Summary
Translesion DNA polymerases are more efficient at bypass of many DNA adducts than replicative polymerases. Previous work with the translesion polymerase Sulfolobus solfataricus Dpo4 showed a decrease in catalytic efficiency during bypass of bulky N2-alkyl deoxyguanosine (G) adducts with N2-isobutylG showing the largest effect, decreasing ∼120-fold relative to unmodified dG (Zhang, H. et al. (2009) J. Biol. Chem. 284, 3563-3576). The effect of adduct size upon individual catalytic steps has not been easy to decipher because of the difficulty of distinguishing early non-covalent steps from phosphodiester bond formation. We developed a mutant with a single Trp (T239W) to monitor fluorescence changes associated with a conformational change that occurs after binding a correct dNTP (Beckman, J.W. et al. (2008) J. Biol. 283, 36711-36723) and, in the present work, utilized this approach to monitor insertion opposite N2-alkylG-modified oligonucleotides. We estimated maximal rates for the forward conformational step, which coupled with measured rates of product formation yielded rate constants for the conformational step (both directions) during insertion opposite several N2-alkylG adducts. With the smaller N2-alkylG adducts, the conformational rate constants were not changed dramatically (< 3-fold), indicating that the more sensitive steps are phosphodiester bond formation and partitioning into inactive complexes. With the larger adducts (≥ (2-naphthyl)methyl) the absence of fluorescence changes suggests impaired ability to undergo an appropriate conformational change, consistent with previous structural work.
Keywords: DNA polymerase, DNA adducts, translesion DNA synthesis, fluorescence, pre-steady-state kinetics
Introduction
The preservation and transmittal of accurate genetic information is dependent upon the fidelity of DNA replication.1 The role of Watson-Crick base pairing in storing genetic information has been recognized since the elucidation of the structure of DNA,2 but energetic considerations argue that the specificity of G:C and A:T pairing is not sufficient to account for the selectivity and the accuracy of the genetic code is instead maintained to a large extent by DNA polymerases through various mechanisms.3
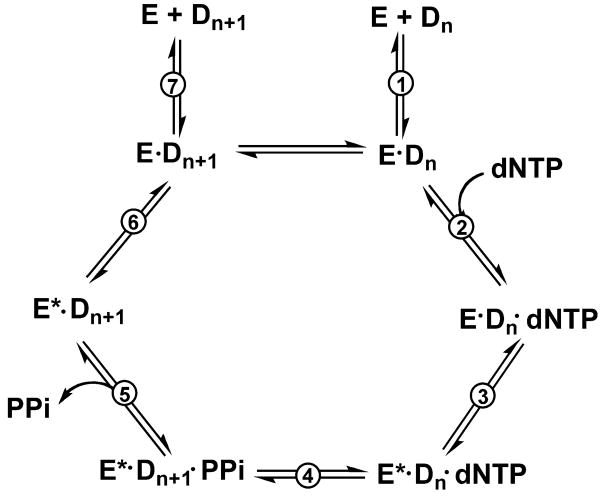
Most DNA polymerases catalyze dNTP incorporation according to a general polymerization mechanism (Fig. 1). First, enzyme and DNA bind to form a binary complex, which then binds dNTP to form a polymerase-DNA-dNTP ternary complex and induces a conformational change to facilitate the formation of a phosphodiester bond. After the reaction, PPi is released and the ternary complex is relaxed to initiate a new cycle. The crystal structures of ternary complexes have revealed large nucleotide-induced conformational changes for some DNA polymerases.4-10 Efficient polymerization results from a conformational change that appropriately aligns the primer 3′-hydroxyl and the α-phosphate group of the incoming dNTP for phosphodiester bond formation. However, in at least one instance (pol β) the large “closing” event observed in the crystal structure was shown to be too fast to be significantly rate-limiting in the kinetic mechanism,11,12 and a more subtle change is considered to be more relevant to discrimination by the DNA polymerases.13 This conformational change step prior to phosphoryl transfer has been considered to be very significant in nucleotide selection and polymerase fidelity.13-15
Fig. 1.
General DNA polymerization mechanism. E: polymerase, Dn: DNA (oligonucleotide) substrate; E*: conformationally-modified polymerase; Dn+1: DNA (oligonucleotide) extended by one base (product); PPi: pyrophosphate. Numbers of steps are associated with nomenclature of the rate constants.
Although crystal structures are important they are static, and dynamic fluorescence approaches can provide kinetic information about conformational changes in a reaction pathway. Several methods have been used with DNA polymerases. 2-AP can be positioned at the site of incorporation or the 5′-next template position and has been used to study conformational changes with Escherichia coli pol I and bacteriophage pol T4,16,17 pol β,18-20 RB69,21,22 and Sulfolobus acidocaldarius Dbh,23 with the fluorescence signal dependent on dNTP-induced protein structure changes. The contribution of local conformational changes in the enzyme structure to the changes observed with 2-AP is not entirely clear.21,24,25 Stopped-flow fluorescence resonance energy transfer methods have been used with E. coli DNA pol I25,26 and Klentaq127-29 by attaching a donor and an acceptor at different positions of the protein or DNA to report changes in protein structure or DNA movement. Covalent attachment of an environmentally sensitive dye to the finger domain has also been used to monitor conformational changes with bacteriophage T7 DNA polymerase (pol T7-),30,31 although the question of whether a local conformational change near the dye or a global movement of the finger subdomain is involved has been raised.25 Single-molecule and ensemble fluorescence measurements32 as well as hydrogen-deuterium exchange methods33 have also been used to study conformational changes in DNA polymerases.
Recently our group reported that Sulfolobus solfataricus Dpo4 T239W and N188W have similar activity to wild-type Dpo4 and can be directly used to monitor conformational changes that occur during DNA polymerization, without the use of dyes and without altering the catalytic efficiency of the enzyme.34 Previous kinetic work with Dpo4 has shown that this model Y-family DNA polymerase shows decreased catalytic efficiency during bypass of N2-alkylG adducts that is dependent on the increasing size of the moiety attached to the N2 atom of guanine.35 The availability of kinetic and structural data related to Dpo4-catalyzed bypass of N2-alkylG adducts provides an excellent opportunity to use kinetic monitoring of conformational changes to understand the effect of bulk upon individual steps in the polymerase catalytic cycle. In order to discriminate between the effects of DNA adducts bulk on conformational change and phosphodiester bond formation, early steps in the incorporation of dCTP opposite N2-alkylG adducts were studied using Dpo4 T239W.
