Abstract
Single-walled carbon nanotubes (SWNT) have unique electronic, mechanical, and structural properties as well as chemical stability that make them ideal nanomaterials for applications in materials science and medicine. Here we report the design and creation of a novel strategy for functionalizing SWNT to systemically silence a target gene in mice by delivering siRNA at doses <1 mg/Kg. SWNT were functionalized with lipids and natural amino acid-based dendrimers (TOT) and complexed to siRNA. Our model study of the silencing efficiency of the TOT-siRNA complex showed that in mice injected at 0.96 mg/kg, an endogenous gene for apoliproprotein B (ApoB) was silenced in liver, plasma levels of ApoB decreased, and total plasma cholesterol decreased. TOT-siRNA treatment was non-toxic and did not induce an immune response. Most (80%) of the RNA trigger molecules assembled with TOT were cleared from the body 48 h after injection, suggesting that the nanotubes did not cause siRNA aggregation or inhibit biodegradation and drug clearance in vivo. These results provide the first evidence that nanotubes can be functionalized with lipids and amino acids to systemically deliver siRNA. This new technology cannot only be used for systemic RNAi, but may also be used to deliver other drugs in vivo.
Keywords: RNAi, nanotubes, siRNA delivery, chemically modified siRNA, therapeutic silencing
Introduction
Single-walled carbon nanotubes (SWNT) have unique electronic, mechanical, and structural properties as well as chemical stability that make them ideal nanomaterials for applications in materials science and medicine. However, native SWNT have poor solubility and biocompatibility, which has stimulated approaches to functionalize them by conjugating molecules such as peptides, nucleic acids and polymers (1, 2). The potential of SWNT has recently been investigated in the cellular delivery of DNA plasmids and of small interfering RNAs (siRNAs) for gene silencing (2, 3). To date, however, the potential of designing, creating, and applying SWNT for systemic delivery of nucleic acids or drugs has not been investigated.
RNA interference (RNAi), a gene-silencing mechanism whereby double-stranded RNA (dsRNA) induces the sequence-specific degradation of homologous mRNA (4), has been widely used to identify gene functions and holds great potential to provide a new class of therapeutic agents (5). During RNAi, long dsRNA is processed by Dicer into short-interfering RNAs (siRNAs), and incorporated into the RNA-induced silencing complex (RISC) (6), a multiturnover enzyme complex that cleaves the target mRNA. The RNAi machinery can also be programmed in cells by introducing duplexes of siRNAs (7, 8) that are assembled into siRISC containing Dicer, Argonautes and other proteins (reviewed in (9)). Therefore, new siRNA-based therapeutic agents could be designed to lower concentrations of disease-causing gene products.
However, the development of such siRNA-based therapies faces two barriers: (1) identification of chemically stable and effective siRNA sequences, and (2) efficient delivery of these sequences to in vivo targets using siRNA amounts that can be translated to clinically feasible doses for humans. Recent advances in understanding the rules for chemically modifying siRNA sequences without compromising their gene-silencing efficiency (10-12) have allowed the design and synthesis of therapeutically effective siRNA molecules that can silence target genes in vivo (13, 14). Furthermore, siRNAs have recently been delivered in vivo to successfully inhibit various gene functions. This delivery has been facilitated by conjugating cholesterol to siRNA (14) or to oligonucleotide inhibitors of miRNA (15), by forming stable nucleic acid-lipid particles (SNALP) of siRNA (13, 16), and by assembling lipid-siRNA complexes (17, 18). In addition, a protamine-antibody fusion protein has been used to deliver siRNAs to HIV-infected cells (19). Recently, application of nanotubes to successfully deliver siRNA in cells have been reported (3, 20-22).
Despite this progress, new chemistry and delivery approaches are greatly needed to systematically silence disease-causing genes in a tissue-specific manner with high efficiencies and at clinically achievable doses. Here we report the design and creation of novel nanotubes (TOT) functionalized with lipids and natural amino acid-based dendrimers. We used these TOT to systemically silence a target gene in mice at siRNA doses <1 mg/Kg. Moreover, this treatment did not induce an immune response and showed favorable pharmacokinetics.
Experimental Procedures
Synthesis of nanotubes functionalized with lipids and lysine dendrimers (TOT)
To create functionalized new nanomaterial, single-walled carbon nanotubes (SWNT) were oxidized and cut by suspending 20 mg of SWNT in 40 ml of a mixture of concentrated H2SO4/HNO3 (3:1) and sonicating at 40 to 50 °C for 3h (23). The resulting suspension was poured into 350 ml water and filtered through a 100 nm pore membrane. The resulting solid was washed repeatedly with water, resuspended in water, and centrifuged (5000 rpm, 10 min) to remove large particles.
Synthesis of Glutamic acid-Lysine Dendrimer Generation 1 (G1L1)
A mixture of H-Glu(OMe)-OMe hydrochlrode (0.50 g, 2.4 mmol), Boc-lysine(Boc)-OH dicyclohexylamine salt (1.24 g, 2.4 mmol) and DIEA (0.80 ml, 4.72 mmol) was suspended in 30 ml DMF under nitrogen atmosphere. After the suspension was cooled to 0 °C in an ice bath, BOP reagent (1.1 g, 2.4 mmol) was added. The reaction was performed at 0 °C for 30 min and then at room temperature for 24 h. Then, the solvent was removed by evaporation, and the resulting syrup was dissolved in ethyl acetate, followed by washing with 5% citric acid, 5% sodium bicarbonate and water. After the solution was dried over anhydrous sodium sulfate, the target compound was purified by silica gel chromatography using ethyl acetate and hexane (4:1) as eluent. The purity of the compound was confirmed by TLC, and G1L1 was obtained as a white powder (1.01g) in 91% yield.
1H NMR (DMSO-d6, ppm): 1.17-1.57 (m, 24H, -C(CH3)3, -CH2-), 2.23-2.31 (m, 4H, -CH2-), 3.01 (m, 2H, -CH2-), 3.84 (m, 2H, -C(NH-)HCH2-), 3.68 (s, 6H, -CH3), 6.61-6.65 (m, 2H, -CONH-), 7.65 (t, 1H, -CONH-). MS (ESI) m/z 526.48 (M + Na)+.
Synthesis of Glutamic acid-Lysine Dendrimer Generation 2 (G1L3)
Deprotection of G1L1: G1L1 was dissolved in a mixture of dichloromethane and trifluoroacetic acid. After vigorously stirring at room temperature for 30min, the solvents were evaporated, and the syrup was washed with anhydrous ethyl ether.
