We determined the environments of polypeptide chains during membrane translocation and integration using site-directed Cys alkylation. Migration of a signal-anchor sequence into the membrane synchronizes with formation of its TM orientation, and the ER translocon can provide the aqueous pathway capable of two hydrophilic chains.
Abstract
In biogenesis of membrane proteins on the endoplasmic reticulum, a protein-conducting channel called the translocon functions in both the membrane translocation of lumenal domains and the integration of transmembrane segments. Here we analyzed the environments of polypeptide chains during the processes by water-dependent alkylation of N-ethylmaleimide at site-directed Cys residues. Using the technique, the region embedded in the hydrophobic portion of the membrane within a signal-anchor sequence and its shortening by insertion of a Pro residue could be detected. When translocation of the N-terminal domain of the signal-anchor was arrested by trapping an N-terminally fused affinity tag sequence, the signal-anchor was susceptible to alkylation, indicating that its migration into the hydrophobic environment was also arrested. Furthermore, when the tag sequence was separated from the signal-anchor by insertion of a hydrophilic sequence, the signal-anchor became inaccessible to alkylation even in the N-terminally trapped state. This suggests that membrane integration of the signal-anchor synchronizes with partial translocation of its N-terminal domain. Additionally, in an integration intermediate of a membrane protein, both of the two translocation-arrested hydrophilic chains were in an aqueous environment flanking the translocon, suggesting that the translocon provides the hydrophilic pathway capable of at least two translocating chains.
INTRODUCTION
In eukaryotic cells, cotranslational integration of membrane proteins into the endoplasmic reticulum (ER) membrane is mediated by a protein-conducting channel formed by the Sec61 complex called the translocon (Johnson and van Waes, 1999; Rapoport, 2007). Findings from electrophysiological (Simon and Blobel, 1991) and site-specific fluorescent labeling (Crowley et al., 1994) studies indicate that the translocon forms an aqueous pore. In addition, studies using site-specific cross-linking techniques demonstrated that Sec61α, a central component of the Sec61 complex, is transiently cross-linked not only with polypeptide chains translocating across the membrane but also with transmembrane (TM) segments of membrane proteins or signal sequences (Mothes et al., 1994, 1997; Martoglio et al., 1995; Do et al., 1996; Heinrich et al., 2000). Evidence suggests that the translocon provides an environment not only for the translocation of secretory proteins, lumenal soluble proteins, and lumenal domains of membrane proteins across the ER membrane, but also for the lateral delivery of hydrophobic sequences into the lipid phase. The Sec61 complex is formed by a heterotrimer that consists of three components: Sec61α, Sec61β, and Sec61γ. Based on studies of crystal structures of archaeal (Van den Berg et al., 2004) and bacterial (Tsukazaki et al., 2008) SecY complexes, which are homologues of the eukaryotic Sec61 complex, one SecY molecule forms an hourglass-like shape across the membrane with a slight opening that faces toward the lipid environment, suggesting that a single SecY complex may be sufficient for both membrane translocation and integration. On the other hand, Sec61 and SecY heterotrimers are observed in oligomeric states by cryo-electron microscopic imaging (Hanein et al., 1996; Beckmann et al., 1997; Mitra et al., 2005) and by biochemical and genetic analyses (Mori et al., 2003; Schaletzky and Rapoport, 2006; Osborne and Rapoport, 2007). Moreover, during integration of multispanning membrane proteins, multiple TM segments or hydrophilic chains can interact simultaneously with the Sec61α subunit (Heinrich and Rapoport, 2003; Sadlish et al., 2005; Kida et al., 2007). These data indicate the possibility that the Sec61 complex also functions as an oligomer. Several ER membrane proteins, such as translocating chain–associated membrane protein or translocon-associated protein complex, also serve with or near the Sec61 complex (Osborne et al., 2005), but their precise functions are not known.
Membrane protein biogenesis begins with the exposure of a hydrophobic signal sequence from a ribosome synthesizing a membrane protein. The signal sequence is recognized by a signal recognition particle and the ribosome with a nascent membrane protein is targeted to the ER. The signal recognition particle is recognized by its receptor on the ER and the signal sequence is transferred to the translocon together with the ribosome. At the translocon, signal sequences form their own TM orientations. TM orientation of signal sequences is influenced mainly by the difference in the number of positively charged residues between both flanking regions and formed by translocation of the side containing the fewest positive charges (Sakaguchi, 1997; Goder and Spiess, 2001). Among the uncleavable signal sequences (the so-called signal-anchor sequences), type I signal-anchor (SA-I) sequences translocate their N-terminal portions and settle in the Nlum/Ccyt orientation (termed the type I orientation), and type II signal-anchor (SA-II) sequences form the opposite Ncyt/Clum orientation, like cleavable signal sequences. Signal sequences are laterally transferred to the lipid environment, and then the following TM segments are sorted into the lipid phase when each TM segment is synthesized and exposed to the translocon. Studies of membrane integration of systematically constructed TM sequences suggest that a direct interaction between TM sequences and the lipid phase may be involved in their recognition at the translocon (Hessa et al., 2005, 2007). Site-specific cross-linking experiments revealed that some signal sequences and TM segments migrate directly from the translocon into the lipid environment (Mothes et al., 1997; Heinrich et al., 2000). Depending on the sequence, however, TM segments are indirectly transferred to the lipid phase via translocating chain–associated membrane protein (Do et al., 1996; Heinrich et al., 2000) or PAT-10 (10 kDa of a protein associated with the ER translocon; Meacock et al., 2002) or are continuously cross-linked with the Sec61α even after the following TM segments enter the translocon (Heinrich and Rapoport, 2003; Sadlish et al., 2005), suggesting that the integration of TM segments is coordinated not only by a single Sec61 complex, but also by its oligomerizaton and/or additional components.
SA-I sequences are integrated into the membrane while translocating their N-terminal domain (N-domain) just after they emerge from the ribosome (Heinrich et al., 2000; Kida et al., 2000). This translocation can be regulated by fusing the dihydrofolate reductase or the streptavidin-binding peptide tag (SBP-tag) sequence to the N-terminus of the N-domain and using their ligands, methotrexate and streptavidin (SAv), respectively (Kida et al., 2005, 2007, 2009). We therefore focused on N-domain translocation and found that the ribosome has a critical role in translocation, whereas ATP, GTP, and a lumenal Hsp70 BiP are not required (Kida et al., 2005). We also fused the 27th immunoglobulin-like domain (I27 domain) of titin protein to the N-domain to evaluate the motive force for the translocation coupling with unfolding of the I27 domain (Kida et al., 2009). Translocations of passenger domains were diminished not only by the insertion of a Pro residue into the SA-I sequence, but also by separating each domain from the SA-I sequence through the insertion of a spacer sequence. We thus concluded that the SA-I sequence itself serves to generate motive force for the N-domain translocation, even though it affects only a limited upstream portion. Moreover, these data led us to hypothesize that motive force is generated when the SA-I sequence associates with and/or migrates into the lipid environment.
