tumors.
Acknowledgements
We would like to thank Dr. Richard A. Morgan, National Cancer Institute, for providing the retroviral vectors and packaging cell lines used in this study.
Footnotes
"This is an author-produced version of a manuscript accepted for publication in The Journal of Immunology (The JI). The American Association of Immunologists, Inc. (AAI), publisher of The JI, holds the copyright to this manuscript. This version of the manuscript has not yet been copyedited or subjected to editorial proofreading by The JI; hence, it may differ from the final version published in The JI (online and in print). AAI (The JI) is not liable for errors or omissions in this author-produced version of the manuscript or in any version derived from it by the U.S. National Institutes of Health or any other third party. The final, citable version of record can be found at www.jimmunol.org."
Disclosures
X. Zhu, W. D. Marcus, W. Xu, H.-I. Lee, K. Han, J. Egan, P. R. Rhode, and H. C. Wong were employees and equity holders of Altor BioScience Corporation at the time of this work.
Abbreviations used in this paper: scTCR, single chain T-cell receptor; γc, common γ chain; hIL, human IL; rhIL, recombinant human IL; mIL, mouse IL
References
- 1.Belmont HJ, Price-Schiavi S, Liu B, Card KF, Lee HI, Han KP, Wen J, Tang S, Zhu X, Merrill J, Chavillaz PA, Wong JL, Rhode PR, Wong HC. Potent antitumor activity of a tumor-specific soluble TCR/IL-2 fusion protein. Clin Immunol. 2006;121:29–39. doi: 10.1016/j.clim.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 2.Wen J, Zhu X, Liu B, You L, Kong L, Lee HI, Han KP, Wong JL, Rhode PR, Wong HC. Targeting activity of a TCR/IL-2 fusion protein against established tumors. Cancer Immunol Immunother. 2008;57:1781–1794. doi: 10.1007/s00262-008-0504-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Waldmann TA. The biology of interleukin-2 and interleukin-15: implications for cancer therapy and vaccine design. Nat Rev Immunol. 2006;6:595–601. doi: 10.1038/nri1901. [DOI] [PubMed] [Google Scholar]
- 4.Ma A, Koka R, Burkett P. Diverse functions of IL-2, IL-15, and IL-7 in lymphoid homeostasis. Annu Rev Immunol. 2006;24:657–679. doi: 10.1146/annurev.immunol.24.021605.090727. [DOI] [PubMed] [Google Scholar]
- 5.Stoklasek TA, Schluns KS, Lefrancois L. Combined IL-15/IL-15Ralpha immunotherapy maximizes IL-15 activity in vivo. J Immunol. 2006;177:6072–6080. doi: 10.4049/jimmunol.177.9.6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dubois S, Patel HJ, Zhang M, Waldmann TA, Muller JR. Preassociation of IL-15 with IL-15R alpha-IgG1-Fc enhances its activity on proliferation of NK and CD8+/CD44high T cells and its antitumor action. J Immunol. 2008;180:2099–2106. doi: 10.4049/jimmunol.180.4.2099. [DOI] [PubMed] [Google Scholar]
- 7.Epardaud M, Elpek KG, Rubinstein MP, Yonekura AR, Bellemare-Pelletier A, Bronson R, Hamerman JA, Goldrath AW, Turley SJ. Interleukin-15/interleukin-15R alpha complexes promote destruction of established tumors by reviving tumor-resident CD8+ T cells. Cancer Res. 2008;68:2972–2983. doi: 10.1158/0008-5472.CAN-08-0045. [DOI] [PubMed] [Google Scholar]
- 8.Kobayashi H, Carrasquillo JA, Paik CH, Waldmann TA, Tagaya Y. Differences of Biodistribution, Pharmacokinetics, and Tumor Targeting between Interleukins 2 and 15. Cancer Res. 2000;60:3577–3583. [PubMed] [Google Scholar]
