Abstract
Post-translational modifications of histones play key roles in the regulation of gene expression and chromatin structure in eukaryotes. Methylation of histone 3 on lysine 27 (H3K27) is one of the most common and well-studied histone post-translational modifications. The vast majority of research on this histone residue, however, has focused on the trimethylated form (H3K27me3). Despite occurring at higher levels than H3K27me3 in animals and plants, the monomethylated form of H3K27 (H3K27me1) remains relatively poorly characterized. The absence of information concerning H3K27me1 is due in large part to the fact that the enzymes catalyzing this epigenetic mark were only recently identified. In this article, we highlight new findings concerning H3K27me1, including the identification of two plant-specific H3K27 monomethyltransferases that are required for gene silencing and heterochromatin condensation. We also discuss the emerging similarities and differences in H3K27 methylation in plant and animal systems.
Keywords: ATXR5, ATXR6, H3K27me1, heterochromatin, gene silencing, chromatin condensation
Introduction
Almost a decade ago, the SET domains of the human and mouse orthologs of SUPPRESSOR OF VARIEGATION 3-9 HOMOLOG 1 were found to catalyze the methylation of histone 3 lysine 9 (H3K9) 1. This discovery had a major impact on the field of epigenetics, as it led to the identification of a large number of methyltransferases that establish and maintain various post-translational modifications (PTM) on histones in eukaryotes 2. Today, the functional analysis of a particular histone PTM still frequently begins with the identification of the enzyme(s) responsible for it. Removal of the enzyme, and hence the PMT, often leads to molecular, cellular, and developmental phenotypes, which can be analyzed to understand the role of the specific PTM. An alternative approach to study the significance of specific histone modifications is to eliminate “reading” proteins, which carry domains capable of specifically interacting with particular PTMs 3. Both approaches, however, are often complicated by genetic redundancy, a lack of knowledge regarding the proteins involved, and/or absence of mutant alleles for these proteins.
Among histone PMTs, histone 3 lysine 27 monomethylation (H3K27me1) can be considered a quintessential “orphan” mark. Although H3K27me1 is found at high levels in most complex eukaryotic systems 4, relatively little is known about the proteins that catalyze and bind H3K27me1 or the biological role of this epigenetic mark. The recent identification of enzymes responsible for H3K27me1 in plants and mammals has yielded a first glimpse into the functions of this histone PTM 5, 6. In this article, we present the new findings concerning H3K27me1 in Arabidopsis and compare these findings with what is known about this epigenetic mark in animals. In addition, we speculate on how H3K27me1 might be maintained during DNA replication in Arabidopsis and its possible biochemical function.
Identification of novel H3K27 monomethyltransferases in Arabidopsis
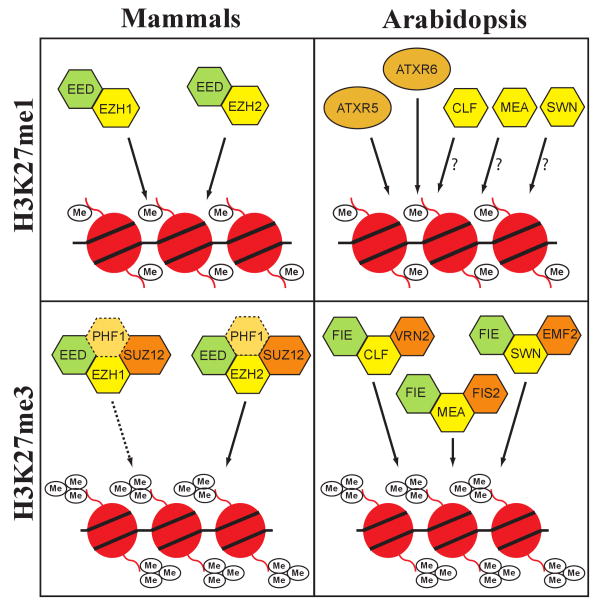