Synthesis of G1L3: deprotected G1L1 was reacted with Boc-lysine(Boc)-OH in a similar method described above to give G1L3 in 88% yield. 1H NMR (DMSO-d6, ppm): 1.17-1.57 (m, 54H, -C(CH3)3, -CH2-), 2.23-2.31 (m, 4H, -CH2-), 3.01 (m, 6H, -CH2-), 3.84 (m, 2H, -C(NH-)HCH2-), 4.14 (m, 2H, -C(NH-)HCH2-), 3.68 (s, 6H, -CH3), 6.61-7.65 (m, 7H, -CONH-). MS (ESI) m/z 982.95 (M + Na)+.
Synthesis of Glutamic acid-Lysine Dendrimer Generation 3 (G1L7)
G1L3 was deprotected, and reacted with Boc-lysine(Boc)-OH in a similar method described above to give G1L7 in 82% yield.
1H NMR (DMSO-d6, ppm): 1.17-1.57 (m, 118H, -C(CH3)3, -CH2-), 2.23-2.31 (m, 4H, -CH2-), 3.01 (m, 14H, -CH2-), 3.84 (m, 4H, -C(NH-)HCH2-), 4.14 (m, 4H, -C(NH-)HCH2-), 3.68 (s, 6H, -CH3), 6.61-7.65 (m, 15H, -CONH-). MS (ESI) m/z 1896.84 (M + Na)+.
Synthesis of Glutamic acid-Lysine Dendrimer Generation 4 (G2L7)
G1L7 (0.36 g, 0.19 mmol) was stirred in a mixture of methanol (15ml) and 1M NaOH solution (15ml) at room temperature. The demethylation process was monitored by TCL (DCM: methanol, 10: 1). After the reaction was complete, the suspension was neutralized by adding dilute hydrochloric acid, followed by evaporation, extraction with chloroform, and washing with dilute HCl solution. Then the solution was dried and the solvent was removed to give demethylated G1L7 as oil.
The oil was dissolved in 5 ml DMF. To this solution was added H-Glu(OMe)-OMe hydrochloride (0.163 g, 0.77 mmol), DIEA (0.164 ml) and BOP reagent (0.34 g, 0.77 mmol). The reaction was performed at 0 °C for 30 min and at room temperature for 24 h. Then, a mixture of ethyl acetate and hexane (4: 1, 100 ml) was poured into the reaction flask, and the resulting precipitate was collected by filtration. The solid was suspended in 10 ml methanol and re-precipitated by adding ethyl acetate and hexane to give a white powder (0.31 g) in 75% yield. 1H NMR (DMSO-d6, ppm): 1.17-1.57 (m, 118H, -C(CH3)3, -CH2-), 2.23-2.31 (m, 12H, -CH2-), 3.01 (m, 14H, -CH2-), 3.84 (m, 6H, -C(NH-)HCH2-), 4.14 (m, 4H, -C(NH-)HCH2-), 3.68 (s, 12H, -CH3), 6.61-7.65 (m, 17H, -CONH-). MS (ESI) m/z 2183.06 (M + Na)+.
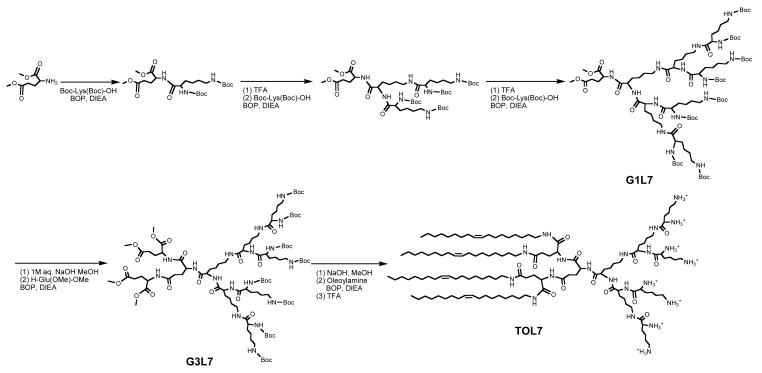
Synthesis of Tetra-Oleoyl Lysine Dendrimer Generation 3 (TOL7)
Demethylated G2L7 (0.09 g, 0.05 mmol), oleoylamine (0.14 g, 0.54 mmol) and DIEA (0.15 ml) was dissolved in DMF. BOP (0.24 g, 0.54 mmol) was added and the solution was stirred at r.t. for 24 hrs. After workup, a white powdery TOL7 (0.12 g) was obtained in 80% yield.
1H NMR (DMSO-d6, ppm): 0.81 (t, 12H, -CH3), 1.21-1.62 (m, 154H, -CH2-), 1.93-1.99 (m, 16H, -CH2CH=CHCH2-), 2.73, 2.96-3.03 (m, 22H, -CH2NH-, -CH2NH2), 3.59, 4.18 (m, 10H, -OCCH(NH-)CH2-), 5.28 (m, 8H, -CH=CH-), 7.75-8.48 (m, 25H, -CONH-, -NH2). MS (MALDI-TOF) m/z 2322.91 (M + Na)+.
Conjugation of TOL7 to oxidized SWNT (TOT)
To a solution of TOL7 (0.5 ml, 24 mg/ml) in water was added SWNT stock solution (2 ml). EDC (80 mg) was added and the suspension was stirred at r.t. After 2 hrs, the mixture was filtered through a polycarbonate filter, rinsed, and re-suspended in water, followed by dialysis against pure water for 3 days. The larger particles were removed by centrifuge at 7,000 rpm for 5 min. The final concentration of TOT was made 0.2mg/ml (24, 25). The concentration of TOL7 on SWNT was determined by quantitative Kaiser assay (26). The structure of TOT was characterized by transmission electron microscopy (Core Electron Microscopy Facility, UMASS Medical School) and dynamic light scattering (Department of Food Science, Umass Amherst).
