Abstract
Diethylstilbestrol (DES) is a human carcinogen, based on sufficient epidemiological evidence. DES is mainly metabolized to its catechol, 3′-hydroxyDES (3′-OH-DES), which can further oxidize to DES-3′,4′-quinone (DES-3′,4′-Q). Similarly to estradiol-3,4-quinone, the reaction of DES-3′,4′-Q with DNA would form the depurinating 3′-OH-DES-6′-N3Ade and 3′-OH-DES-6′-N7Gua adducts. To prove this hypothesis, synthesis of DES-3′,4′-Q by oxidation of 3′-OH-DES with Ag2O was tried; this failed due to instantaneous formation of a spiro-quinone. Oxidation of 3′-OH-DES by lactoperoxidase or tyrosinase in the presence of DNA led to the formation of 3′-OH-DES-6′-N3Ade and 3′-OH-DES-6′-N7Gua adducts. These adducts were tentatively identified by LC-MS/MS as 3′-OH-DES-6′-N3Ade, m/z = 418 [M+H]+, and 3′-OH-DES-6′-N7Gua, m/z = 434 [M+H]+. Demonstration of their structures derived from their oxidation by MnO2 to the DES quinone adducts and subsequent tautomerization to the dienestrol (DIES) catechol adducts, which are identical to the standard 3′-OH-DIES-6′-N3Ade, m/z = 416 [M+H]+, and 3′-OH-DIES-6′-N7Gua, m/z = 432 [M+H]+, adducts. The reaction of DIES-3′,4′-Q or lactoperoxidase-activated 3′-OH-DIES with DNA did not produce any depurinating adducts, due to the dienic chain being perpendicular to the phenyl planes, which impedes the intercalation of DIES into the DNA. Enzymic oxidation of 3′-OH-DES suggests that the catechol of DES intercalates into DNA and is then oxidized to its quinone to yield N3Ade and N7Gua adducts. These results suggest that the common denominator of tumor initiation by the synthetic estrogen DES and the natural estrogen estradiol is formation of their catechol quinones, which react with DNA to afford the depurinating N3Ade and N7Gua adducts.
Introduction
Diethylstilbestrol (DES), a potent synthetic estrogen, was obtained in 1938 and thereafter used to prevent spontaneous abortions and premature deliveries [1]. In 1971 an association was found between prenatal exposure to DES and clear cell adenocarcinoma of the vagina and cervix in young women [2]. Evidence for a causal relationship between transplacental exposure to DES and clear cell adenocarcinoma is conclusive [3–5]. Furthermore, it was later reported that women who took DES during pregnancy had a higher incidence of breast cancer [6,7]. More recently, it was found that women exposed to DES prenatally also have a higher incidence of breast cancer after age 40 [8,9].
The natural estrogens estradiol (E2) and estrone (E1) are metabolically oxidized to their respective catechols, and the catechols are subsequently oxidized to their quinones, in particular the E1(E2)-3,4-quinones (Q). We have proposed and demonstrated that reaction of the E1(E2)-3,4-Q with DNA is the first critical step in the initiation of cancer [10–13]. The depurinating adducts 4-OHE1(E2)-1-N3Ade and 4-OHE1(E2)-1-N7Gua, as well as the resulting apurinic sites in the DNA, are formed in this reaction. These apurinic sites generate mutations that we think can lead to cancer initiation [13–15].
The same mechanism of metabolic activation and reaction with DNA has been demonstrated for the synthetic estrogen hexestrol (HES) [16,17]. The catechol quinone of HES has chemical and biochemical properties similar to those of E1(E2)-3,4-Q; namely, it forms analogous depurinating N3Ade and N7Gua adducts after reaction with DNA. In addition, depurination of the N7Gua adduct occurs rather slowly, analogously to the respective adducts of the natural E1(E2)-3,4-Q [16,17]. HES is the derivative of DES hydrogenated at the ethylenic bond.
The major metabolites of HES and DES are their catechols [18–21]. The natural estrogens E1 and E2, as well as the synthetic HES and DES, are carcinogenic in the kidney of male Syrian golden hamsters [22]. In this article, we report that the human carcinogen DES is activated similarly to the natural estrogens and the synthetic estrogen HES. The catechol quinone of DES reacts with DNA to form N3Ade and N7Gua adducts analogously to the quinones of E1, E2 and HES.
Materials and methods
Chemicals, reagents and enzymes
DES, lactoperoxidase (LP, from bovine milk, 78 units/mg solid), H2O2, methemoglobin, and tyrosinase (Tyr, 2, 130 units/mg solid) were purchased from Sigma-Aldrich (St. Louis, Mo). Prostaglandin H synthase (PHS, ovine, 5K) and arachidonic acid were purchased from Cayman Chemical Co. (Ann Arbor, MI). E,E-Dienestrol (E,E-DIES) was purchased from Steraloids (Newport, RI). Z,Z-DIES was synthesized from DES as reported [23]. 3′-OH-DES, 3′-OH-E,E-DIES, 3′-OH-Z,Z-DIES and 3′-OH-4″-OCH3-DES were synthesized as reported previously [24]. Ag2O, Ade, 2-iodoxybenzoic acid (IBX), trifluoroacetic acid (TFA), DMSO-d6 and acetone-d6 were purchased from Aldrich Chemical Co. (Milwaukee, WI).
Instrumentation
UV
The UV spectra were obtained during HPLC by using the photodiode array detector (Waters 996, Milford, MA) for all compounds synthesized.
NMR
1H NMR spectra of all new compounds were recorded in deuterated solvents on a Varian Inova 500 instrument at 499.562 MHz at 25 °C. Labile protons on oxygen and nitrogen atoms were detected by recording spectra after shaking the sample with one drop of D2O. Chemical shifts are reported relative to DMSO (2.50 ppm) or acetone (2.04 ppm) and coupling constants are given in hertz (Hz).
HPLC
Preparative HPLC was conducted on a Waters 600E solvent delivery system equipped with a 996 photodiode array detector and a Luna C-18(2) column (10 μm 100 Å, 250 × 21.2 mm) (Phenomenex, Torrance, CA). A gradient elution system began with 10% CH3CN/90% H2O (containing 0.4% acetic acid) and then linearly increased the proportion of CH3CN up to 50% in 30 min and 100% in the next 5 min at a flow rate of 9 mL/min. Eluting compounds were monitored at 270 nm. Analysis of reaction mixtures was conducted on a Waters 2690 (Alliance) separation module equipped with Waters 996 photodiode array detector interfaced to a digital Venturis Fx 5100 computer. A 50-μL aliquot of a reaction mixture was injected and compounds were separated on a Luna C-18 (2) column (5 μm, 100 Å, 250 × 4.6 mm, Phenomenex) by using a gradient elution system starting from 10% CH3CN/90% H2O (containing 0.25% TFA) and changing linearly to 100% CH3CN in 30 min at a flow rate of 1 mL/min. Analytical HPLC using electrochemical detection was conducted on a system equipped with dual ESA Model 580 solvent delivery modules, an ESA Model 540 autosampler and a 12-channel CoulArray detector (ESA, Chelmsford, MA). Depurinating adducts from the reaction of chemically formed quinones or enzyme-activated catechols with DNA were separated on a reverse phase Luna C-18 (2) column (5 μm, 100 Å, 250 × 4.6 mm) with two mobile phases, A [CH3CN/CH3OH/H2O/buffer, pH 3.7 (15:5:70:10)] and B [CH3CN/CH3OH/H2O/buffer, pH 3.7(50:20:20:10)]. The buffer was a 1:1 mixture of 1 M NH4OAc and 1 M citric acid and pH (3.7) was adjusted with conc NH4OH. Elution was started with 90%A/10%B and the proportion of B was increased linearly up to 35% in 25 min, eluting isocratically for 5 min and, finally, raising the proportion of B linearly up to 90% in 20 min with a flow rate of 0.5 mL/min at 35 °C. The serial array of 12 coulometric electrodes was set at potentials between −10 and 440 mV (−10, 80, 150, 200, 230, 260, 290, 320, 350, 380, 410, and 440). The system was controlled and data were acquired and processed using the CoulArray software package. Peaks were identified by both retention time and peak height ratios between the dominant peak and the peaks in the two adjacent channels. The depurinating adducts were quantified by comparison of peak response ratios with known amounts of standards. Triplicate samples were analyzed for each data point.
MS
Fast atom bombardment tandem mass spectrometry (FAB-MS) was conducted at the Nebraska Center for Mass Spectrometry (University of Nebraska-Lincoln) using a MicroMass (Manchester, England) AutoSpec high resolution magnetic sector mass spectrometer. Xenon was admitted to the collision cell at a level to attenuate the precursor ion signal by 75%. Data acquisition and processing were accomplished using OPUS software that was provided by the manufacturer (Microcasm). Samples were dissolved in 5–10 μL of methanol; 1-μL aliquots were placed on the sample probe tip along with 1 μL of a 1:1 mixture of glycerol/thioglycerol.
