Abstract
Signaling by the epidermal growth factor receptor requires an allosteric interaction between the kinase domains of two receptors, whereby one activates the other. We show that the intracellular juxtamembrane segment of the receptor, known to potentiate kinase activity, is able to dimerize the kinase domains. The C-terminal half of the juxtamembrane segment latches the activated kinase domain to the activator, and the N-terminal half of this segment further potentiates dimerization, most likely by forming an antiparallel helical dimer that engages the transmembrane helices of the activated receptor. Our data are consistent with a mechanism in which the extracellular domains block the intrinsic ability of the transmembrane and cytoplasmic domains to dimerize and activate, with ligand binding releasing this block. The formation of the activating juxtamembrane latch is prevented by the C-terminal tails in a new structure of an inactive kinase domain dimer, suggesting how alternative dimers can prevent ligand-independent activation.
Introduction
Inter-cellular signaling in animals relies on receptor tyrosine kinases that consist of an extracellular ligand binding domain and a cytoplasmic kinase domain (Hubbard and Miller, 2007). A distinct sub-family of these receptors includes the epidermal growth factor receptor (EGFR, also known as ErbB1 or Her1) and its three homologs in humans: ErbB2/Her2 and ErbB4/Her4, which are kinase-active, and ErbB3/Her3, which is not (Landau and Ben-Tal, 2008; Schlessinger, 2002; Yarden and Sliwkowski, 2001). The ligand-dependent dimerization of various combinations of EGFR family members results in phosphorylation of their C-terminal tail segments, which is crucial for cellular proliferation and survival.
Activation of the catalytic domain of EGFR family members is controlled primarily by an allosteric interaction between two protein kinase domains in an asymmetric dimer, rather than by phosphorylation (Gotoh et al., 1992; Stamos et al., 2002; Zhang et al., 2006). The kinase domain of one receptor molecule (the activator) plays a role analogous to that of a cyclin bound to a cyclin-dependent protein kinase, and activates the kinase domain of a second receptor (the receiver) (Zhang et al., 2006) (Figure 1A). The formation of the asymmetric dimer appears to underlie the activation of all EGFR family members (Qiu et al., 2008; Zhang et al., 2006).
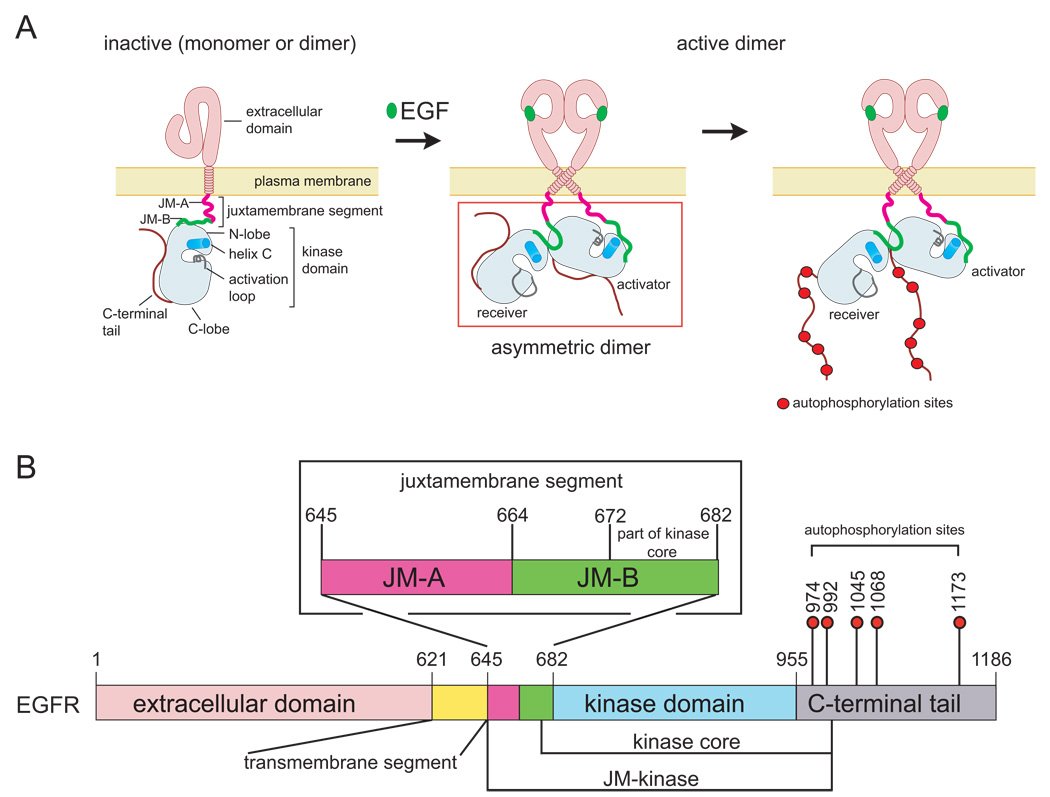
Figure 1. Schematic Diagrams of EGFR.
(A) Activation of EGFR by EGF results in the formation of an asymmetric kinase domain dimer. (B) Domains of EGFR. Residue numbering corresponds to human EGFR, excluding the signal sequence.
Upon binding ligand, the extracellular domains of EGFR family members dimerize such that their C-terminal ends are brought close together at the junction with the transmembrane segments (Burgess et al., 2003). The transmembrane segments connect to the cytoplasmic juxtamembrane segments of the receptor. The role of the juxtamembrane segment of EGFR family members is distinct from that of typical receptor tyrosine kinases because it activates, rather than inhibits, the kinase domain (Thiel and Carpenter, 2007). The nature of this coupling between the juxtamembrane segment and the kinase domain is not understood.
The juxtamembrane segment of human EGFR spans residues 645 to 682, and we refer to the N-terminal half (residues 645 to 663) as JM-A, and the C-terminal half (residues 664 to 682) as JM-B (Figure 1B). An examination of crystal lattice contacts in a previously reported structure of the Her4 kinase domain (Wood et al., 2008) reveals that the JM-B portion of the juxtamembrane segment forms a clamp that reaches across from the N-terminal lobe of the receiver kinase domain in an asymmetric dimer to engage the C-terminal lobe of the activator kinase domain. The significance of this interaction has not been noted previously, but we demonstrate that this interface involving JM-B, which we refer to as the “juxtamembrane latch,” is crucial for receptor activation.
We show that JM-A segments on the receiver and the activator are both required for dimerization and activation, and we propose that the two JM-A segments in an asymmetric kinase domain dimer form short α-helices that are likely to interact in an antiparallel manner and connect to the C-terminal ends of the dimeric form of the transmembrane helices. This allows a model for the entire activated receptor to be built, in which ligand engagement by the extracellular domains stabilizes the formation of the JM-A helical dimer which, in turn, stabilizes the asymmetric kinase domain dimer, resulting in activation.
We have determined a new structure of the EGFR kinase core in which formation of the juxtamembrane latch is blocked by the C-terminal tails of the receptor. This structure forms a symmetrical dimer of inactive kinase domains, and suggests a potential mechanism whereby alternative dimers can prevent ligand-independent activation.
Results and Discussion
The Juxtamembrane Segment Activates the Kinase Domain in Solution
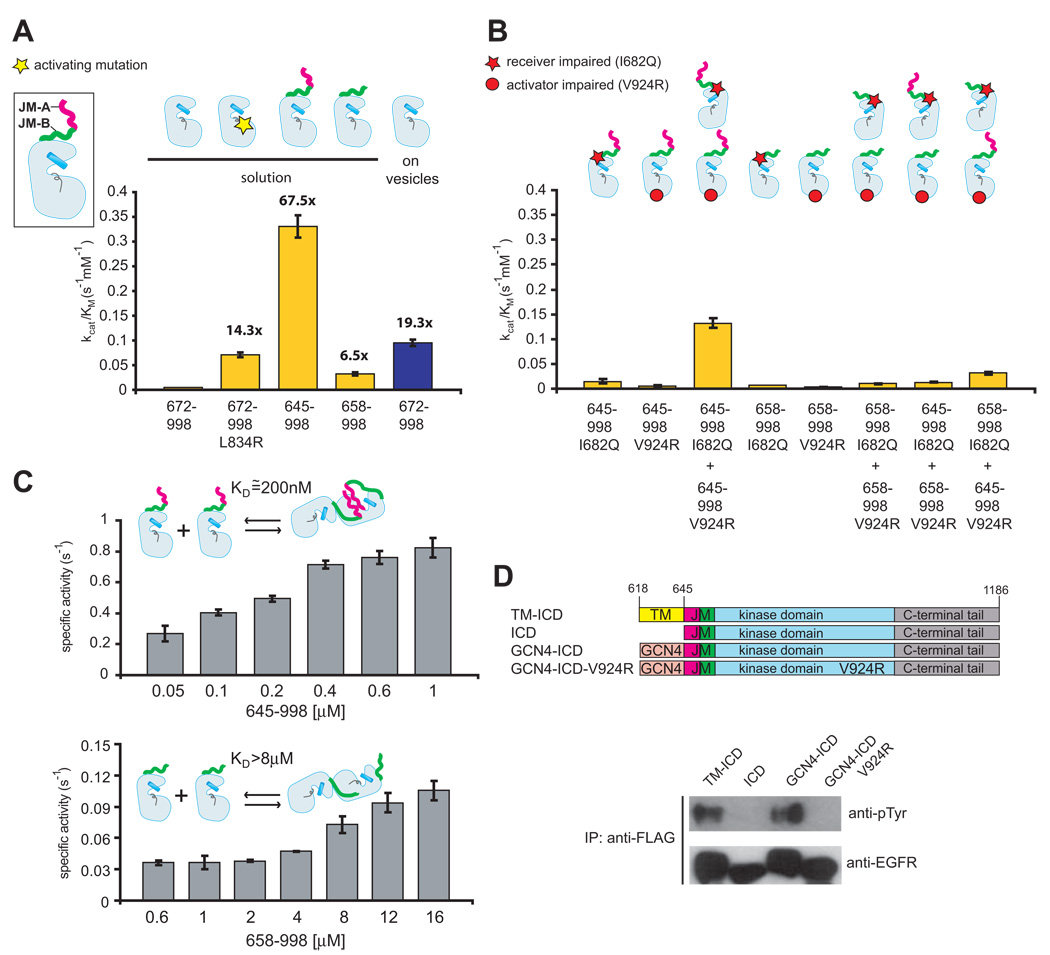
The core kinase domain has low activity, when measured in solution using purified protein (the catalytic efficiency, kcat/KM, is 0.0049 ± 0.0005 s−1mM−1, Figure 2A). Introduction of the L834R mutation, commonly found in lung cancer patients, increases the activity of the kinase core by ~14-fold, consistent with previous results (Yun et al., 2007; Zhang et al., 2006) (Figure 2A). Attachment of the juxtamembrane segment to the wild type kinase domain results in substantially greater activity. The value of kcat/KM for the JM-kinase construct (0.33 ± 0.02 s−1mM−1) is ~70-fold greater than for the kinase core alone. The activity of the kinase core is increased ~20-fold by concentrating it on lipid vesicles (Zhang et al., 2006), but the addition of the juxtamembrane segment results in greater catalytic efficiency in solution, without concentration on vesicles (Figure 2A).