Sequence analysis of the paralogous proteins ARABIDOPSIS TRITHORAX-RELATED PROTEIN 5 (ATXR5) and ATXR6 shows that they contain divergent SET domains. Although most SET domains function to methylate specific lysine residues of H3 1, some have been shown to catalyze the methylation of non-histone substrates 7-10. Thus, it remained unclear for some time whether ATXR5 and ATXR6 actually play a role in histone methylation, as phylogenetic analysis did not indicate strong similarity with functionally-characterized SET domains known to methylate H3 11. The in vitro characterization of the enzymatic activity of ATXR5 and ATXR6 by our group has shown that these enzymes are in fact H3K27 methyltransferases 5. More specifically, ATXR5 and ATXR6 are H3K27 monomethyltransferases, meaning that they convert H3K27me0 to H3K27me1, but not to H3K27me2 or H3K27me3 (Fig. 1). The Arabidopsis genome encodes three other H3K27 methyltransferases: CURLY LEAF (CLF), SWINGER (SWN), and MEDEA (MEA) 12. These three enzymes are homologs of the Drosophila protein ENHANCER OF ZESTE (E(z)). Like E(z), CLF, SWN, and MEA are the catalytic subunits of Polycomb Repressive Complex 2 (PRC2)-like complexes in plants (Fig. 1), which are responsible for the deposition of H3K27me3 13. The presence of a new class of H3K27 methyltransferases in Arabidopsis was somewhat expected, as previous experiments had shown that CLF, SWN, and MEA were not required for the establishment of H3K27me1 14. ATXR5 and ATXR6 are the first eukaryotic H3K27 methyltransferases to be identified that are not orthologous to E(z). It should be noted, however, that the viral protein vSET from the Paramecium bursaria chlorella virus 1 is also a non-E(z)-type H3K27 methyltransferase 15, 16.
Figure 1.
Proteins involved in mono- and trimethylation of H3K27 in mammals and Arabidopsis. PRC2 core components and other associated proteins are shown in hexagons bounded by solid lines and dashed lines, respectively. Non-PRC2 proteins are shown in ovals. Homologous proteins are color-coded.
Further work on ATXR5 and ATXR6 has revealed that these enzymes are responsible for the enrichment of H3K27me1 at chromocenters (constitutive heterochromatin) in Arabidopsis 5. Also, ATXR5 and ATXR6 appear to function redundantly in maintaining the condensed and transcriptionally-silent state of heterochromatin in Arabidopsis, as atxr5 atxr6 double mutants show both chromatin decondensation and loss of gene silencing. Interestingly, a reduction in H3K27me1 does not affect H3K27me2/3. This last result strongly suggests that the phenotypes observed in atxr5 atxr6 mutants are due specifically to lower levels of H3K27me1 and that H3K27me2/3 does not depend on H3K27me1 as an intermediate for their catalysis.
Many questions arise from the discovery of the enzymes responsible for H3K27me1 in Arabidopsis. For example, what is the mechanism by which the spatial distribution of H3K27me1 is regulated in Arabidopsis? Apart from their SET domains, ATXR5 and ATXR6 also contain a plant homeodomain (PHD) domain. Because some PHD domains have been shown to bind specific PTMs on H3 proteins 17, 18, one possibility would be that the PHD domains of ATXR5 and ATXR6 could control the distribution of H3K27me1 by interacting only with certain forms of H3. Analysis of the spatial distribution of H3K27me1 in Arabidopsis shows that it is enriched at the chromocenters 14, 19. If the binding and catalytic activity of ATXR5 and ATXR6 are linked, we can hypothesize that the PHD domains of these proteins might interact with an epigenetic mark that is also enriched in heterochromatin.