To investigate this hypothesis, we assessed the environment (water-accessible or water-free) of the SA-I sequence during its N-domain translocation. In previous studies (Kimura-Someya et al., 1998; Kuwabara et al., 2004; Koide et al., 2007), Cys-scanning mutants of membrane proteins were constructed and the local environment surrounding each Cys position was analyzed based on the reactivity with maleimide reagents because a water molecule is required to function as a proton acceptor for alkylation of the SH groups with maleimide reagents. By adopting the technique, we determined the actual region embedded in the hydrophobic interior of the membrane within an SA-I sequence and also observed that the region was shortened by inserting a Pro residue. When the N-domain was stalled in the cytosol by trapping the N-terminally fused SBP-tag, the SA-I sequence was susceptible to alkylation, indicating that its migration into the hydrophobic environment was also arrested. When the SA-I sequence was separated from the SBP-tag by insertion of a spacer sequence, it became inaccessible to alkylation even in the N-terminally trapped state. This suggests that membrane integration of the SA-I sequence synchronizes with partial translocation of its N-terminally flanking portion. These findings support the hypothesis that motive force is generated by an interaction between the signal-anchor sequence and lipid. Furthermore, fusion of an SA-II sequence following the SBP-tagged N-domain and SA-I sequence forms a complex integration intermediate of a multispanning membrane protein when both N-terminal and C-terminal translocation are arrested (Kida et al., 2007). Our site-directed alkylation analysis indicated that two translocation-arrested hydrophilic chains of the intermediate remained in the aqueous environment whereas two signal-anchor sequences were released into the water-free environment. Also, the two hydrophilic sequences and one signal-anchor sequence were cross-linked with the Sec61α. We thus suggest that the Sec61-based translocon provides the hydrophilic pathway capable of at least two translocating chains.
MATERIALS AND METHODS
Constructs
The plasmid expressing SBP-Syt200 protein, consisting of the SBP-tag (MDEKTTGWRGGHVVEGLAGELEQLRARLEHHPQGQREP), a glycosylation probe sequence (KLNSTAT), and Arg2-Arg200 of mouse synaptotagmin II (SytII; an AflII site was generated just downstream of Arg200 for synthesis of truncated mRNAs), as described previously (Kida et al., 2007). Substitution of all seven Cys residues within SBP-Syt200 by Ala (as shown in Figure 1) and introduction of a Cys residue into each position in the SA-I sequence were performed by site-directed mutagenesis using either Kunkel's method (Kunkel, 1985) or the QuickChange procedure (Stratagene, La Jolla, CA). A Pro residue was also inserted by site-directed mutagenesis as shown in Figure 4. To lengthen the linker between the SBP-tag and SA-I sequence, PCR fragments encoding 10 residues (T627YTQKLSV634EF), 20 residues (T627YTQKLSVPDGFKVSNSA644EF), 30 residues (T627YTQKLSVPDGFKVSNSAARGWVIHPLG654EF), and 38 residues (T627YTQKLSVPDGFKVSNSAARGWVIHPLGLRSEFPIW662EF; the mutated point for silencing glycosylation and two residues derived from a restriction enzyme site are underlined and boldface, respectively) from human anion exchanger 1 were inserted between the glycosylation probe sequence and SytII.
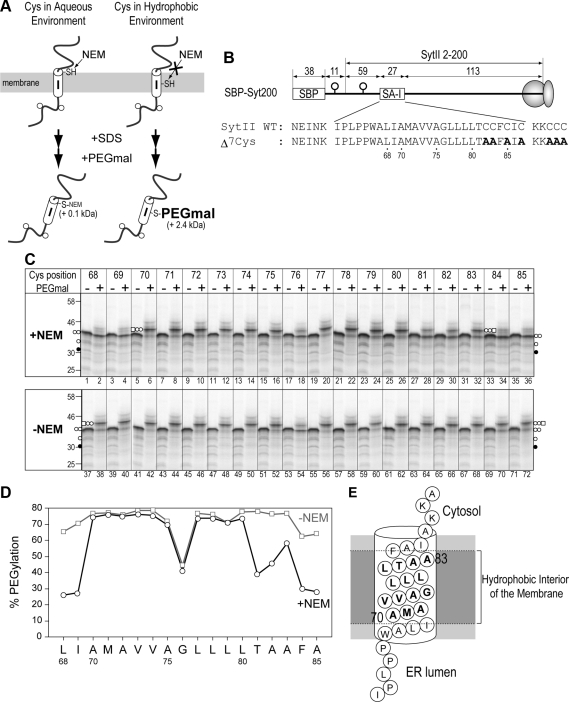
Figure 1.
Membrane-embedded region within a signal-anchor sequence can be determined by site-directed Cys alkylation. (A) Scheme of two-step Cys alkylation with N-ethylmaleimide (NEM) and (methyl-PEG12)3-PEG4-maleimide (PEGmal). A Cys residue in an aqueous environment is reactive with NEM, which blocks the subsequent PEGmal modification. A Cys residue in a hydrophobic environment is not reactive with NEM so that the PEGmal modification can occur after solubilization with SDS. PEGmal-conjugated molecules are identified by their decreased mobility on SDS-PAGE gels. (B) The SBP-tag and N-glycosylation sequence were fused to the N-terminus of SytII. Potential N-glycosylation sites are indicated by the open circles. mRNAs truncated at Arg200 of SytII were used for cell-free translation. Numbers indicate the amino acid residues within the indicated regions. All seven Cys residues within the SBP-Syt200 protein were substituted with Ala residues (bold letters). Each residue between the Leu68 and Ala85 of the Δ7Cys mutant was exchanged for a Cys residue. (C) Single-Cys derivatives of SBP-Syt200 were synthesized in the presence of rough microsomal membranes (RM) and then applied to the two-step alkylation. In the second step, the reaction was performed in the presence of PEGmal (+) or its solvent only (−). Nonglycosylated (•), monoglycosylated (○), diglycosylated (○ ○), and diglycosylated and PEGmal-conjugated (○ ○ and □) forms are observed. (D) The PEGmal conjugation efficiency (% PEGylation) of the diglycosylated forms at each position in C was quantified. (E) The region roughly between Ala70 and Ala83 within the SA-I sequence is highly likely to be in the hydrophobic interior of the membrane.