To verify the presence and identity of adducts, the reaction mixtures were analyzed by direct infusion tandem mass spectrometry (MS/MS) using a MicroMass QuattroMicro triple quadrupole mass spectrometer (Waters). A 100-μL aliquot of the reaction mixture was diluted 1:1 in triple distilled H2O then, passed through a 0.22 μm syringe filter prior to infusion into the mass spectrometer. Flow from the syringe was 5 μL/min directed to an interface at 120 °C using nitrogen as both the desolvation and auxiliary gas. Reaction components were ionized in positive mode with the capillary voltage at 3.5 kV and cone voltage at 25 V. Full scan spectra were obtained between 20 and 700 amu. MS/MS conditions were optimized for each individual peak identified in the full-scan spectra. Argon was used as the collision gas.
For analysis of depurinating adducts, samples were run on a Waters Acquity UPLC equipped with a MicroMass QuattroMicro triple stage quadrupole system (Waters). The 10-μL injections were carried out on a Waters Acquity UPLC™ BEHC18 column (1.7 μm, 1 × 100 mm). The instrument was operated in positive electrospray ionization mode. All aspects of system operation, data acquisition and processing were controlled using QuanLynx v4.0 software (Waters). The column was eluted starting with 20% CH3CN in H2O (0.1% formic acid) for 1 min at a flow rate of 75 μL/min; raised to 35% CH3CN in H2O (0.1% formic acid) in 5 min and then to 80% in 1 min. Ionization was achieved by using the following settings: capillary voltage 3 kV; cone voltage 30–66 V; source temperature 100 °C; desolvation temperature 200 °C with a nitrogen flow of 400 L/hr. MS/MS conditions were optimized for each compound using pure standard adducts and argon as the collision gas. Triplicate samples were analyzed for each data point.
Syntheses
Synthesis of 3′-OH-Z,Z-DIES
Synthesis of 3′-OH-Z,Z-DIES was achieved by reacting Z,Z-DIES with IBX, as described previously [24]. Yield 75%; 1H NMR (DMSO-d6): δ 9.35 (s, 1H, Ar-OH, exchangeable with D2O), 8.81 (s, 1H, Ar-OH, exchangeable with D2O), 8.75 (s, 1H, Ar-OH, exchangeable with D2O), 7.12 (d, J = 8.79 Hz, 2 H, H-2″,6″), 6.71 (d, J = 1.9, 1H, H-2′), 6.63-6.57 (m, 4 H, H-5′,6′,3″,5″), 6.13 (q, J = 6.34 Hz, 1 H, CH), 6.08 (q, J = 6.82 Hz, 1 H, CH), 1.60 (bt, J = 7.32 Hz, 6 H, 2 × CH3).
Synthesis of DIES-3′,4′-Q
To a stirred solution of 3′-OH-Z,Z DIES or 3′-OH-E,E DIES (15.4 mg, 0.05 mmol) in acetone (2 mL) was added Ag2O (76.6 mg, 0.3 mmol) at room temperature. After 15–20 min, the orange-colored solution of quinone was filtered and used as such for further reactions.
Z,Z-DIES-3′,4′-Q
Yield 95%, 1H NMR (acetone-d6): δ 8.40 (s, 1H, Ar-OH, exchangeable with D2O), 7.75 (dd, J = 2.4, 10.7 Hz, 1 H, H-6′), 7.24 (d, J = 8.3 Hz, 2 H, H-2″,6″), 7.02 (q, J = 7.32 Hz, 1 H, CH), 6.78 (d, J = 8.30 Hz, H-3″,5″), 6.42 (d, J = 10.7 Hz, 1 H, H-5′), 6.33 (q, J = 6.83 Hz, 1 H, CH), 6.11 (d, J = 2.0 Hz, 1 H, H-2′), 1.84 (d, J = 6.83 Hz, 3 H, CH3), 1.67 (d, J = 6.83 Hz, 3 H, CH3).
E,E-DIES-3′,4′-Q
Yield 99%, 1H NMR (acetone-d6): δ 8.46 (s, 1H, Ar-OH, exchangeable with D2O), 7.04 (dd, J = 2.4, 10.2 Hz, 1 H, H-6′), 7.00 (d, J = 8.3 Hz, 2 H, H-2″,6″), 6.86 (d, J = 8.30 Hz, H-3″,5″), 6.36 (d, J = 10.2 Hz, 1 H, H-5′), 6.17 (d, J = 1.5 Hz, 1 H, H-2′), 5.75 (q, J = 6.84 Hz, 1 H, CH), 5.50 (q, J = 6.84 Hz, 1 H, CH), 1.76 (d, J = 7.32 Hz, 3 H, CH3), 1.58 (d, J = 6.83 Hz, 3 H, CH3).
Synthesis of standard depurinating adducts
3′-OH-Z,Z-/E,E-DIES-6′-N3Ade
A freshly prepared solution of Z,Z- or E,E-DIES-3′,4′-Q (0.05 mmol) in acetone (1 mL) was reacted with a solution of Ade (67.5 mg, 0.5 mmol) in DMF/H2O/acetic acid (1:1:1, 5 mL) at room temperature for 24 h. The reaction mixture was filtered and solvent was removed at low pressure. The residue was re-dissolved in 2 mL of DMF/CH3OH (1:1) and analyzed by HPLC as described above. Separation of the reaction mixture was carried out by preparative HPLC, as described above.
3′-OH-Z,Z-DIES-6′-N3Ade
Yield 25%. 1H NMR (DMSO-d6): δ 9.50 (bs, 1H, Ar-OH, exchangeable with D2O), 9.40 (bs, 2 H, Ar-OH, exchangeable with D2O), 8.00 (bs, 2 H, NH2-Ade), 7.67 (s, 1 H, H-2/8-Ade), 7.62 (s, 1 H, H-2/8-Ade), 6.85 (d, J = 8.30 Hz, 2 H, H-2″,6″), 6.82 (s, 1 H, H-5′/2′), 6.60 (d, J = 8.30 Hz, H-3″,5″), 6.58 (s, 1 H, H-5′/2′), 5.55 (bs, 2 H, 2 × CH), 1.87 (d, J = 7.32 Hz, 3 H, CH3), 1.39 (d, J = 6.83 Hz, 3 H, CH3). FAB-MS: m/z 416.1699 ([M+H]+ C23H21N5O3, calc. 416.1644).
3′-OH-E,E-DIES-6′-N3Ade
Yield 20%. 1H NMR (DMSO-d6): δ 9.58 (bs, 1H, Ar-OH, exchangeable with D2O), 9.35 (bs, 2 H, Ar-OH, exchangeable with D2O), 7.96 (bs, 2 H, NH2-Ade), 7.84 (s, 1 H, H-2/8-Ade), 7.66 (s, 1 H, H-2/8-Ade), 7.03 (s, 1 H, H-5′/2′), 6.63 (d, J = 8.30 Hz, 2 H, H-2″,6″), 6.63 (s, 1 H, H-5′/2′), 6.39 (d, J = 8.30 Hz, H-3″,5″), 5.10 (q, J = 6.83 Hz, 1 H, CH), 5.04 (q, J = 6.83 Hz, 1 H, CH), 1.39 (d, J = 7.32 Hz, 3 H, CH3), 1.04 (d, J = 6.83 Hz, 3 H, CH3). FAB-MS: m/z 416.1735 ([M+H]+ C23H21N5O3, calc. 416.1644).
3′-OH-Z,Z-/E,E-DIES-6′-N7Gua
A freshly prepared solution of Z,Z- or E,E-DIES-3′,4′-Q (0.05 mmol) in acetone (1 mL) was reacted with a solution of dG (142.5 mg, 0.5 mmol) in DMF/H2O/acetic acid (1:1:1, 5 mL) at room temperature for 24 h. The reaction mixture was filtered and solvent was removed at low pressure. The residue was redissolved in 2 mL of DMF/MeOH (1:1) and analyzed by HPLC as described above. Separation of the reaction mixture was carried out by preparative HPLC as described above.
3′-OH-Z,Z-DIES-6′-N7Gua
Yield 30%. 1H NMR (DMSO-d6): δ 10.68 (s, 1 H, NH-Gua, exchangeable with D2O), 9.36 (s, 1 H, Ar-OH, exchangeable with D2O), 9.32 (s, 1 H, Ar-OH, exchangeable with D2O), 9.24 (s, 1 H, Ar-OH, exchangeable with D2O), 7.31 (s, 1 H, H-8-Gua), 6.88 (d, J = 8.8 Hz, 2 H, H-2″,6″), 6.69 (s, 1 H, H-5′/2′), 6.60 (d, J = 8.30 Hz, 2 H, H-3″,5″), 6.46 (s, 1 H, H-5′/2′), 6.09 (bs, 2 H, NH2-Gua), 5.64 (bs, 1 H, CH), 5.55 (bs, 1 H, CH), 1.45 (d, J = 7.32 Hz, 6 H, 2 × CH3). FAB-MS: m/z 432.1678 ([M+H]+ C23H21N5O4, calc. 432.1594).