Preparation of TOT-siRNA complexes
All siRNAs used in these studies were chemically synthesized using silyl ethers to protect 5′-hydroxyls and acid-labile orthoesters to protect 2′-hydroxyls (2′-ACE) (Dharmacon, Lafayette, CO, USA). After deprotection and purification, siRNA strands were annealed as described (11): ApoB siRNA (ORF position 10049-10071): Unmodified (UM) sense 5′-GUCAUCACACUGAAUACCAAU-3′, antisense 5′-AUUGGUAUUCAGUGUGAUGACAC-3′; chemically modified (CM): sense 5′-GSUFCFAUFCFACACUGAAUACFSCFAASUF-propylamine-3′, antisense 5′-PAUFUFGGUAUUCAGUGUGAUFGACFSASC; UM mismatch (mm) siRNA: sense 5′-GUGAUCAGACUCAAUACGAAU propylamine-3′, antisense 5-’AUUCGUAUUGAGUCUGAUCACAC-3′; CM mm: sense 5′-GSUFGAUFCFAGACUCAAUACFGAASUF propylamine-3′, antisense 5′-AUFUFCGUAUUGAGUCUGAUFCACFSASC. The superscript letters F and S represent 2′-O-F and HS-backbone modifications, respectively. TOT-siRNA complexes were prepared by mixing siRNA and TOT in Hepes saline or Opti-MEM culture medium (Invitrogen) and incubating at room temperature for 20 min (see below).
in vitro Assay for RNAi activity of TOT-siRNA
FL83B (mouse hepatocyte) cells were maintained at 37°C with 5% CO2 in F12 Khangians modified culture medium (ATCC, USA) supplemented with 10% fetal bovine serum (FBS), 100U/ml penicillin and 100 μg/ml streptomycin. Cells were regularly passaged and plated in 96-well and 6 well-culture plates 16 h before transfection at 70% confluency. Cells were transfected with 1ml/well of complex (siRNA: TOT) for 2.5 h at 37°C. Medium was removed and replaced with complete medium without antibiotics and incubated for an additional 24 h. Cell viability was assessed using a CellTiter 96® AQueous One Solution cell proliferation assay by colorimetric analysis of the MTS tetrazolium compound according to the manufacturer’s instructions (Promega, USA). Total RNA was extracted using RNeasy mini spin columns and treated with DNase I (Qiagen, USA) before quantitation. To assess levels of ApoB mRNA, real time quantitative PCR (qPCR) was performed using SYBR Green (Qiagen USA) with forward (5′-TTCCAGCCATGGGCAACTTTACCT-3′) and reverse (5′-TACTGCAGGGCGTCAGTGACAAAT-3′) ApoB primers. ApoB mRNA levels were normalized against the housekeeping gene GAPDH using forward (5′-ATCAAGAAGGTGGTGAAGCAGGCA-3′) and reverse (5′-TGGAAGAGTGGGAGTTGCTGTTGA-3′) GAPDH primers.
in vivo Assay for RNAi activity of TOT-siRNA
Six- to eight-week-old male C57BL/6 mice (Charles River laboratories, USA) were maintained under a 12 h light/dark cycle in a pathogen-free animal facility. Mice were injected via the lateral tail vein with phosphate buffered saline pH 7.4 (PBS) or TOT-siRNA complexes of chemically modified (CM) ApoB siRNA or its mismatched (mm) siRNA. At 48 h after injection, liver tissue levels of ApoB mRNA, plasma levels of ApoB protein, and total plasma cholesterol were measured. For pharmacokinetic studies, liver tissue levels of ApoB mRNA were analyzed at various time intervals (48-144 h) after the injection.
Measurement of in vivo ApoB mRNA and protein levels
To determine ApoB mRNA levels in liver tissue after siRNA treatment, small uniform tissue samples were collected from 3 regions of the liver. Total RNA was extracted with Trizol and treated with DNase I before quantification. ApoB mRNA levels were determined by qPCR as described above. ApoB protein levels were determined by western blot using a polyclonal goat anti-ApoB100/48 antibody (Santa Cruz, USA). ApoB protein levels were then detected by enhanced chemiluminescence (PerkinElmer Life Sciences, USA). As a control, fibronectin was visualized by immunoblot using a polyclonal rabbit anti-fibronectin antibody (Sigma, USA).
Measurement of plasma ApoB and total cholesterol levels
Plasma cholesterol was measured by a commercial enzyme assay according to the manufacturer’s instructions (Biodesign International, USA).
In vivo interferon induction
To assess for any nonspecific immune response to injected TOT-containing siRNA, mouse liver tissue was assessed for expression of two interferon (IFN)-inducible genes, IFN-induced protein with tetratricopeptide repeats 1 (IFIT1), and signal transducer and activator of transcription 1 (STAT 1). Liver tissue was obtained from mice treated with PBS (control) and TOT-ApoB siRNA. Expression of IFIT1 and STAT1 were measured by qPCR as described above using the following primers: IFIT1 forward: 5′-AAACCCTGAGTACAACGCTGGCTA-3′ IFIT1 reverse: 5′-AAACCCTGAGTACAACGCTGGCTA-3′; STAT 1: forward: 5′-CAGCTGCAAAGCTGGTTCACCATT-3′ STAT 1 reverse: 5′-AGGTTCGATCTGACAACACCTGCT-3′. IFIT1 and STAT 1 mRNA levels were normalized against the housekeeping gene GAPDH. In addition, plasma IFN-α levels were quantified 24 h after the final injection using sandwich ELISA according to the manufacturer’s instructions (PBL biomedical Laboratories, USA). As a positive control for both assays, C57BL/6 mice were injected via the lateral tail vein with 250 μg polyinosinic-polycytidylic acid (Poly IC, total volume 0.125 ml). Six h after the injection, liver and plasma samples were collected.