Results
Steady-state kinetics of dCTP incorporation opposite G and N2-alkylG adducts by Dpo4 T239W
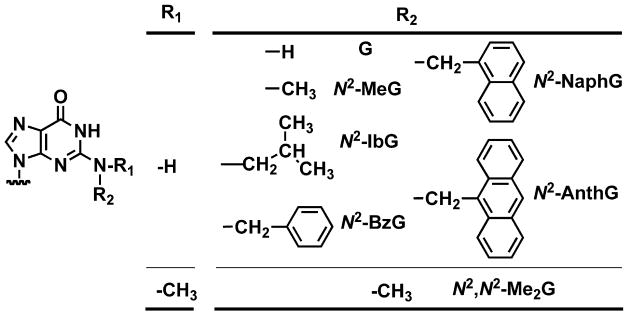
A 13-mer primer/18-mer template duplex (Table 1) containing G or various N2-alkylG adducts (N2-MeG, N2-IbG, N2-BzG, N2-NaphG, N2-AnthG, or N2,N2-Me2G, Fig. 2) was used to measure steady-state kinetic parameters (kcat and Km) for the incorporation of dCTP opposite G or these adducts by Dpo4 T239W, varying the concentration of dCTP (Table 2). As reported for the wild-type enzyme,35,36 the incorporation efficiencies were similar for N2-MeG and G and decreased as much as 80-fold for other N2-alkylG adducts and 160,000-fold for N2,N2-Me2G, due to both the decrease in kcat and increase in Km,dCTP. As noted previously,36 the trend was partially reversed with the bulky N2-AnthG.
Table 1.
Oligodeoxynucleotides used in this study
| 13Cdd-mer | 5′-GGGGGAAGGATTCdd- 3′ |
| 13C-mer | 5′-GGGGGAAGGATTC - 3′ |
| 18-mer | 3′-CCCCCTTCCTAAGG*CACT- 5′ |
G*: G, N2-MeG, N2-IbG, N2-BzG, N2-NaphG, N2-AnthG, or N2,N2-Me2G. Cdd: terminal 2′,3′-dideoxycytidine.
Fig. 2.
Structures of N2-alkylG adducts used in this study.
Table 2.
Steady-state kinetic parameters for dCTP incorporation by Dpo4 T239Wa
| Template base |
kcat s-1 |
Km,dCTP μM |
kcat/Km,dCTP mM-1 s-1 |
kcat/Km,dCTP relative to G |
|---|---|---|---|---|
| G | 0.057 ± 0.001 | 5.9 ± 0.5 | 9.7 | 1.0 |
| N2-MeG | 0.078 ± 0.003 | 9.7 ± 1.8 | 8.0 | 0.82 |
| N2-IbG | 0.011 ± 0.001 | 105 ± 30 | 0.10 | 0.010 |
| N2-BzG | 0.0061 ± 0.0003 | 36 ± 6 | 0.17 | 0.018 |
| N2-NaphG | 0.015 ± 0.001 | 124 ± 16 | 0.12 | 0.012 |
| N2-AnthG | 0.0031 ± 0.001 | 5.1 ± 0.6 | 0.61 | 0.062 |
| N2,N2-Me2G | (11 ± 0.1) × 10-6 | 190 ± 44 | 5.8 × 10-5 | 5.9 × 10-6 |
The extent of conversion of primer to the product was kept < 20% by adjustment of the enzyme concentration and reaction time.
Determination of kpol and Kd,dCTP for dCTP incorporation by Dpo4 T239W
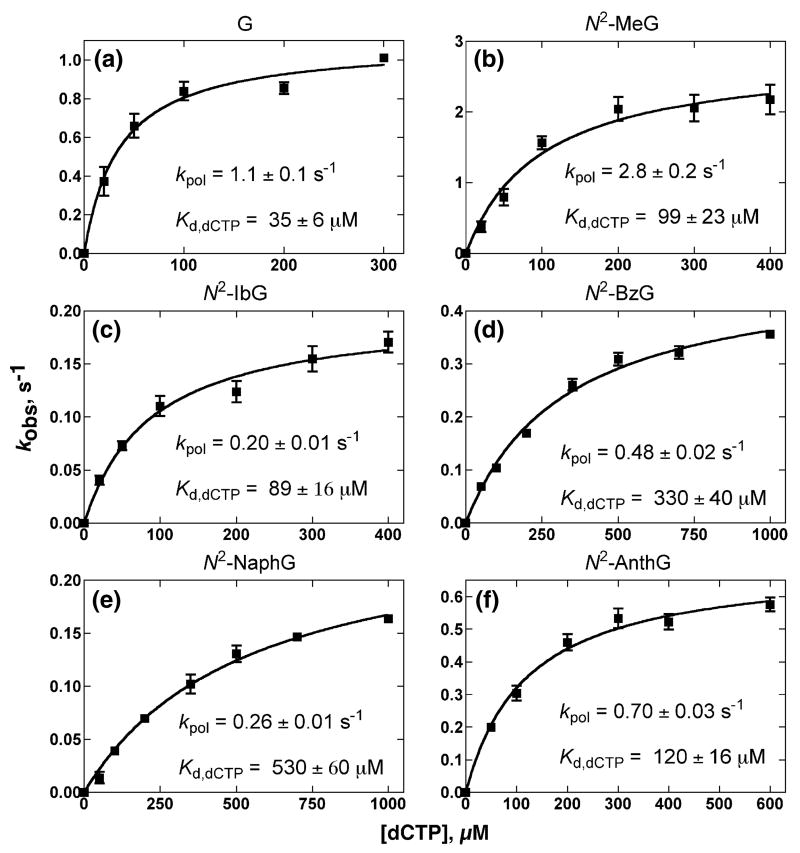
Dpo4 T239W showed burst kinetics for the incorporation of dCTP opposite G or N2-alkylG but no burst phase for incorporation opposite N2,N2-Me2G, as observed with the wild-type enzyme35,36 (results not shown). Analysis of the changes in rates as a function of dCTP concentration (under conditions of excess Dpo4 T239W over oligonucleotide) yielded maximal rates of nucleotide incorporation (kpol) and apparent dCTP binding affinity (Kd,dCTP)37,38 (Fig. 3, Table 3, Supplementary Data Fig. S1). The incorporation efficiency (kpol/Kd,dCTP) was decreased as a function of the bulk of the N2-alkylG adduct, with the exception of N2-AnthG (also noted in the steady-state measurements) (Table 2), similar to results observed with 24-mer/36-mer oligonucleotides and wild-type Dpo4.36
Fig. 3.
Estimation of kpol and apparent Kd,dCTP for the incorporation of dCTP opposite G and N2-alkylG adducts in 13C-mer/18-mer oligonucleotide pairs catalyzed by Dpo4 T239W. Dpo4 T239W (200 nM) was incubated with 10 nM 13C-mer/18-mer complexes in a rapid quench-flow instrument and then mixed with varying concentrations of dCTP (plus 5 mM Mg2+) to initiate reactions. Plots of burst rates (kobs) versus [dCTP] were fit to a hyperbolic equation to yield kpol and apparent Kd,dCTP (values shown in figures).
Table 3.