Different mechanisms are involved in the deposition of H3K27me1 in plants and animals
The analysis of histone PTMs in a broad spectrum of eukaryotes has revealed both conservation and divergence in their spatial distributions and biological roles 4, 20. For H3K27me1, one clear difference between animals and plants is the molecular mechanism by which this PTM is deposited onto histones. Homologs of ATXR5 and ATXR6 are present in land plants, but not in metazoans. Thus, different proteins are used to monomethylate H3K27 in animals. In Drosophila, H3K27me1is completely dependent on E(z) 21. Similarly, two homologs of E(z), ENHANCER OF ZESTE HOMOLOG 1 (EZH1) and EZH2, function redundantly in mammals to control global levels of H3K27me1 6. The mammalian PRC2 subunit EMBRYONIC ECTODERM DEVELOPMENT (EED) was also demonstrated to be required for H3K27me1 22. Taken together, these results suggest that PRC2-type complexes are required for H3K27me1 in mammals (Fig. 1).
E(z) and its homologs have been implicated in both H3K27 mono- and trimethylation. How these enzymes regulate the level of H3K27 methylation, however, remains to be determined. The answer to this question may lie in the particular protein composition of the different PRC2-like complexes. For example, SUPPRESSOR OF ZESTE 12 (SUZ12) has been found to be essential for H3K27me3, but dispensable for H3K27me1 (Fig.1) 23. In the same manner, the mammalian protein PHD FINGER PROTEIN 1 (PHF1), a homologue of the Drosophila protein POLYCOMBLIKE (PCL), was shown to associate with PRC2 components and stimulate formation of H3K27me3, but not H3K27me1 or H3K27me2, in vitro and in vivo 24-26. Although the Arabidopsis genome contains three SUZ12-related proteins (VERNALIZATION2 (VRN2), EMBRYONIC FLOWER2 (EMF2)), and FERTILIZATION-INDEPENDENT SEED 2 (FIS2)) 13, no PHF1/PCL homologs are found in Arabidopsis. This suggests that both conserved and plant/animal-specific mechanisms may regulate the histone methyltransferase activity of E(z)/PRC2-like complexes. It remains to be determined whether the plant E(z) homologs also contribute to H3K27me1. Only a 22 percent decrease in H3K27me1 was observed in atxr5 atxr6 mutants, which suggests that other H3K27 methyltransferases can also catalyze this epigenetic mark 5. However, a definitive conclusion still cannot be drawn, as the atxr6 allele used in the study is not a null.
Aside from being catalyzed by non-homologous enzymes, there are striking similarities between the global distribution of H3K27me1 in plants and animals. Immunostaining experiments have shown centromeric and pericentromeric enrichments for H3K27me1 in mammals and Arabidopsis 14, 19, 27. More precise mapping of this histone PTM in mammals has revealed that H3K27me1 is actually widely distributed in the genome, being only excluded from the region surrounding the promoter of active genes 28. Similarly, the distribution of H3K27me1 was found recently to be largely homogenous over the entire length of the chromosomes in two gymnosperms and maize 29, 30. It should be noted that the compact nature of the constitutive heterochromatin in both plants and animals may have the effect of enhancing the apparent heterochromatic enrichment of H3K27me1 in immunolocalization experiments. The similar genomic distributions in plants and animals suggests that the role of H3K27me1 might be at least partially conserved in eukaryotes.
Epigenetic inheritance of H3K27me1 in plants
One of the most interesting questions concerning epigenetic marks like H3K27me1 is the mechanism by which they are maintained through DNA replication. Although DNA replication is a challenge for chromatin homeostasis, it also provides the cell with a window of opportunity for chromatin remodeling 31, 32. Transcription factors and other bound proteins that might otherwise restrict access to chromatin are displaced, allowing passage of the replication fork. In addition, the newly-synthesized histones incorporated on both daughter strands can be given new PTMs that can alter the epigenetic status of the associated locus. A few SET-domain proteins in animals have already been shown to function during DNA replication 33, 34.