Results and Discussion
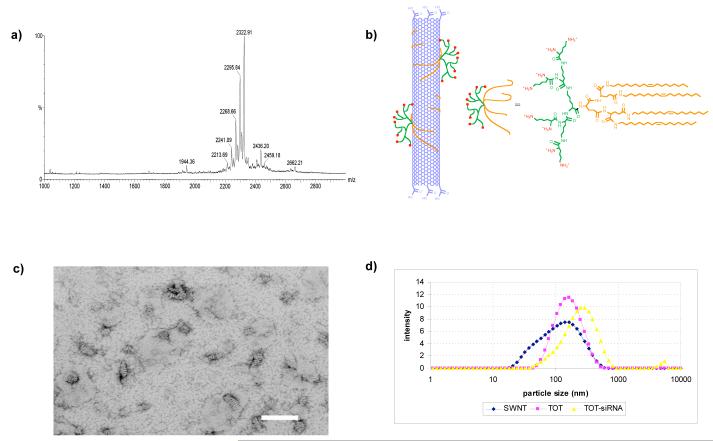
To create a novel SWNT-based nanomaterial for efficient systemic siRNA delivery, we hypothesized that SWNT should be functionalized with at least two moieties: (1) a positive-charge for siRNA binding, and (2) lipid chains to modify the hydrophilic properties of siRNA-SWNT and make contacts with cellular membranes (Fig 1a,b). Thus, we constructed TOL7, a lysine-based dendrimer with covalently attached oleoyl lipid chains. The MADLI-TOF mass spectrum (Fig 1a) shows a major peak at 2322.91, which matches the calculated molecular mass of TOL7 (2300.47 + Na+). Although the spectrum also shows some other minor peaks due to the lower purity of starting material (oleoylamine contains 60% C18 chain), nonetheless, the MS and 1H NMR confirms the successful synthesis of the target molecule. The lipid moieties on TOL7 bind to the hydrophobic wall of the nanotube upon mixing a solution of TOL7 and oxidized SWNT. However, we found that adding EDC to the mixture during the coupling increases the loading of TOL7 to the nanotube, suggesting that a covalent attachment occured through amide bonds between carboxyl groups of SWNT and amines on the surface of TOL7, which facilitated the TOL7-SWNT interaction. Quantitative analysis by performing Kaiser assay showed that in the presence of EDC the loading was 1.35 mg TOL7 to 1.0 mg SWNT, higher than in the absent of EDC (1.0 mg TOL7 to 1.0 mg SWNT). SWNT modified with TOL7 (see Experimental Procedures for details) is denoted here as TOT (Fig 1). Electron microscopic images of TOL7 assembled on the surface of SWNT reveal regular lateral striations on the surface of 80- to 150-nm nanotubes (Fig 1c). These striations suggest that TOL7 molecules are coupled to SWNT by forming half-cylinders on the nanotube surface, consistent with a previous report (27) showing supramolecular self-assembly of lipid derivatives on carbon nanotubes.
Figure 1. Structural analysis of lysine dendrimers TOL7 and functionalized nanotube TOT.
(a) MALDI-TOF MS spectrum of TOL7. (b) Chemical structure of TOL7 and schematic illustration of TOT. (c) Electron micrograph of TOT complex and (d) Particle size distribution of oxidized single walled nanotube (blue), TOT (pink), and TOT-siRNA (yellow) complex by dynamic light scattering.
To test the efficacy of TOT for systemic RNAi, we chose as model system the endogenous mouse gene, apolipoprotein B (ApoB). ApoB, a large protein that exists in two forms, ApoB100 and ApoB48 (28), is a ligand for the low-density lipoprotein (LDL) receptor and is involved in cholesterol metabolism. Mouse ApoB contains 4,515 amino acids and is predominantly expressed in the liver and intestine. In mice, both ApoB100 and ApoB48 are expressed in the liver, whereas ApoB48 is predominantly expressed in the intestine. Heterozygous knockout mouse models for ApoB show a 20% decrease in serum cholesterol levels and resist diet-induced hypercholesterolemia (29).
In humans, serum levels of ApoB have been strongly correlated with increased risk of coronary artery disease (28, 30). Because ApoB is a large protein for which no three-dimensional structural data are available, it is not a suitable target for small molecules development to treat hypercholesterolemia. An alternative therapy that has recently been explored in a mouse model is to silence ApoB with siRNA chemically modified by conjugation to cholesterol (14, 31). These studies were very encouraging in providing proof of the concept for developing RNAi-based therapeutics (14), but the amounts of siRNA used (50 mg/kg) would translate to unfeasible doses for clinical application in humans (31). Furthermore, the delivery of siRNA required cholesterol conjugation, which might increase risk factors in patients who cannot tolerate higher levels of cholesterol. Lower doses of siRNAs have recently been used by encapsulating them in SNALP to efficiently silence ApoB in non-human primates (16), but this approach requires a sophisticated SNAPL assembly. Therefore, we planned to develop new nanomaterial that can be used to easily assemble siRNA for systemic delivery. To test our approach for systemic siRNA delivery and to achieve ApoB silencing with siRNA doses in animals that are clinically relevant for RNAi-based therapy in humans, we chose ApoB silencing in mice as an animal model system.
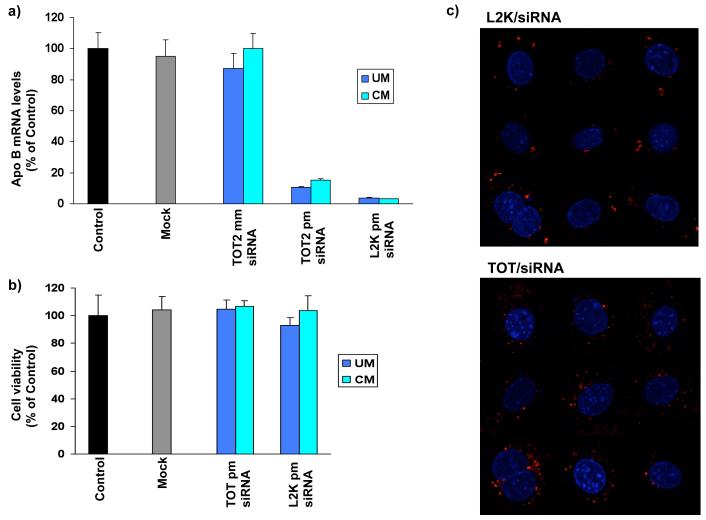
To determine whether TOT could deliver active siRNA to its target and silence ApoB mRNA levels in vitro, we treated FL83B (mouse hepatocyte) cells with TOT-siRNA complexes and analyzed ApoB mRNA levels by quantitative PCR (qPCR). TOT-siRNA containing ApoB siRNA almost completely silenced ApoB mRNA expression (>80%) in FL83B cells relative to that in controls or cells treated with TOT containing mismatched siRNA (Fig 2a). Notably, the efficiency of ApoB mRNA silencing using TOT to transport siRNA was similar to that of cells transfected with Lipofectamine 2000 complexed to siRNA (Fig 2a), although we observed slightly lower RNAi with TOT as compared with L2K. Similar to unmodified siRNA, TOT containing our chemically modified siRNA directed against ApoB efficiently silenced ApoB mRNA (Fig 2a). These results therefore show that our modifications to siRNA did not negatively influence its RNAi activity (Fig 2a). Furthermore, these reduced levels of ApoB mRNA levels in FL83B cells were not due to TOT-siRNA induced cell toxicity, as confirmed by cell viability assays (Fig 2b). Taken together, these results demonstrate that TOT-siRNA is non-toxic and efficiently transports siRNA into cells.
Figure 2. in vitro silencing of ApoB by TOT-siRNA.