3′-OH-E,E-DIES-6′-N7Gua
Yield 28%. 1H NMR (DMSO-d6): δ 10.68 (s, 1 H, NH-Gua, exchangeable with D2O), 9.36 (bs, 3 H, Ar-OH, exchangeable with D2O), 7.46 (s, 1 H, H-8-Gua), 6.82 (s, 1 H, H-5′/2′), 6.67 (d, J = 8.8 Hz, 2 H, H-2″,6″), 6.54 (s, 1 H, H-5′/2′), 6.50 (d, J = 8.30 Hz, 2 H, H-3″,5″), 6.03 (bs, 2 H, NH2-Gua), 5.16 (q, J = 6.83 Hz, 1 H, CH), 5.02 (q, J = 6.83 Hz, 1 H, CH), 1.41 (d, J = 7.32 Hz, 3 H, CH3), 1.17 (d, J = 7.32 Hz, 3 H, CH3). FAB-MS: m/z 432.1647 ([M+H]+ C23H21N5O4, calc. 432.1594).
3′-OH-4″-OCH3-DES-6′-N3Ade
A freshly prepared solution of 4″-OCH3-DES-3′,4′-Q2 (0.05 mmol) in CH3CN (1 mL) was reacted with a solution of Ade (67.5 mg, 0.5 mmol) in DMF/H2O/acetic acid (1:1:1, 5 mL) at room temperature for 24 h. The reaction mixture was filtered and solvent was removed at low pressure. The residue was redissolved in 2 mL of DMF/CH3OH (1:1) and analyzed by HPLC as described before. Separation of the reaction mixture was carried out by preparative HPLC as described before. Yield 35%, 1H NMR (DMSO-d6): δ 9.74 (bs, 1 H, Ar-OH, exchangeable with D2O), 9.62 (bs, 1 H, Ar-OH, exchangeable with D2O), 9.54 (bs, 1 H, Ar-OH, exchangeable with D2O), 8.91 (s, 1 H, H-2/8-Ade), 8.54 (bs, 2 H, NH2-Ade), 7.05 (s, 1 H, H-2/8-Ade), 6.88-6.70 (m, 6 H, H-5′,2′,2″,3″,5″,6″), 3.71 (s, 3 H, OCH3), 2.23 (m, 2 H, CH2), 1.74 (m, 2 H, CH2), 0.61 (m, 6 H, 2 × CH3). FAB-MS: m/z 432.1965 ([M+H]+ C24H25N5O3, calc. 432.1957).
3′-OH-4″-OCH3-DES-6′-N7Gua
A freshly prepared solution of 4″-OCH3-DES-3′,4′-Q [24] (0.05 mmol) in CH3CN (1 mL)was reacted with a solution of dG (142.5 mg, 0.5 mmol) in DMF/H2O/acetic acid (1:1:1, 5 mL) at room temperature for 24 h. The reaction mixture was filtered and solvent was removed at low pressure. The residue was re-dissolved in 2 mL of DMF/CH3OH (1:1) and analyzed by HPLC as described before. Separation of the reaction mixture was carried out by preparative HPLC as described before. Yield 32%, 9.43 (bs, 3 H, Ar-OH, NH-Gua), 7.87 (s, 1 H, H-8-Gua), 6.87 (s, 4 H, H-2″,3″,5″,6″), 6.80 (s, 1 H, H-2′/5′), 6.63 (s, 1 H, H-2′/5′), 6.47 (bs, 2 H, NH2-Gua), 3.72 (s, 3 H, OCH3), 2.20 (m, 2 H, CH2), 1.79 (m, 2 H, CH2), 0.61 (t, J = 7.32 Hz, 6 H, 2 × CH3). FAB-MS: m/z 448.1978 ([M+H]+ C24H25N5O4, calc. 448.1907).
Covalent binding of quinones to DNA
Z,Z- or E,E-DIES-3′,4′-Q (2.5 mg/500 μL of acetone), or 4″-OCH3-DES-3′,4′-Q (2.5 mg/500 μL of CH3CN) was mixed with a 10-mL solution of calf thymus DNA (3 mM) in 67 mM sodium-potassium phosphate buffer (pH 7.0) and incubated at 37 °C. After 10 h, DNA was precipitated with two volumes of ethanol, and processed for measurement of stable adducts by the 32P-postlabeling technique [25]. The supernatant, containing depurinating adducts, was dried under low pressure, the residue redissolved in CH3OH/DMF (500 μL), filtered and semipurified by preparative HPLC. Samples of standard depurinating adducts were first injected in the preparative HPLC to determine the exact retention times of the adducts. The DNA samples were then injected under the same conditions and the fractions that had the retention times of the standards were collected and dried under vacuum for a second HPLC analysis, which was conducted by using an analytical HPLC equipped with a series of electrochemical detectors and the UPLC-MS/MS technique, as described above.
Covalent binding of enzyme-activated catechols to DNA
3′-OH-DES, 3′-OH-Z,Z-DIES or 3′-OH-E,E-DIES was incubated with DNA in the presence of Tyr, LP or PHS. The reaction volume in each experiment was 10 ml. In the Tyr-catalyzed reaction, the mixture containing 3mM calf thymus DNA in 67 mM sodium-potassium phosphate (pH 7.0), 3′-OH-DES (2.5 mg/500μL DMSO) and 1 mg of enzyme (2000 units) was incubated at 37 °C for 10 h. For the LP-catalyzed reaction, the mixture containing 3mM calf thymus DNA, 3′-OH-DES (2.5 mg/500μL DMSO), H2O2 (0.5 mM) and 1 mg of enzyme (78 units) was incubated at 37 °C for 10 h. For the PHS-catalyzed reaction, the mixture containing 3 mM calf thymus DNA, 3′-OH-DES (2.5 mg/500μL DMSO), 1 ml of methemoglobin (2.95 mg/ml in 75 mM KH2PO4, pH 7.5), 1 ml of arachidonic acid (50 mM in ethanol) and 800 μL of PHS (400 units) was incubated at 37 °C for 10 h. After incubation, DNA was precipitated with 2 volumes of ethanol and processed for 32P-postlabeling analysis of stable adducts [25]. The supernatant, containing depurinating adducts, was evaporated, resuspended in CH3CN, filtered and treated with 10 mg of solid MnO2 to convert the DES adducts to DIES adducts. After stirring at room temperature for 10 min, the reaction mixture was filtered to remove solid material, CH3CN was evaporated and the residue was redissolved in 200 μL of CH3OH/H2O. The solution was passed through a 5000 MW cut-off filter and analyzed on UPLC-MS/MS by comparing with synthesized depurinating DIES adducts. Control reactions were carried out under identical conditions either with no enzyme or no cofactor. 3′-OH-4″-OCH3-DES, 3′-OH-Z,Z-DIES, and 3′-OH-E,E-DIES were reacted with DNA under the same conditions, but without treating the supernatant with MnO2.
Results
Synthesis of quinones
Due to the presence of the double bond between C3 and C4 of 3′-OH-DES (Fig. 1), its chemical oxidation afforded a highly unstable DES-3′,4′-Q, which cyclized instantaneously to spiro-catechol and then to spiro-quinone (Fig. 2) [26]. Use of different solvents and reaction conditions did not help to stabilize DES-3′,4′-Q.
Figure 1.
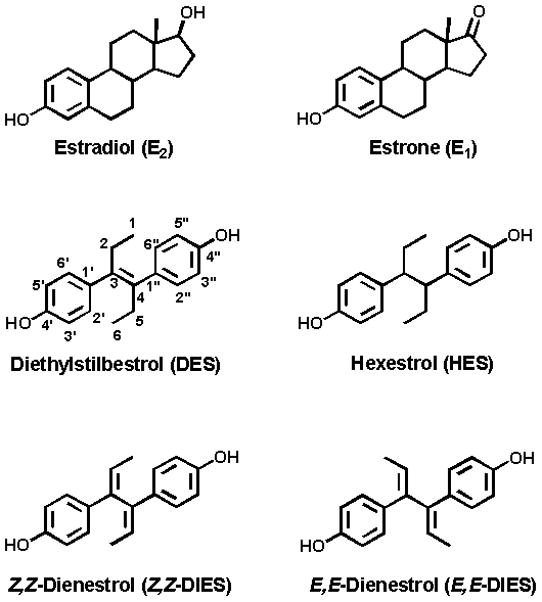
Structures of natural and synthetic estrogens.
Figure 2.