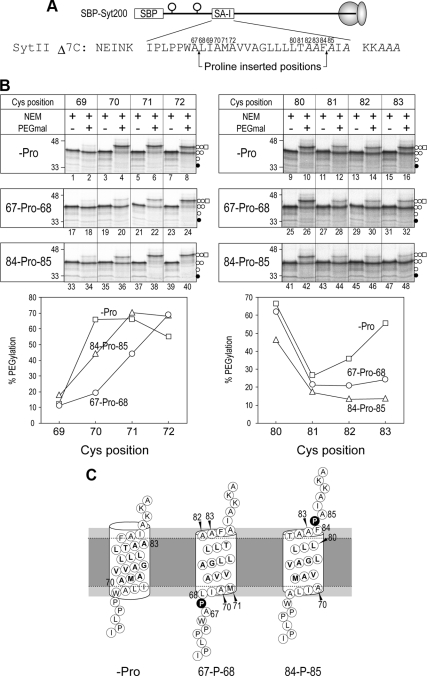
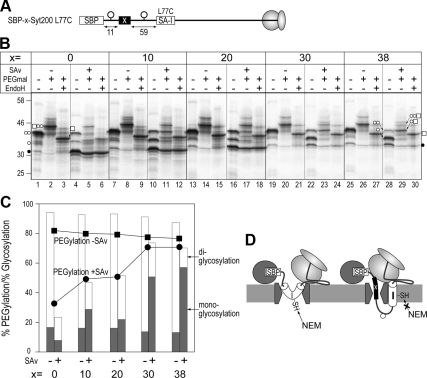
Figure 4.
Insertion of a Pro residue into the SA-I sequence shortens the membrane-embedded region. (A) A Pro residue was inserted between positions 67 and 68 or positions 84 and 85 within the SA-I sequence, and then the regions from position 69–72 and from position 80–83 were scanned with a Cys residue. (B) After synthesis in the presence of RM, the molecules were applied to two-step Cys alkylation as described in Figure 1. PEGylation efficiency of the diglycosylated forms of each protein (−Pro, □; 67-Pro68, ○; 84-Pro85, ▵) was quantified. (C) When a Pro residue was inserted between positions 67 and 68, positions 70 and 83 were exposed to the aqueous environment and the location of positions 71 and 82 were a little affected. The insertion of a Pro residue between positions 84 and 85 also exposed positions 70 and 83 to an aqueous environment and the environment of position 80 was significantly changed.
The plasmid expressing S-38-I-II fusion protein, consisting of the SBP-tag, a glycosylation probe sequence, SytII (Arg2-Thr160), TM3 of human Na+/H+ exchanger isoform 6 (R97FLHETGLAMIYGLLVGLVLRYGIHVPSDVNNV129), another glycosylation probe sequence, and bovine prolactin (Thr31-Cys229), was also previously described (Kida et al., 2007). Disruption of three glycosylation sites of S-38-I-II by exchange of Thr in glycosylation consensus sequences with Ala, substitution of all 10 Cys residues between Met1 and His368 (just upstream of BspHI site) of S-38-I-II to Ala, and introduction of 1 Cys residue into the positions indicated in Figure 5 was performed by site-directed mutagenesis.
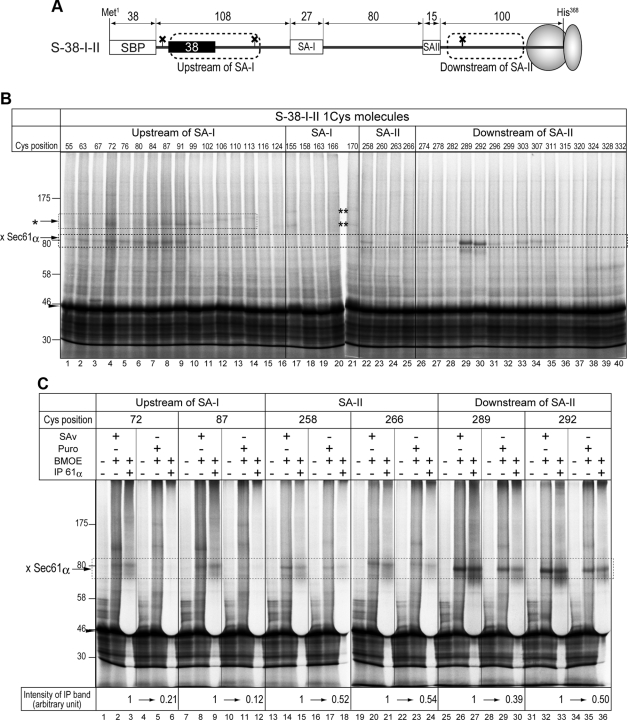
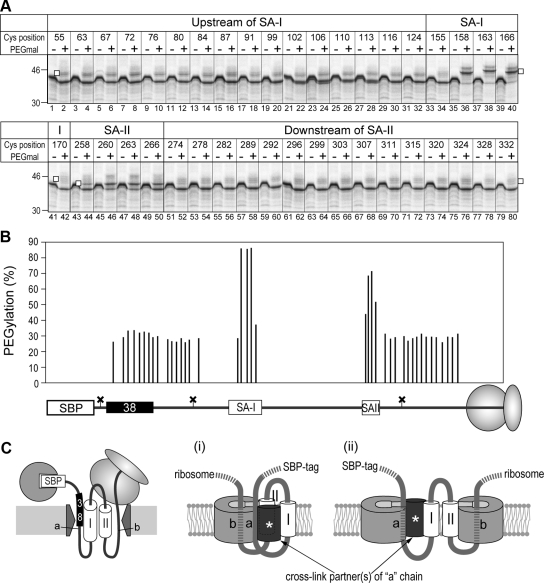
Figure 5.
Two translocation-arrested chains and one signal-anchor sequence are simultaneously cross-linked with Sec61α. (A) A fusion protein consisting of the SBP-tag, a 38-residue spacer sequence, and an SA-I sequence, as shown in Figure 3, was further followed by the SA-II and hydrophilic sequences to construct the S-38-I-II protein. To simplify band patterns on SDS-PAGE gels, three glycosylation sites of the original S-38-I-II were silenced (shown as crosses). Four regions, upstream of the SA-I sequence, within the SA-I and SA-II sequences, and downstream of the SA-II sequence, of the Cys-less mutant of S-38-I-II were scanned with a Cys residue. (B) Truncated mRNAs of single-Cys derivatives of S-38-I-II were translated in the presence of RM and streptavidin (SAv), and then the products were treated with a bifunctional cross-linker, bis-maleimidoethane (BMOE). Numbers at the top of the lanes indicate Cys positions from the N-terminus. The map of the positions is illustrated in Figure 6B. The former hydrophilic chain was also cross-linked with another molecule(s) (asterisk). In addition, two cross-linked products between the SA-I sequence and proteins but Sec61α was detected (double-asterisk). (C) Molecules with a Cys residue at the indicated positions were synthesized in the presence of SAv to form the intermediate state of the translocation of two hydrophilic chains (SAv +) or synthesized in the absence of SAv and the translation was terminated with puromycin (Puro) to translocate both chains (SAv − and Puro +). After cross-linking with BMOE, aliquots were immunoprecipitated with anti-Sec61α antiserum. Immunoprecipitated forms were quantified and the reduction rates caused by releasing both chains from translocation arrest were calculated (bottom of the panel).