(a) TOT-siRNA complex specifically silences ApoB in mouse hepatocyte cells. FL83B cells were treated for 24 h with TOT without siRNA (Mock) or with TOT-siRNA complex containing unmodified (UM) siRNA, chemically modified (CM) siRNA, or their respective mismatches (mm). As a control, Lipofectamine 2000 (L2K) was used to transfect chemically modified or unmodified siRNA. ApoB mRNA levels are expressed as percent of control (no transfection). Each value represents the mean ± SD of duplicate cultures from two representative experiments. (b) FL83B cells remain viable after 24 h treatment with TOT-siRNA complex. Cells were treated with TOT without siRNA (Mock) or with TOT-siRNA complex containing unmodified (UM) or chemically modified (CM) siRNA. Cell toxicity levels are expressed as percent of control (no transfection). Each value represents the mean ± SD of duplicate cultures from two representative experiments. (c) Localization of siRNA. Cells were transfected with Cy3-labeled siRNA using Lipofectamine 2000 (L2K) or with siRNA-TOT complex. Localization of the duplex siRNA after 24 h was monitored by confocal microscopy. Overlay images of siRNA and nuclear DNA (4′,6-diamidino-2-phenylindole [DAPI] stained) are shown.
To determine the effect of TOT on the uptake and localization of siRNA in FL83B cells, we used Lipofectamine 2000 (L2K) or TOT as described above to deliver Cy3-labeled siRNA to cells. The localization of Cy3-SS/AS-3′N3 siRNA was first established by transfecting it into cells using L2K. In both live and 4′,6-diamidino-2-phenylindole (DAPI)-stained transfected cells (32), Cy3-labeled siRNA delivered by L2K was localized to the cytoplasm around the periphery of the nucleus (Fig 2c). To determine the effect of TOT on localization of Cy3-SS/AS-3′N3 siRNA, cells were transfected with the Cy3-labeled siRNA-TOT complex. Similar to the siRNA delivered into cells by L2K, siRNA delivered by TOT localized to perinuclear regions of the cytoplasm (Fig 2c). This localization pattern is typical for siRNA transfection because siRNA duplexes are first localized to cytoplasm, which are not P bodies, and then released to form RISC assembly for RNAi functions and localization to P bodies together with argonaute proteins (9, 32, 33). Due to low fluorescence intensity and higher backgrounds, it is quite difficult to quantify the amount of RNA released and subsequent assembly into RISC in cells. Nonetheless, this result indicates that siRNA delivery by TOT did not affect cellular localization of the RNA and did not cause any aggregate formation or nuclear translocation of siRNA.
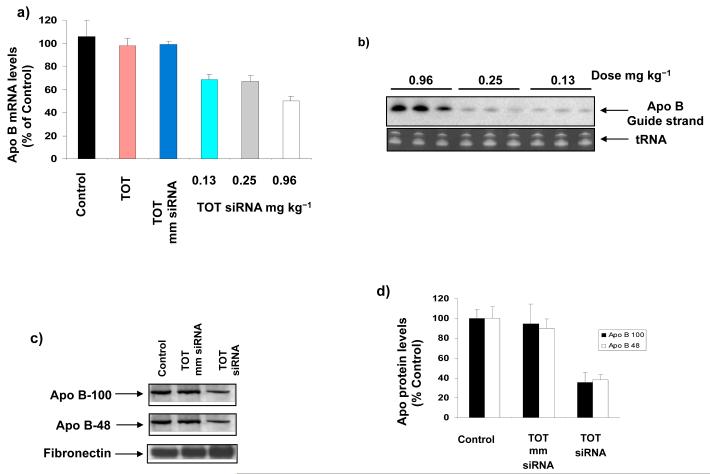
We next determined the ability of TOT to deliver siRNA to its target and to silence ApoB expression in vivo. To determine if chemical modifications in the siRNA sequence are necessary for in vivo RNAi, we verified that injecting mice with cholesterol-conjugated unmodified siRNA did not significantly knockdown ApoB expression (data not shown), consistent with a previous report (14). Therefore, our in vivo experiments focused on TOT containing chemically modified siRNA. After mice were injected via tail vein with TOT complexed to either chemically modified ApoB-targeted siRNA or its mismatch, samples of liver and plasma were analyzed (see Experimental Procedures for details). To quantify ApoB mRNA silencing throughout the liver, separate regions were analyzed for ApoB mRNA levels. ApoB mRNA decreased in a dose-dependent manner in liver tissue from mice treated with TOT-siRNA relative to that in liver tissue from control mice and mice treated with TOT containing mismatched siRNA (Fig 3a).
Figure 3. In vivo silencing of ApoB by TOT-siRNA.
(a) ApoB is specifically silenced in livers of mice injected with TOT-siRNA complex. ApoB mRNA levels were reduced in livers of mice treated 48 h earlier with varying doses of TOT complexed to chemically modified (CM) siRNA or its mismatch (mm). Values represent the mean ± SD of tissue samples from 3 liver regions (n= 3-5 animals). Data are expressed as percent of control. (b) ApoB siRNA content in mouse liver varies with TOT-siRNA dose. Total RNA was isolated from livers of mice treated with varying doses (a) of TOT complexed to unmodified (UM) or chemically modified (CM) siRNA, and was identified by northern blot analysis. A representative northern blot shows the presence of the ApoB guide strand siRNA in mouse liver 48 h after the treatment. tRNA was identified as a positive control. (c) ApoB protein plasma levels are reduced in mice injected with TOT-siRNA. Mice were injected with TOT-siRNA containing chemically modified (CM) siRNA or its mismatch (mm), and 48 h later plasma was analyzed for ApoB100 and ApoB48 protein levels by western blot analysis with anti-ApoB100 or anti-ApoB48. Total protein loading was confirmed by assessing plasma fibronectin levels. (d) Quantification of reduced ApoB plasma levels after TOT-siRNA treatment. Western blots for mouse plasma levels of ApoB100 and ApoB48 (d) were analyzed by densitometry. Data are expressed as percent of control (n = 3-5 animals).
The maximum silencing effect of TOT-siRNA was reached at 0.96 mg/kg, resulting in 56 ± 4% reduction in ApoB mRNA (n = 5 animals). Increasing the siRNA dose did not enhance the in vivo RNAi efficiency (data not shown). ). We reasoned that applying multiple doses would not enhance the silencing efficiency of TOT-siRNA, since our previous work showed that a nanoparticle created from similar dendrimer (iNOP) did not increase the RNAi efficiency when injected multiple times in animals (34). To quantify the amount of ApoB siRNA in livers of mice injected with varying doses of TOT-siRNA, total RNA was isolated from liver 48 h after treatment and guide strand ApoB siRNA was identified by northern blot analysis. Interestingly, the amount of guide strand in mouse liver decreased with decreasing doses of injected siRNA (Fig 3b). It was not clear why higher concentrations of TOT-siRNA did not increase RNAi efficiency. Nonetheless, these observations show a clear correlation between in vivo RNAi efficiency and amount of guide strand delivered.