Figure 2. The Effect of the Juxtamembrane Segment on Activity.
(A) Catalytic efficiency (kcat/KM) of the kinase core (residues 672–998) in solution (yellow) and on vesicles (blue), compared to the catalytic efficiency in solution of constructs that include the full juxtamembrane segment (JM-A and JM-B, residues 645–998) or only JM-B (residues 658–998). The values of kcat/KM were obtained from the linear dependence of reaction velocity on substrate concentration at low substrate concentration, and the error bars are derived from the linear fit (Zhang et al., 2006). (B) The activity of constructs that are either receiver-impaired (restricted to serve as activators, with the I682Q mutation) or activator-impaired (restricted to serve as receivers, with the V924R mutation). (C) Concentration-dependent change in specific activity, in solution, for the JM-kinase construct (containing both JM-A and JM-B, residues 645–998) and a construct containing JM-B but lacking JM-A (residues 658–998). Data shown are mean values from two independent experiments ± STD. (D) EGFR constructs were immunoprecipitated from cell lysates using an anti-FLAG antibody and EGFR autophosphorylation was examined by immunobloting using an anti-phosphotyrosine antibody (anti-pTyr).
Dimerization and Activation Requires Intact Juxtamembrane Segments on both the Receiver and the Activator Kinase Domains
Deletion of the JM-A segment reduces catalytic efficiency by ten-fold, to 0.032 ± 0.003 s−1mM−1 (Figure 2A), showing that an intact juxtamembrane segment is required for full activation. In order to determine if the JM-A segment is required on both the activator and receiver, we made mutant kinases that can only take the activator or the receiver position. We refer to kinases with an I682Q mutation, which can take only the activator position, as “receiver-impaired” (Zhang et al., 2006). Kinases with a V924R mutation cannot be activated, and are referred to as “activator-impaired”. Both the activator-impaired and receiver-impaired JM-kinase constructs have low activity alone, but mixing these constructs recovers ~50% of the activity of wild type JM-kinase constructs in solution, as observed in cell transfection studies (Thiel and Carpenter, 2007) (Figure 2B). In contrast, activity is reduced significantly if the JM-A segment on either the activator-impaired or the receiver-impaired kinase is missing (Figure 2B). Thus, both JM-A and JM-B segments are required on both the activator and the receiver kinases for robust stimulation of activity.
The core kinase domain is a monomer, based on static light scattering at a concentration of 150 µM, whereas the JM-kinase construct is predominantly a dimer with no detectable monomer (Figure S1). The specific activity of the JM-kinase construct increases with concentration, and the dissociation constant (KD) for dimerization is estimated to be ~200 nM (Figure 2C). Deletion of the JM-A segment increases the midpoint value of the concentration dependence curve to at least ~8 µM (Figure 2C). The >40 fold increase in the estimated values for KD for dimerization upon deletion of the JM-A segment provides strong evidence for a role of the JM-A segment in dimerization.
Activation by the Transmembrane and Juxtamembrane Segments Is Comparable to the Effects of Enforced Dimerization
We used cell-based assays to compare the activities of various constructs containing the JM-kinase portion of the receptor. One construct includes the transmembrane and intracellular domains of EGFR (TM-ICD). The other construct is a fusion of the intracellular domains to 29 residues of the coiled-coil portion of the transcriptional activator GCN4 (O'Shea et al., 1991), which is expected to enforce constitutive dimerization on the intracellular domains.
We transfected COS7 cells with a ~5-fold lower level of DNA than used in the previous study (Thiel and Carpenter, 2007) (Figure 2D). Under these conditions, the intracellular domains alone display very low levels of autophosphorylation in an immunoprecipitation assay. In contrast, a construct that contains the transmembrane segment, but without the extracellular segment (TM-ICD), shows high activity. The activity of the TM-ICD construct is about the same as that for the GCN4-ICD construct, indicating that the effect of the juxtamembrane segment, when fused to the transmembrane domain and localized to membranes, is comparable to that of enforced dimerization. This result is consistent with previous experiments showing that removal of the extracellular domains activates the kinase domains of EGFR (Chantry, 1995; Nishikawa et al., 1994; Zhu et al., 2003).
Many kinases are activated by imposed dimerization, where the effect could simply be due to enhancement of trans-phosphorylation. Introduction of the V924R mutation in the GCN4-ICD construct, which is expected to disrupt the asymmetric dimer interface, results in a complete loss of activity (Figure 2D). Thus, even when dimerization is enforced by GCN4, formation of the asymmetric dimer appears to be essential for activity.
A Structure of the Her4 Kinase Domain Suggests a Latching Function for the JM-B Segment of the Juxtamembrane Domain
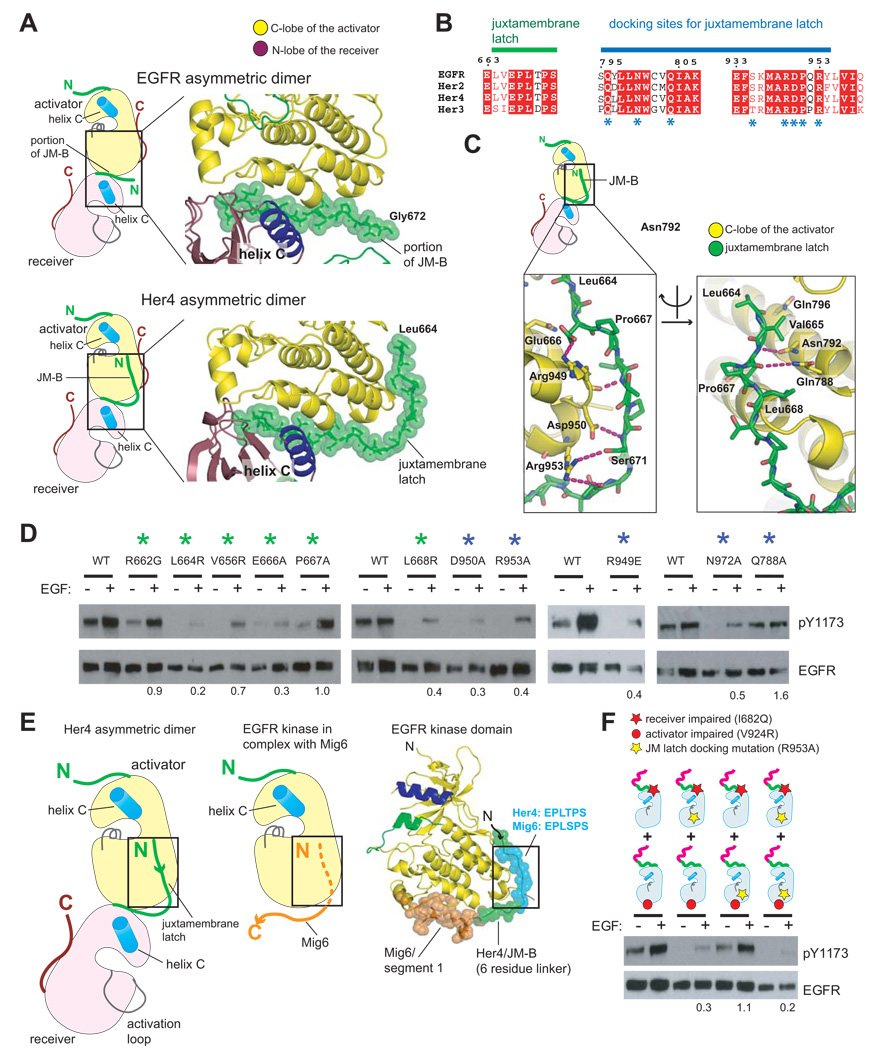
A crystal structure of the Her4 kinase domain bound to a covalently linked inhibitor (PDB ID: 2R4B) (Wood et al., 2008) provides insight into the structural basis for the role of the juxtamembrane segment. Crystallization of the Her4–inhibitor complex inhibitor utilized a construct that includes the JM-B portion of the juxtamembrane segment (Wood et al., 2008). Although the authors do not comment on this fact, the kinase domains in their crystal structure form an asymmetric dimer that is similar in general terms to that seen in other crystal structures for active forms of EGFR and Her4. The kinase domain of Her4 in this complex is in an inactive conformation, which is required for accommodation of the covalently bound inhibitor (Wood et al., 2008). Nevertheless, the crystal lattice generates a “daisy chain” of kinase domains in which each molecule in the chain is docked on the next one, as in crystals of the active kinase domain.