Sequences of all constructs were confirmed with DNA sequencing. Information for all of the oligo DNAs used in this study are available from the authors.
In Vitro Transcription and Translation
For synthesis of truncated mRNAs coding SBP-Syt200 and its derivatives, plasmids were linearized by AflII at Arg200 of SytII. For truncated mRNAs coding S-38-I-II and its derivatives, plasmids were linearized by BspHI at His368 of S-38-I-II. Template DNAs were transcribed with T7 RNA polymerase (Takara, Tokyo, Japan) as previously described (Sakaguchi et al., 1992). mRNAs were translated in a reticulocyte lysate cell-free system for 1 h at 25°C in the presence of rough microsomal membrane (RM). Preparation of RM (Walter and Blobel, 1983) and rabbit reticulocyte lysate (Jackson and Hunt, 1983) was performed as previously described. RM was extracted with EDTA and treated with Staphylococcus aureus nuclease (Roche Chemical, Indianapolis, IN) as described previously (Walter and Blobel, 1983). The translation reaction contained 100 mM potassium acetate (KOAc), 1.0 mM magnesium acetate (Mg(OAc)2), 32% reticulocyte lysate, and 20 kBq/μl EXPRESS protein-labeling mix (PerkinElmer, Norwalk, CT). Where indicated, 1 mg/ml SAv (Wako, Osaka, Japan) was included in the translation reaction. For the translocation chase in the presence of biotin (Sigma-Aldrich, Tokyo, Japan), translation was terminated by incubation in the presence of 2 mM cycloheximide (Sigma-Aldrich) for 10 min at 25°C and then further incubated at 25°C for 1 h in the presence of 0.2–0.4 mM biotin.
Two-Step Cys Alkylation of One-Cys Molecules
For modification with N-ethylmaleimide (NEM) after translation was terminated with cycloheximide, the mixtures were incubated in the absence or presence of 10 mM NEM (Wako) at 5°C for 1 h. NEM treatment was quenched by addition of a 10-fold volume of physiological salt buffer (30 mM HEPES, pH 7.4, 150 mM KOAc, and 2.5 mM Mg(OAc)2) containing 2 mM dithiothreitol (DTT) and further incubation for 10 min. To detect whether each Cys residue was blocked with NEM, membrane fractions were sedimented by centrifugation at 100,000 × g for 5 min at 4°C, solubilized with lysis buffer (50 mM Tris/HCl, pH 8.5, 1% SDS, 2 mM Tris[2-carboxyethyl]phosphine hydrochloride), and incubated in the absence or presence of 8 mM (methyl-PEG12)3-PEG4-maleimide (PEGmal; Thermo Scientific, Waltham, MA). After PEGmal modification, aliquots were incubated with 20 mM DTT and then treated with endoglycosidase H (EndoH; New England Biolabs, Ipswich, MA) at 37°C for 1 h, in accordance with the manufacturer's instructions. All the alkylation experiments were carried out at least twice and the results were reproducible.
Chemical Cross-Linking and Immunoprecipitation
For chemical cross-linking, single Cys derivatives of S-38-I-II were synthesized in the presence of RM and in the presence or absence of SAv. Translation in the presence of SAv was terminated by 10-min incubation with cycloheximide. The translation mixtures in the absence of SAv were supplemented with 2 mM puromycin (Sigma-Aldrich) and incubated at 25°C for 60 min to allow for translocation of the downstream portion of the SA-II sequence. These mixtures were then treated with 6–10 mM bis-maleimidoethane (BMOE; Thermo Scientific) or its solvent DMSO (where indicated as a minus-cross-linker) on ice for 60 min. Cross-linking reactions were quenched by dilution with a 10-fold volume of high-salt buffer (30 mM HEPES. pH 7.4, 500 mM KOAc, 2.5 mM Mg(OAc)2) containing 3 mM DTT and incubated for 15 min on ice. For immunoprecipitation, membrane fractions were isolated by centrifugation at 100,000 × g for 10 min, solubilized with 1% SDS for 5 min at 95°C, and then diluted with a 20-fold volume of immunoprecipitation buffer (1% Triton X-100, 50 mM Tris/HCl, pH 7.5, 150 mM NaCl). The solutions were incubated for 30 min with protein A-Sepharose (GE Healthcare, Waukesha, WI) alone to remove materials nonspecifically bound to the resin. The unbound fractions were incubated for 2 h with anti-Sec61α antiserum and for another 2 h with protein A-Sepharose. The resin was washed twice with 0.5 ml immunoprecipitation buffer and then extracted with sample buffer for SDS-PAGE.
SDS-PAGE and Image Analysis
Radiolabeled proteins were analyzed by SDS-PAGE using 7–12% slab gels and visualized on a Bioimage analyzer BAS-1800 (Fuji Film, Tokyo, Japan). Quantification was performed using Image Gauge software (v4.0; Fuji Film).
RESULTS
Environment of the SA-I Sequence in the Intermediate of N-domain Translocation
To determine the environment of the SA-I sequence during its N-domain translocation, we first examined the actual region embedded in the hydrophobic interior of the membrane within an SA-I sequence. Each amino acid residue from position 68–85 of the Cys-less mutant of SBP-Syt200 protein, which consists of the SBP-tag and N-terminal 200 residues of SytII (Syt200; a model protein possessing an SA-I sequence), was sequentially replaced with a Cys residue (Figure 1). mRNAs truncated at Arg200 of SytII were synthesized in vitro and translated in a cell-free system in the presence of RM. Translation was completed with puromycin, which releases a nascent polypeptide from the ribosome, and the mixtures were treated with 10 mM NEM, a membrane-permeable maleimide reagent, at 5°C for 1 h. In this step, the SH groups of Cys residues in an aqueous environment were alkylated with NEM. After quenching NEM with DTT, membrane fractions were isolated by centrifugation, solubilized with SDS, and then treated with 8 mM PEGmal to modify the Cys residues that were not blocked with NEM. Because the molecular mass of PEGmal is ∼2.4 kDa, in contrast to the ∼0.1-kDa molecular weight of NEM, Cys residues located in the hydrophobic environment are modified with PEGmal and the conjugated molecule is detectable as a band with lower mobility on SDS-PAGE.
After synthesis in the presence of RM, all single-Cys derivatives of SBP-Syt200 were glycosylated at both N-glycosylation sites in the N-domain (Figure 1C, PEGmal (−) lanes), indicating that these mutations in the SA-I sequence did not affect membrane orientation. The membrane-integrated proteins were applied to sequential Cys modification with NEM followed by PEGmal. When the first NEM treatment was skipped, diglycosylated molecules of all single-Cys proteins except one at position 76 displayed slower mobility on SDS-PAGE, and therefore PEGmal was efficiently reactive with almost all Cys positions examined here in the absence of NEM treatment (Figure 1C, bottom panel, PEGmal + lanes). Because position 76 shows low reactivity with PEGmal irrespective of NEM treatment, we did not take this position into account. Treatment with NEM, however, remarkably diminished the PEGmal modification efficiency at positions 68, 69, 84, and 85 (Figure 1C, +NEM, lanes 2, 4, 34, and 36), indicating that these positions were located in an aqueous environment. PEGmal modification at most of the positions between 70 and 83 were affected little by prior NEM treatment (Figure 1C, lanes 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, and 32), in contrast to positions 81 and 82 (Figure 1C, lanes 28, and 30). It is possible that these two positions are partially exposed to the aqueous environment by inclination of the SA-I sequence across the membrane or partial perturbation of the cytosolic leaflet of the lipid bilayer. These results indicate that the region between positions 70 and 83 within the SA-I sequence is embedded in the hydrophobic interior of the membrane.