To determine if silencing of ApoB mRNA correlated with reduced plasma levels of ApoB protein, we used immunoblot analysis to measure levels of ApoB100 and ApoB48. Both ApoB100 and ApoB48 serum levels decreased to >60% of control (Figs 3c and d) in mice injected with 0.96 mg/kg TOT-siRNA, while fibronectin levels were unaffected. These results show that TOT-siRNA complex efficiently silenced ApoB expression in vivo. Remarkably, this TOT-mediated silencing required only 0.96 mg/kg siRNA, a clinically feasible dose for RNAi therapeutic applications.
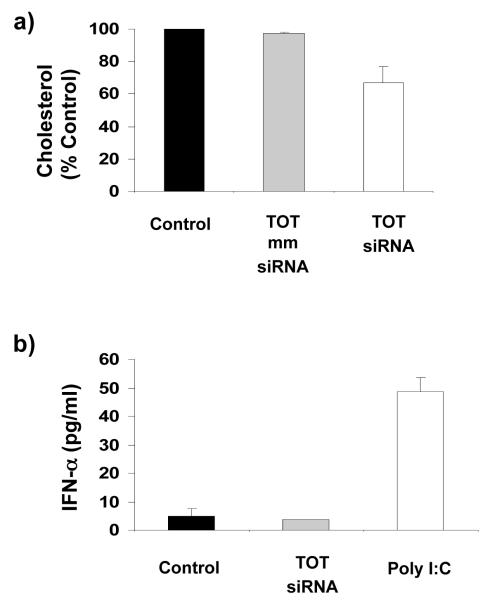
To investigate the physiological effects of ApoB mRNA silencing on cholesterol metabolism, we measured total plasma cholesterol levels in mice 48 h after a single injection of 0.96 mg/kg TOT-siRNA. As shown in Fig 4a, TOT-siRNA-mediated silencing of ApoB expression in liver and plasma samples was correlated with a reduction of total cholesterol (33 ± 10%). However, cholesterol levels were unchanged in mice treated with PBS TOT (control) or with TOT containing mismatched siRNA (Fig 4a). Together, these findings demonstrate that TOT-siRNA-mediated targeting of ApoB could provide a clinically significant new approach to reducing cholesterol levels in patients with hypercholesterolemia.
Figure 4. (a) TOT-siRNA therapeutically reduces cholesterol levels.
Plasma cholesterol levels were measured 48 h after injecting mice with 0.96 mg/kg TOT-siRNA complex, (b) TOT-siRNA treatment does not induce interferon response in mice. IFN-α plasma levels were measured 48 h after injecting mice with 0.96 mg/kg TOT without siRNA (Mock) or with 0.96 mg/kg TOT-CM siRNA. As a positive control, IFN-α plasma levels were measured after Injecting mice with 250 μg Poly IC. Each value represents the mean ± SD of 5 plasma samples from each treatment group.
siRNA-based therapies could have at least two major side effects: (1) activation of an immune response (35, 36), and (2) toxic effects. To address the concern that a non-specific immune response could be elicited by injecting mice with TOT-CM siRNA, their liver tissue RNA was assessed by qPCR for induction of the interferon (IFN)-inducible genes, IFN-induced protein with tetratricopeptide repeats 1 (IFIT1) and signal transducer and activator of transcription 1 (STAT 1), and plasma IFN-α levels were measured by ELISA. Our results show that injecting mice with TOT-siRNA did not alter the expression of IFLT1 and STAT 1 genes in the liver (data not shown), nor did it induce the release of IFN-α in plasma relative to controls (Fig 4b). To address concerns about TOT-siRNA toxicity, all mice were monitored daily for overall health, food intake and weight changes. At the end of TOT-siRNA treatment, mice were sacrificed and necropsied. Histological sections of liver, our target tissue for ApoB silencing, were prepared and independently examined for toxic effects by a board-certified animal pathologist. No histological differences were noted between tissues from control (no treatment or TOT only) mice and from those treated with TOT-siRNA complex (data not shown). These results demonstrate that TOT-siRNA treatment may not induce an immune response in animals and caused no apparent toxic effects.
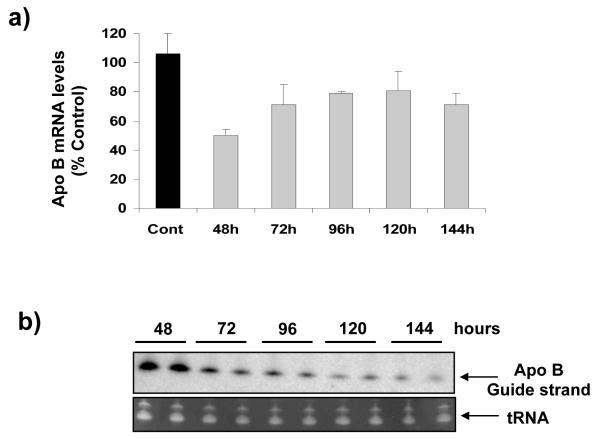
One concern about applying nanotechnology in medicine is its safety. Nanomaterials used to deliver a drug could cause undesired effects by inhibiting the drug’s biodegradation and clearance, thus prolonging its half-life in vivo. To address this question, we determined the in vivo pharmacokinetics of TOT-siRNA by injecting mice with 0.96 mg/kg of TOT-siRNA via tail vein, isolating liver tissues at various times after injection, and analyzing different regions for ApoB mRNA levels. The maximum knockdown of ApoB mRNA was observed 48 h after injecting TOT-siRNA and the ApoB mRNA levels increased to ~80% after 72h and remained at that level until 144 h (Fig 5a). To determine the amount of ApoB siRNA in mouse liver, total RNA was isolated from livers at various times after injecting mice with TOT-siRNA, and guide strand ApoB siRNA was identified by northern blot analysis. The amount of guide strand found in mouse liver was greatest at 48 h after TOT-siRNA injection and decreased with time (up to 144 h) (Fig 5b). Taken together, these results show a correlation between in vivo RNAi efficiency at various times after TOT-siRNA treatment and amount of guide strand present in mouse liver. Although these findings do not directly demonstrate the half-life of nanomaterial in vivo, however, these results suggest that the RNA trigger molecules in the siRNA-TOT complexes were not trapped in liver and were cleared quite rapidly from the body.