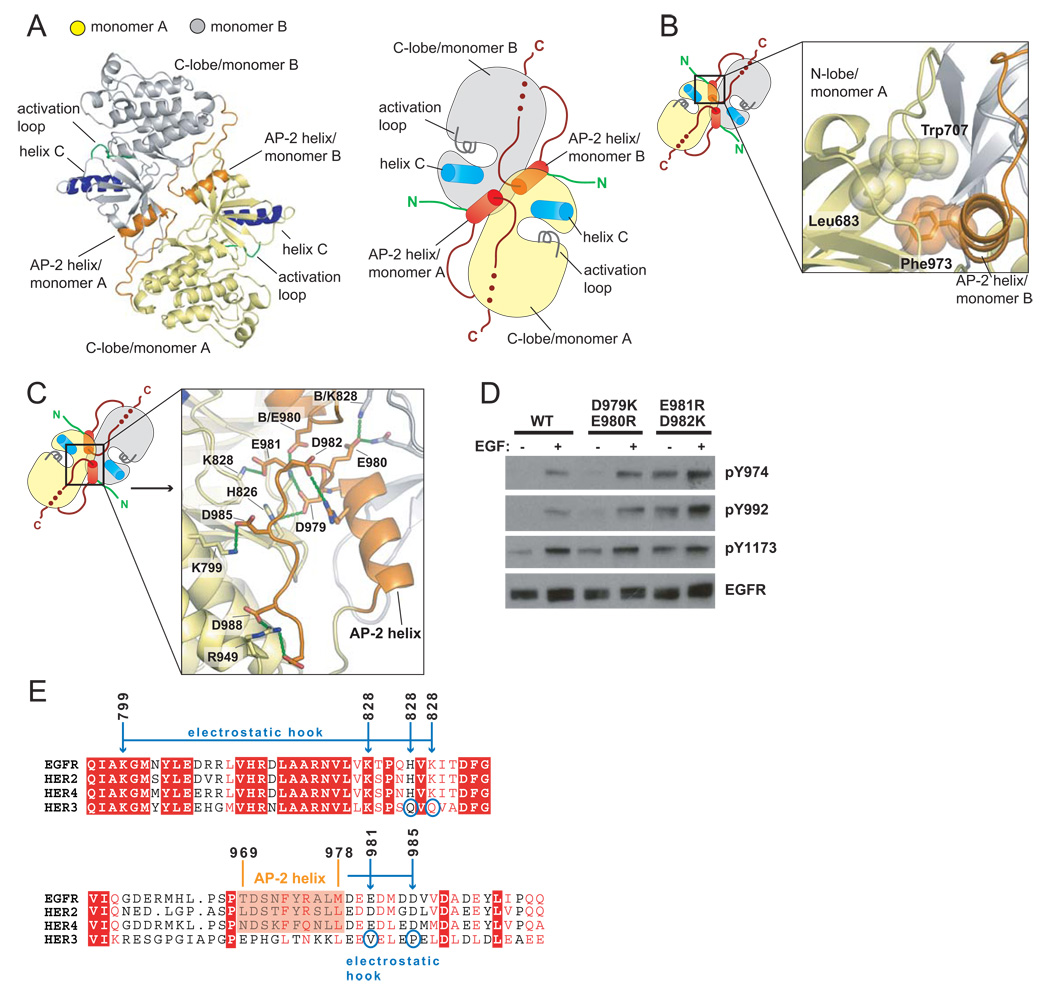
The last ten residues of JM-B (corresponding to Gly 672 to Ile 682 in EGFR) have been visualized previously at the asymmetric dimer interface, and are critical for EGFR activation (Zhang et al., 2006). What is new in the Her4-inhibitor complex is that the rest of the JM-B segment, provided by the kinase domain in the receiver position, latches two kinase domains together by running along the surface of the C-lobe of the activator kinase domain (Figure 3A).
Figure 3. Role of the Juxtamembrane Latch in Activation of EGFR.
(A) Comparison of the structures of asymmetric dimers of kinase domains for EGFR (PDB ID: 2GS6), (Zhang et al., 2006) and Her4 (PDB ID: 2R4B), (Wood et al., 2008). Residues are identified using EGFR numbering. (B) Sequence conservation in the juxtamembrane latch/C-lobe interface. Residues interacting with the juxtamembrane latch are indicated by asterisks. (C) Detailed view of the structure of the juxtamembrane latch in the Her4 structure (PDB ID: 2R4B), with residues identified by EGFR numbering. (D) Effect of mutating residues involved in formation of the juxtamembrane latch. The level of EGF-stimulated phosphorylation on Tyr 1173 relative to wild type, after normalizing for EGFR levels, is shown below each lane. (E) Comparison of the juxtamembrane latch with the docking of the EGFR inhibitor, Mig6 (PDB ID: 2RFE). (F) The effect of a mutation that prevents docking of the juxtamembrane latch (R953A). The results of co-transfection experiments using full length EGFR receptor variants that are receiver-impaired (I682Q) or activator-impaired (V924R) are shown. The level of EGF-stimulated phosphorylation relative to I682Q and V924R co-transfection in the wild type background, after normalizing for EGFR levels, is shown below each lane.
Formation of the juxtamembrane latch involves residues in the receiver and activator kinases that are conserved in EGFR family members (Figure 3B). The interaction involves several hydrogen bonds and hydrophobic contacts (see Figure 3C). Mutations in C-lobe residues that anchor the JM-B region (e.g., Asn 972, Arg 949, Asp 950, Arg 953 in the C-lobe) have substantial inhibitory effects on EGFR autophosphorylation in cell-based assays (Figure 3D). Mutation of Glu 666 in the JM-B region, which forms an ion pair with Arg 949, is also inhibitory. Three hydrophobic residues in the JM-B segment (Leu 664, Val 656, Leu 668) are also essential for EGFR activation (Figure 3D). Leu 668 packs against Pro 951 in the C-lobe of the kinase domain. Leu 664 and Val 665 are located near the junction of JM-B with the N-terminal JM-A segment.
A segment spanning residues 315–374 of the EGFR inhibitor Mig6 blocks asymmetric dimer formation (Zhang et al., 2007). A six residue motif (EPLTPS) in the juxtamembrane latch is almost identical in sequence with six residues in Mig6 (residues 323–327, with sequence EPLSPS). This sequence motif is located six residues upstream of the first ordered residue in the crystal structure containing Mig6, and so was not seen in that complex. The EPLSPS motif of Mig6 can be docked onto the C-lobe of the kinase domain based on the corresponding motif in the juxtamembrane latch, while allowing the rest of Mig6 to block the interface between the activator and receiver kinase domains (Figure 3E). This indicates that part of the function of Mig6 is to prevent formation of the juxtamembrane latch.
The Juxtamembrane Latch Involves the C-lobe of the Activator Kinase Domain, and not that of the Receiver
A crucial assumption in our subsequent modeling is that the JM-kinase portion of the receptor forms a closed dimer, in which the two juxtamembrane segments interact with each other rather than repeating identical interactions in a daisy chain. We used cell transfection assays to confirm that the juxtamembrane latch is engaged only by the activator and not by the receiver in activated full length receptors.
When EGFR variants that are receiver-impaired or activator-impaired are co-expressed, robust EGF-dependent activity is observed (Figure 3F). We introduced a mutation in the C-lobe of the kinase domain (R953A) that disrupts the juxtamembrane latch (see Figure 3F). If the mutation is introduced only in the receiver-impaired EGFR construct, which presumably serves only as an activator, activity is reduced in co-transfection experiments. There is essentially no effect on EGFR activity if the R953A mutation is introduced in the activator-impaired kinase domain. These results are consistent with the formation of a closed dimer and imply that the juxtamembrane segment of the activator may be free to interact with the receiver.
The JM-A Segment is Likely to be Helical
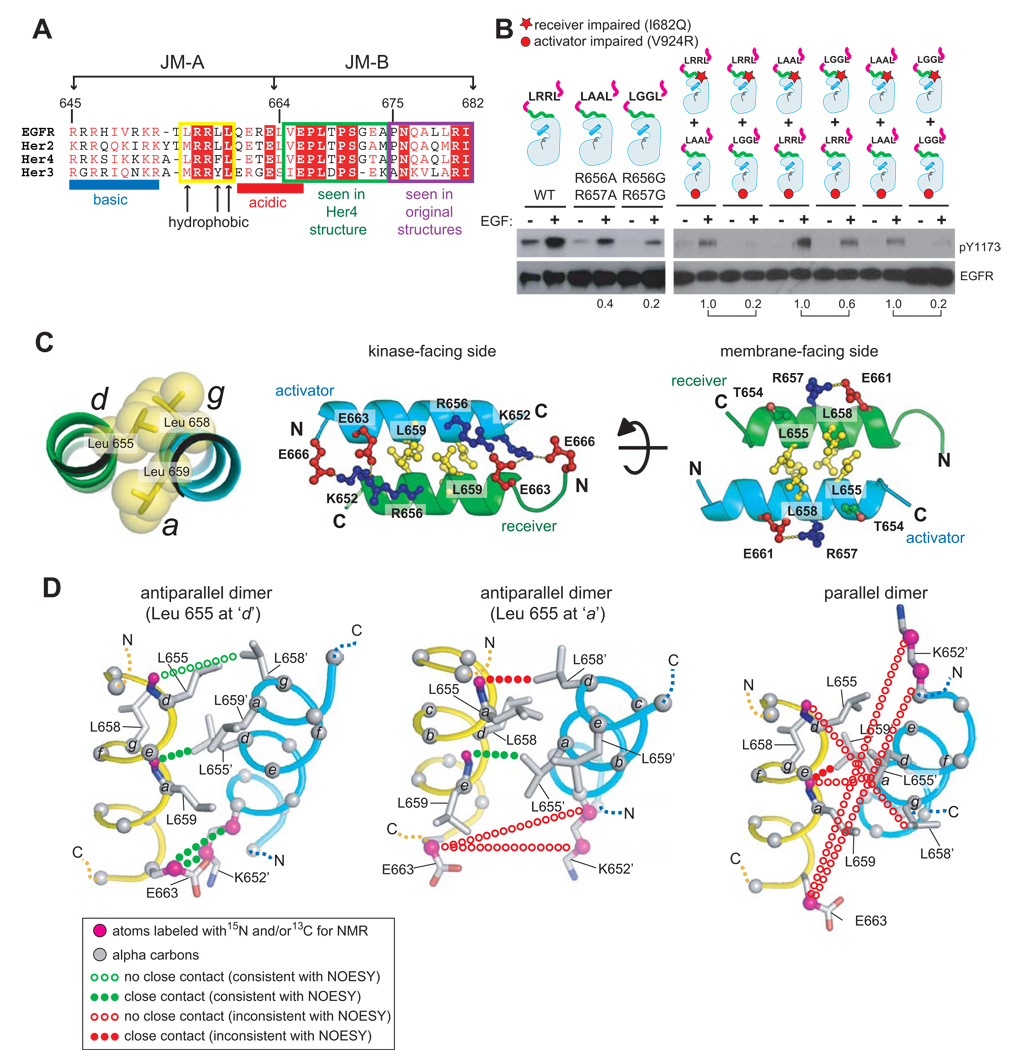
The JM-A segments have three elements that are conserved among EGFR family members (Figure 4A). The N-terminal region contains 5–7 basic residues, including several arginines. At least some of these residues are likely to interact with phosphate groups in the membrane bilayer (McLaughlin et al., 2005). The central portion contains hydrophobic residues in an i, i+3, i+4 pattern within an LRRLL motif in EGFR. The C-terminal region contains acidic residues.