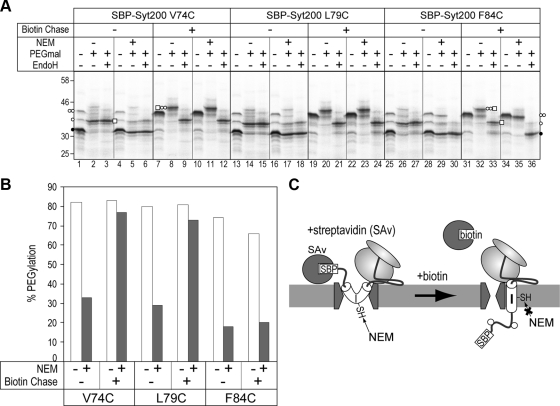
We then examined the environment of the SA-I sequence in the intermediate of the N-domain translocation. N-domain translocation of SBP-Syt200 is arrested in the presence of SAv and restored by the addition of biotin. We performed two-step Cys modification in the translocation-arrested state or after translocation resumed (Figure 2, A and B). To quantify PEGmal-modification efficiency, EndoH treatment was performed after the PEGmal reaction, because the mobility shift caused by PEGmal conjugation is similar to that produced by N-glycosylation and the band patterns are rather complex. According to Figure 1, positions 74 and 79 are in the interior and position 84 is in the exterior of the membrane-embedded region within the SA-I sequence. In the arrested state with SAv, mainly nonglycosylated molecules were detected (Figure 2A, lanes 1, 4, 13, 16, 25, and 28), and they were diglycosylated after incubation in the presence of biotin (Figure 2A, lanes 7, 10, 19, 22, 31, and 34). In the absence of NEM treatment, both translocated and translocation-arrested molecules of three single-Cys proteins were efficiently (∼80%) modified with PEGmal (Figure 2A, lanes 2, 3, 8, 9, 14, 15, 20, 21, 26, and 27, and Figure 2B), so the glycosylation states of the N-domain did not affect the PEGmal modification. By resuming translocation with biotin, positions 74 and 79, but not 84, were efficiently (>70%) modified with PEGmal after two-step Cys alkylation (Figure 2A, lanes 11, 12, 23, 24, 35, and 36, and Figure 2B), suggesting that the SA-I sequence was normally integrated into the membrane. In the translocation-arrested state, however, PEGmal-modification efficiency at positions 74 and 79 was rather low (∼30%) because NEM blocked the modification (Figure 2A, lanes 5, 6, 17, and 18, and 2B). These results indicate that the SA-I sequence in the translocation-arrested state remains in an aqueous environment, and that it migrates into the hydrophobic environment, presumably the lipid phase, after the resumption of the translocation. In addition, the membrane-embedded region of the SA-I sequence and the migration into the hydrophobic environment could be quantitatively observed using this method. Thus, this method was validated for assessing the local environment of membrane proteins.
Figure 2.
SA-I sequence migration into the hydrophobic environment is coupled with N-domain translocation. (A) Three single-Cys derivatives (positions 74, 79, and 84) of SBP-Syt200 were synthesized in the presence of streptavidin (SAv) to arrest the N-domain translocation and then treated with biotin (biotin chase) to release the translocation arrest. Before (−) or after (+) biotin chase, products were applied to two-step Cys alkylation. Each reaction was performed in the absence (−) or presence (+) of each maleimide reagent. To quantify PEGylation efficiency, aliquots following PEGmal reaction were treated with endoglycosidase H (EndoH). ○ ○ and □, glycosylation and PEGylation, respectively. (B) PEGylation efficiencies in the EndoH + lanes of Figure 2A were quantified. (C) When the N-domain translocation was arrested, Cys residues in the central part of the SA-I sequence were reactive to NEM. In the intermediate state, the SA-I sequence of SBP-Syt200 is in an aqueous environment.
Membrane Integration of the SA-I Sequence Synchronizes with the Formation of Its TM Orientation and Partial Translocation of the N-domain
As we previously reported, when the SA-I sequence was separated from the SBP-tag by the insertion of a hydrophilic sequence, a portion in the N-domain just flanking the SA-I sequence partially reached the luminal space in the N-terminally trapped state (Kida et al., 2007). Therefore, we next analyzed the SA-I sequence in the N-terminally trapped state when the linker sequence was elongated. To this end, the SA-I sequence was stepwise separated from the SBP-tag by the insertion of successively longer hydrophilic sequences derived from a lumenal loop region of human anion exchanger 1 (Figure 3). When synthesized in the absence of SAv, all proteins shown in Figure 3 were efficiently diglycosylated (Figure 3B, lanes 1, 7, 13, 19, and 25) and each position 77 was conjugated with PEGmal even after NEM treatment (Figure 3B, lanes 2, 8, 14, 20, and 26), indicating that spacer sequences alone did not affect membrane translocation and integration. On the other hand, when synthesized in the presence of SAv, glycosylation of the molecule without a spacer was inhibited (Figure 3B, lane 4) and position 77 was hardly reactive with PEGmal because of masking by NEM (Figure 3B, lanes 5 and 6), similar to positions 74 and 79 in Figure 2. When molecules containing spacer sequences were synthesized in the presence of SAv, the signals of the monoglycosylated forms became stronger as the spacer sequence was elongated, and the monoglycosylation efficiencies of molecules containing 30- and 38-residue spacers increased to 50 and 57%, respectively (Figure 3B, lanes 10, 16, 22, and 28, and 3C, filled bars). After two-step Cys modification, not only diglycosylated but also monoglycosylated forms of molecules with spacer sequences were efficiently modified with PEGmal, whereas nonglycosylated bands showed little change (Figure 3B, lanes 11, 17, 23, and 29). Moreover, the sum of the monoglycosylation and diglycosylation modification efficiency rates was almost identical to that achieved with PEGmal (Figure 3C). These results indicate that the SA-I sequence migrated into the hydrophobic environment, even when its upstream domain was still translocating in the translocon and was partially exposed to the ER lumen. Thus, it is highly likely that membrane integration of the SA-I sequence synchronizes with the formation of its own TM orientation. According to our previous data, the translocation motive force affecting the N-terminally fused SBP-tag or I27 domain was decreased by increasing the spacing between the SA-I and each sequence, and we thus concluded that the SA-I sequence directly drives N-domain translocation within a limited upstream portion (Kida et al., 2009). Together, the previous and present data suggest that the SA-I sequence provides the motive force for the translocation simultaneously with the formation of TM orientation and migration into the membrane.