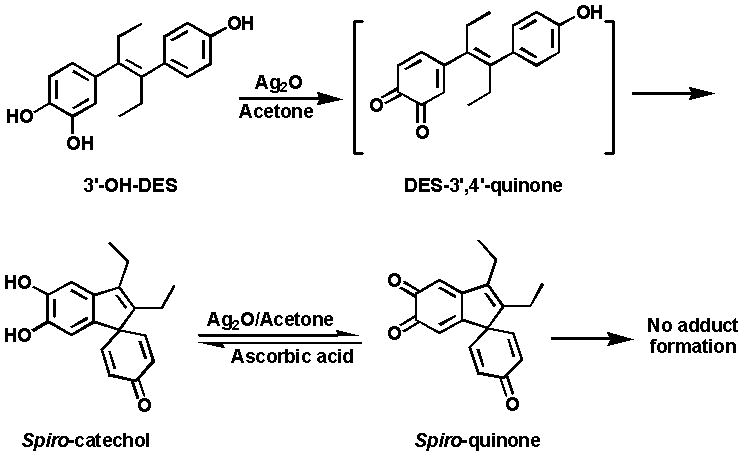
Formation of spiro-quinone by oxidation of 3′-OH-DES.
On the other hand, the two dehydrogenated derivatives of 3′-OH-DES, 3′-OH-Z,Z-DIES and 3′-OH-E,E-DIES, were easily converted into their respective DIES-3′,4′-Q, when reacted with silver oxide (Ag2O) in acetone (Figs. 3 and 4). The structure of the quinones was confirmed by 1H NMR, which along with HPLC analysis, indicated quantitative conversion of the catechols to the corresponding ortho-quinones.
Figure 3.
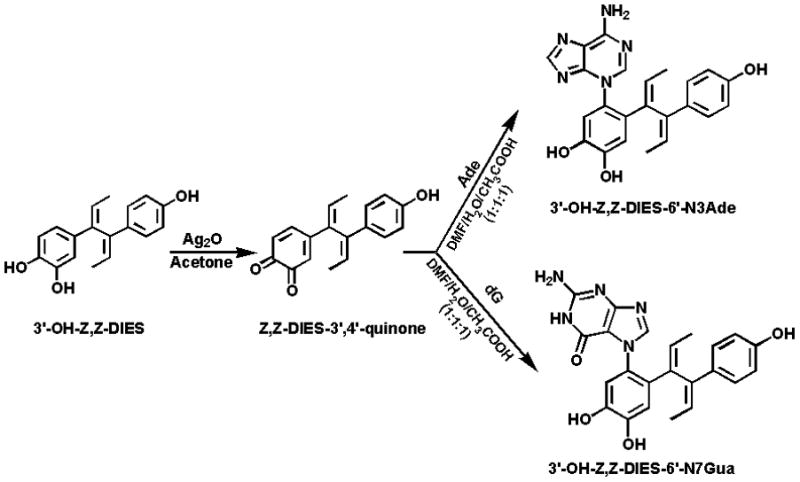
Oxidation of 3′-OH-Z,Z-DIES to Z,Z-DIES-3′,4′-Q and reaction of the quinone with Ade or dG to form N3Ade or N7Gua adducts.
Figure 4.
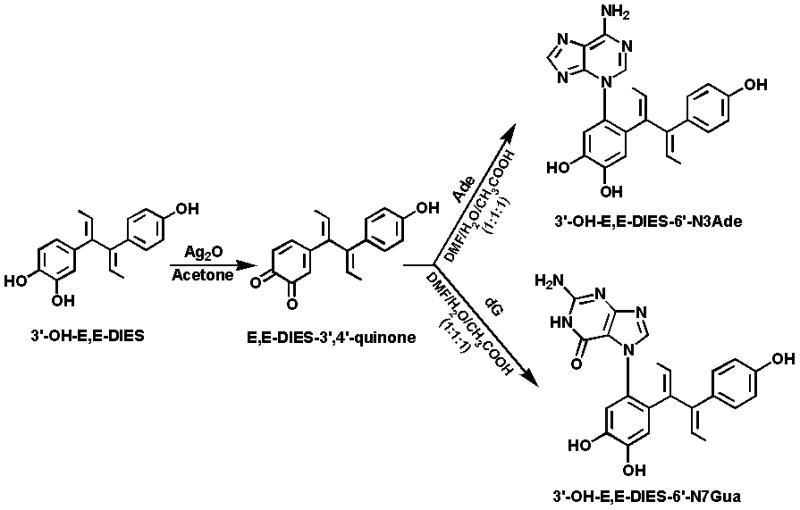
Oxidation of 3′-OH-E,E-DIES to E,E-DIES-3′,4′-Q and reaction of the quinone with Ade or dG to form N3Ade or N7Gua adducts.
Synthesis and structure determination of standard adducts
Reaction of Z,Z-DIES-3′,4′-Q (Fig. 3) or E,E-DIES-3′,4′-Q (Fig. 4) with Ade or dG led to formation of the N3Ade or N7Gua adduct.
3′-OH-DIES-6′-N3Ade
Reaction of Z,Z-DIES-3′,4′-Q or E,E-DIES-3′,4′-Q with Ade afforded one major product with some unknown minor impurities, as observed by analytical HPLC. The reaction mixture was purified by using preparative HPLC as described above and structure elucidation was carried out with the help of spectroscopic techniques.
FAB-MS of the adducts showed a protonated molecular ion [M+H]+ at m/z 416, which was consistent with addition of the DIES moiety to an Ade base. Furthermore, the presence of the DIES moiety in the 1H NMR spectrum was easily recognized due to their characteristic correlation patterns of two aliphatic methyl groups as two doublets resonating between δ 1.00 to 1.90 ppm and two olefinic methine protons resonating as a quartet between δ 5.00 to 5.65 ppm, in the 1H NMR spectra. In general, the chemical shifts of a particular proton attached to the Z,Z-isomer were found to be upfield with respect to those attached to the E,E-isomer. Characteristic signals of H-2′ (d), H-5′ (d), and H-6′ (dd) in the starting catechol [24] were replaced by two sharp singlets around δ 7.03 and 6.63 ppm, which were assigned to H-5′ and H-2′, respectively, indicating that substitution had taken place at C-6′ of the DIES moiety. On the other hand, the presence of two additional sharp singlets around δ 7.84 and 7.66 ppm, assigned to H-8 (Ade) and H-2 (Ade), indicated binding of Ade to DIES. The presence of a broad signal, with integration of two protons around δ 7.95 ppm ruled out the possibility of an exocyclic NH2 adduct. The bond of Ade via the N3 position was established by using detailed HMQC and HMBC analyses (data not shown).
3′-OH-DIES-6′-N7Gua
The reaction of Z,Z-DIES-3′,4′-Q or E,E-DIES-3′,4′-Q with dG yielded initially two peaks in HPLC analysis (data not shown). Interestingly, the parent ion scan by direct infusion in mass spectrometry of the reaction mixture after 2 h showed the presence of a molecular ion [M+H]+ at m/z 548, indicating formation of the 3′-OH-DIES-6′-N7dG adduct. The concentration of this adduct initially increased and then it started to decrease after 5 h and became a trace after 24 h. At the same time, the concentration of the depurinating adduct, 3′-OH-DIES-6′-N7Gua, formed after the loss of deoxyribose from the initial adduct, increased continuously and became constant after 24 h. Formation of dG adducts was already reported when E2-3,4-Q or HES-3′,4′-Q was reacted with dG [16]. HPLC analysis after 24 h indicated the presence of one major compound with some unknown less polar impurities. The reaction mixture was then separated by using preparative HPLC, as described above.
1H NMR and MS techniques were used for structure elucidation of the N7Gua adduct. FAB-MS analysis of the compounds showed an [M+H]+ ion at m/z 432, corresponding to the proposed elemental composition. Again, assignments of chemical shifts corresponding to the DIES moiety were easily accomplished by their characteristic coupling patterns. The most upfield chemical shift around δ 10.68 ppm, assigned to NH-1, as well as two proton resonances around δ 6.00 ppm, assigned to the exocyclic NH2, were used as diagnostic signals for the formation of the N7Gua adducts [11,17,27]. Moreover, the presence of a one-proton sharp singlet at δ 7.31 (Z,Z-isomer) or 7.46 (E,E-isomer), assigned to C-8 of Gua, further ruled out the possibility of the formation of adducts with the DIES moiety bound to the C-8 or NH2 of Gua. Hence, binding of the DIES moiety to Gua occurred via the N7 position. Furthermore, the binding site of the DIES moiety at C-6′ was confirmed by the absence of the characteristic d, d, and dd pattern of 3′-OH-DIES [24] (see above), and the appearance of two sharp singlets around δ 6.82 and 6.54 ppm in the 1H NMR spectra of the adducts.