Results from previous work indicate that ATXR5 and ATXR6 may be involved in chromatin modification during DNA synthesis. ATXR5 and ATXR6 are expressed at highest levels during DNA synthesis and physically interact with PROLIFERATING CELL NUCLEAR ANTIGEN (PCNA) through a PCNA-interacting protein box motif 35. These two findings indicate that ATXR5 and ATXR6 are well positioned to modify histones during DNA replication. Newly-synthesized histones could be a substrate for monomethylation by ATXR5 and ATXR6, before and/or after deposition on the chromatin. Modification of these histones would prevent dilution of H3K27me1 by restoring pre-replication levels of this mark. Interestingly, it has been speculated that H3K27me1 might also be the default state of newly assembled chromatin following DNA replication in mammals 28. PCNA seems to remain associated with the newly-replicated daughter strands, at least in animal cells, for approximately 20 minutes 36. If this also happens in plants, it would represent a temporal window for PCNA-binding factors, like ATXR5 and ATXR6, to perform their activity.
What is the role of H3K27me1?
In addition to increasing our understanding of the mechanisms that maintain or modify chromatin during DNA replication, further studies of ATXR5 and ATXR6 may also shed light on the biochemical function of H3K27me1. Our work strongly suggests that H3K27me1 plays a critical role in maintaining heterochromatic chromatin condensation and gene silencing in Arabidopsis 5. It remains unknown, however, how this histone modification signals such events. One possibility is that H3K27me1 simply acts as a recruitment signal for an H3K27me1-binding protein. Alternatively, monomethylation of H3K27 (and possibly H3K27me2 and H3K27me3) may act in a negative manner. This could occur by either preventing H3K27me0-interacting proteins from binding chromatin or by blocking alternative histone modifications from being made on H3K27.
If an H3K27me1-binding protein exists, what might we expect its function to be? Based on our results, this protein would likely promote chromatin condensation, either directly or as part of a complex. Chromatin-remodeling enzymes are good candidates for this activity, as they use the energy stored in ATP to promote nucleosome sliding, remodeling, and dissociation 37. DECREASE IN DNA METHYLATION1 (DDM1) is a chromatin-remodeling protein in Arabidopsis that could potentially be linked to H3K27me1. Similar to atxr5 atxr6 double mutants, ddm1 plants show chromatin decondensation and reactivation of repressed heterochromatic elements (Fransz 2006). These effects of ddm1 mutations, however, are associated with decreased levels of DNA methylation, and our work clearly shows that atxr5 atxr6 has no impact on DNA methylation 5, 38. Another chromatin-remodeling protein that could be part of an H3K27me1 pathway is MORPHEUS' MOLECULE 1 (MOM1) 39. The MOM1 protein is responsible for silencing heterochromatic elements, and it does so, like ATXR5 and ATXR6, without affecting DNA methylation 39-42. However, unlike atxr5 atxr6 mutants, compaction of the heterochromatin is unaffected in mom1 40. More work will have to be done in order to identify the factor(s) working downstream of H3K27me1.
Concluding Remarks
For various reasons, our understanding of H3K27me1 has lagged behind that of other epigenetic marks. With the identification of the enzymes responsible for this mark in animals and plants, however, the near future should reveal many exciting discoveries about the function of this mark in eukaryotic cells. It will be particularly interesting to see the degree of similarity and divergence that exists between plants and animals regarding the deposition of H3K27me1 and its biological function.
Acknowledgments
This work was supported by a grants to S.D.M. from the NIH (GM075060) and to Y.J. from NATEQ (Fonds québécois de recherche sur la nature et les technologies).