Figure 3.
Migration of the SA-I sequence into the hydrophobic environment synchronizes with partial translocation of the N-domain. (A) The SA-I sequence was separated from the SBP-tag by insertion of a hydrophilic sequence of the lengths indicated by the “x =” row in B. Leu77 within the membrane-embedded region was exchanged for a Cys residue. (B) Molecules with spacer sequences (0, 10, 20, 30, or 38 residues) were synthesized in the absence (−) or presence (+) of streptavidin (SAv) and applied to two-step Cys alkylation and endoglycosidase H (EndoH) treatment, as in the above figures. (C) Monoglycosylated and diglycosylated efficiencies (filled and open columns, respectively) in the cases with (+) or without (−) SAv were quantified from PEGmal (−) lanes in B. PEGylation efficiencies in the presence (•) or absence (■) of SAv were also quantified from the EndoH + lanes. (D) As the spacer (x) was elongated, the N-domain partially reached the lumenal space even when the N-terminal SBP-tag was trapped in the cytosol. In this situation, the SA-I sequence formed the TM orientation and migrated into the hydrophobic interior of the membrane.
Pro insertion into the SA-I Sequence Shortens the Membrane-embedded Region
The insertion of one Pro residue into the SA-I sequence diminishes the translocation of N-terminally fused passenger sequences, the SBP-tag, and the I27 domain (Kida et al., 2009). To further explore the role of the SA-I sequence in generating the motive force for translocation, we examined the intramembrane environments of the Pro containing SA-I sequences. A Pro residue was inserted between either positions 67 and 68 or positions 84 and 85, and the environments at the lumenal and cytosolic boundary positions of the SA-I sequence were examined by site-directed Cys modification. All proteins mutated in the SA-I sequence (Figure 4A) were correctly inserted into the membrane and diglycosylated when synthesized in the presence of RM (Figure 4B, PEGmal lanes). When a Pro residue was not introduced, positions 70 and 83 were the lumenal and cytosolic ends embedded in the membrane, respectively (Figures 1 and 4B, lanes 4 and 16). When a Pro residue was inserted between positions 67 and 68, PEGmal modification at positions 70 and 71 was decreased (Figure 4B, lanes 20 and 22), suggesting that these positions were exposed to the aqueous environment by the Pro residue insertion. In addition, reactivity at position 83 was influenced by the same Pro insertion (Figure 4B, lane 32). In the case of Pro insertion between positions 84 and 85, NEM reactivity at positions 83 and 70 also increased (Figure 4B, lanes 36 and 48), indicating that the Pro residue affected water accessibility of not only the close end but also the distal end of the membrane-embedded region. These findings suggest that the membrane-embedded region within the SA-I sequence was shortened by the insertion of a Pro residue. Based on the results presented in Figures 3 and 4 and previous results, the conformational change of the SA-I sequence caused by Pro insertion probably influences the membrane-integration process.
Translocation-arrested N-domain in Aqueous Site Flanking the Sec61α Does Not Affect Integration of the Signal-Anchor Sequence That Follows
When an SA-II sequence is fused following the SA-I sequence of SBP-38-Syt protein (x = 38 in Figure 3A), it is integrated into the membrane with translocation of its downstream chain, irrespective of the translocation-arrested N-domain (Kida et al., 2007). When the N-terminal SBP-tag of the S-38-I-II chimera protein is trapped by SAv and the C-terminus is also trapped in a ribosome by translating the truncated mRNA, the two translocation-arrested hydrophilic chains penetrating the membrane flank Sec61α. Therefore, we proposed that one translocon could accommodate at least two translocating hydrophilic chains (Kida et al., 2007). In the present study, we examined the environment surrounding each of four membrane-penetrating chains by the site-directed Cys alkylation. A Cys residue was scanned within four regions (as shown in Figure 5A) of the S-38-I-II protein. Three glycosylation sites in the original S-38-I-II protein were silenced to more easily detect the PEGmal-modified forms. Before the two-step Cys modification analysis, single-Cys derivatives were applied to chemical cross-linking using the bifunctional cross-linker, BMOE. After synthesis in the presence of RM and SAv, BMOE was added and the cross-linking reaction was performed. S-38-I-II proteins were detected as ∼45-kDa bands on an SDS-PAGE gel, and cross-linked products of ∼80 kDa were detected on the molecules possessing a single Cys within the region upstream of the SA-I sequence, the SA-II sequence, and the region downstream of the SA-II sequence (Figure 5B). Bands of ∼80 kDa were immunoprecipitated with anti-Sec61α antiserum (Figure 5C, lanes 3, 9, 15, 21, 27, and 33), and therefore these bands contained cross-linked products between S-38-I-II and Sec61α proteins. Cross-linking with the region upstream of the SA-I sequence completely disappeared in the absence of SAv (Figure 5C, lanes 6 and 12), and cross-linking with the SA-II sequence and its downstream portion decreased after the C-terminus was released from the ribosome by puromycin (Figure 5C, lanes 18, 24, 30, and 36), indicating that these regions are close to the Sec61 channel in each translocation-arrested situation. Furthermore, the upstream hydrophilic chain was cross-linked with another protein(s), and cross-linked products of ∼110 kDa were detected (Figure 5B). The band seems to shift gradually to ∼120 kDa from position 99–113 (Figure 5B, lanes 10–14). Although the cross-link partner(s) was not identified, this suggests that the former and latter hydrophilic chains were located in or near Sec61α but under somewhat different situations. Also, the SA-I sequence was cross-linked at positions 155 and 170, and two cross-linked products were detected. The SA-I sequence may be located near other factors after it leaves the Sec61 channel.
The same proteins were then analyzed by two-step Cys modification (Figure 6). After sequential treatment with NEM followed by PEGmal, all positions in regions upstream of the SA-I sequence and downstream of the SA-II sequence were hardly modified with PEGmal (Figure 6B, lanes 1–32 and 51–80), whereas the central positions of the SA-I and SA-II sequences were highly modified with PEGmal (lanes 36, 38, 40, 46, and 48). Thus, the two translocation-arrested chains were located in an aqueous environment and reactive to NEM, although the SA-I and SA-II sequences migrated into the hydrophobic environment even at such an intermediate state. Unmodified molecules showed a little higher mobility upon PEGmal treatment. Some contaminations within the PEGmal reagent may affect the mobility on SDS-PAGE. These results suggest the flexible nature of the translocon: 1) The translocon forms an aqueous environment that can accommodate two hydrophilic chains, and 2) the preceding hydrophilic chain penetrating via an aqueous pathway flanking the Sec61α does not inhibit integration and translocation of the following chains.