Figure 4. A Helical Dimer in the JM-A Segment.
(A) Alignment of the sequences of the juxtamembrane segments of EGFR family members. (B) Comparison of the effects of alanine and glycine substitutions in the JM-A segment. The first three panels compare activity of the full length wild type receptor with that for variants in which Arg 656 and Arg 657 are replaced by alanine and glycine. The next six panels shows results of co-transfection experiments using activator-impaired and receiver-impaired variants of the receptor, and compare the results of alanine and glycine substitutions in each variant. (C) The modeled antiparallel JM-A helical dimer, with Leu655 at the d position of a heptad motif in one helix and Leu658 and Leu659 at the g and a positions in the second helix. (D) Models for antiparallel (left and middle, with Leu 655 at the d and a positions, respectively) and parallel (right, with Leu 655 at the d position). The dotted lines indicate interatomic contacts that are either consistent or inconsistent with NMR data for a peptide containing two tandem repeats of the JM-A segment (see Supplemental Data).
The i, i+3, i+4 pattern of hydrophobic residues in JM-A suggests an α-helical structure. The structure of a micelle-bound peptide containing the juxtamembrane segment of EGFR has been determined by NMR (Choowongkomon et al., 2005). The relevance of this structure, in which the JM-A segment is helical, is uncertain because one portion of this peptide is normally integrated into the folded structure of the kinase domain core as a β strand, but instead adopts an α-helical conformation in this micelle-bound peptide.
We analyzed an isolated 15 residue peptide spanning the JM-A segment by solution NMR. These data provide evidence for the transient formation of an α-helix spanning the length of the peptide, and also for a concentration-dependent interaction between peptides (see Figure S2 and Supplemental Data). We used cell transfection experiments to probe the conformation of the JM-A segment in the full length receptor, taking advantage of the fact that glycine residues weaken an α-helix but alanine residues do not (Pace and Scholtz, 1998). We examined the activity of full length EGFR constructs with arginine residues in the 655LRRLL659 motif in JM-A replaced by glycines. This led to a significant reduction in EGFR activity (Figure 4B and Figure S3A). To account for the effects of charge removal, as opposed to helix weakening, we compared the effects of the glycine substitutions to that of alanine substitutions. The effect of alanine substitutions is smaller than that of glycine substitutions, consistent with a helical conformation for the JM-A segment (Figure 4B and Figure S3A).
The results of co-transfection experiments using activator-impaired and receiver-impaired kinases show that replacement of Arg 656 and Arg 657 by glycines on either the activator or receiver kinase individually also results in a reduction of EGFR activity relative to alanine substitutions (Figure 4B and Figure S3A). These data indicate that a helical conformation for the JM-A segment is important for both the activator and the receiver.
A Potential Antiparallel Helical Dimer Interaction in the Juxtamembrane Segment
Mutation of the three leucine residues in the LRRLL motif (residues 655, 658 and 659) to either alanine or aspartic acid attenuates the activity of the receptor (Figure S3B). This led us to consider models for helical dimers in which the JM-A segments are packed closely together with a hydrophobic interface, in either an antiparallel or a parallel arrangement. We based our models on the structures of coiled coils (Woolfson, 2005), but because we are considering only a short helical segment the predicted intermolecular contacts are similar for tightly packed straight helices. For each choice of orientation, parallel or antiparallel, there are two choices of sequence register, corresponding to whether the first leucine or the fifth one is at the a position of a heptad sequence motif (Woolfson, 2005).
If the orientation were parallel, several basic residues would be brought close together at the N-terminal ends of the two helices, which is likely to be energetically unfavorable. Only one inter-molecular ion pair is predicted for the parallel arrangement, and it involves Arg 662 and a glutamate sidechain (Glu 661 in one register and Glu 663 in the other; Figure S4A). The parallel arrangement puts two such ion pairs close together by symmetry, as shown in Figure S4A. Mutation of Arg 662 to glutamate would therefore place four glutamate sidechains in close proximity in the parallel dimer (Figure S4A) which should destabilize the dimer. Mutation of Arg 662 to glutamate has little or no effect on EGFR activity (Figures S3B and S4B), arguing against a parallel arrangement.
A parallel helical dimer is also inconsistent with the formation of heterodimers between the JM-A segments of EGFR family members. A single amino acid deletion in the JM-A segment of Her4 (Figure 4A) results in Glu 693 of Her4 being directly apposed to another glutamate residue in modeled parallel heterodimers involving either Her2 or EGFR, in both possible sequence registers (Figure S4C). In addition, no favorable intermolecular ion pairs are formed in these parallel heterodimers.
In an antiparallel coiled coil, residues at the a and d positions of the heptad repeat in one helix interact with the residues at the d and a positions, respectively, in the other (Woolfson, 2005). Additional inter-helical interactions can be made by the sidechains at the e or g positions. The LxxLL motif in EGFR JM-A fits into this pattern if the first leucine is at either the a position or the d position, with the second two leucines at the d and e positions or the g and a positions, respectively. We favor placement of the first leucine at the d position because it provides a role for both arginine residues in the LRRLL motif, consistent with the strong effects of mutating these residues (Figure 4B and Figure S3B). In this pairing, Leu 658 and Leu 659 at the g and a positions in each helix form a V-shaped crevice into which the sidechain of Leu 655 at the d position from the other helix is inserted (Figure 4C).
An antiparallel dimer leads naturally to inter-helical ion pairing between arginine or lysine sidechains at the N-terminal end of each helix with the acidic sidechains located at the C-terminal end of the partner helix, and so we favor this arrangement for the helical dimer over a parallel one (Figure 4C and Figure S5). The pattern of inter-helical ion pairs predicted in this way for EGFR is also consistent with the formation of relevant heterodimers pairs (Figure S6).
Additional support for the antiparallel model is provided by NMR measurements on a 35 residue peptide containing two copies of the JM-A segment of EGFR with a five residue flexible spacer (see Supplemental Data and Figure S7A). The first and last leucine residues in the LRRLL motif in the first JM-A segment (Leu 655 and Leu 659) are labeled with 15N. The second glutamate in the first segment (Glu 663) is labeled with 15N and 13C. As for the 15 residue JM-A peptide, NMR data for the tandemly linked JM-A segments provide evidence for transient rather than stable adoption of helical structure in both JM-A segments under the conditions of the NMR experiment (see Supplemental Data). Despite the transient nature of these helices, the NMR data demonstrate that the two JM-A segments in the peptide interact with each other.
Data from Nuclear Overhauser Effect (NOE) experiments are consistent with an antiparallel rather than parallel orientation of two JM-A helices. This is not surprising, since an antiparallel interaction could occur in an intramolecular fashion within this peptide, whereas a parallel interaction would require dimerization. Nevertheless, the significance of these NMR data arises from the evidence that it provides for a specific register in the antiparallel interaction, in which the first leucine of the LRRLL motif is at the d position of a heptad repeat, with the second two leucines at the g and a positions, respectively (Figure 4D and Supplemental Data).