3′-OH-4″-OCH3-DES-6′-N3Ade and 3′-OH-4″-OCH3-DES-6′-N7Gua
The rapid conversion of DES-3′,4′-Q to a spiro-quinone was due to the presence of the ethylenic double bond between the two rings (Fig. 2), which rendered the DES-3′,4′-Q extremely unstable. To impede rearrangement of the quinone to the spiro-quinone, the 4″-OH was methylated (Fig. 5). In this case, oxidation of the catechol to the quinone provided a stable compound that could react with Ade or dG in a 1,4-Michael addition.
Figure 5.
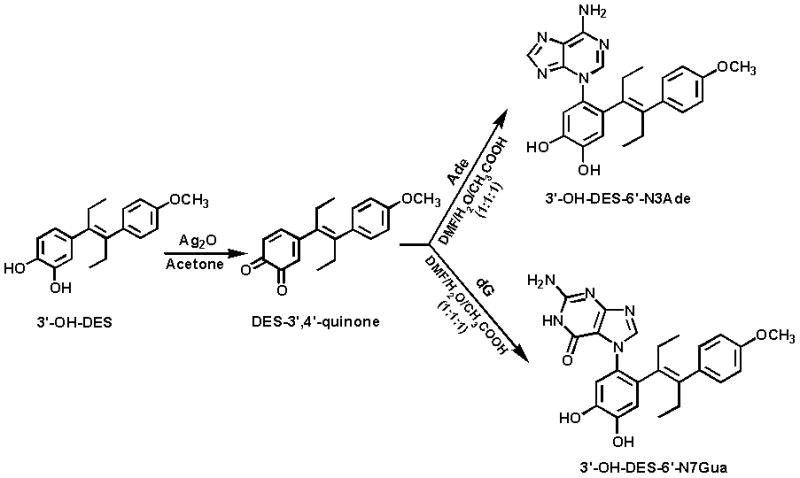
Oxidation of 3′-OH-4″-OCH3-DES to 4″-OCH3-DES-3,4-Q and reaction of the quinone with Ade or dG to form N3Ade or N7Gua adducts.
Formation of the standard depurinating adducts 3′-OH-4″-OCH3-DES-6′-N3Ade and 3′-OH-4″-OCH3-DES-6′-N7Gua (Fig. 5) was accomplished by reacting 4″-OCH3-DES-3′,4′-Q with Ade or dG, respectively. Structure elucidation was carried out by using NMR and MS techniques as described above. Assignment of the chemical shifts and confirmation of proposed structures of the adducts were accomplished by following the same spectroscopic analyses as described above.
Covalent binding of quinones to DNA
DES-3′,4′-Q instantaneously formed a spiro-quinone. When the spiro-quinone was incubated with DNA, it was recovered unreacted with no adduct present in the supernatant (data not shown).
Reaction of Z,Z- DIES-3′,4′-Q or E,E-DIES-3′,4′-Q with DNA yielded no depurinating adducts (Table 1). The precipitated DNA was analyzed for stable adduct formation by 32P-postlabeling [25] and stable adducts were observed as 1.10 μmol/mol DNA-P with Z,Z-DIES-3′,4′-Q, and 0.89 μmol/mol DNA-P with E,E-DIES-3′,4′-Q (Table 2).
Table 1.
Formation of depurinating adducts after incubation of DNA with quinones or enzyme-activated catechols
| Depurinating adducts, μmol/mol DNA-P |
||||||
|---|---|---|---|---|---|---|
| Reactant | 3′-OH-Z,Z-DIES- 6′-N3Ade |
3′-OH-Z,Z-DIES- 6′-N7Gua |
3′-OH-E,E-DIES- 6′-N3Ade |
3′-OH-E,E-DIES- 6′-N7Gua |
4″-OCH3-3′-OH- DES-6′-N3Ade |
4″-OCH3-3′-OH- DES-6′-N7Gua |
| Quinones | ||||||
| Z,Z-DIES-3′,4′-Q | <0.5a | <0.5 | ||||
| E,E-DIES-3′,4′-Q | <0.5 | <0.5 | ||||
| 4″-OCH3-DES-3′,4′-Q | 17.7 ± 10.2 | 32.3 ± 10.6 | ||||
| Catechol/enzymes | ||||||
| 3′-OH- DES/Tyr | 3.7 ± 2.0b | 5.8 ± 1.7 | 1.7 ± 0.2 | 2.8 ± 1.5 | ||
| 3′-OH- DES/LP | 16.7 ± 2.0 | 23.8 ± 5.7 | 11.7 ± 3.2 | 15.8 ± 3.8 | ||
| 3′-OH- DES/PHS | <0.5c | <0.5 | <0.5 | <0.5 | ||
| 3′-OH-Z,Z- DIES/Tyr | <0.5 | <0.5 | ||||
| 3′-OH-Z,Z-DIES/LP | <0.5 | <0.5 | ||||
| 3′-OH-Z,Z-DIES/PHS | <0.5 | <0.5 | ||||
| 3′-OH-E,E- DIES/Tyr | <0.5 | <0.5 | ||||
| 3′-OH-E,E-DIES/LP | <0.5 | <0.5 | ||||
| 3′-OH-E,E-DIES/PHS | <0.5 | <0.5 | ||||
Less than the limit of detection of the instrument.
Based on LC-Ms/MS analysis of the reaction mixture, N3Ade and N7Gua adducts of DES were tentatively identified. Their structures were definitely demonstrated after treatment of supernatant with MnO2 to convert DES adducts to DIES adducts that were compared with the synthesized depurinating adducts.
The major compound found in this experiment was spiro-catechol which after oxidation with MnO2 was observed as spiro-quinone.
Table 2.
Formation of stable adducts in DNA after incubation with quinones or enzyme-activated catechols
| Reactant | Stable adducts, μmol/mol DNA-P |
|---|---|
| Quinones | |
| Z,Z-DIES-3′,4′-Q | 1.10 |
| E,E-DIES-3′,4′-Q | 0.89 |
| 4″-OCH3-DES-3′,4′-Q | 0.59 |
| Catechol/enzymes | |
| 3′-OH- DES/Tyr | 0.71 |
| 3′-OH- DES/LP | 1.00 |
| 3′-OH- DES/PHS | 0.80 |
| 3′-OH-Z,Z- DIES/Tyr | 0.32 |
| 3′-OH-Z,Z-DIES/LP | 0.45 |
| 3′-OH-Z,Z-DIES/PHS | 0.44 |
| 3′-OH-E,E- DIES/Tyr | 0.35 |
| 3′-OH-E,E-DIES/LP | 0.47 |
| 3′-OH-E,E-DIES/PHS | 0.46 |
When 4″-OCH3-DES-3′,4′-Q was reacted with DNA, two depurinating adducts, 3′-OH-4″-OCH3-DES-6′-N3Ade and 3′-OH-4″-OCH3-DES-6′-N7Gua were formed (Fig. 5 and Table 1). The amount of 3′-OH-4″-OCH3-DES-6′-N7Gua was 32.3 ± 10.6 μmol/mol DNA-P, which was almost double the amount of 3′-OH-4″-OCH3-DES-6′-N3Ade, 17.7 ± 10.2 μmol/mol DNA-P. Precipitated DNA was then analyzed for stable adducts and the level of unknown adduct formation was 0.59 μmol/mol DNA-P. When the level of stable adducts was compared with the level of depurinating adducts, about 99% of the adducts were depurinating adducts.
Covalent binding of enzyme-activated catechols to DNA
3′-OH-DES was enzymatically activated by PHS, LP or Tyr in the presence of DNA at 37 °C for 10 h. After processing the supernatant as described in the Methods section, the level of depurinating adducts was measured by UPLC-MS/MS. Activation with PHS did not yield any detectable depurinating adducts, and the only product identified was the spiro-catechol, observed as spiro-quinone (Fig. 2) after oxidation with MnO2.
Activation with Tyr yielded a very low level of 3′-OH-DIES adducts (5.4 ± 2.2μmol/mol DNA-P for the N3Ade adducts and 8.6 ± 5.5 μmol/mol DNA-P for the N7Gua) compared to activation carried out with LP (28.4 ± 5.2 μmol/mol DNA-P for the N3Ade adducts and 39.6 ± 9.5 μmol/mol DNA-P for the N7Gua), as shown in Table 1. Precipitated DNA was then analyzed for stable adducts, and the level of unknown adducts was 0.71 μmol/mol DNA-P with Tyr-activation, 1.00 μmol/mol DNA-P with LP-activation, and 0.80 μmol/mol DNA-P with PHS-activation. Again, the level of depurinating adduct formation was 95–99% higher with Tyr and LP compared to the level of stable adducts (Table 2).
Incubation of both 3′-OH-Z,Z-DIES and 3′-OH-E,E-DIES with DNA in the presence of Tyr, LP or PHS produced none of the depurinating adducts 3′-OH-DIES-6′-N7Gua and 3′-OH-DIES-6′-N3Ade (Table 1). However, analysis of the DNA yielded various low amounts of unknown stable adducts, as shown in Table 2.