Pre-steady-state kinetic analysis of dCTP incorporation by Dpo4 T239W
| Template base |
kpol s-1 |
Kd,dCTP μM |
kpol/Kd,dCTP mM-1 s-1 |
kpol/Kd,dCTP relative to G |
|---|---|---|---|---|
| G | 1.1 ± 0.1 | 35 ± 6 | 31 | 1.0 |
| N2-MeG | 2.8 ± 0.2 | 99 ± 23 | 28 | 0.90 |
| N2-IbG | 0.20 ± 0.01 | 89 ± 16 | 2.2 | 0.071 |
| N2-BzG | 0.48 ± 0.02 | 330 ± 40 | 1.5 | 0.15 |
| N2-NaphG | 0.26 ± 0.01 | 530 ± 60 | 0.49 | 0.016 |
| N2-AnthG | 0.70 ± 0.03 | 120 ± 16 | 5.8 | 0.19 |
Rapid fluorescence changes associated with dCTP binding to Dpo4 T239W-oligonucleotide complexes
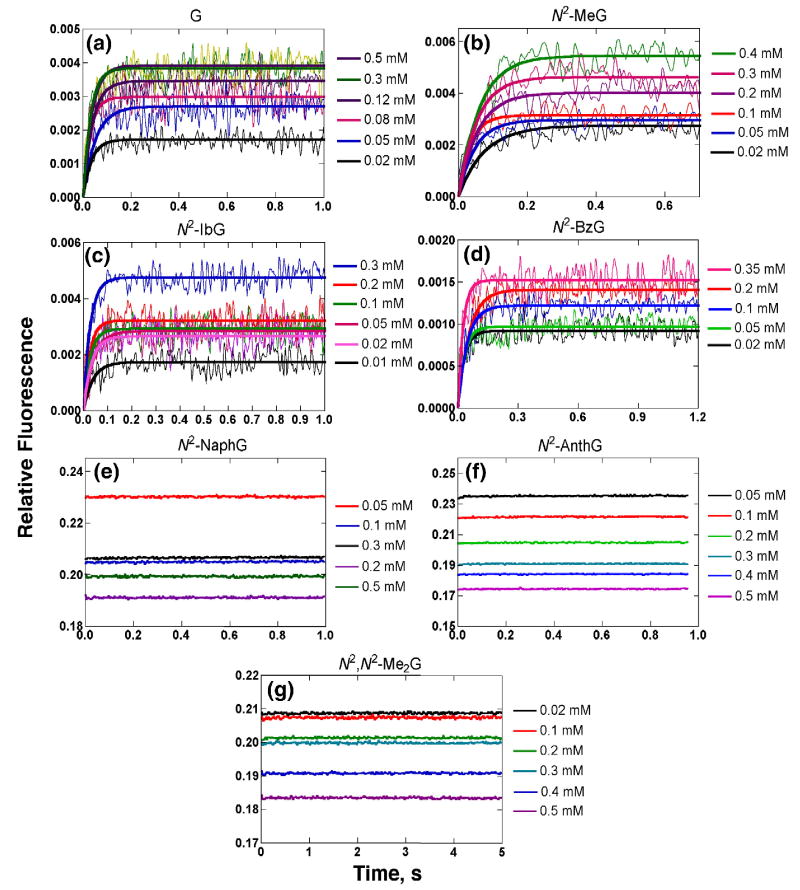
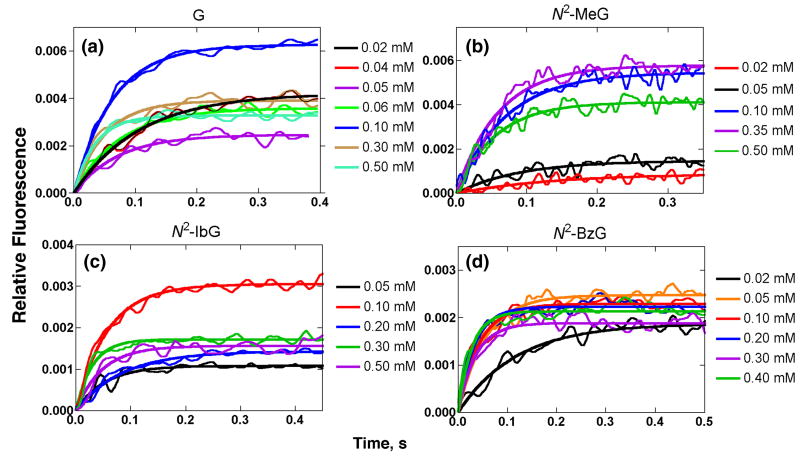
Fluorescence changes were observed upon mixing dCTP opposite templates containing G and N2-alkylG adducts at varying dCTP concentrations (Fig. 4), consonant with previous results.34 The use of 2′,3′-dideoxy sugars precluded the formation of phosphodiester bonds, providing kinetic information prior to the step in which the phosphodiester bond is formed (Fig. 1, step 4). Addition of dCTP opposite G, N2-MeG, N2-IbG, and N2-BzG showed fast increases (Fig. 4a-d). Such changes have previously been attributed to conformational changes in the protein,34 and rates were obtained by fitting the change of the fluorescence signal to a single exponential equation (Eq. 3) at each dCTP concentration. No detectable fluorescence changes were observed for the addition of dCTP opposite N2-NaphG, N2-AnthG, or N2,N2-Me2G (Fig. 4e-g). (We checked the effect of the N2-NaphG and N2-AnthG oligomers on the steady-state (Trp) fluorescence of Dpo4 T239W, under the conditions used in Fig. 4, and found that none of the fluorescence (350 nm) was lost, similar to previous results with unmodified oligomers34 (the N2-AnthG oligomer had major bands at 400-420 nm, in addition, but these did not interfere).
Fig. 4.
Stopped-flow fluorescence changes for the addition of dCTP opposite 13Cdd-mer/18-mer oligonucleotide pair containing G or N2-alkylG adducts by Dpo4 T239W. The reactions included a complex of 1 μM Dpo4 T239W–13Cdd-mer/18-mer oligonucleotide pair, 5 mM Mg2+, and varying concentrations of dCTP at 37 °C. dCTP concentrations are shown at the right of each part of the figure.
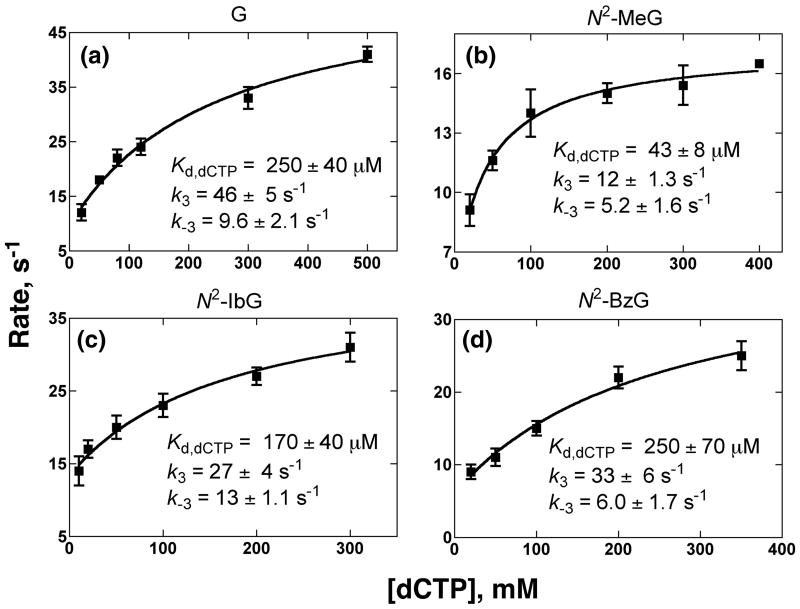
The dCTP ground dissociation constants (Kd,dCTP,ground) and dCTP-induced conformational change rate constants were estimated by fitting the observed rates (from the experiments with the 2′, 3′-dideoxy oligonucleotides) against dCTP concentrations based on Eq. 4 in the following model34 (Fig. 5 and Table 4)
Fig. 5.