Abbreviations
- ATXR5
ARABIDOPSIS TRITHORAX-RELATED PROTEIN 5
- ATXR6
ARABIDOPSIS TRITHORAX-RELATED PROTEIN 6
- CLF
CURLY LEAF
- DDM1
DECREASE IN DNA METHYLATION1
- EED
EMBRYONIC ECTODERM DEVELOPMENT
- EMF2
EMBRYONIC FLOWER2
- E(z)
ENHANCER OF ZESTE
- EZH1
ENHANCER OF ZESTE HOMOLOG 1
- EZH2
ENHANCER OF ZESTE HOMOLOG 2
- FIS2
FERTILIZATION-INDEPENDENT SEED 2
- H3
histone 3
- K
lysine
- MEA
MEDEA
- me
methyl
- MOM1
MORPHEUS' MOLECULE 1
- PHF1
PHD FINGER PROTEIN 1
- PHD
plant homeodomain
- PRC2
Polycomb Repressive Complex 2
- PCL
POLYCOMBLIKE
- PTM
post-translational modifications
- PCNA
PROLIFERATING CELL NUCLEAR ANTIGEN
- SUZ12
SUPPRESSOR OF ZESTE 12
- SWN
SWINGER
- VRN2
VERNALIZATION2
Works Cited
- 1.Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, Jenuwein T. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–9. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 2.Lachner M, Jenuwein T. The many faces of histone lysine methylation. Curr Opin Cell Biol. 2002;14:286–98. doi: 10.1016/s0955-0674(02)00335-6. [DOI] [PubMed] [Google Scholar]
- 3.Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol. 2007;14:1025–40. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fuchs J, Demidov D, Houben A, Schubert I. Chromosomal histone modification patterns--from conservation to diversity. Trends Plant Sci. 2006;11:199–208. doi: 10.1016/j.tplants.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 5.Jacob Y, Feng S, Leblanc CA, Bernatavichute YV, Stroud H, Cokus S, Johnson LM, Pellegrini M, Jacobsen SE, Michaels SD. ATXR5 and ATXR6 are H3K27 monomethyltransferases required for chromatin structure and gene silencing. Nat Struct Mol Biol. 2009 doi: 10.1038/nsmb.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shen X, Liu Y, Hsu YJ, Fujiwara Y, Kim J, Mao X, Yuan GC, Orkin SH. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell. 2008;32:491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chuikov S, Kurash JK, Wilson JR, Xiao B, Justin N, Ivanov GS, McKinney K, Tempst P, Prives C, Gamblin SJ, Barlev NA, Reinberg D. Regulation of p53 activity through lysine methylation. Nature. 2004;432:353–60. doi: 10.1038/nature03117. [DOI] [PubMed] [Google Scholar]
- 8.Huang J, Perez-Burgos L, Placek BJ, Sengupta R, Richter M, Dorsey JA, Kubicek S, Opravil S, Jenuwein T, Berger SL. Repression of p53 activity by Smyd2-mediated methylation. Nature. 2006;444:629–32. doi: 10.1038/nature05287. [DOI] [PubMed] [Google Scholar]
- 9.Shi X, Kachirskaia I, Yamaguchi H, West LE, Wen H, Wang EW, Dutta S, Appella E, Gozani O. Modulation of p53 function by SET8-mediated methylation at lysine 382. Mol Cell. 2007;27:636–46. doi: 10.1016/j.molcel.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trievel RC, Beach BM, Dirk LM, Houtz RL, Hurley JH. Structure and catalytic mechanism of a SET domain protein methyltransferase. Cell. 2002;111:91–103. doi: 10.1016/s0092-8674(02)01000-0. [DOI] [PubMed] [Google Scholar]
- 11.Springer NM, Napoli CA, Selinger DA, Pandey R, Cone KC, Chandler VL, Kaeppler HF, Kaeppler SM. Comparative analysis of SET domain proteins in maize and Arabidopsis reveals multiple duplications preceding the divergence of monocots and dicots. Plant Physiol. 2003;132:907–25. doi: 10.1104/pp.102.013722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baumbusch LO, Thorstensen T, Krauss V, Fischer A, Naumann K, Assalkhou R, Schulz I, Reuter G, Aalen RB. The Arabidopsis thaliana genome contains at least 29 active genes encoding SET domain proteins that can be assigned to four evolutionarily conserved classes. Nucleic Acids Res. 2001;29:4319–33. doi: 10.1093/nar/29.21.