Discussion
In vitro and in vivo data generated from our laboratory over several years [10–15], as well as our recent studies on human urine [28–31], have clearly demonstrated the relevance of the formation of depurinating natural estrogen-DNA adducts to the initiation of breast and prostate cancer. Because the synthetic estrogen DES shares many similarities with the natural estrogen E2, i.e., it is a potent estrogen and initiates cancer, it is plausible to hypothesize that the initiation of cancer by DES occurs by formation of depurinating adducts. Toward this goal, the first hindrance was the synthesis of the standard depurinating adducts of DES.
Previously, we synthesized depurinating adducts of the natural estrogen E2 and synthetic estrogen HES, by chemical oxidation of the catechol estrogens to their respective electrophilic quinones, followed by reaction with Ade or dG [11,17,27]. Under similar conditions, chemical oxidation of 3′-OH-DES with Ag2O in acetone afforded a highly unstable DES-3′,4′-Q, which underwent novel cyclization to spiro-quinone (Fig. 2) [26]. Efforts to synthesize the standard depurinating adducts of DES suffered because of the unavailability of DES-3′,4′-Q to react with Ade or dG.
Cyclization of DES-3′,4′-Q to spiro-compound takes place in a state where the molecule is free to undergo various rotations with a high degree of freedom. We hypothesized that if free rotations of these chemical bonds were blocked, for instance by a physical complex with DNA, then there would be a chance that the DES-3′,4′-Q might be sufficiently stabilized to react with the DNA. To test this hypothesis, 3′-OH-DES was enzymatically activated in the presence of DNA. Under these conditions the quinone DES-3′4′-Q was stable enough to react specifically at the N3 position of Ade and N7 position of Gua, thus forming the depurinating adducts. This result can only be explained by intercalation of the 3′-OH-DES in the DNA, followed by enzymatic oxidation to its quinone. The N3Ade and N7Gua adducts could be identified by rapidly determining their molecular weight by MS (Fig. 6) before the adducts autoxidized to their corresponding DIES adducts. For further demonstration that these adducts were formed, the freshly obtained DES adducts were oxidized with MnO2 and converted to their DIES adducts (Fig. 6). The structures of these DIES adducts were elucidated by comparison with the synthesized standard DIES adducts.
Figure 6.
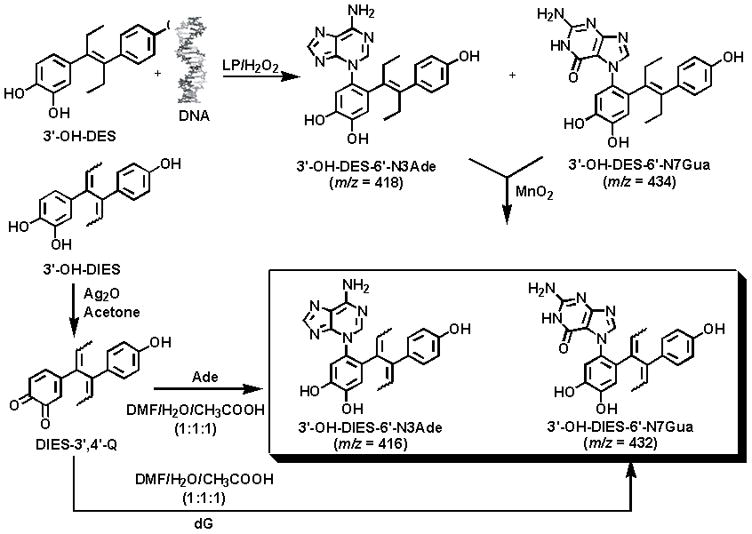
Formation of depurinating N3Ade and N7Gua adducts of DES following enzymic activation of 3′-OH-DES in the presence of DNA. The DES adducts were oxidized to the corresponding DIES adducts.
To confirm that the DIES-DNA adducts were formed from DES and not from DIES, we treated DNA with DIES-3′,4′-Q or 3′-OH-DIES after enzymatic activation. No depurinating adducts were observed in these experiments (Table 1), which indicated that the DIES-DNA adducts observed following reaction of enzyme-activated 3′-OH-DES with DNA were solely formed from DES and not from DIES.
The lack of formation of depurinating adducts in the reaction of DIES-3′,4′-Q with DNA derives from the chemical structures of these compounds. Data from X-ray crystallography of DES and DIES derivatives indicate that the orientation of the dienic chain of the DIES derivatives is perpendicular to the phenyl planes [32], whereas it is almost linear with the phenyl planes in DES [33]. Thus, the perpendicular dienic chain of DIES derivatives impedes the appropriate intercalation of DIES, but not DES, into DNA. Hence, the quinones of DES give rise to the formation of depurinating adducts, whereas the quinones of DIES do not.
It was surprising to observe the formation of stable adducts even with DIES metabolites. Although DIES metabolites are not suitable candidates for intercalation in DNA, they can generate free radicals. Kalyanaraman and Sealy [34] have studied the formation of free radicals from stilbene catechol estrogens. The DIES radicals generated under the quinone-semiquinone-catechol system may damage DNA, thus giving rise to lesions observed as stable adducts.
Unified mechanism of depurinating estrogen-DNA adduct formation
The totality of experiments on estrogen metabolism [35–41], formation of DNA adducts [10–13,28,30,42,43], mutagenicity [13–15,44,45], cell transformation [46–49] and carcinogenicity [50–53] have led to the hypothesis that certain estrogen metabolites, predominantly catechol estrogen-3,4-quinones, react with DNA to cause mutations that lead to the initiation of cancer [13]. Catechol estrogen-3,4-quinones can react with DNA to form the depurinating adducts, 4-OHE1(E2)-1-N3Ade and 4-OHE1(E2)-1-N7Gua, generating apurinic sites in the DNA (Fig. 7). Various types of evidence indicate that error-prone repair of these apurinic sites to mutations that can initiate breast cancer [13–15,44,45].
Figure 7.
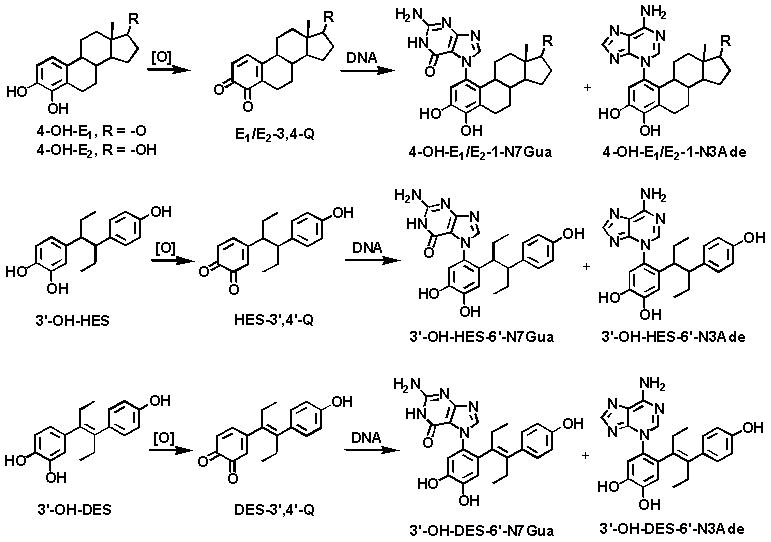
Unified mechanism of metabolic activation and reaction with DNA of the natural estrogens and the synthetic estrogens HES and DES.
The more abundant formation of depurinating N3Ade and N7Gua adducts obtained from E1(E2)-3,4-Q versus E1(E2)-2,3-Q [12] is in line with other studies that indicate the important role of E1(E2)-3,4-Q in the initiation of cancer [13]. In fact, in animal models 4-OHE1(E2) is more carcinogenic than the extremely weak carcinogen 2-OHE1(E2) [50–52]. The catechol of DES, when oxidized to its corresponding catechol quinone and reacted with DNA, yields N3Ade and N7Gua adducts analogous to those formed by E1(E2)-3,4-Q and HES-3′,4′-Q [16,17].
This common mechanism of metabolic activation for natural estrogens and DES (Fig. 7) is extremely important because DES is a proved human carcinogen [2–9]. Furthermore, through the studies reported here, we have demonstrated that intercalation of the catechol estrogen in DNA is the necessary preliminary step to generate specificity in the formation of the depurinating adducts.
Acknowledgments
This research was supported by U.S. Public Health Service grant P01 CA49210 from the National Cancer Institute and the U.S. Army Breast Cancer Research Program grant DAMD 17-03-1-0229. Core support at the Eppley Institute was provided by grant P30 CA36727 from the National Cancer Institute.