Estimation of forward and reverse conformational change rate constants (k3 and k-3) and ground state Kd,dCTP for the incorporation of dCTP opposite G or N2-alkylG adducts in 13Cdd-mer/18-mer oligonucleotide pairs by fitting the observed fluorescence change rates (Fig. 4) vs. dCTP concentrations (Eq. 4).
Table 4.
Pre-steady-state rate constants for conformational changes of Dpo4 T239W as detected by stopped-flow fluorescence
| Template base | Basis of 2′,3′-dideoxy oligonucleotide | Basis of normal (3′-OH) oligonucleotide | ||||
|---|---|---|---|---|---|---|
| k3 (s-1) | k-3 (s-1) | k3/k-3 | Kd (μM) | k3′ (s-1) | k-3′ (s-1) | |
| G | 46 | 9.6 | 4.8 | 250 | 29 | 9.2 |
| N2-MeG | 12 | 5.2 | 2.3 | 43 | 11 | 5.2 |
| N2-IbG | 27 | 13 | 2.1 | 170 | 15 | 11 |
| N2-BzG | 33 | 6 | 5.5 | 250 | 8.9 | 7.6 |
(where “DNA” indicates the primer-template oligonucleotide complex). For the incorporation of dCTP opposite unmodified G, the ground-state dissociation constant (Kd,dCTP,ground = 250 ± 40 μM,), a forward conformational change rate (k3 = 46 ± 5 s-1), and a reverse conformational change rate (k-3 = 9.6 ± 2.1 s-1) were all similar to previous results.34 The addition of dCTP opposite N2-MeG, N2-IbG, or N2-BzG by Dpo4 T239W yielded similar kinetic parameters for dCTP binding and the induced conformational change (Table 4).
As shown previously with Dpo4 T239W using a normal primer (with 3′-OH),34 the fluorescence signal (Trp) showed a rapid increase (within 0.5 s) and then slowly decreased for incorporation. The fast phase (increasing fluorescence) was previously concluded to correspond to a dCTP-induced conformation change (step 3 in Fig. 1) (following very rapid binding, step 2) and was observed with G, N2-MeG, N2-IbG, and N2-BzG but not any of the bulkier lesions (Fig. 6). The rates of dCTP–induced conformational change increased as a function of the dCTP concentration (Fig. 7), and fitting of the observed fast phase rates against dCTP concentrations according to Eq. 4 gave estimates of the fast phase rate constant (k3′) and its reverse rate constant (k-3′) by an independent method. The kinetic parameters obtained using this primer were similar to those obtained using the 13Cdd-mer/18-mer oligonucleotide pair (Table 4), with one exception (k3 for N2-BzG), indicating that the primer 3′-OH had little effect on dCTP binding and the conformational change of Dpo4 T239W.
Fig. 6.
Stopped-flow fluorescence changes (fast phase) for the addition of dCTP opposite G or N2-alkylG adducts in 13C-mer/18-mer oligonucleotide pairs by Dpo4 T239W. The conditions included 1 μM Dpo4 T239W–13C-mer/18-mer oligonucleotide complex, 5 mM Mg2+, and varying concentrations of dCTP at 37 °C. dCTP concentrations are shown at the right of each part of the figure.
Fig. 7.
Estimation of rate constants for conformational change and Kd,dCTP following the addition of dCTP opposite G and N2-alkyl G adducts in 13C-mer/18-mer oligonucleotides. The rate constants, designated k3′and k-3′ here (to distinguish from k3 and k-3 estimated with dideoxy oligonucleotide, Figs. 4, 5), were estimated by fitting the observed fluorescence change rates against dCTP concentration.
The addition of dTTP opposite a 13Cdd-mer/18-mer oligonucleotide pair containing G or an N2-alkylG adduct did not yield an increase in the fluorescence signal (Supplementary Data Fig. S2a), in contrast to the (correct) dCTP incorporation (Fig. 4a-d). The misincorporation of dTTP opposite G or any of the N2-alkylG adducts showed no kinetic burst phase of product formation (Supplementary Data Fig. S2b). Misincorporation of dTTP opposite G or any N2-alkylG adduct in a 13C-mer/18-mer oligonucleotide pair (with free 3′-OH) also showed no increase in fluorescence (Supplementary Data Fig. S2c).
Discussion
N2-AlkylG adducts have been extensively studied with the replicative DNA polymerases bacteriophage pol T7- and HIV-1 transcriptase and the translesion polymerases human DNA pols η, ι, κ, and Rev1, as well as Dpo4.35,36,39-43 Small lesions, even N2-MeG and N2-EtG, strongly block pol T7- and HIV-1 reverse transcriptase,39 in stark contrast to the relatively minor (or non-existent) inhibition of translesion polymerases. The Y-family DNA polymerases η, ι, κ, and Rev1 can all bypass the bulkier N2-alkylG adducts but the incorporation efficiency does decrease for the larger modifications.40-43 We have shown that wild-type Dpo4 is strongly blocked by N2,N2-Me2G35 and bypasses N2-alkylG adducts, with the incorporation efficiency lowered by about ∼120-fold.36
Recently we developed the Dpo4 mutants T239W and N188W to monitor conformational changes associated with dNTP binding.34 In this work Dpo4 T239W was used to analyze conformational changes associated with the addition of dCTP opposite N2-alkylG adducts (Fig. 2). A fluorescent signal that we attribute to a catalytically-relevant conformational change34 was observed with G and adducts ranging in size up to N2-BzG. No fluorescence increases associated with conformational changes were observed for the N2-alkylG adducts bulkier than N2-BzG (N2-NaphG, N2-AnthG) or N2,N2-Me2G, consistent with abnormal crystal structures we have reported for N2-NaphG and N2,N2-Me2G.35,36 The incorporation of dCTP opposite G (and possibly the smaller N2-alkylG adducts) via standard W-C base pairing44 induces a fast conformational change in Dpo4 T239W.34 However, the crystal structures of Dpo4 with oligonucleotide duplexes containing N2-NaphG or N2,N2-Me2G showed disrupted, non-W-C base paring.35,36 The fluorescent signal observed in the stopped-flow experiment is considered to be a result of the majority of Dpo4 molecules undergoing the same or at least a similar change in the orientation of Trp-239 upon binding of dCTP. The multiple binding orientations observed with N2-NaphG and N2,N2-Me2G35,36 are in agreement with the lack of a fluorescence signal (Fig. 4e-g) reflecting the conformational change. As alternative abnormal binding modes are accommodated in the polymerase active site in solution, the amplification of the fluorescence signal will be diminished because these non-productive binding modes will not elicit the appropriate change in the conformation of Dpo4.