A Model for the Juxtamembrane Segment in the Context of the Asymmetric Kinase Domain Dimer
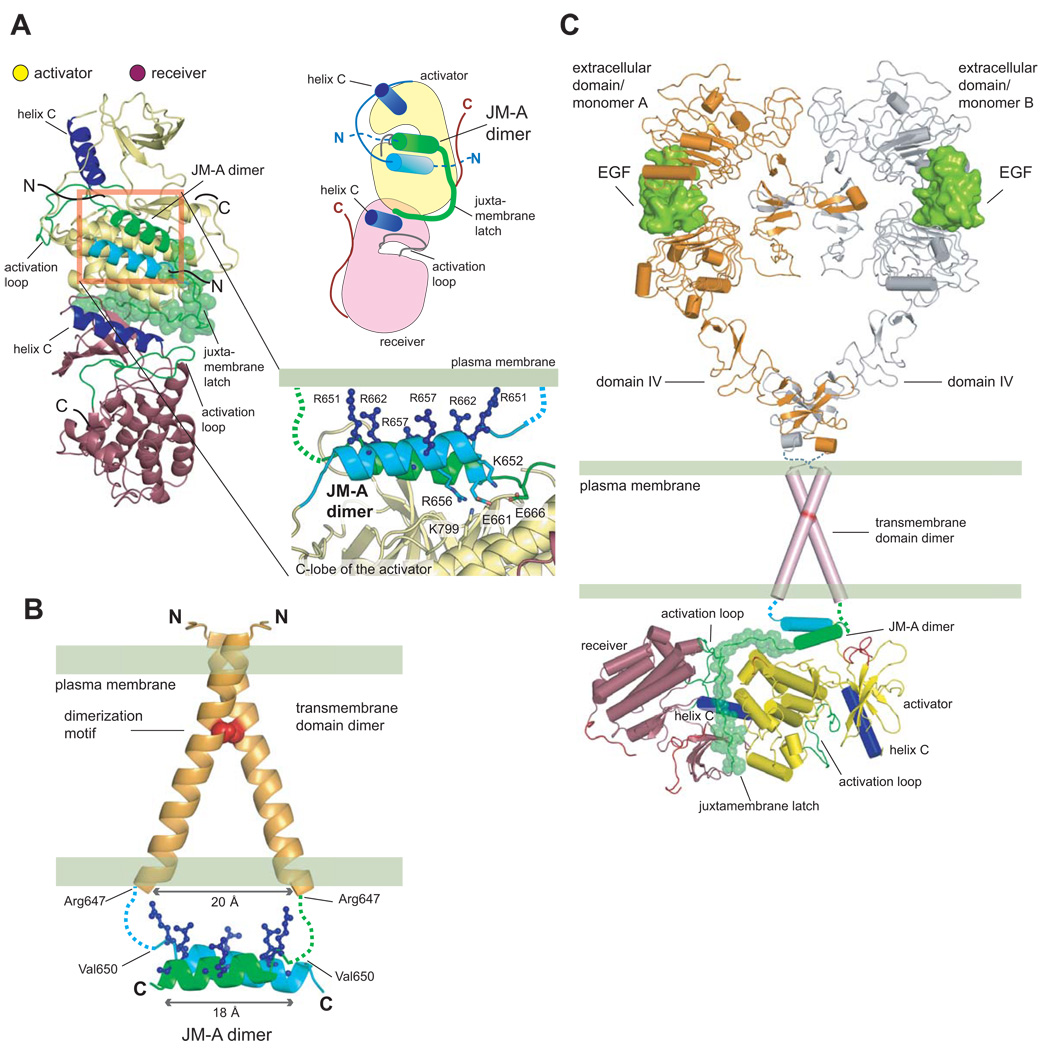
We joined the last residue in the modeled JM-A α-helix of the receiver kinase (the JM-A/receiver helix) to the first residue of the JM-B segment of the receiver kinase (the JM-B/receiver segment), while preserving the JM-B/receiver segment as seen in the Her4 structure (Wood et al., 2008) (Figure 5A). Although the precise orientation of the JM-A/receiver helix with respect to the kinase domain is uncertain, we chose to orient this helix such that the sidechain of Glu 663 in the receiver helix points towards the sidechain of Lys 799 in the activator kinase domain, a residue that is crucial for activity (Zhang et al., 2006). Another attractive feature of this orientation is that the face of the dimer that is likely to pack against the phospholipid bilayer has several arginine side chains, which could interact favorably with the membrane (Figure 5A). Mutation of many of these residues (e.g. Arg 651, Arg 657) results in greater reduction in EGFR activity than mutation of glutamate residues in JM-A, indicating that they are important for other functions in addition to dimerization. Particularly interesting is the presence of Thr 654 on this face of the dimer. Phosphorylation of Thr 654 attenuates EGFR signaling (Hunter et al., 1984), and the model suggests that the addition of a phosphate group to this side of the helix dimer might weaken interaction with membranes, as shown for other membrane-interacting peptide segments (Thelen et al., 1991).
Figure 5. Structural Coupling Between the Extracellular and Intracellular Domains in Active EGFR.
(A) A model for the JM-A helical dimer in the context of the asymmetric dimer of kinase domains. In the exploded view, arginine sidechains that face the membrane are shown. (B) Structure of the transmembrane domain dimer of Her2 (PDB ID: 2JWA), (Bocharov et al., 2008) and the modeled JM-A dimer. Positively charged sidechains that face the membrane are in blue. (C) A model for the activated EGF receptor. Two liganded EGFR extracellular domains are shown in an active dimeric assembly (PDB ID: 1IVO) (Ogiso et al., 2002), with domains IV based on the structure of the inactive EGFR extracellular domain (PDB ID: 1NQL) (Ferguson et al., 2003). This arrangement is compatible with the transmembrane domain dimer and couples to the asymmetric kinase domain dimer via the dimeric JM-A helices and the juxtamembrane latch.
Given the orientation of the JM-A/receiver helix, the antiparallel geometry naturally places the JM-A/activator helix between it and the first residue of the activator kinase domain core. In order to connect the activator kinase core to the JM-A/activator helix we modeled a 12 residue loop that connects Gln 677 of the activator kinase to Val 665 the JM-A/activator helix. We do not attach any particular significance to our modeled loop, except to note that the connection does not appear to impose unreasonable stereochemical constraints. We verified that the model we have constructed is without unnatural energetic strain by generating multiple molecular dynamics trajectories to relax the structure and noting that the dimeric structure is stable (see Supplemental Data and Figure S8).
Potential Coupling Between the Juxtamembrane and Transmembrane Segments
The NMR-derived structure of the Her2 transmembrane helices (Bocharov et al., 2008) shows that the transmembrane helices dimerize through a conserved glycine-containing motif (Burke et al., 1997; Fleishman et al., 2002; Sternberg and Gullick, 1989). Alignment of the EGFR sequence onto the Her-2 NMR structure shows that the Cα atoms of the residues corresponding to Arg 647 in EGFR are ~20 Å apart. The distance between the Cα atoms of the N-terminal residues in our model for the two antiparallel JM-A helices (Arg 651) is 18 Å, and the three residues that bridge the gap between the juxtamembrane and transmembrane segments can do so readily (Figure 5B). The convergence between the transmembrane helical dimer and the ends of our modeled JM-A antiparallel dimer suggests that distortions in the relative orientations of the transmembrane helices could weaken the coupling to the juxtamembrane segments. Such misalignment might explain the position-sensitive effects of crosslinking or mutation in the transmembrane segments on EGFR activity (Bell et al., 2000; Burke and Stern, 1998; Moriki et al., 2001).
Crystal structures of the extracellular domains of EGFR show that the active dimer brings the two C-terminal ends of the extracellular domains into close proximity (Burgess et al., 2003). We propose that this conformation brings the transmembrane helices close enough together to dimerize via their N-termini, thus supporting the active juxtamembrane and kinase domain dimers. Our model therefore specifies how the activating signal is transmitted across the membrane (Figure 5C).
Crystal Structure of an Inactive Kinase Domain In a Dimer Form
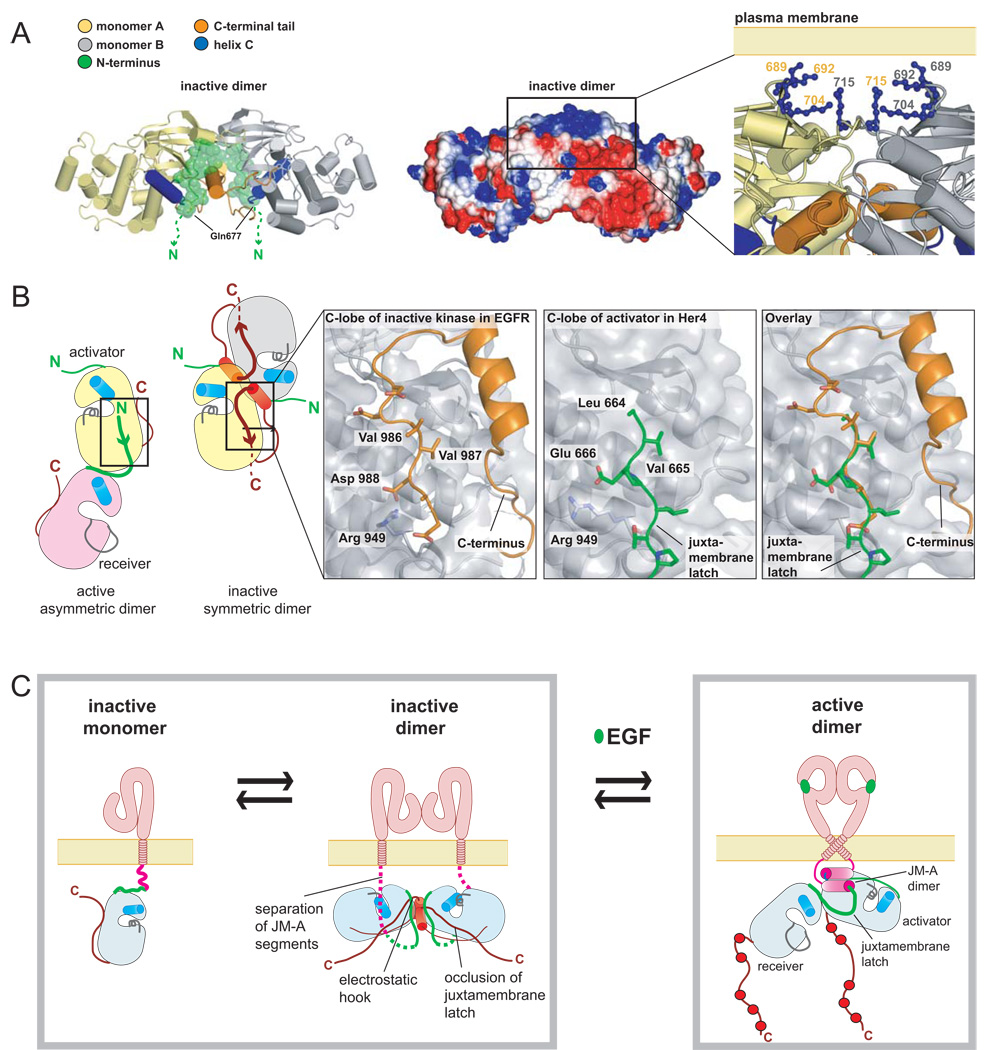
We have obtained a crystal structure, at 3Å resolution, of an EGFR kinase domain variant inactivated by the V924R mutation. The kinase domains in this structure form a symmetrical dimer (Figure 6A and Table S1). Crystal structures of inactive forms of the EGFR kinase domain that have been determined previously have the kinase domain in essentially the same conformation as in our structure, but do not show extensive contacts within symmetrical dimers (Wood et al., 2004; Xu et al., 2008; Zhang et al., 2006). These structures were determined in the presence of salts that might disrupt the electrostatic interactions that are at the center of the dimer interface of our crystal form, which is obtained under low salt conditions (see Experimental Procedures). A symmetrical EGFR dimer described previously has the kinase domains in an active conformation (Landau et al., 2004).