Figure 6.
Two translocation-arrested chains are located in an aqueous environment flanking the Sec61α. (A) Single-Cys derivatives of S-38-I-II were synthesized in the presence of RM and SAv and applied to two-step Cys alkylation. PEGylated forms are indicated with open squares. (B) PEGylation efficiencies in the panel were quantified. Each value was placed in each position on the map. (C) Distribution models of four membrane-penetrating chains and prospective cross-link partner(s) with one (i) or two (ii) Sec61 channels. In the integration intermediate of the S-38-I-II protein, two hydrophilic chains (a and b) and the SA-II sequence (II) flank the Sec61 channel. Although two hydrophilic chains are located in the aqueous environment, only the former “a” chain is cross-linked with another cross-link partner(s) and thus these two chains may be in different situations.
DISCUSSION
In the present study, we determined the environments of the SA-I sequence and polypeptide chains during membrane translocation and integration using site-directed Cys alkylation. The findings indicated, first, that ∼14 residues in the central region of the SA-I sequence comprise the actual region embedded in the hydrophobic portion of the membrane. Second, the SA-I sequence of the SBP-Syt200 molecule remains in the aqueous environment in the intermediate state of the N-domain translocation and migrates into the hydrophobic environment immediately after the translocation resumes. Third, separation of the SA-I sequence from the SBP-tag releases the SA-I sequence into the hydrophobic environment irrespective of trapping the N-terminal tag in the cytosol. Fourth, insertion of a Pro residue into the SA-I sequence shortens its membrane-embedded region. Fifth, in the integration intermediate of the S-38-I-II chimera protein, two translocation-arrested hydrophilic chains remain in the aqueous environment flanking Sec61α, whereas two signal-anchor sequences have migrated into the water-free environment. We here suggest that migration of the SA-I sequence into the membrane synchronizes with the formation of its own TM orientation, and it is likely that this environmental transition supplies the motive force for N-domain translocation. We also suggest that the translocon can provide the aqueous pathway capable of at least two translocating chains.
Membrane Integration of the Signal-Anchor Sequence Coupled with Translocation of its Lumenal Domain
In the present study, the SA-I sequence remained in the aqueous environment when the N-terminally fused SBP-tag was trapped in the cytosol, and it migrated into the hydrophobic environment by separating between the SBP-tag and the SA-I sequence. Because N-terminally fused passenger sequences closer to the SA-I sequence are translocated more efficiently (Kida et al., 2009), migration of the SA-I sequence into the hydrophobic environment may synchronize with movement of the passengers into the translocon pore. Findings from site-specific cross-linking experiments demonstrated that highly hydrophobic sequences enter the lipid environment directly from the Sec61 channel (Mothes et al., 1997; Heinrich et al., 2000). Assessments of membrane insertion of systematically constructed stop-transfer TM sequences also indicated that direct interaction between hydrophobic sequences and lipids seems to be involved in recognition at the translocon (Hessa et al., 2005; Hessa et al., 2007). Thus, it is likely that the SA-I sequence associates with and/or migrates into the lipid environment in an early stage during the topogenesis. We also observed that insertion of a Pro residue into the SA-I sequence shortened its membrane-embedded region, indicating that the Pro residue causes a conformation change of the SA-I sequence within the membrane, presumably by its helix-breaking propensity. The same mutation reduces its N-domain translocation (Kida et al., 2009). Because a Pro residue within a hydrophobic sequence shows inhibitory effect on the membrane integration (Hessa et al., 2005), the inserted Pro residue may attenuate migration of the SA-I sequence into the lipid environment and consequently lead to a lower efficiency of N-domain translocation. It is possible that the Pro residue diminishes the hydrophobic interaction between the SA-I sequence and the lipid and/or decelerates the release from the translocon into the lipid phase. We thus propose that membrane integration of the SA-I sequence provides the motive force for its N-domain translocation.
In addition, our translocation-trapping system can arrest migration of the SA-I sequence into the hydrophobic environment at various intermediate stages by increasing the distance between the SA-I sequence and the SBP-tag. Biophysical analyses of these intermediates, for example by using an environment-sensitive fluorescent probe for live detection of the environment (Alder et al., 2008), may provide additional insight into the dynamics of the migration of hydrophobic sequences into the membrane.
Environment Provided by the Translocon during Membrane Protein Integration
Data from the site-specific BMOE cross-linking showed that two translocation-arrested chains and the SA-II sequence of the S-38-I-II chimera protein in the integration intermediate state were simultaneously flanking the Sec61α. SH groups located in the hydrophobic environment should not be used for BMOE cross-linking because of the water-dependent reactivity of maleimide groups. The positions exposed partially to the aqueous environment within the SA-II sequence, however, were cross-linked with Sec61α, whereas those within the SA-I sequence were not. This finding suggested that the SA-II sequence, but not the SA-I sequence, is proximal to the Sec61 channel in the intermediate state. Furthermore, various Cys positions in both translocation-arrested hydrophilic chains were proximal to the Sec61α and were located in an aqueous environment. This strongly suggests that two hydrophilic chains penetrated the membrane via the aqueous pathway in or near the Sec61 channel. According to a previous study (Brambillasca et al., 2006), several tail-anchored TM sequences can mediate the translocation of the downstream sequences of more than 80 residues through the protein-free liposomal membrane. In this study, however, two chains were cross-linked with Sec61α as long as their translocations were arrested and therefore they should penetrate the membrane through an aqueous environment flanking Sec61α. Only the chain upstream of the SA-I sequence contacted another protein(s) and showed lower cross-linking efficiency with Sec61α when using bis-maleimidohexane, an analogue of BMOE with a longer linker (data not shown). Hence, these two chains are thought to be in different situations and maybe the former chain is closer to the rim of the Sec61 channel. The results obtained by the two techniques suggest that the Sec61 translocon simultaneously interacts with three polypeptide chains and provides an aqueous environment that accommodates at least two hydrophilic chains. Figure 6C shows two cartoons of our model of each polypeptide chain around the Sec61 channel. In the model with one Sec61 channel (Figure 6C, i), the former hydrophilic chain should move from the central pore of the channel to a secondary pathway near or outside the lateral exit crack. In this case, an additional hydrophilic spot formed by the gathered Sec61 channels or with other components may be required. An alternative model is that two hydrophilic chains are accommodated in distinct channels (Figure 6C, ii). Considering the cross-linked pattern, the former hydrophilic chain may locate near the lateral exit site of the channel and be accessible to another protein.