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kohler C, Villar CB. Programming of gene expression by Polycomb group proteins. Trends Cell Biol. 2008;18:236–43. doi: 10.1016/j.tcb.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 14.Lindroth AM, Shultis D, Jasencakova Z, Fuchs J, Johnson L, Schubert D, Patnaik D, Pradhan S, Goodrich J, Schubert I, Jenuwein T, Khorasanizadeh S, Jacobsen SE. Dual histone H3 methylation marks at lysines 9 and 27 required for interaction with CHROMOMETHYLASE3. EMBO J. 2004;23:4286–96. doi: 10.1038/sj.emboj.7600430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manzur KL, Farooq A, Zeng L, Plotnikova O, Koch AW, Sachchidanand, Zhou MM. A dimeric viral SET domain methyltransferase specific to Lys27 of histone H3. Nat Struct Biol. 2003;10:187–96. doi: 10.1038/nsb898. [DOI] [PubMed] [Google Scholar]
- 16.Mujtaba S, Manzur KL, Gurnon JR, Kang M, Van Etten JL, Zhou MM. Epigenetic transcriptional repression of cellular genes by a viral SET protein. Nat Cell Biol. 2008 doi: 10.1038/ncb1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, Carney D, Pena P, Lan F, Kaadige MR, Lacoste N, Cayrou C, Davrazou F, Saha A, Cairns BR, Ayer DE, Kutateladze TG, Shi Y, Cote J, Chua KF, Gozani O. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–9. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, Wu C, Allis CD. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 19.Mathieu O, Probst AV, Paszkowski J. Distinct regulation of histone H3 methylation at lysines 27 and 9 by CpG methylation in Arabidopsis. EMBO J. 2005;24:2783–91. doi: 10.1038/sj.emboj.7600743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcia BA, Hake SB, Diaz RL, Kauer M, Morris SA, Recht J, Shabanowitz J, Mishra N, Strahl BD, Allis CD, Hunt DF. Organismal differences in post-translational modifications in histones H3 and H4. J Biol Chem. 2007;282:7641–55. doi: 10.1074/jbc.M607900200. [DOI] [PubMed] [Google Scholar]
- 21.Ebert A, Schotta G, Lein S, Kubicek S, Krauss V, Jenuwein T, Reuter G. Su(var) genes regulate the balance between euchromatin and heterochromatin in Drosophila. Genes Dev. 2004;18:2973–83. doi: 10.1101/gad.323004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Montgomery ND, Yee D, Chen A, Kalantry S, Chamberlain SJ, Otte AP, Magnuson T. The murine polycomb group protein Eed is required for global histone H3 lysine-27 methylation. Curr Biol. 2005;15:942–7. doi: 10.1016/j.cub.2005.04.051. [DOI] [PubMed] [Google Scholar]
- 23.Pasini D, Bracken AP, Jensen MR, Lazzerini Denchi E, Helin K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 2004;23:4061–71. doi: 10.1038/sj.emboj.7600402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cao R, Wang H, He J, Erdjument-Bromage H, Tempst P, Zhang Y. Role of hPHF1 in H3K27 methylation and Hox gene silencing. Mol Cell Biol. 2008;28:1862–72. doi: 10.1128/MCB.01589-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Margueron R, Li G, Sarma K, Blais A, Zavadil J, Woodcock CL, Dynlacht BD, Reinberg D. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol Cell. 2008;32:503–18. doi: 10.1016/j.molcel.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sarma K, Margueron R, Ivanov A, Pirrotta V, Reinberg D. Ezh2 requires PHF1 to efficiently catalyze H3 lysine 27 trimethylation in vivo. Mol Cell Biol. 2008;28:2718–31. doi: 10.1128/MCB.02017-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peters AH, Kubicek S, Mechtler K, O'Sullivan RJ, Derijck AA, Perez-Burgos L, Kohlmaier A, Opravil S, Tachibana M, Shinkai Y, Martens JH, Jenuwein T. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell. 2003;12:1577–89. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- 28.Vakoc CR, Sachdeva MM, Wang H, Blobel GA. Profile of histone lysine methylation across transcribed mammalian chromatin. Mol Cell Biol. 2006;26:9185–95. doi: 10.1128/MCB.01529-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fuchs J, Jovtchev G, Schubert I. The chromosomal distribution of histone methylation marks in gymnosperms differs from that of angiosperms. Chromosome Res. 2008;16:891–8. doi: 10.1007/s10577-008-1252-4. [DOI] [PubMed] [Google Scholar]