Earlier studies with the replicative DNA polymerases pol T7- and E. coli pol I have been interpreted to indicate that the rate-limiting step is the conformational change for correct incorporations but changes to phosphodiester bond formation for misincorporations.13,37,45-47 With a modified pol T7-, the conformational change acts as a molecular switch to discriminate matched dNTP from mismatched dNTPs.15 The directions of fluorescence changes (reflecting conformational changes) differed for correct and incorrect dNTP binding with coumarin-tagged pol T7-.30 Thus, binding of the “wrong” dNTP is proposed to induce pol T7- into a state that has higher energy barriers for proceeding to catalysis, thus favoring reversal of equilibrium events that would lead to phosphodiester bond formation. The results of computational calculations with pol β have been interpreted to suggest that correct incorporation is through a transition state with a closed conformation but that misincorporation occurs with one or several partially open conformations.48 With E. coli pol I selection against mismatched dNTP has been proposed to occur in an “earlier” pathway (open state), and amismatched dNTP is proposed to accelerate DNA dissociation.26
Our results suggest that the behavior of Dpo4 (T239W) may be different, probably because it is a translesion DNA polymerase (although pol β should not really be considered a replicative polymerase, in that it is primarily a gap filler). With Dpo4, small (N2-alkylG) DNA adducts that reduce catalytic efficiency by 60- to 80-fold (Tables 2, 3) still appear to produce the fluorescence changes associated with normal W-C pairing and replication (Figs. 4, 6) and the estimated rate constants are very similar to those measured with (unmodified) G (Figs. 5, 7 and Table 4). Thus, there seems to be little discrimination against the modification at G in the earlier stages of the mechanism (i.e., through step 3 in Fig. 1). This result appears to differ from the conclusion advanced for E. coli pol I, pol β, and pol T7- (vide supra, although the point should be made that the focus with these polymerases is misincorporation). We presume the discrimination that lowers catalytic efficiency (Tables 2, 3) occurs in either step 4 (Fig.1) (i.e. phosphodiester bond formation) or, alternatively, in the an altered partitioning into an inactive ternary complex, which we have proposed to account for the partial kinetic bursts observed (with wild type Dpo4).36 In our earlier kinetic modeling studies36 we could fit the varying kinetic bursts observed for dCTP incorporation opposite the N2-alkylG adducts by varying k4 and holding k3, k-3, and the ratio for partitioning into an inactive complex nearly constant with the N2-MeG, N2-EtG, N2-IbG, and N2-BzG adducts. At this point we are not able to further refine the relevant rate constants (other than k3 and k-3, Table 4) on the basis of experimental data. One caveat about our earlier modeling with wild-type Dpo436 is that the values of k3 and k-3 we used there are much lower than the newer rate constants we have estimated with the T239W mutant,34 and the values should probably be considered inappropriate.
With N2-alkylG adducts as large as N2-NaphG (or with N2,N2-Me2G) no fluorescence changes were observed upon addition of dCTP (Fig. 4) and presumably the correct conformational change is blocked or a least very slow. This result is of interest in that the difference in catalytic efficiency of dCTP incorporation opposite N2-BzG and N2-NaphG was small (Tables 2, 3).
With some adducts the anti/syn conformation about template nucleosides or dNTPs can explain catalytic selectivity, e.g. with Dpo4 and 8-oxoG. In that case hydrogen bonding to the C8 oxygen (of 8-oxoG) drives the anti/syn equilibrium and accordingly the fidelity.49-51 However, the conformational changes considered here are probably more complex. Crystal structures of N2-NaphG36 and N2,N2-·Me2G35 have shown both anti and syn conformations with no real correlation of either with catalytic activity.
Another point that should be made, relevant to the comparisons of Dpo4 (T239W) with E. coli pol I, pol β, and pol T7-, is that the focus of our work has been on the fluorescence/conformational changes observed with “normal” incorporation (of dCTP opposite G derivatives). For the addition of dTTP opposite any of the adducts there was no fluorescence change observed (Supplemental Data Fig. S2), and little can be said other than the correct conformational change appears not to occur. Whether an alternate (but silent) change occurs such as with pol T7-15,30 is unknown.
In summary, we have used fluorescent Dpo4 mutant T239W to show that a 2-order of magnitude loss of in catalytic efficiency of dCTP incorporation due to alkylation of the N2 atom of G by small moieties does not appear to involve changes in the rate or extent of dCTP-induced conformational changes, as judged by protein fluorescence measurements. Larger alkyl groups (N2-NaphG, N2-AnthG, and N2,N2-Me2G) blocked the fluorescence change, indicating that the conformation changes were strongly perturbed. Thus, modifications of different sizes at a single atom have multiple mechanisms of slowing DNA polymerization. In all of the cases examined, misincorporation (of a nucleoside triphosphate other than dCTP) is not preferred36 and no signals were observed for fluorescence related to any conformational changes.
Materials and Methods
Materials
Unlabeled dNTPs were purchased from New England Biolabs (Beverly, MA), and [γ-32P]ATP (specific activity 3 × 103 Ci mmol-1) was from PerkinElmer Life Sciences (Boston, MA). T4 polynucleotide kinase and restriction endonucleases were purchased from New England Biolabs (Beverly, MA). Bio-spin columns were obtained from Bio-Rad (Hercules, CA). Other reagents were of the highest quality commercially available.
Dpo4 mutant T239W was expressed in E. coli and purified to electrophoretic homogeneity as described previously,34,52 except that the 80 °C heat step used for the wild-type enzyme was omitted because of the possibility of decreased mutant stability. Purified Dpo4 T239W was stored in small aliquots at -80 °C in 50 mM Tris-HCl buffer (pH 7.7 at 22 °C) containing 50 mM NaCl, 1 mM dithiothreitol, and 50% glycerol (v/v).
Oligonucleotides
Oligonucleotide complexes containing a 13-mer primer and an 18-mer template were used in this study (Table 1, Fig. 1). For analysis of fluorescence changes in the absence of phosphodiester bond formation, the 3′-end of the primer strand was rendered incapable of phosphoryl transfer by substituting a 2′,3′-dideoxy analog (labeled “dd” in the text). For the analysis of fluorescence changes under conditions where phosphodiester bond formation can occur and the steady-state and pre-steady-state reactions in which oligonucleotide products were measured, the 13-mer primer contained the normal 3′-OH group.
The unmodified 13-mer and 18-mer and two 18-mer oligonucleotides containing N2-MeG or N2-IbG (Table 1) were synthesized and purified using HPLC by Midland Certified Reagent Co. (Midland, TX). The other four 18-mers, each containing an N2-alkylG adduct (N2-BzG, N2-NaphG, N2-AnthG, or a N2,N2-Me2G (Fig. 1) were prepared as previously described, purified by electrophoresis, and characterized by capillary gel electrophoresis and matrix-assisted laser desorption ionization/time-of-flight mass spectrometry.35,36,39-43 The extinction coefficients for the oligonucleotides, estimated by the Borer method,53 were: 13-mer, ε260 = 112 mM-1 cm-1; and 18-mer, ε260 = 157 mM-1 cm-1.35
The 5′ end of the 13-mer primer was labeled with [γ-32P]ATP using T4 polynucleotide kinase at 37 °C for 30 min. After removal of excess ATP using a Bio-Spin 6 column (Bio-Rad, Hercules, CA), the labeled primer and the template (molar ratio 1:1) were heated at 95 °C for 5 min and then slowly cooled to room temperature to form the 13-mer/18-mer primer/template duplexes, which were used for steady-state and pre-steady-state kinetic experiments.