Figure 6. A Symmetric Inactive Dimer of the EGFR Kinase Domain.
(A) Overview of the crystal structure of the symmetric inactive dimer. (B) Detailed view of the hydrophobic packing between the C-terminal AP-2 helix of monomer B and the N-lobe of monomer A. (C) Exploded view of the electrostatic hook formed between the C-terminal tail (residues 979–990) of EGFR and the hinge region in the kinase domain. (D) Effect of mutations in the electrostatic hook on autophosphorylation of full length EGFR in COS7 cells. (E) Alignment of the sequences of EGFR family members in the C-terminal tail regions encompassing residues in the electrostatic hook and AP-2 helix.
Dimer formation in this new crystal form is mediated principally by the C-terminal tail of the kinase core (Figure 6A). There are four independent molecules in the asymmetric unit, designated A, B, C and D, which form two nearly identical dimers (A:B and C:D). In one molecule (A), electron density for the C-terminal segment is visualized up to Asp 990. The residues between Ser 967 and Met 978 form an α-helix (the AP-2 helix, see below), which is followed by a five residue turn spanning Asp 979 to Met 983. The last seven residues in the structural model, Asp 984 to Asp 990, run along the surface of the C-lobe of the kinase domain in an extended conformation. In molecule D there is no electron density for the extended strand (residues 984 to 990), and this region is blocked by crystal contacts. Electron density for the extended strand is present but weak in molecules B and C. Subsequent discussion of the dimer interactions is focused on the A:B dimer. The portion of the C-terminal tail (residues 967 to 983) that is of interest here is mainly disordered in structures of the active conformation of the EGFR kinase domain (Stamos et al., 2002) and is partially ordered but in a different conformation in the other inactive structures (Wood et al., 2004; Zhang et al., 2007).
The Inactive Kinase Domain Dimer May Suppress Activity Prior to EGF Binding
Several studies have shown that EGFR dimerizes prior to EGF binding (Clayton et al., 2008; Sako et al., 2000). Although these preformed dimers are likely to involve the intracellular domain (Yu et al., 2002), the orientation of the kinase domains in these preformed dimers is unknown. Our new inactive dimer has several features, which suggest it could play a role in the inhibition of kinase activity.
Each AP-2 helix in the C-terminal tail of one kinase subunit interacts with the other subunit, burying ~1400Å2 of surface area at each interface. The helix encompasses residues 973 to 977, which form the recognition element in EGFR for the AP-2 clathrin adapter protein (Sorkin et al., 1996). The recognition of AP-2 by EGFR is dependent on activation by EGF (Sorkin and Carpenter, 1993), and our structure shows how an inactive form of EGFR can sequester the AP-2 recognition motif. The interactions made by the AP-2 helix are reminiscent of those made by the SH2-kinase linker in inactive Src family kinases (Figure 6B) (Sicheri et al., 1997; Xu et al., 1997). In particular, the engagement of the N-lobe of the adjacent kinase domain by the sidechain of Phe 973 in the C-terminal tail of EGFR is analogous to interactions made by the sidechain of Leu 255 in the SH2-kinase linker of c-Src (Xu et al., 1997).
Acidic sidechains (Asp979, Glu 980, Glu 981) in the turn following the AP-2 helix form ion pairs with residues in the kinase domain (His 749, His 826, Lys 828 and Lys 822) (Figure 6C). The turn, referred to as an “electrostatic hook”, is located near the hinge region of the kinase domain and near the αC/β4 loop. In ZAP-70 and in certain other tyrosine kinases the formation of a hydrogen bonded network similar to that seen here in EGFR has been correlated with the inhibition of kinase activity (Chen et al., 2007; Deindl et al., 2007).
The conformation of the portion of the C-terminal tail that follows the electrostatic hook (residues 982 to 990) and runs along the surface of the C-lobe of the kinase mimics the manner in which the JM-B segment of the receiver kinase domain engages the same surface of the activator kinase domain when forming the juxtamembrane latch (Figure 7B). Thus, formation of the inactive dimer blocks formation of the activating juxtamembrane latch, in a manner similar to that postulated for Mig6 (Figure 3E).
Figure 7. Proposed Role of the Inactive Dimer in EGFR Autoinhibition.
(A) The surface electrostatic potential of the inactive dimer, with positively and negatively charged regions in blue and red, respectively. The exploded view shows the proposed docking at the plasma membrane. (B) Schematic diagram comparing the juxtamembrane latch in the active asymmetric dimer and the docking of the C-terminal tail in the inactive symmetric dimer. (C) A schematic representation of the activation mechanism of EGFR.
The key residues in the AP-2 helix, the electrostatic hook and the region of the C-terminal tail that interacts with the JM-B binding interface, as well as the basic residues that interact with the electrostatic hook, are conserved between EGFR and its kinase-active homologs (Her2 and Her4) (Figure 6E). The residues that form the electrostatic hook are absent in Her3, as are two of the three basic residues in the kinase domain. Presumably, the lack of kinase activity in Her3 renders inhibition of kinase activity unnecessary.
The surface electrostatic potential of the inactive dimer is strongly polarized (Figure 7A and S9). Eight lysine residues (residues 689, 692, 704, 715 in each subunit) are clustered together on the face of the dimer that is opposite to the internally engaged position of the C-terminal tail, which is a region of negative electrostatic potential. The lysine residues are conserved among EGFR family members. We speculate that the inactive dimer might be oriented with respect to the membrane such that the lysine residues can interact with negatively charged lipid head groups. The juxtamembrane segments would then be located on the far side of the dimer with respect to the membrane and dimerization of the JM-A segments would be prevented (Figure 7A).
Effect of Mutations in the C-terminal Tail on Activity of EGFR
Cell transfection experiments show that mutations in the electrostatic hook result in substantial activation of EGFR in the absence of EGF, as well an increased response to EGF, consistent with a role for this region in inhibiting EGFR kinase activity (Figure 6D). Notably, several insertions in exon 20 that drive constitutive EGFR activation are detected in lung cancer patients (Shigematsu et al., 2005)((Greulich et al., 2005). These insertions are in the β4/αC loop and are likely to disturb the electrostatic hook. Mutation of Lys 828, located within the electrostatic hook, increases the basal level of EGFR autophosphorylation (Zhang et al., 2006).
Surprisingly, deletion of the AP-2 helix or mutations of residues within this helix resulted consistently in impaired EGFR function in the cell-based assay (Figure S10A and S10B). These mutations result in activation in vitro (Figure S10C), demonstrating that the AP-2 helix is not required for the integrity of the catalytic domain. Mutation of residues in the AP-2 recognition motif does not lead to an obvious change in kinetics of EGFR internalization, presumably because of redundancy in endocytic targeting signals (Sorkin et al., 1996). The observed reduction in activity in cell-based assays suggests that the AP-2 helix plays a role in the activation mechanism, perhaps by presenting the C-terminal tails for phosphorylation.
Conclusions
We propose a structural mechanism for the activation of the EGFR receptor by integrating our model for the juxtamembrane segment with the separately determined structures of the transmembrane and extracellular domains (Figure 5C). In this model, the proximity of the C-terminal ends of the activated extracellular domains (Burgess et al., 2003) stabilizes the dimer of transmembrane helices observed in the Her2 NMR structure (Bocharov et al., 2008). This provides the proper geometry for the formation of the helical JM-A dimer and the juxtamembrane latch, which activate the kinase domain by stabilizing the asymmetric dimer.
Our model suggests that an important role for the extracellular domain is to inhibit the formation of the activated kinase domain dimer prior to ligand binding, as proposed earlier (Chantry, 1995; Nishikawa et al., 1994; Zhu et al., 2003). Our model also provides an explanation for why the insertion of a flexible linker between the extracellular domain and the transmembrane segment activates EGFR in the absence of ligand (Sorokin, 1995), since such a linker would decouple the conformation of the extracellular domains from the transmembrane helices. It appears that merely allowing the transmembrane segments to interact in a manner consistent with dimeric engagement of the JM-A helices suffices for activation.
In the absence of ligand the extracellular domains of EGFR adopt a compact conformation that converts to an extended conformation upon ligand binding (Burgess et al., 2003). Conversion of the extracellular domain to the active conformation without dimerization is by itself insufficient for activation (Mattoon et al., 2004). It appears that the extracellular segments either prevent interaction in the absence of ligand, or else dimerize in a way that keeps their C-terminal ends at a distance that is sufficient to prevent the transmembrane segments from stabilizing the dimeric interaction between the juxtamembrane helices. Separation of the C-terminal ends of the extracellular domains in an inactive dimer would be consistent with coupling to the inactive kinase domain dimer that we have described here.