Figure 5. Pharmacokinetics of TOT-siRNA.
(a) Mice were injected with 0.96 mg/kg of TOT-siRNA and ApoB mRNA levels were measured in liver at various times after injection. Values represent the mean ± SD of tissue samples from 3-5 animals, (b) ApoB siRNA decreases with time in livers of mice injected with TOT-siRNA. Mice were injected with TOT-siRNA (a), total RNA was isolated from their livers at various times after injection, and ApoB siRNA guide strand was identified by northern blot analysis. A representative northern blot shows that the amount of ApoB guide strand siRNA decreased over the time range shown. tRNA was Identified as a positive control.
Nanomedicine has recently emerged as a new branch of science that explores applications of nanotechnology for diagnosing, monitoring, treating, and controlling biological functions (37). However, the questions of physiological toxicity and favorable pharmacology of nanomaterials remains unanswered. Here we present evidence that a new approach to designing nanotubes can be used to create nanomaterials for in vivo delivery of siRNA to silence endogenous disease-related genes at clinically acceptable and therapeutically affordable doses. Injecting mice with only 0.96 mg/kg siRNA in complex with TOT (SWNT functionalized with lipids and natural amino acid-based dendrimers) silenced ApoB mRNA and reduced protein levels in liver, decreased plasma levels of ApoB, and lowered total plasma cholesterol. Moreover, TOT treatment was non-toxic and did not induce an immune response. Most of the RNA molecules (80%) assembled with TOT were cleared from the body 48 h after treatment, indicating that the nanotubes did not inhibit siRNA biodegradation and clearance, ensuring a short half-life for siRNA in vivo. These results provide the first evidence that nanotubes can be safely and effectively used to systemically deliver siRNA, thus offering a new technology for systemic RNAi and potentially for delivering other drugs in vivo.
Scheme 1.
Synthesis of Tetra-Oleoyl Lysine Dendrimer Generation 3 (TOL7)
Acknowledgments
We thank Michael Czech, Craig Mello and members of the Rana lab for helpful discussions and Dr. David Garlick for histological examination of slides. We also thank Dr. Gregory Hendricks and his group at the Core Electron Microscopy Facility, UMASS Medical School for their advice and help with the electron microscopy experiments. This work was supported in part by a grant from the NIH to T.M.R.
References
- (1).Star A, Stoddart JF, Steuerman D, Diehl M, Boukai A, Wong EW, Yang X, Chung SW, Choi H, Heath JR. Preparation and Properties of Polymer-Wrapped Single-Walled Carbon Nanotubes. Angew Chem Int Ed Engl. 2001;40:1721–1725. doi: 10.1002/1521-3773(20010504)40:9<1721::aid-anie17210>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- (2).Singh R, Pantarotto D, McCarthy D, Chaloin O, Hoebeke J, Partidos CD, Briand JP, Prato M, Bianco A, Kostarelos K. Binding and condensation of plasmid DNA onto functionalized carbon nanotubes: toward the construction of nanotube-based gene delivery vectors. J Am Chem Soc. 2005;127:4388–96. doi: 10.1021/ja0441561. [DOI] [PubMed] [Google Scholar]
- (3).Kam NW, Liu Z, Dai H. Functionalization of carbon nanotubes via cleavable disulfide bonds for efficient intracellular delivery of siRNA and potent gene silencing. J Am Chem Soc. 2005;127:12492–3. doi: 10.1021/ja053962k. [DOI] [PubMed] [Google Scholar]
- (4).Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–11. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- (5).Hannon GJ, Rossi JJ. Unlocking the potential of the human genome with RNA interference. Nature. 2004;431:371–8. doi: 10.1038/nature02870. [DOI] [PubMed] [Google Scholar]
- (6).Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–6. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- (7).Caplen NJ, Parrish S, Imani F, Fire A, Morgan RA. Specific inhibition of gene expression by small double-stranded RNAs in invertebrate and vertebrate systems. Proc Natl Acad Sci U S A. 2001;98:9742–7. doi: 10.1073/pnas.171251798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–8. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- (9).Rana TM. Illuminating the silence: understanding the structure and function of small RNAs. Nat Rev Mol Cell Biol. 2007;8:23–36. doi: 10.1038/nrm2085. [DOI] [PubMed] [Google Scholar]
- (10).Amarzguioui M, Holen T, Babaie E, Prydz H. Tolerance for mutations and chemical modifications in a siRNA. Nucleic Acids Res. 2003;31:589–95. doi: 10.1093/nar/gkg147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Chiu YL, Rana TM. siRNA function in RNAi: a chemical modification analysis. Rna. 2003;9:1034–48. doi: 10.1261/rna.5103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Chiu YL, Rana TM. RNAi in human cells: basic structural and functional features of small interfering RNA. Mol Cell. 2002;10:549–61. doi: 10.1016/s1097-2765(02)00652-4. [DOI] [PubMed] [Google Scholar]
- (13).Morrissey DV, Lockridge JA, Shaw L, Blanchard K, Jensen K, Breen W, Hartsough K, Machemer L, Radka S, Jadhav V, Vaish N, Zinnen S, Vargeese C, Bowman K, Shaffer CS, Jeffs LB, Judge A, MacLachlan I, Polisky B. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat Biotechnol. 2005;23:1002–7. doi: 10.1038/nbt1122. [DOI] [PubMed] [Google Scholar]