Reaction Conditions for Dpo4 T239W Assays and Product Analysis Methods
Standard oligonucleotide polymerization reactions with Dpo4 T239W included 50 mM Tris-HCl buffer (pH 7.5 at 25 °C) containing 50 mM NaCl, 5 mM dithiothreitol, and 5% (v/v) glycerol) at 37 °C.34 Although bovine serum albumin is often used in DNA polymerase assays, it was not added here because it contains two Trp residues that could interfere with the fluorescence of Dpo4 mutant T239W.34 Removal of albumin was previously found not to affect the rate of DNA polymerization. All reactions were initiated by mixing dNTP-MgCl2 (final MgCl2 concentration of 5 mM) solution to preincubated enzyme-DNA mixtures. After reaction, 5 μl aliquots were quenched with EDTA-formamide solution (50 μl of 20 mM EDTA in 95% formamide (v/v), with 0.5% bromphenol blue (w/v) and 0.05% xylene cyanol (w/v)). Products were resolved using 20% polyacrylamide (w/v) denaturing gel electrophoresis (containing 8 M urea) and visualized and quantitated by phosphorimaging analysis using a Bio-Rad Molecular Imager FX instrument and Quantity One software.
Steady-state Kinetic Assays
Steady-state single-base incorporation experiments were performed by adding a single dNTP at varying concentrations (twelve points) (plus MgCl2) to the Dpo4 T239W-oligonucleotide complexes, incubated in the reaction buffer. The molar ratio of Dpo4 to oligonucleotide was < 10%, and primer conversion to product was kept < 20% by adjusting the Dpo4 T239W concentration and incorporation time.54 Reactions were quenched and products were analyzed and quantitated; graphs of the incorporation rates versus dNTP concentration were fit to a hyperbolic equation to yield kcat and Km values using nonlinear regression in GraphPad Prism Version 3.0 (GraphPad, San Diego, CA). The corresponding errors were also obtained to indicate the goodness of fit.
Pre-steady-state Kinetic Analysis
Rapid quench experiments were performed in a model RQF-3 KinTek Quench Flow Apparatus (KinTek, Austin, TX) with a 50 mM Tris-HCl (pH 7.4) aqueous solution in the drive syringes. Reactions with excess Dpo4 T239W were initiated by mixing 200 nM Dpo4/10 nM oligonucleotide mixtures (12.5 μl) with dCTP (varying concentrations)-5 mM MgCl2 complex (10.9 μl). After the reactions were quenched by the addition of 0.6 M EDTA from the central syringe line after varying times, the products were analyzed and quantitated. Reactions with excess Dpo4 T239W were fit to Eq. 1 (where P is product and t is time) to obtain a burst amplitude A and burst rate kp:
| (Eq. 1) |
Plots of kp versus dCTP concentration were fit to a hyperbolic equation to estimate kpol and Kd,dCTP,app:
| (Eq. 2) |
where kpol is the maximal rate of nucleotide incorporation and Kd,dCTP,app is the apparent equilibrium dissociation constant for dCTP in the activated form of polymerase-oligonucleotide-dNTP complex.37,38 All nonlinear regression analysis used GraphPad Prism Version 3.0, with corresponding errors included to indicate the goodness of fit.
Stopped-flow fluorescence measurements
An OLIS RSM-1000 spectrofluorimeter was used in the measurement of transient fluorescent assays. For optimal signal/noise ratios for observing changes in Trp fluorescence, 3.16 mm slits (corresponding to a 20 nm bandwidth) were employed along with both 335 nm long-pass (CVI Laser Corp., Albuquerque, NM) and 355 nm bandpass filters (Newport, Irvine, CA) attached to the sample photomultiplier tube in series. Fluorescence signals were collected in the ratio mode (sample/reference photomultiplier). MgCl2 (5 mM) was included in both syringes. In typical experiments for measuring the effect of dNTP binding on fluorescence of Dpo4 T239W, one syringe included Dpo4 T239W and the oligonucleotide in 50 mM Tris buffer (pH 7.5 at 25 °C) containing 5 mM dithiothreitol and 5% glycerol (v/v), and the second syringe contained various concentrations of the correct dNTP in the same Tris buffer. After rapid mixing, the final concentration of Dpo4 T239W:oligonucleotide complex was 1.0 μM. In all cases, standard assays were performed including all components except the reagent producing a change (e.g., dNTP in the case of fluorescence change dependence on dNTPs). The fluorescence traces shown in the figures are averages of eight independent mixing events.
Data analysis of stopped-flow fluorescence measurements
The rate of conformational change at different dCTP concentration was analyzed by non-linear regression fitting the fluorescence increase signal against time using the equation:
| (Eq. 3) |
where y is the fluorescence signal, A is the amplitude of the signal, kobs is the observed rate, and t is time. kobs values were plotted against the dCTP concentration using a hyperbolic equation (Eq. 4) to estimate the ground state dissociation constant of dCTP (Kd,dCTP,ground), maximal forward conformational change rate constant (k3), and reverse conformational change rate constant (k-3) (Fig. 1):
| (Eq. 4) |
All nonlinear regression analysis used the OLIS RSM-1000 software and GraphPad Prism Version 3.0, with corresponding errors indicating the goodness of fit.
Supplementary Material
Acknowledgments
We thank I. D. Kozekoff for synthesis of the oligonucleotides containing N2-BzG and N2-AnthG, R. L. Eoff for comments on the manuscript, and K. Trisler for assistance in preparation of the manuscript,. This research was supported, in part, by National Institutes of Health grants NIH R01 ES010375 and P30 ES000267 (to F.P.G.).
Abbreviations used
- Anth
(9-anthracenyl)methyl
- Bz
benzyl
- Dbh
DinB homologue of Sulfolobus acidocaldarius (Dpo4 ortholog)
- Dpo4
Sulfolobus solfataricus P2 DNA polymerase IV
- dNTP
2′-deoxyribonucleoside triphosphate
- Et
ethyl
- G
guanine
- Ib
isobutyl
- Me
methyl
- Naph
(1-naphthyl)methyl
- PPi
pyrophosphate
- pol
(DNA) polymerase
- W-C
Watson-Crick (base pair). For simplicity the N2-guanyl adducts are usually collectively referred to as “alkyl” although technically some are aralkyl
Footnotes
Supplementary Data: Supplementary Data associated with this article can be found, in the online version, at doi….