- 9.Zamai L, Ponti C, Mirandola P, Gobbi G, Papa S, Galeotti L, Cocco L, Vitale M. NK cells and cancer. J Immunol. 2007;178:4011–4016. doi: 10.4049/jimmunol.178.7.4011. [DOI] [PubMed] [Google Scholar]
- 10.Lawyer FC, Stoffel S, Saiki RK, Chang SY, Landre PA, Abramson RD, Gelfand DH. High-level expression, purification, and enzymatic characterization of full-length Thermus aquaticus DNA polymerase and a truncated form deficient in 5' to 3' exonuclease activity. PCR Methods Appl. 1993;2:275–287. doi: 10.1101/gr.2.4.275. [DOI] [PubMed] [Google Scholar]
- 11.Hughes MS, Yu YY, Dudley ME, Zheng Z, Robbins PF, Li Y, Wunderlich J, Hawley RG, Moayeri M, Rosenberg SA, Morgan RA. Transfer of a TCR gene derived from a patient with a marked antitumor response conveys highly active T-cell effector functions. Hum Gene Ther. 2005;16:457–472. doi: 10.1089/hum.2005.16.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang S, Rosenberg SA, Morgan RA. Clinical-scale lentiviral vector transduction of PBL for TCR gene therapy and potential for expression in less-differentiated cells. J Immunother. 2008;31:830–839. doi: 10.1097/CJI.0b013e31818817c5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu X, Belmont HJ, Price-Schiavi S, Liu B, Lee HI, Fernandez M, Wong RL, Builes J, Rhode PR, Wong HC. Visualization of p53264-272/HLA-A*0201 Complexes Naturally Presented on Tumor Cell Surface by a Multimeric Soluble Single-Chain T Cell Receptor. J Immunol. 2006;176:3223–3232. doi: 10.4049/jimmunol.176.5.3223. [DOI] [PubMed] [Google Scholar]
- 14.Ruckert R, Brandt K, Hofmann U, Bulfone-Paus S, Paus R. IL-2-IgG2b fusion protein suppresses murine contact hypersensitivity in vivo. J Invest Dermatol. 2002;119:370–376. doi: 10.1046/j.1523-1747.2002.01849.x. [DOI] [PubMed] [Google Scholar]
- 15.Kaspar M, Trachsel E, Neri D. The antibody-mediated targeted delivery of interleukin-15 and GM-CSF to the tumor neovasculature inhibits tumor growth and metastasis. Cancer Res. 2007;67:4940–4948. doi: 10.1158/0008-5472.CAN-07-0283. [DOI] [PubMed] [Google Scholar]
- 16.Chirifu M, Hayashi C, Nakamura T, Toma S, Shuto T, Kai H, Yamagata Y, Davis SJ, Ikemizu S. Crystal structure of the IL-15-IL-15Ralpha complex, a cytokine-receptor unit presented in trans. Nat Immunol. 2007;8:1001–1007. doi: 10.1038/ni1492. [DOI] [PubMed] [Google Scholar]
- 17.Olsen SK, Ota N, Kishishita S, Kukimoto-Niino M, Murayama K, Uchiyama H, Toyama M, Terada T, Shirouzu M, Kanagawa O, Yokoyama S. Crystal Structure of the interleukin-15.interleukin-15 receptor alpha complex: insights into trans and cis presentation. J Biol Chem. 2007;282:37191–37204. doi: 10.1074/jbc.M706150200. [DOI] [PubMed] [Google Scholar]
- 18.Johnston JA, Bacon CM, Finbloom DS, Rees RC, Kaplan D, Shibuya K, Ortaldo JR, Gupta S, Chen YQ, Giri JD, et al. Tyrosine phosphorylation and activation of STAT5, STAT3, and Janus kinases by interleukins 2 and 15. Proc Natl Acad Sci U S A. 1995;92:8705–8709. doi: 10.1073/pnas.92.19.8705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meazza R, Basso S, Gaggero A, Detotero D, Trentin L, Pereno R, Azzarone B, Ferrini S. Interleukin (IL)-15 induces survival and proliferation of the growth factor-dependent acute myeloid leukemia M-07e through the IL-2 receptor beta/gamma. Int J Cancer. 1998;78:189–195. doi: 10.1002/(sici)1097-0215(19981005)78:2<189::aid-ijc12>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 20.Russell SM, Johnston JA, Noguchi M, Kawamura M, Bacon CM, Friedmann M, Berg M, McVicar DW, Witthuhn BA, Silvennoinen O, et al. Interaction of IL-2R beta and gamma c chains with Jak1 and Jak3: implications for XSCID and XCID. Science. 1994;266:1042–1045. doi: 10.1126/science.7973658. [DOI] [PubMed] [Google Scholar]