References
- 1.Dodds EC, Goldberg L, Lawson W, Robinson R. Estrogenic activity of certain synthetic compounds. Nature. 1938;141:247–8. [Google Scholar]
- 2.Herbst AL, Ulfelder H, Poskanzer DC. Adenocarcinoma of the vagina. Association of maternal stilbestrol therapy with tumor appearance in young women. N Engl J Med. 1971;284:878–81. doi: 10.1056/NEJM197104222841604. [DOI] [PubMed] [Google Scholar]
- 3.Swan SH. Diethylstilbestrol and clear cell vaginal carcinoma. Am J Med. 1987;83:372–4. doi: 10.1016/0002-9343(87)90728-5. [DOI] [PubMed] [Google Scholar]
- 4.Noller KL, O’Brien PC. Diethylstilbestrol and clear cell vaginal carcinoma. Am J Med. 1987;83:374. doi: 10.1016/0002-9343(87)90728-5. [DOI] [PubMed] [Google Scholar]
- 5.Herbst AL. Diethylstilbestrol and clear cell vaginal carcinoma. Am J Med. 1987;83:375–6. doi: 10.1016/0002-9343(87)90728-5. [DOI] [PubMed] [Google Scholar]
- 6.Greenberg ER, Barnes AB, Resseguie L, Barrett JA, Burnside S, Lanza LL, Neff RK, Stevens M, Young RH, Colton T. Breast cancer in mothers given diethylstilbestrol in pregnancy. N Engl J Med. 1984;311:1393–8. doi: 10.1056/NEJM198411293112201. [DOI] [PubMed] [Google Scholar]
- 7.Colton T, Greenberg ER, Noller K, Resseguie L, Van Bennekom C, Heeren T, Zhang Y. Breast cancer in mothers prescribed diethylstilbestrol in pregnancy. Further follow-up. Jama. 1993;269:2096–100. [PubMed] [Google Scholar]
- 8.Palmer JR, Wise LA, Hatch EE, Troisi R, Titus-Ernstoff L, Strohsnitter W, Kaufman R, Herbst AL, Noller KL, Hyer M, Hoover RN. Prenatal diethylstilbestrol exposure and risk of breast cancer. Cancer Epidemiol Biomarkers Prev. 2006;15:1509–14. doi: 10.1158/1055-9965.EPI-06-0109. [DOI] [PubMed] [Google Scholar]
- 9.Troisi R, Hatch EE, Titus-Ernstoff L, Hyer M, Palmer JR, Robboy SJ, Strohsnitter WC, Kaufman R, Herbst AL, Hoover RN. Cancer risk in women prenatally exposed to diethylstilbestrol. Int J Cancer. 2007;121:356–60. doi: 10.1002/ijc.22631. [DOI] [PubMed] [Google Scholar]
- 10.Cavalieri EL, Stack DE, Devanesan PD, Todorovic R, Dwivedy I, Higginbotham S, Johansson SL, Patil KD, Gross ML, Gooden JK, Ramanathan R, Cerny RL, et al. Molecular origin of cancer: catechol estrogen-3,4-quinones as endogenous tumor initiators. Proc Natl Acad Sci U S A. 1997;94:10937–42. doi: 10.1073/pnas.94.20.10937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li KM, Todorovic R, Devanesan P, Higginbotham S, Kofeler H, Ramanathan R, Gross ML, Rogan EG, Cavalieri EL. Metabolism and DNA binding studies of 4-hydroxyestradiol and estradiol-3,4-quinone in vitro and in female ACI rat mammary gland in vivo. Carcinogenesis. 2004;25:289–97. doi: 10.1093/carcin/bgg191. [DOI] [PubMed] [Google Scholar]
- 12.Zahid M, Kohli E, Saeed M, Rogan E, Cavalieri E. The greater reactivity of estradiol-3,4-quinone vs estradiol-2,3-quinone with DNA in the formation of depurinating adducts: implications for tumor-initiating activity. Chem Res Toxicol. 2006;19:164–72. doi: 10.1021/tx050229y. [DOI] [PubMed] [Google Scholar]
- 13.Cavalieri E, Chakravarti D, Guttenplan J, Hart E, Ingle J, Jankowiak R, Muti P, Rogan E, Russo J, Santen R, Sutter T. Catechol estrogen quinones as initiators of breast and other human cancers: implications for biomarkers of susceptibility and cancer prevention. BBA Reviews on Cancer. 2006;1766:63–78. doi: 10.1016/j.bbcan.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 14.Chakravarti D, Mailander PC, Li KM, Higginbotham S, Zhang HL, Gross ML, Meza JL, Cavalieri EL, Rogan EG. Evidence that a burst of DNA depurination in SENCAR mouse skin induces error-prone repair and forms mutations in the H-ras gene. Oncogene. 2001;20:7945–53. doi: 10.1038/sj.onc.1204969. [DOI] [PubMed] [Google Scholar]
- 15.Mailander PC, Meza JL, Higginbotham S, Chakravarti D. Induction of A.T to G. C mutations by erroneous repair of depurinated DNA following estrogen treatment of the mammary gland of ACI rats. J Steroid Biochem Mol Biol. 2006;101:204–15. doi: 10.1016/j.jsbmb.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 16.Saeed M, Zahid M, Gunselman SJ, Rogan E, Cavalieri E. Slow loss of deoxyribose from the N7deoxyguanosine adducts of estradiol-3,4-quinone and hexestrol-3′,4′-quinone. Implications for mutagenic activity. Steroids. 2005;70:29–35. doi: 10.1016/j.steroids.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 17.Saeed M, Gunselman SJ, Higginbotham S, Rogan E, Cavalieri E. Formation of the depurinating N3adenine and N7guanine adducts by reaction of DNA with hexestrol-3′,4′-quinone or enzyme-activated 3-hydroxyhexestrol. Implication for a unifying mechanism of tumor initiation by natural and syntetic estrogens. Steroids. 2005;70:37–45. doi: 10.1016/j.steroids.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 18.Liehr JG, Ballatore AM, Dague BB, Ulubelen AA. Carcinogenicity and metabolic activation of hexestrol. Chem Biol Interact. 1985;55:157–76. doi: 10.1016/s0009-2797(85)80125-3. [DOI] [PubMed] [Google Scholar]
- 19.Haaf H, Metzler M. In vitro metabolism of diethylstilbestrol by hepatic, renal and uterine microsomes of rats and hamsters. Effects of different inducers. Biochem Pharmacol. 1985;34:3107–15. doi: 10.1016/0006-2952(85)90155-8. [DOI] [PubMed] [Google Scholar]
- 20.Blaich G, Gottlicher M, Cikryt P, Metzler M. Effects of various inducers on diethylstilbestrol metabolism, drug-metabolizing enzyme activities and the aromatic hydrocarbon (Ah) receptor in male Syrian golden hamster liver. J Steroid Biochem. 1990;35:201–4. doi: 10.1016/0022-4731(90)90275-w. [DOI] [PubMed] [Google Scholar]
- 21.Metzler M, McLachlan JA. Oxidative metabolism of the synthetic estrogens hexestrol and dienestrol indicates reactive intermediates. Adv Exp Med Biol. 1981;136(Pt A):829–37. doi: 10.1007/978-1-4757-0674-1_65. [DOI] [PubMed] [Google Scholar]
- 22.Li JJ, Li SA, Klicka JK, Parsons JA, Lam LK. Relative carcinogenic activity of various synthetic and natural estrogens in the Syrian hamster kidney. Cancer Res. 1983;43:5200–4. [PubMed] [Google Scholar]
- 23.Oda T, Murai T, Sato Y. Carbon-13 nuclear magnetic resonance study of meso-hexestrol and its derivatives. Chem Pharm Bull. 1988;36:1534–9. doi: 10.1248/cpb.36.1534. [DOI] [PubMed] [Google Scholar]
- 24.Saeed M, Zahid M, Rogan E, Cavalieri E. Synthesis of the catechols of natural and synthetic estrogens by using 2-iodoxybenzoic acid (IBX) as the oxidizing agent. Steroids. 2005;70:173–8. doi: 10.1016/j.steroids.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 25.Bodell WJ, Devanesan PD, Rogan EG, Cavalieri EL. 32P-postlabeling analysis of benzo[a]pyrene-DNA adducts formed in vitro and in vivo. Chem Res Toxicol. 1989;2:312–5. doi: 10.1021/tx00011a008. [DOI] [PubMed] [Google Scholar]
- 26.Saeed M, Rogan E, Cavalieri E. Novel spiro-quinone formation from 3-hydroxydiethylstilbestrol after oxidation with silver oxide. Tetrahedron Lett. 2005;46:4449–51. [Google Scholar]
- 27.Stack DE, Byun J, Gross ML, Rogan EG, Cavalieri EL. Molecular characteristics of catechol estrogen quinones in reaction with deoxyribonucleosides. Chem Res Toxicol. 1996;9:851–9. doi: 10.1021/tx960002q. [DOI] [PubMed] [Google Scholar]