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. 2nd. ASM Press; Washington, DC: 2006. [Google Scholar]
- 2.Watson JD, Crick FHC. Genetical implications of the structure of deoxyribonucleic acid. Nature. 1953;171:964–967. doi: 10.1038/171964b0. [DOI] [PubMed] [Google Scholar]
- 3.Goodman MF. Hydrogen bonding revisited: geometric selection as a principal determinant of DNA replication fidelity. Proc Natl Acad Sci USA. 1997;94:10493–10495. doi: 10.1073/pnas.94.20.10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Franklin MC, Wang J, Steitz TA. Structure of the replicating complex of a pol α family DNA polymerase. Cell. 2001;105:657–667. doi: 10.1016/s0092-8674(01)00367-1. [DOI] [PubMed] [Google Scholar]
- 5.Doublié S, Sawaya MR, Ellenberger T. An open and closed case for all polymerases. Structure Fold Design. 1999;7:R31–R35. doi: 10.1016/S0969-2126(99)80017-3. [DOI] [PubMed] [Google Scholar]
- 6.Doublié S, Tabor S, Long AM, Richardson CC, Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 Å resolution. Nature. 1998;391:251–257. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 7.Huang H, Chopra R, Verdine GL, Harrison SC. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: Implications for drug resistance. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- 8.Pelletier H, Sawaya MR, Wolfle W, Wilson SH, Kraut J. A structural basis for metal ion mutagenicity and nucleotide selectivity in human DNA polymerase β. Biochemistry. 1996;35:12762–77. doi: 10.1021/bi9529566. [DOI] [PubMed] [Google Scholar]
- 9.Li Y, Korolev S, Waksman G. Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: Structural basis for nucleotide incorporation. EMBO J. 1998;17:7514–7525. doi: 10.1093/emboj/17.24.7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sawaya MR, Prasad R, Wilson SH, Kraut J, Pelletier H. Crystal structures of human DNA polymerase β complexed with gapped and nicked DNA: evidence for an induced fit mechanism. Biochemistry. 1997;36:11205–11215. doi: 10.1021/bi9703812. [DOI] [PubMed] [Google Scholar]
- 11.Showalter AK, Tsai MD. A reexamination of the nucleotide incorporation fidelity of DNA polymerases. Biochemistry. 2002;41:10571–10576. doi: 10.1021/bi026021i. [DOI] [PubMed] [Google Scholar]
- 12.Kim SJ, Beard WA, Harvey J, Shock DD, Knutson JR, Wilson SH. Rapid segmental and subdomain motions of DNA polymerase β. J Biol Chem. 2003;278:5072–5081. doi: 10.1074/jbc.M208472200. [DOI] [PubMed] [Google Scholar]
- 13.Joyce CM, Benkovic SJ. DNA polymerase fidelity: kinetics, structure, and checkpoints. Biochemistry. 2004;43:14317–14324. doi: 10.1021/bi048422z. [DOI] [PubMed] [Google Scholar]
- 14.Johnson KA. Conformational coupling in DNA polymerase fidelity. Annu Rev Biochem. 1993;62:685–713. doi: 10.1146/annurev.bi.62.070193.003345. [DOI] [PubMed] [Google Scholar]
- 15.Johnson KA. Role of induced-fit in enzyme specificity: A molecular forward/reverse switch. J Biol Chem. 2008;283:26297–26301. doi: 10.1074/jbc.R800034200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frey MW, Sowers LC, Millar DP, Benkovic SJ. The nucleotide analog 2-aminopurine as a spectroscopic probe of nucleotide incorporation by the Klenow fragment of Escherichia coli polymerase I and bacteriophage T4 DNA polymerase. Biochemistry. 1995;34:9185–9192. doi: 10.1021/bi00028a031. [DOI] [PubMed] [Google Scholar]
- 17.Hariharan C, Bloom LB, Helquist SA, Kool ET, Reha-Krantz LJ. Dynamics of nucleotide incorporation: Snapshots revealed by 2-aminopurine fluorescence studies. Biochemistry. 2006;45:2836–2844. doi: 10.1021/bi051644s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bakhtina M, Roettger MP, Kumar S, Tsai MD. A unified kinetic mechanism applicable to multiple DNA polymerases. Biochemistry. 2007;46:5463–5472. doi: 10.1021/bi700084w. [DOI] [PubMed] [Google Scholar]
- 19.Bakhtina M, Roettger MP, Tsai MD. Contribution of the reverse rate of the conformational step to polymerase β fidelity. Biochemistry. 2009;48:3197–208. doi: 10.1021/bi802119f. [DOI] [PubMed] [Google Scholar]
- 20.Roettger MP, Bakhtina M, Tsai MD. Mismatched and matched dNTP incorporation by DNA polymerase beta proceed via analogous kinetic pathways. Biochemistry. 2008;47:9718–9727. doi: 10.1021/bi800689d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee HR, Wang M, Konigsberg W. The reopening rate of the fingers domain is a determinant of base selectivity for RB69 DNA polymerase. Biochemistry. 2009;48:2087–2098. doi: 10.1021/bi8016284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang H, Cao W, Zakharova E, Konigsberg W, De La Cruz EM. Fluorescence of 2-aminopurine reveals rapid conformational changes in the RB69 DNA polymerase-primer/template complexes upon binding and incorporation of matched deoxynucleoside triphosphates. Nucleic Acids Res. 2007;35:6052–6062. doi: 10.1093/nar/gkm587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DeLucia AM, Grindley ND, Joyce CM. Conformational changes during normal and error-prone incorporation of nucleotides by a Y-family DNA polymerase detected by 2-aminopurine fluorescence. Biochemistry. 2007;46:10790–10803. doi: 10.1021/bi7006756. [DOI] [PubMed] [Google Scholar]
- 24.Rachofsky EL, Osman R, Ross JBA. Probing structure and dynamics of DNA with 2-aminopurine: effects of local environment on fluorescence. Biochemistry. 2001;40:946–956. doi: 10.1021/bi001664o. [DOI] [PubMed] [Google Scholar]
- 25.Stengel G, Gill JP, Sandin P, Wilhelmsson LM, Albinsson B, Norden B, Millar D. Conformational dynamics of DNA polymerase probed with a novel fluorescent DNA base analogue. Biochemistry. 2007;46:12289–12297. doi: 10.1021/bi700755m. [DOI] [PubMed] [Google Scholar]
- 26.Joyce CM, Potapova O, Delucia AM, Huang X, Basu VP, Grindley ND. Fingers-closing and other rapid conformational changes in DNA polymerase I (Klenow fragment) and their role in nucleotide selectivity. Biochemistry. 2008;47:6103–6116. doi: 10.1021/bi7021848. [DOI] [PubMed] [Google Scholar]
- 27.Rothwell PJ, Mitaksov V, Waksman G. Motions of the fingers subdomain of Klentaq1 are fast and not rate limiting: implications for the molecular basis of fidelity in DNA polymerases. Mol Cell. 2005;19:345–355. doi: 10.1016/j.molcel.2005.06.032. [DOI] [PubMed] [Google Scholar]
- 28.Rothwell PJ, Waksman G. A pre-equilibrium before nucleotide binding limits fingers subdomain closure by Klentaq1. J Biol Chem. 2007;282:28884–28892. doi: 10.1074/jbc.M704824200. [DOI] [PubMed] [Google Scholar]