- 21.Van Parijs L, Refaeli Y, Lord JD, Nelson BH, Abbas AK, Baltimore D. Uncoupling IL-2 signals that regulate T cell proliferation, survival, and Fas-mediated activation-induced cell death. Immunity. 1999;11:281–288. doi: 10.1016/s1074-7613(00)80103-x. [DOI] [PubMed] [Google Scholar]
- 22.Wang X, Rickert M, Garcia KC. Structure of the quaternary complex of interleukin-2 with its alpha, beta, and gammac receptors. Science. 2005;310:1159–1163. doi: 10.1126/science.1117893. [DOI] [PubMed] [Google Scholar]
- 23.Stauber DJ, Debler EW, Horton PA, Smith KA, Wilson IA. Crystal structure of the IL-2 signaling complex: paradigm for a heterotrimeric cytokine receptor. Proc Natl Acad Sci U S A. 2006;103:2788–2793. doi: 10.1073/pnas.0511161103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pettit DK, Bonnert TP, Eisenman J, Srinivasan S, Paxton R, Beers C, Lynch D, Miller B, Yost J, Grabstein KH, Gombotz WR. Structure-Function Studies of Interleukin 15using Site-specific Mutagenesis, Polyethylene Glycol Conjugation, and Homology Modeling. J. Biol. Chem. 1997;272:2312–2318. doi: 10.1074/jbc.272.4.2312. [DOI] [PubMed] [Google Scholar]
- 25.Bernard J, Harb C, Mortier E, Quemener A, Meloen RH, Vermot-Desroches C, Wijdeness J, van Dijken P, Grotzinger J, Slootstra JW, Plet A, Jacques Y. Identification of an interleukin-15alpha receptor-binding site on human interleukin-15. J Biol Chem. 2004;279:24313–24322. doi: 10.1074/jbc.M312458200. [DOI] [PubMed] [Google Scholar]
- 26.Quemener A, Bernard J, Mortier E, Plet A, Jacques Y, Tran V. Docking of human interleukin-15 to its specific receptor alpha chain: correlation between molecular modeling and mutagenesis experimental data. Proteins. 2006;65:623–636. doi: 10.1002/prot.21103. [DOI] [PubMed] [Google Scholar]
- 27.Studier FW, Rosenberg AH, Dunn JJ, Dubendorff JW. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 28.Bamford RN, Grant AJ, Burton JD, Peters C, Kurys G, Goldman CK, Brennan J, Roessler E, Waldmann TA. The interleukin (IL) 2 receptor beta chain is shared by IL-2 and a cytokine, provisionally designated IL-T, that stimulates T-cell proliferation and the induction of lymphokine-activated killer cells. Proc Natl Acad Sci U S A. 1994;91:4940–4944. doi: 10.1073/pnas.91.11.4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giron-Michel J, Fogli M, Gaggero A, Ferrini S, Caignard A, Brouty-Boye D, Baouz S, Le Bousse-Kerdiles MC, Peault B, van Dijk M, Bulfone-Paus S, Durali D, Chouaib S, Azzarone B. Detection of a functional hybrid receptor gammac/GM-CSFRbeta in human hematopoietic CD34+ cells. J Exp Med. 2003;197:763–775. doi: 10.1084/jem.20020150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rubinstein MP, Kovar M, Purton JF, Cho JH, Boyman O, Surh CD, Sprent J. Converting IL-15 to a superagonist by binding to soluble IL-15R{alpha} Proc Natl Acad Sci U S A. 2006;103:9166–9171. doi: 10.1073/pnas.0600240103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mortier E, Quemener A, Vusio P, Lorenzen I, Boublik Y, Grotzinger J, Plet A, Jacques Y. Soluble interleukin-15 receptor alpha (IL-15R alpha)-sushi as a selective and potent agonist of IL-15 action through IL-15R beta/gamma. Hyperagonist IL-15 x IL-15R alpha fusion proteins. J Biol Chem. 2006;281:1612–1619. doi: 10.1074/jbc.M508624200. [DOI] [PubMed] [Google Scholar]