- 28.Markushin Y, Gaikwad N, Zhang H, Kapke P, Rogan EG, Cavalieri EL, Trock BJ, Pavlovich C, Jankowiak R. Potential biomarker for early risk assessment of prostate cancer. Prostate. 2006;66(14):1565–71. doi: 10.1002/pros.20484. [DOI] [PubMed] [Google Scholar]
- 29.Yang L, Gaikwad N, Meza J, Cavalieri E, Muti P, Trock B, Rogan E. Novel biomarkers for risk of prostate cancer. Results from a case-control study. Prostate. 2008 doi: 10.1002/pros.20850. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gaikwad NW, Yang L, Muti P, Meza JL, Pruthi S, Ingle JN, Rogan EG, Cavalieri EL. The molecular etiology of breast cancer: evidence from biomarkers of risk. Int J Cancer. 2008;122(9):1949–57. doi: 10.1002/ijc.23329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gaikwad N, Yang L, Pruthi S, Ingle J, Meza J, Sandhu N, Suman V, Rogan E, Cavalieri E. Validation of urine biomarkers of risk in the molecular etiology of breast cancer. Int J Cancer. 2008 doi: 10.4137/bcbcr.s2112. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fornies-Marquina JM, Courseille C, Busetta B, Hospital M. Structure crystalline et moléculaire du cycladiène (dièneoestrol) Acta Crystallogr B. 1972;28:655–6. [Google Scholar]
- 33.Weeks CM, Cooper A, Norton DA. The crystal and molecular structure of diethylstilbestrol. Acta Crystallogr B. 1970;26:429–33. doi: 10.1107/s0567740870002509. [DOI] [PubMed] [Google Scholar]
- 34.Kalyanaraman B, Sealy RC. Characterization of semiquinone free radicals formed from stilbene catechol estrogen: an ESR spin stabilization and spin trapping study. J Biol Chem. 1989;264:11014–9. [PubMed] [Google Scholar]
- 35.Lu F, Zahid M, Saeed M, Cavalieri EL, Rogan EG. Estrogen metabolism and formation of estrogen-DNA adducts in estradiol-treated MCF-10F cells. The effects of 2,3,7,8-tetrachlorodibenzo-p-dioxin induction and catechol-O-methyltransferase inhibition. J Steroid Biochem Mol Biol. 2007;105:150–8. doi: 10.1016/j.jsbmb.2006.12.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zahid M, Saeed M, Lu F, Gaikwad N, Rogan E, Cavalieri EL. Inhibition of catechol-O-methyltransferase increases estrogen DNA-adduct formation. Free Radic Biol Med. 2007;43:1534–40. doi: 10.1016/j.freeradbiomed.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu BT, Conney AH. Functional role of estrogen metabolism in target cells: Review and perspectives. Carcinogenesis. 1998;19:1–27. doi: 10.1093/carcin/19.1.1. [DOI] [PubMed] [Google Scholar]
- 38.Cavalieri EL, Kumar S, Todorovic R, Higginbotham S, Badawi AF, Rogan EG. Imbalance of estrogen homeostasis in kidney and liver of hamsters treated with estradiol: Implications for estrogen-induced initiation of renal tumors. Chem Res Toxicol. 2001;14:1041–50. doi: 10.1021/tx010042g. [DOI] [PubMed] [Google Scholar]
- 39.Devanesan P, Santen RJ, Bocchinfuso WP, Korach KS, Rogan EG, Cavalieri EL. Catechol estrogen metabolites and conjugates in mammary tumors with hyperplastic tissue from estrogen receptor-α knock-out (ERKO)/Wnt-1 Mice: Implications for initiation of mammary tumors. Carcinogenesis. 2001;22:1573–6. doi: 10.1093/carcin/22.9.1573. [DOI] [PubMed] [Google Scholar]
- 40.Cavalieri EL, Devanesan P, Bosland MC, Badawi AF, Rogan EG. Catechol estrogen metabolites and conjugates in different regions of the prostate of Noble rats treated with 4-hydroxyestradiol: implications for estrogen-induced initiation of prostate cancer. Carcinogenesis. 2002;23:329–33. doi: 10.1093/carcin/23.2.329. [DOI] [PubMed] [Google Scholar]
- 41.Rogan EG, Badawi AF, Devanesan PD, Meza JL, Edney JA, West WW, Higginbotham SM, Cavalieri EL. Relative imbalances in estrogen metabolism and conjugation in breast tissue of women with carcinoma: Potential biomarkers of susceptibility to cancer. Carcinogenesis. 2003;24:697–702. doi: 10.1093/carcin/bgg004. [DOI] [PubMed] [Google Scholar]
- 42.Markushin Y, Zhong W, Cavalieri EL, Rogan EG, Small GJ, Yeung ES, Jankowiak R. Spectral characterization of catechol estrogen quinone (CEQ)-derived DNA adducts and their identification in human breast tissue extract. Chem Res Toxicol. 2003;16:1107–17. doi: 10.1021/tx0340854. [DOI] [PubMed] [Google Scholar]
- 43.Saeed M, Rogan E, Fernandez SV, Sheriff F, Russo J, Cavalieri E. Formation of depurinating N3Adenine and N7Guanine adducts by MCF-10F cells cultured in the presence of 4-hydroxyestradiol. Int J Cancer. 2007;120:1821–4. doi: 10.1002/ijc.22399. [DOI] [PubMed] [Google Scholar]
- 44.Zhao Z, Kosinska W, Khmelnitsky M, Cavalieri EL, Rogan EG, Chakravarti D, Sacks PG, Guttenplan JB. Mutagenic activity of 4-hydroxyestradiol, but not 2-hydroxyestradiol, in BB rat2 embryonic cells, and the mutational spectrum of 4-hydroxyestradiol. Chem Res Toxicol. 2006;19:475–9. doi: 10.1021/tx0502645. [DOI] [PubMed] [Google Scholar]
- 45.Fernandez SV, Russo IH, Russo J. Estradiol and its metabolites 4-hydroxyestradiol and 2-hydroxyestradiol induce mutations in human breast epithelial cells. Int J Cancer. 2006;118:1862–8. doi: 10.1002/ijc.21590. [DOI] [PubMed] [Google Scholar]
- 46.Russo J, Lareef MH, Balogh G, Guo S, Russo IH. Estrogen and its metabolites are carcinogenic agents in human breast epithelial cells. J Steroid Biochem Mol Biol. 2003;87:1–25. doi: 10.1016/s0960-0760(03)00390-x. [DOI] [PubMed] [Google Scholar]
- 47.Lareef MH, Garber J, Russo PA, Russo IH, Heulings R, Russo J. The estrogen antagonist ICI-182–780 does not inhibit the transformation phenotypes induced by 17-beta-estradiol and 4-OH estradiol in human breast epithelial cells. Int J Oncol. 2005;26:423–9. [PubMed] [Google Scholar]
- 48.Russo J, Fernandez SV, Russo PA, Fernbaugh R, Sheriff FS, Lareef HM, Garber J, Russo IH. 17-Beta-estradiol induces transformation and tumorigenesis in human breast epithelial cells. Faseb J. 2006;20:1622–34. doi: 10.1096/fj.05-5399com. [DOI] [PubMed] [Google Scholar]
- 49.Venugopal D, Zahid M, Mailander PC, Meza JL, Rogan EG, Cavalieri EL, Chakravarti D. Reduction of estrogen-induced transformation of mouse mammary epithelial cells by N-acetylcysteine. J Steroid Biochem Mol Biol. 2008;109:22–30. doi: 10.1016/j.jsbmb.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liehr JG, Fang WF, Sirbasku DA, Ari-Ulubelen A. Carcinogenicity of catecholestrogens in Syrian hamsters. J Steroid Biochem. 1986;24:353–6. doi: 10.1016/0022-4731(86)90080-4. [DOI] [PubMed] [Google Scholar]
- 51.Li JJ, Li SA. Estrogen carcinogenesis in Syrian hamster tissue: Role of metabolism. Fed Proc. 1987;46:1858–63. [PubMed] [Google Scholar]
- 52.Newbold RR, Liehr JG. Induction of uterine adenocarcinoma in CD-1 mice by catechol estrogens. Cancer Res. 2000;60:235–7. [PubMed] [Google Scholar]
- 53.Yue W, Santen RJ, Wang JP, Li Y, Verderame MF, Bocchinfuso WP, Korach KS, Devanesan P, Todorovic R, Rogan EG, Cavalieri EL. Genotoxic metabolites of estradiol in breast: potential mechanism of estradiol induced carcinogenesis. J Steroid Biochem Mol Biol. 2003;86:477–86. doi: 10.1016/s0960-0760(03)00377-7. [DOI] [PubMed] [Google Scholar]