- 29.Allen WJ, Rothwell PJ, Waksman G. An intramolecular FRET system monitors fingers subdomain opening in Klentaq1. Prot Sci. 2008;17:401–408. doi: 10.1110/ps.073309208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsai YC, Johnson KA. A new paradigm for DNA polymerase specificity. Biochemistry. 2006;45:9675–9687. doi: 10.1021/bi060993z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsai YC, Jin Z, Johnson KA. Site-specific labeling of T7 DNA polymerase with a conformationally sensitive fluorophore and its use in detecting single-nucleotide polymorphisms. Anal Biochem. 2009;384:136–44. doi: 10.1016/j.ab.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luo G, Wang M, Konigsberg WH, Xie XS. Single-molecule and ensemble fluorescence assays for a functionally important conformational change in T7 DNA polymerase. Proc Natl Acad Sci USA. 2007;104:12610–12615. doi: 10.1073/pnas.0700920104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eoff RL, Sanchez-Ponce R, Guengerich FP. Conformational changes during nucleotide selection by Sulfolobus solfataricus DNA polymerase Dpo4. J Biol Chem. 2009;284:21090–21099. doi: 10.1074/jbc.M109.009506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beckman JW, Wang Q, Guengerich FP. Kinetic analysis of nucleotide insertion by a Y-family DNA polymerase reveals conformational change both prior to and following phosphodiester bond formation as detected by tryptophan fluorescence. J Biol Chem. 2008;283:36711–36723. doi: 10.1074/jbc.M806785200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang H, Eoff RL, Egli M, Kozekov ID, Rizzo CJ, Guengerich FP. Structure-function relationships in miscoding by Sulfolobus solfataricus DNA polymerase Dpo4. Guanine N2,N2-dimethyl substitution produces inactive and miscoding polymerase complexes. J Biol Chem. 2009;284:17687–17699. doi: 10.1074/jbc.M109014274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang H, Eoff RL, Egli M, Guengerich FP. Versatility of Y-family Sulfolobus solfataricus DNA polymerase Dpo4 in transletion synthesis past bulky N2-alkylguanine adducts. J Biol Chem. 2009;284:3563–3576. doi: 10.1074/jbc.M807778200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patel SS, Wong I, Johnson KA. Pre-steady-state kinetic analysis of processive DNA replication including complete characterization of an exonuclease-deficient mutant. Biochemistry. 1991;30:511–525. doi: 10.1021/bi00216a029. [DOI] [PubMed] [Google Scholar]
- 38.Johnson KA. Rapid quench kinetic analysis of polymerases, adenosine triphosphatases, and enzyme intermediates. Methods Enzymol. 1995;249:38–61. doi: 10.1016/0076-6879(95)49030-2. [DOI] [PubMed] [Google Scholar]
- 39.Choi YJ, Guengerich FP. Analysis of the effect of bulk at N2-alkylguanine DNA adducts on catalytic efficiency and fidelity of the processive DNA polymerase T7 exonuclease− and HIV-1 reverse transcriptase. J Biol Chem. 2004;279:19217–19229. doi: 10.1074/jbc.M313759200. [DOI] [PubMed] [Google Scholar]
- 40.Choi JY, Guengerich FP. Adduct size limits efficient and error-free bypass across bulky N2-guanine DNA lesions by human DNA polymerase η. J Mol Biol. 2005;352:72–90. doi: 10.1016/j.jmb.2005.06.079. [DOI] [PubMed] [Google Scholar]
- 41.Choi JY, Guengerich FP. Kinetic evidence for efficient and error-prone bypass across bulky N2-guanine DNA adducts by human DNA polymerase ι. J Biol Chem. 2006;281:12315–12324. doi: 10.1074/jbc.M600112200. [DOI] [PubMed] [Google Scholar]
- 42.Choi JY, Angel KC, Guengerich FP. Translesion synthesis across bulky N2-alkylguanine DNA adducts by human DNA polymerase κ. J Biol Chem. 2006;2006:21062–21072. doi: 10.1074/jbc.M602246200. [DOI] [PubMed] [Google Scholar]
- 43.Choi JY, Guengerich FP. Kinetic analysis of translesion synthesis opposite bulky N2- and O6-alkylguanine DNA adducts by human DNA polymerase REV1. J Biol Chem. 2008;283:23645–23655. doi: 10.1074/jbc.M801686200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ling H, Boudsocq F, Woodgate R, Yang W. Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell. 2001;107:91–102. doi: 10.1016/s0092-8674(01)00515-3. [DOI] [PubMed] [Google Scholar]
- 45.Wong I, Patel SS, Johnson KA. An induced-fit kinetic mechanism for DNA replication fidelity: direct measurement by single-turnover kinetics. Biochemistry. 1991;30:526–537. doi: 10.1021/bi00216a030. [DOI] [PubMed] [Google Scholar]
- 46.Dahlberg ME, Benkovic SJ. Kinetic mechanism of DNA polymerase I (Klenow fragment): identification of a second conformational change and evaluation of the internal equilibrium constant. Biochemistry. 1991;30:4835–4843. doi: 10.1021/bi00234a002. [DOI] [PubMed] [Google Scholar]
- 47.Eger BT, Benkovic SJ. Minimal kinetic mechanism for misincorporation by DNA polymerase I (Klenow fragment) Biochemistry. 1992;31:9227–9236. doi: 10.1021/bi00153a016. [DOI] [PubMed] [Google Scholar]
- 48.Xiang Y, Goodman MF, Beard W, Wilson S, Warshal A. Exploring the role of large conformational changes in the fidelity of DNA polymerase β. Proteins. 2007;70:231–247. doi: 10.1002/prot.21668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eoff RL, Irimia A, Angel K, Egli M, Guengerich FP. Hydogen bonding of 7,8-dihydro-8-oxodeoxyguanosine with a charged residue in the little finger domain determines miscoding events in Sulfolobus solfataricus DNA polymerase Dpo4. J Biol Chem. 2007;282:19831–19843. doi: 10.1074/jbc.M702290200. [DOI] [PubMed] [Google Scholar]
- 50.Zang H, Irminia A, Choi JY, Angel KC, Loukachevitch LV, Egli M, Guengerich FP. Efficient and high fidelity incorporation of dCTP opposite 7,8-dihydro-8-oxo-deoxyguanosine by Sulfolobus solfataricus DNA polymerase Dpo4. J Biol Chem. 2006;281:2358–2372. doi: 10.1074/jbc.M510889200. [DOI] [PubMed] [Google Scholar]
- 51.Rechkoblit O, Malinina L, Cheng Y, Kuryavyi V, Broyde S, Geacintov NE, Patel DJ. Stepwise translocation of Dpo4 polymerase during error-free bypass of an oxoG lesion. PLoS Biol. 2006;4:e11. doi: 10.1371/journal.pbio.0040011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zang H, Goodenough AK, Choi JY, Irminia A, Loukachevitch LV, Kozekov ID, Angel KC, Rizzo CJ, Egli M, Guengerich FP. DNA adduct bypass polymerization by Sulfolobus solfataricus DNA polymerase Dpo4. Analysis and crystal structures of multiple base-pair substitution and frameshift product with the adduct 1,N2-ethenoguanine. J Biol Chem. 2005;280:29750–29764. doi: 10.1074/jbc.M504756200. [DOI] [PubMed] [Google Scholar]
- 53.Borer PN. In: Handbook of Biochemistry and Molecular Biology. 3rd. Fasman GD, editor. CRC Press; Cleveland, OH: 1975. pp. 589–590. [Google Scholar]
- 54.Boosalis MS, Petruska J, Goodman MF. DNA polymerase insertion fidelity: gel assay for site-specific kinetics. J Biol Chem. 1987;262:14689–14696. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.