Our structure of an inactive kinase domain dimer reveals that the C-terminal tails are central to the extensive inactive dimer interface that blocks formation of the juxtamembrane latch. Analysis of EGFR mutations in cancer patients has shown that deletion of the C-terminal tail can drive constitutive EGFR activation (Frederick et al., 2000). The formation of pre-formed EGFR dimers has been documented (Clayton et al., 2008; Sako et al., 2000), and the intracellular domain is implicated in formation of these dimers (Yu et al., 2002). Thus, inactive kinase dimers may constitute a key autoinhibitory mechanism that prevents ligand-independent activation. The extracellular domains determine the balance between inactive and active dimeric states of the kinase domain, with inactive monomers and dimers predominating prior to EGF stimulation, and active dimers, or higher order assemblies, predominating after stimulation (Clayton et al., 2008) (Figure 7C). In this model both the extracellular domains and the inactive dimer work synergistically to prevent the high local concentration of receptors at the membrane from generating spurious signals through uncontrolled trans-phosphorylation. Determining the mechanism by which this balance is achieved remains an important focus for further study.
Experimental Procedures
Protein Expression and Purification
Human EGFR kinase core constructs (residues 672:998) were expressed and purified as described (Zhang et al., 2006). JM-kinase constructs (residues 645–998 and 658–998) expressed and purified using the same protocol. All proteins constructs contained N-terminal 6His-tags (Zhang et al., 2006). Mutations were introduced using the Quick-Change kit (Stratagene) and confirmed by DNA sequencing.
In Vitro Kinase Assays
Kinase activity was measured using a continuous enzyme-coupled kinase assay in solution and on vesicles as described (Zhang et al., 2006) using the substrate peptide corresponding to the Tyr 1173 phosphorylation site in EGFR (TAENAEYLRVAPQ). The details of the reaction conditions are given in Table S2. The concentration dependence of activity was measured using poly Glu:Tyr as a substrate, at a substrate concentration significantly above the KM value (3mg/ml).
Crystallization and Structure Determination
The crystal form containing the EGFR inactive symmetric dimer was obtained during an attempt to crystallize a complex of the EGFR kinase core (V924R) with the kinase domain of Her3. Crystals (space group P21, a = 61.9 Å, b = 72.5 Å, c = 143.5 Å, β=101.7°) were obtained from solutions containing a 1:1 mixture of the EGFR and Her3 kinase domains at 6mg/ml, 5mM AMP-PNP, 2mM MgCl2, 0.1M Bis-Tris pH5.5, 0.1M ammonium acetate, 17% w/v PEG 10,000. Refinement of the structure to 3 Å usinging PHENIX (Adams et al., 2002) showed unambiguously that the structure was that of an EGFR dimer, and that the crystals did not contain Her3. We then crystallized the EGFR kinase domain alone, obtaining the same crystal form (space group P21, a = 62.6 Å, b = 73.3 Å, c = 144.5 Å, β=102.0°), containing the same pair of dimers, with essentially the same structure. The resolution limit for these data is slightly lower (3.2 Å), and our analysis is based on the structure obtained at higher resolution. Data collection and refinement statistics are in Supplemental Material.
Mammalian Cell-Based Assays
DNA encoding the full-length human EGFR gene with an N-terminal FLAG tag in pcDNA3.1 mammalian expression vector was used in cell-based assays, as described (Zhang et al., 2006). The AP-2 helix deletion was constructed by deleting residues 973–978 in full length EGFR. The JM-kinase and TM-JM-kinase constructs were cloned into pcDNA3.1 (Invitrogen) and a FLAG tag was inserted N-terminal to the EGFR sequence. The GCN4-JM-kinase construct was generated by inserting the GCN4 sequence (RVKQLEDKVEELLSKNAHLENEVARLKKL) N-terminal to the JM-kinase constructs.
COS7 cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum (FBS) and streptomycin/penicillin. Cells were transiently transfected using Fugene 6 (Roche) and cultured for 24h after transfection, followed by 12h serum starvation. Cells were treated with EGF (50 ng/ml, PeproTech) for 5 min at 37°C and lysed in Lysis Buffer (50 mM TrispH7.5, 150 mM NaCl, 1 mM EDTA, 1 mMa Na3VO4, 1 mM NaF, 1% Triton X-100 and a protease inhibitor cocktail (Roche)). Cell lysates were subjected to Western blot analyses using the following primary antibodies: anti-EGFR, sc-03, polyclonal (Santa Cruz), anti-phosphoTyr 4G10 (Upstate) and EGFR site-specific phosphorylation antibodies, Tyr1173 (Santa Cruz), Tyr974 (Cell Signaling), Tyr992 (Cell Signaling), Tyr1045 (Cell Signaling) and Tyr1068 (Cell Signaling). Secondary antibodies used were either goat anti-mouse (GE Healthcare) or goat anti-rabbit (Cell Signaling) coupled to horseradish peroxidase. For immunoprecipitation assay, cells were lysed in the Lysis Buffer, cleared by centrifugation and incubated with anti-FLAG monoclonal antibody M2 (Sigma-Aldrich) followed by incubation with 1:1 protein A-sepharose slurry (Sigma-Aldrich). The immunoprecipitates were analysed by Western blotting with the indicated antibodies.
Supplementary Material
Acknowledgments
We thank Deborah Makino for help with structure determination, Xiaoxian Cao for Sf9 cell culture, David King for peptide synthesis and Jeffrey Pelton for help with NMR experiments. We thank Tony Hunter, Nir Ben-Tal, Alexander Sorkin, Susan Marqusee and members of the Kuriyan laboratory for helpful discussions. This work is supported in part by grant from the National Cancer Institute to J. K. (RO1 CA96504-06). N. E. is a fellow of the Leukemia and Lymphoma Society. We thank the staff at the Advanced Light Source, which is supported by U.S Department of Energy under Contract DE-AC03-76SF00098 at the Lawrence Berkeley National Laboratory. NMR instrumentation and operation were supported by NIH-GM 68933
References
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Bell CA, Tynan JA, Hart KC, Meyer AN, Robertson SC, Donoghue DJ. Rotational coupling of the transmembrane and kinase domains of the Neu receptor tyrosine kinase. Mol Biol Cell. 2000;11:3589–3599. doi: 10.1091/mbc.11.10.3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocharov EV, Mineev KS, Volynsky PE, Ermolyuk YS, Tkach EN, Sobol AG, Chupin VV, Kirpichnikov MP, Efremov RG, Arseniev AS. Spatial structure of the dimeric transmembrane domain of the growth factor receptor ErbB2 presumably corresponding to the receptor active state. J Biol Chem. 2008;283:6950–6956. doi: 10.1074/jbc.M709202200. [DOI] [PubMed] [Google Scholar]
- Burgess AW, Cho HS, Eigenbrot C, Ferguson KM, Garrett TP, Leahy DJ, Lemmon MA, Sliwkowski MX, Ward CW, Yokoyama S. An open-and-shut case? Recent insights into the activation of EGF/ErbB receptors. Mol Cell. 2003;12:541–552. doi: 10.1016/s1097-2765(03)00350-2. [DOI] [PubMed] [Google Scholar]
- Burke CL, Lemmon MA, Coren BA, Engelman DM, Stern DF. Dimerization of the p185neu transmembrane domain is necessary but not sufficient for transformation. Oncogene. 1997;14:687–696. doi: 10.1038/sj.onc.1200873. [DOI] [PubMed] [Google Scholar]
- Burke CL, Stern DF. Activation of Neu (ErbB-2) mediated by disulfide bond-induced dimerization reveals a receptor tyrosine kinase dimer interface. Mol Cell Biol. 1998;18:5371–5379. doi: 10.1128/mcb.18.9.5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chantry A. The kinase domain and membrane localization determine intracellular interactions between epidermal growth factor receptors. J Biol Chem. 1995;270:3068–3073. [PubMed] [Google Scholar]
- Chen H, Ma J, Li W, Eliseenkova AV, Xu C, Neubert TA, Miller WT, Mohammadi M. A molecular brake in the kinase hinge region regulates the activity of receptor tyrosine kinases. Mol Cell. 2007;27:717–730. doi: 10.1016/j.molcel.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choowongkomon K, Carlin CR, Sonnichsen FD. A structural model for the membrane-bound form of the juxtamembrane domain of the epidermal growth factor receptor. J Biol Chem. 2005;280:24043–24052. doi: 10.1074/jbc.M502698200. [DOI] [PubMed] [Google Scholar]
- Clayton AH, Orchard SG, Nice EC, Posner RG, Burgess AW. Predominance of activated EGFR higher-order oligomers on the cell surface. Growth Factors. 2008:1. doi: 10.1080/08977190802442187. [DOI] [PubMed] [Google Scholar]
- Deindl S, Kadlecek TA, Brdicka T, Cao X, Weiss A, Kuriyan J. Structural basis for the inhibition of tyrosine kinase activity of ZAP-70. Cell. 2007;129:735–746. doi: 10.1016/j.cell.2007.03.039. [DOI] [PubMed] [Google Scholar]
- Ferguson KM, Berger MB, Mendrola JM, Cho HS, Leahy DJ, Lemmon MA. EGF activates its receptor by removing interactions that autoinhibit ectodomain dimerization. Mol Cell. 2003;11:507–517. doi: 10.1016/s1097-2765(03)00047-9. [DOI] [PubMed] [Google Scholar]