- (14).Soutschek J, Akinc A, Bramlage B, Charisse K, Constien R, Donoghue M, Elbashir S, Geick A, Hadwiger P, Harborth J, John M, Kesavan V, Lavine G, Pandey RK, Racie T, Rajeev KG, Rohl I, Toudjarska I, Wang G, Wuschko S, Bumcrot D, Koteliansky V, Limmer S, Manoharan M, Vornlocher HP. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173–8. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- (15).Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–9. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- (16).Zimmermann TS, Lee AC, Akinc A, Bramlage B, Bumcrot D, Fedoruk MN, Harborth J, Heyes JA, Jeffs LB, John M, Judge AD, Lam K, McClintock K, Nechev LV, Palmer LR, Racie T, Rohl I, Seiffert S, Shanmugam S, Sood V, Soutschek J, Toudjarska I, Wheat AJ, Yaworski E, Zedalis W, Koteliansky V, Manoharan M, Vornlocher HP, MacLachlan I. RNAi-mediated gene silencing in non-human primates. Nature. 2006;441:111–4. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]
- (17).Palliser D, Chowdhury D, Wang QY, Lee SJ, Bronson RT, Knipe DM, Lieberman J. An siRNA-based microbicide protects mice from lethal herpes simplex virus 2 infection. Nature. 2006;439:89–94. doi: 10.1038/nature04263. [DOI] [PubMed] [Google Scholar]
- (18).Santel A, Aleku M, Keil O, Endruschat J, Esche V, Fisch G, Dames S, Loffler K, Fechtner M, Arnold W, Giese K, Klippel A, Kaufmann J. A novel siRNA-lipoplex technology for RNA interference in the mouse vascular endothelium. Gene Ther. 2006 doi: 10.1038/sj.gt.3302777. [DOI] [PubMed] [Google Scholar]
- (19).Song E, Zhu P, Lee SK, Chowdhury D, Kussman S, Dykxhoorn DM, Feng Y, Palliser D, Weiner DB, Shankar P, Marasco WA, Lieberman J. Antibody mediated in vivo delivery of small interfering RNAs via cell-surface receptors. Nat Biotechnol. 2005;23:709–17. doi: 10.1038/nbt1101. [DOI] [PubMed] [Google Scholar]
- (20).Krajcik R, Jung A, Hirsch A, Neuhuber W, Zolk O. Functionalization of carbon nanotubes enables non-covalent binding and intracellular delivery of small interfering RNA for efficient knock-down of genes. Biochem Biophys Res Commun. 2008;369:595–602. doi: 10.1016/j.bbrc.2008.02.072. [DOI] [PubMed] [Google Scholar]
- (21).Liu Z, Winters M, Holodniy M, Dai H. siRNA delivery into human T cells and primary cells with carbon-nanotube transporters. Angew Chem Int Ed Engl. 2007;46:2023–7. doi: 10.1002/anie.200604295. [DOI] [PubMed] [Google Scholar]
- (22).Wang X, Ren J, Qu X. Targeted RNA interference of cyclin A2 mediated by functionalized single-walled carbon nanotubes induces proliferation arrest and apoptosis in chronic myelogenous leukemia K562 cells. ChemMedChem. 2008;3:940–5. doi: 10.1002/cmdc.200700329. [DOI] [PubMed] [Google Scholar]
- (23).Liu J, Rinzler AG, Dai H, Hafner JH, Bradley RK, Boul PJ, Lu A, Iverson T, Shelimov K, Huffman CB, Rodriguez-Macias F, Shon YS, Lee TR, Colbert DT, Smalley RE. Fullerene pipes. Science. 1998;280:1253–6. doi: 10.1126/science.280.5367.1253. [DOI] [PubMed] [Google Scholar]
- (24).Baigude H, Katsuraya K, Okuyama K, Tokunaga S, Uryu T. Synthesis of Sphere-Type Monodispersed Oligosaccharide-Polypeptide Dendrimers Macromolecules. 2003;36:7100–7106. [Google Scholar]
- (25).Shao J, Tam JP. Unprotected Peptides as Building Blocks for the Synthesis of Peptide Dendrimers with Oxime, Hydrazone, and Thiazolidine Linkages. J. Am. Chem. Soc. 1995;117:3893–3899. [Google Scholar]
- (26).Gao L, Nie L, Wang T, Qin Y, Guo Z, Yang D, Yan X. Carbon nanotube delivery of the GFP gene into mammalian cells. Chembiochem. 2006;7:239–42. doi: 10.1002/cbic.200500227. [DOI] [PubMed] [Google Scholar]
- (27).Richard C, Balavoine F, Schultz P, Ebbesen TW, Mioskowski C. Supramolecular self-assembly of lipid derivatives on carbon nanotubes. Science. 2003;300:775–8. doi: 10.1126/science.1080848. [DOI] [PubMed] [Google Scholar]
- (28).Olofsson SO, Boren J. Apolipoprotein B: a clinically important apolipoprotein which assembles atherogenic lipoproteins and promotes the development of atherosclerosis. J Intern Med. 2005;258:395–410. doi: 10.1111/j.1365-2796.2005.01556.x. [DOI] [PubMed] [Google Scholar]
- (29).Farese RV, Jr., Veniant MM, Cham CM, Flynn LM, Pierotti V, Loring JF, Traber M, Ruland S, Stokowski RS, Huszar D, Young SG. Phenotypic analysis of mice expressing exclusively apolipoprotein B48 or apolipoprotein B100. Proc Natl Acad Sci U S A. 1996;93:6393–8. doi: 10.1073/pnas.93.13.6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Brown MS, Goldstein JL. A receptor-mediated pathway for cholesterol homeostasis. Science. 1986;232:34–47. doi: 10.1126/science.3513311. [DOI] [PubMed] [Google Scholar]
- (31).Rossi JJ. Medicine: a cholesterol connection in RNAi. Nature. 2004;432:155–6. doi: 10.1038/432155a. [DOI] [PubMed] [Google Scholar]
- (32).Chiu YL, Ali A, Chu CY, Cao H, Rana TM. Visualizing a correlation between siRNA localization, cellular uptake, and RNAi in living cells. Chem Biol. 2004;11:1165–75. doi: 10.1016/j.chembiol.2004.06.006. [DOI] [PubMed] [Google Scholar]
- (33).Chu CY, Rana TM. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol. 2006;4:e210. doi: 10.1371/journal.pbio.0040210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Baigude H, McCarroll J, Yang CS, Swain PM, Rana TM. Design and creation of new nanomaterials for therapeutic RNAi. ACS Chem Biol. 2007;2:237–41. doi: 10.1021/cb7000582. [DOI] [PubMed] [Google Scholar]
- (35).Marques JT, Williams BR. Activation of the mammalian immune system by siRNAs. Nat Biotechnol. 2005;23:1399–405. doi: 10.1038/nbt1161. [DOI] [PubMed] [Google Scholar]
- (36).Robbins MA, Rossi JJ. Sensing the danger in RNA. Nat Med. 2005;11:250–1. doi: 10.1038/nm0305-250. [DOI] [PubMed] [Google Scholar]
- (37).Wagner V, Dullaart A, Bock AK, Zweck A. The emerging nanomedicine landscape. Nat Biotechnol. 2006;24:1211–7. doi: 10.1038/nbt1006-1211. [DOI] [PubMed] [Google Scholar]