- 30.Shi J, Dawe RK. Partitioning of the maize epigenome by the number of methyl groups on histone H3 lysines 9 and 27. Genetics. 2006;173:1571–83. doi: 10.1534/genetics.106.056853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aligianni S, Varga-Weisz P. Chromatin-remodelling factors and the maintenance of transcriptional states through DNA replication. Biochem Soc Symp. 2006:97–108. doi: 10.1042/bss0730097. [DOI] [PubMed] [Google Scholar]
- 32.Probst AV, Dunleavy E, Almouzni G. Epigenetic inheritance during the cell cycle. Nat Rev Mol Cell Biol. 2009;10:192–206. doi: 10.1038/nrm2640. [DOI] [PubMed] [Google Scholar]
- 33.Esteve PO, Chin HG, Smallwood A, Feehery GR, Gangisetty O, Karpf AR, Carey MF, Pradhan S. Direct interaction between DNMT1 and G9a coordinates DNA and histone methylation during replication. Genes Dev. 2006;20:3089–103. doi: 10.1101/gad.1463706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sarraf SA, Stancheva I. Methyl-CpG binding protein MBD1 couples histone H3 methylation at lysine 9 by SETDB1 to DNA replication and chromatin assembly. Mol Cell. 2004;15:595–605. doi: 10.1016/j.molcel.2004.06.043. [DOI] [PubMed] [Google Scholar]
- 35.Raynaud C, Sozzani R, Glab N, Domenichini S, Perennes C, Cella R, Kondorosi E, Bergounioux C. Two cell-cycle regulated SET-domain proteins interact with proliferating cell nuclear antigen (PCNA) in Arabidopsis. Plant J. 2006;47:395–407. doi: 10.1111/j.1365-313X.2006.02799.x. [DOI] [PubMed] [Google Scholar]
- 36.Sporbert A, Gahl A, Ankerhold R, Leonhardt H, Cardoso MC. DNA polymerase clamp shows little turnover at established replication sites but sequential de novo assembly at adjacent origin clusters. Mol Cell. 2002;10:1355–65. doi: 10.1016/s1097-2765(02)00729-3. [DOI] [PubMed] [Google Scholar]
- 37.Jerzmanowski A. SWI/SNF chromatin remodeling and linker histones in plants. Biochim Biophys Acta. 2007;1769:330–45. doi: 10.1016/j.bbaexp.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 38.Jeddeloh JA, Stokes TL, Richards EJ. Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat Genet. 1999;22:94–7. doi: 10.1038/8803. [DOI] [PubMed] [Google Scholar]
- 39.Amedeo P, Habu Y, Afsar K, Mittelsten Scheid O, Paszkowski J. Disruption of the plant gene MOM releases transcriptional silencing of methylated genes. Nature. 2000;405:203–6. doi: 10.1038/35012108. [DOI] [PubMed] [Google Scholar]
- 40.Probst AV, Fransz PF, Paszkowski J, Mittelsten Scheid O. Two means of transcriptional reactivation within heterochromatin. Plant J. 2003;33:743–9. doi: 10.1046/j.1365-313x.2003.01667.x. [DOI] [PubMed] [Google Scholar]
- 41.Steimer A, Amedeo P, Afsar K, Fransz P, Mittelsten Scheid O, Paszkowski J. Endogenous targets of transcriptional gene silencing in Arabidopsis. Plant Cell. 2000;12:1165–78. doi: 10.1105/tpc.12.7.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vaillant I, Schubert I, Tourmente S, Mathieu O. MOM1 mediates DNA-methylation-independent silencing of repetitive sequences in Arabidopsis. EMBO Rep. 2006;7:1273–8. doi: 10.1038/sj.embor.7400791. [DOI] [PMC free article] [PubMed] [Google Scholar]