- Fleishman SJ, Schlessinger J, Ben-Tal N. A putative molecular-activation switch in the transmembrane domain of erbB2. Proc Natl Acad Sci U S A. 2002;99:15937–15940. doi: 10.1073/pnas.252640799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick L, Wang XY, Eley G, James CD. Diversity and frequency of epidermal growth factor receptor mutations in human glioblastomas. Cancer Res. 2000;60:1383–1387. [PubMed] [Google Scholar]
- Gotoh N, Tojo A, Hino M, Yazaki Y, Shibuya M. A highly conserved tyrosine residue at codon 845 within the kinase domain is not required for the transforming activity of human epidermal growth factor receptor. Biochem Biophys Res Commun. 1992;186:768–774. doi: 10.1016/0006-291x(92)90812-y. [DOI] [PubMed] [Google Scholar]
- Greulich H, Chen TH, Feng W, Janne PA, Alvarez JV, Zappaterra M, Bulmer SE, Frank DA, Hahn WC, Sellers WR, Meyerson M. Oncogenic transformation by inhibitor-sensitive and -resistant EGFR mutants. PLoS Med. 2005;2:e313. doi: 10.1371/journal.pmed.0020313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard SR, Miller WT. Receptor tyrosine kinases: mechanisms of activation and signaling. Curr Opin Cell Biol. 2007;19:117–123. doi: 10.1016/j.ceb.2007.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T, Ling N, Cooper JA. Protein kinase C phosphorylation of the EGF receptor at a threonine residue close to the cytoplasmic face of the plasma membrane. Nature. 1984;311:480–483. doi: 10.1038/311480a0. [DOI] [PubMed] [Google Scholar]
- Landau M, Ben-Tal N. Dynamic equilibrium between multiple active and inactive conformations explains regulation and oncogenic mutations in ErbB receptors. Biochim Biophys Acta. 2008;1785:12–31. doi: 10.1016/j.bbcan.2007.08.001. [DOI] [PubMed] [Google Scholar]
- Landau M, Fleishman SJ, Ben-Tal N. A putative mechanism for downregulation of the catalytic activity of the EGF receptor via direct contact between its kinase and C-terminal domains. Structure. 2004;12:2265–2275. doi: 10.1016/j.str.2004.10.006. [DOI] [PubMed] [Google Scholar]
- Mattoon D, Klein P, Lemmon MA, Lax I, Schlessinger J. The tethered configuration of the EGF receptor extracellular domain exerts only a limited control of receptor function. Proc Natl Acad Sci U S A. 2004;101:923–928. doi: 10.1073/pnas.0307286101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin S, Smith SO, Hayman MJ, Murray D. An electrostatic engine model for autoinhibition and activation of the epidermal growth factor receptor (EGFR/ErbB) family. J Gen Physiol. 2005;126:41–53. doi: 10.1085/jgp.200509274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriki T, Maruyama H, Maruyama IN. Activation of preformed EGF receptor dimers by ligand-induced rotation of the transmembrane domain. J Mol Biol. 2001;311:1011–1026. doi: 10.1006/jmbi.2001.4923. [DOI] [PubMed] [Google Scholar]
- Nishikawa R, Ji XD, Harmon RC, Lazar CS, Gill GN, Cavenee WK, Huang HJ. A mutant epidermal growth factor receptor common in human glioma confers enhanced tumorigenicity. Proc Natl Acad Sci U S A. 1994;91:7727–7731. doi: 10.1073/pnas.91.16.7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogiso H, Ishitani R, Nureki O, Fukai S, Yamanaka M, Kim JH, Saito K, Sakamoto A, Inoue M, Shirouzu M, Yokoyama S. Crystal structure of the complex of human epidermal growth factor and receptor extracellular domains. Cell. 2002;110:775–787. doi: 10.1016/s0092-8674(02)00963-7. [DOI] [PubMed] [Google Scholar]
- O'Shea EK, Klemm JD, Kim PS, Alber T. X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science. 1991;254:539–544. doi: 10.1126/science.1948029. [DOI] [PubMed] [Google Scholar]
- Pace CN, Scholtz JM. A helix propensity scale based on experimental studies of peptides and proteins. Biophys J. 1998;75:422–427. doi: 10.1016/s0006-3495(98)77529-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu C, Tarrant MK, Choi SH, Sathyamurthy A, Bose R, Banjade S, Pal A, Bornmann WG, Lemmon MA, Cole PA, Leahy DJ. Mechanism of activation and inhibition of the HER4/ErbB4 kinase. Structure. 2008;16:460–467. doi: 10.1016/j.str.2007.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sako Y, Minoghchi S, Yanagida T. Single-molecule imaging of EGFR signalling on the surface of living cells. Nat Cell Biol. 2000;2:168–172. doi: 10.1038/35004044. [DOI] [PubMed] [Google Scholar]
- Schlessinger J. Ligand-induced, receptor-mediated dimerization and activation of EGF receptor. Cell. 2002;110:669–672. doi: 10.1016/s0092-8674(02)00966-2. [DOI] [PubMed] [Google Scholar]
- Shigematsu H, Takahashi T, Nomura M, Majmudar K, Suzuki M, Lee H, Wistuba II, Fong KM, Toyooka S, Shimizu N, et al. Somatic mutations of the HER2 kinase domain in lung adenocarcinomas. Cancer Res. 2005;65:1642–1646. doi: 10.1158/0008-5472.CAN-04-4235. [DOI] [PubMed] [Google Scholar]
- Sicheri F, Moarefi I, Kuriyan J. Crystal structure of the Src family tyrosine kinase Hck. Nature. 1997;385:602–609. doi: 10.1038/385602a0. [DOI] [PubMed] [Google Scholar]
- Sorkin A, Carpenter G. Interaction of activated EGF receptors with coated pit adaptins. Science. 1993;261:612–615. doi: 10.1126/science.8342026. [DOI] [PubMed] [Google Scholar]
- Sorkin A, Mazzotti M, Sorkina T, Scotto L, Beguinot L. Epidermal growth factor receptor interaction with clathrin adaptors is mediated by the Tyr974-containing internalization motif. J Biol Chem. 1996;271:13377–13384. doi: 10.1074/jbc.271.23.13377. [DOI] [PubMed] [Google Scholar]
- Sorokin A. Activation of the EGF receptor by insertional mutations in its juxtamembrane regions. Oncogene. 1995;11:1531–1540. [PubMed] [Google Scholar]
- Stamos J, Sliwkowski MX, Eigenbrot C. Structure of the epidermal growth factor receptor kinase domain alone and in complex with a 4-anilinoquinazoline inhibitor. J Biol Chem. 2002;277:46265–46272. doi: 10.1074/jbc.M207135200. [DOI] [PubMed] [Google Scholar]
- Sternberg MJ, Gullick WJ. Neu receptor dimerization. Nature. 1989;339:587. doi: 10.1038/339587a0. [DOI] [PubMed] [Google Scholar]
- Thelen M, Rosen A, Nairn AC, Aderem A. Regulation by phosphorylation of reversible association of a myristoylated protein kinase C substrate with the plasma membrane. Nature. 1991;351:320–322. doi: 10.1038/351320a0. [DOI] [PubMed] [Google Scholar]
- Thiel KW, Carpenter G. Epidermal growth factor receptor juxtamembrane region regulates allosteric tyrosine kinase activation. Proc Natl Acad Sci U S A. 2007;104:19238–19243. doi: 10.1073/pnas.0703854104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood ER, Shewchuk LM, Ellis B, Brignola P, Brashear RL, Caferro TR, Dickerson SH, Dickson HD, Donaldson KH, Gaul M, et al. 6-Ethynylthieno[3,2-d]- and 6-ethynylthieno[2,3-d]pyrimidin-4-anilines as tunable covalent modifiers of ErbB kinases. Proc Natl Acad Sci U S A. 2008;105:2773–2778. doi: 10.1073/pnas.0708281105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood ER, Truesdale AT, McDonald OB, Yuan D, Hassell A, Dickerson SH, Ellis B, Pennisi C, Horne E, Lackey K, et al. A unique structure for epidermal growth factor receptor bound to GW572016 (Lapatinib): relationships among protein conformation, inhibitor off-rate, and receptor activity in tumor cells. Cancer Res. 2004;64:6652–6659. doi: 10.1158/0008-5472.CAN-04-1168. [DOI] [PubMed] [Google Scholar]
- Woolfson DN. The design of coiled-coil structures and assemblies. Adv Protein Chem. 2005;70:79–112. doi: 10.1016/S0065-3233(05)70004-8. [DOI] [PubMed] [Google Scholar]
- Xu G, Searle LL, Hughes TV, Beck AK, Connolly PJ, Abad MC, Neeper MP, Struble GT, Springer BA, Emanuel SL, et al. Discovery of novel 4-amino-6-arylaminopyrimidine-5-carbaldehyde oximes as dual inhibitors of EGFR and ErbB-2 protein tyrosine kinases. Bioorg Med Chem Lett. 2008;18:3495–3499. doi: 10.1016/j.bmcl.2008.05.024. [DOI] [PubMed] [Google Scholar]
- Xu W, Harrison SC, Eck MJ. Three-dimensional structure of the tyrosine kinase c-Src. Nature. 1997;385:595–602. doi: 10.1038/385595a0. [DOI] [PubMed] [Google Scholar]
- Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- Yu X, Sharma KD, Takahashi T, Iwamoto R, Mekada E. Ligandin-dependent dimer formation of epidermal growth factor receptor (EGFR) is a step separable from ligand-induced EGFR signaling. Mol Biol Cell. 2002;13:2547–2557. doi: 10.1091/mbc.01-08-0411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun CH, Boggon TJ, Li Y, Woo MS, Greulich H, Meyerson M, Eck MJ. Structures of lung cancer-derived EGFR mutants and inhibitor complexes: mechanism of activation and insights into differential inhibitor sensitivity. Cancer Cell. 2007;11:217–227. doi: 10.1016/j.ccr.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125:1137–1149. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
- Zhang X, Pickin KA, Bose R, Jura N, Cole PA, Kuriyan J. Inhibition of the EGF receptor by binding of MIG6 to an activating kinase domain interface. Nature. 2007;450:741–744. doi: 10.1038/nature05998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu HJ, Iaria J, Orchard S, Walker F, Burgess AW. Epidermal growth factor receptor: association of extracellular domain negatively regulates intracellular kinase activation in the absence of ligand. Growth Factors. 2003;21:15–30. doi: 10.1080/0897719031000096424. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.