- 32.Bouchaud G, Garrigue-Antar L, Sole V, Quemener A, Boublik Y, Mortier E, Perdreau H, Jacques Y, Plet A. The exon-3-encoded domain of IL-15ralpha contributes to IL-15 high-affinity binding and is crucial for the IL-15 antagonistic effect of soluble IL-15Ralpha. J Mol Biol. 2008;382:1–12. doi: 10.1016/j.jmb.2008.07.019. [DOI] [PubMed] [Google Scholar]
- 33.Eisenman J, Ahdieh M, Beers C, Brasel K, Kennedy MK, Le T, Bonnert TP, Paxton RJ, Park LS. Interleukin-15 interactions with interleukin-15 receptor complexes: characterization and species specificity. Cytokine. 2002;20:121–129. doi: 10.1006/cyto.2002.1989. [DOI] [PubMed] [Google Scholar]
- 34.Cornish GH, Sinclair LV, Cantrell DA. Differential regulation of T-cell growth by IL-2 and IL-15. Blood. 2006;108:600–608. doi: 10.1182/blood-2005-12-4827. [DOI] [PubMed] [Google Scholar]
- 35.LaPorte SL, Juo ZS, Vaclavikova J, Colf LA, Qi X, Heller NM, Keegan AD, Garcia KC. Molecular and structural basis of cytokine receptor pleiotropy in the interleukin-4/13 system. Cell. 2008;132:259–272. doi: 10.1016/j.cell.2007.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lindemann MJ, Benczik M, Gaffen SL. Anti-apoptotic signaling by the interleukin-2 receptor reveals a function for cytoplasmic tyrosine residues within the common gamma (gamma c) receptor subunit. J Biol Chem. 2003;278:10239–10249. doi: 10.1074/jbc.M209471200. [DOI] [PubMed] [Google Scholar]
- 37.Gaffen SL. Signaling domains of the interleukin 2 receptor. Cytokine. 2001;14:63–77. doi: 10.1006/cyto.2001.0862. [DOI] [PubMed] [Google Scholar]
- 38.Cheever MA. Twelve immunotherapy drugs that could cure cancers. Immunol Rev. 2008;222:357–368. doi: 10.1111/j.1600-065X.2008.00604.x. [DOI] [PubMed] [Google Scholar]
- 39.Chapoval AI, Fuller JA, Kremlev SG, Kamdar SJ, Evans R. Combination chemotherapy and IL-15 administration induce permanent tumor regression in a mouse lung tumor model: NK and T cell-mediated effects antagonized by B cells. J Immunol. 1998;161:6977–6984. [PubMed] [Google Scholar]
- 40.Wysocka M, Benoit BM, Newton S, Azzoni L, Montaner LJ, Rook AH. Enhancement of the host immune responses in cutaneous T-cell lymphoma by CpG oligodeoxynucleotides and IL-15. Blood. 2004;104:4142–4149. doi: 10.1182/blood-2004-03-1190. [DOI] [PubMed] [Google Scholar]
- 41.Klebanoff CA, Finkelstein SE, Surman DR, Lichtman MK, Gattinoni L, Theoret MR, Grewal N, Spiess PJ, Antony PA, Palmer DC, Tagaya Y, Rosenberg SA, Waldmann TA, Restifo NP. IL-15 enhances the in vivo antitumor activity of tumor-reactive CD8+ T cells. Proc Natl Acad Sci U S A. 2004;101:1969–1974. doi: 10.1073/pnas.0307298101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Teague RM, Sather BD, Sacks JA, Huang MZ, Dossett ML, Morimoto J, Tan X, Sutton SE, Cooke MP, Ohlen C, Greenberg PD. Interleukin-15 rescues tolerant CD8+ T cells for use in adoptive immunotherapy of established tumors. Nat Med. 2006;12:335–341. doi: 10.1038/nm1359. [DOI] [PubMed] [Google Scholar]
- 43.Evans R, Fuller JA, Christianson G, Krupke DM, Troutt AB. IL-15 mediates anti-tumor effects after cyclophosphamide injection of tumorbearing mice and enhances adoptive immunotherapy: the potential role of NK cell subpopulations. Cell Immunol. 1997;179:66–73. doi: 10.1006/cimm.1997.1132. [DOI] [PubMed] [Google Scholar]