Crystal structures of archaeal and bacterial SecY complexes reveal that SecY monomers form a reasonable shape for membrane translocation and integration (Van den Berg et al., 2004; Tsukazaki et al., 2008), and positions in the constricted site of the SecY hourglass are cross-linked with a translocation-arrested chain (Cannon et al., 2005). Also, according to a recent image of the solubilized ribosome-Sec61 channel complex, a ribosome may bind firmly with only one Sec61 channel (Menetret et al., 2008). On the other hand, the results of various studies suggest that the Sec channel also serves as an oligomer. Oligomeric states of the Sec61 and SecY complexes were observed by electron microscopy (Hanein et al., 1996; Mitra et al., 2005) and biochemical analyses (Mori et al., 2003; Schaletzky and Rapoport, 2006). According to site-specific fluorescent labeling experiments, the translocon may form a large pore with a >40-Å diameter and may be regulated by BiP (binding immunoglobulin protein; Hamman et al., 1997; 1998). Additionally, several studies using site-specific cross-linking showed that multiple TM segments or translocating hydrophilic sequences are simultaneously associated with the Sec channel during the integration of multispanning membrane proteins (Heinrich and Rapoport, 2003; Sadlish et al., 2005; Kida et al., 2007). Moreover, recent data analyzed with blue-native PAGE shows that oligomeric states of the SecY channel may change depending on the substrate (Boy and Koch, 2009). On the basis of the findings of the present study, we propose the formation of the hydrophilic pathway capable of at least two translocating chains. How the translocon forms the pathway is the next question.
ACKNOWLEDGMENTS
This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan and the Sumitomo Foundation.
Abbreviations used:
- BMOE
bis-maleimidoethane
- I27 domain
27th immunoglobulin-like domain of titin
- N-domain
N-terminal domain
- NEM
N-ethylmaleimide
- PEGmal
(methyl-PEG12)3-PEG4-maleimide
- RM
rough microsomal membrane
- SA-I
type I signal-anchor
- SA-II
type II signal-anchor
- SAv
streptavidin
- SBP-tag
streptavidin-binding peptide tag
- SytII
synaptotagmin II
- TM
transmembrane.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-08-0738) on December 2, 2009.
REFERENCES
- Alder N. N., Jensen R. E., Johnson A. E. Fluorescence mapping of mitochondrial TIM23 complex reveals a water-facing, substrate-interacting helix surface. Cell. 2008;134:439–450. doi: 10.1016/j.cell.2008.06.007. [DOI] [PubMed] [Google Scholar]
- Beckmann R., Bubeck D., Grassucci R., Penczek P., Verschoor A., Blobel G., Frank J. Alignment of conduits for the nascent polypeptide chain in the ribosome-Sec61 complex. Science. 1997;278:2123–2126. doi: 10.1126/science.278.5346.2123. [DOI] [PubMed] [Google Scholar]
- Boy D., Koch H. G. Visualization of distinct entities of the SecYEG translocon during translocation and integration of bacterial proteins. Mol. Biol. Cell. 2009;20:1804–1815. doi: 10.1091/mbc.E08-08-0886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brambillasca S., Yabal M., Makarow M., Borgese N. Unassisted translocation of large polypeptide domains across phospholipid bilayers. J. Cell Biol. 2006;175:767–777. doi: 10.1083/jcb.200608101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon K. S., Or E., Clemons W. M., Jr, Shibata Y., Rapoport T. A. Disulfide bridge formation between SecY and a translocating polypeptide localizes the translocation pore to the center of SecY. J. Cell Biol. 2005;169:219–225. doi: 10.1083/jcb.200412019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley K. S., Liao S., Worrell V. E., Reinhart G. D., Johnson A. E. Secretory proteins move through the endoplasmic reticulum membrane via an aqueous, gated pore. Cell. 1994;78:461–471. doi: 10.1016/0092-8674(94)90424-3. [DOI] [PubMed] [Google Scholar]
- Do H., Falcone D., Lin J., Andrews D. W., Johnson A. E. The cotranslational integration of membrane proteins into the phospholipid bilayer is a multistep process. Cell. 1996;85:369–378. doi: 10.1016/s0092-8674(00)81115-0. [DOI] [PubMed] [Google Scholar]
- Goder V., Spiess M. Topogenesis of membrane proteins: determinants and dynamics. FEBS Lett. 2001;504:87–93. doi: 10.1016/s0014-5793(01)02712-0. [DOI] [PubMed] [Google Scholar]
- Hamman B. D., Chen J. C., Johnson E. E., Johnson A. E. The aqueous pore through the translocon has a diameter of 40–60 A during cotranslational protein translocation at the ER membrane. Cell. 1997;89:535–544. doi: 10.1016/s0092-8674(00)80235-4. [DOI] [PubMed] [Google Scholar]
- Hamman B. D., Hendershot L. M., Johnson A. E. BiP maintains the permeability barrier of the ER membrane by sealing the lumenal end of the translocon pore before and early in translocation. Cell. 1998;92:747–758. doi: 10.1016/s0092-8674(00)81403-8. [DOI] [PubMed] [Google Scholar]
- Hanein D., Matlack K. E., Jungnickel B., Plath K., Kalies K. U., Miller K. R., Rapoport T. A., Akey C. W. Oligomeric rings of the Sec61p complex induced by ligands required for protein translocation. Cell. 1996;87:721–732. doi: 10.1016/s0092-8674(00)81391-4. [DOI] [PubMed] [Google Scholar]
- Heinrich S. U., Mothes W., Brunner J., Rapoport T. A. The Sec61p complex mediates the integration of a membrane protein by allowing lipid partitioning of the transmembrane domain. Cell. 2000;102:233–244. doi: 10.1016/s0092-8674(00)00028-3. [DOI] [PubMed] [Google Scholar]
- Heinrich S. U., Rapoport T. A. Cooperation of transmembrane segments during the integration of a double-spanning protein into the ER membrane. EMBO J. 2003;22:3654–3663. doi: 10.1093/emboj/cdg346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hessa T., Kim H., Bihlmaier K., Lundin C., Boekel J., Andersson H., Nilsson I., White S. H., von Heijne G. Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature. 2005;433:377–381. doi: 10.1038/nature03216. [DOI] [PubMed] [Google Scholar]
- Hessa T., Meindl-Beinker N. M., Bernsel A., Kim H., Sato Y., Lerch-Bader M., Nilsson I., White S. H., von Heijne G. Molecular code for transmembrane-helix recognition by the Sec61 translocon. Nature. 2007;450:1026–1030. doi: 10.1038/nature06387. [DOI] [PubMed] [Google Scholar]
- Jackson R. J., Hunt T. Preparation and use of nuclease-treated rabbit reticulocyte lysates for the translation of eukaryotic messenger RNA. Methods Enzymol. 1983;96:50–74. doi: 10.1016/s0076-6879(83)96008-1. [DOI] [PubMed] [Google Scholar]
- Johnson A. E., van Waes M. A. The translocon: a dynamic gateway at the ER membrane. Annu. Rev. Cell Dev. Biol. 1999;15:799–842. doi: 10.1146/annurev.cellbio.15.1.799. [DOI] [PubMed] [Google Scholar]
- Kida Y., Mihara K., Sakaguchi M. Translocation of a long amino-terminal domain through ER membrane by following signal-anchor sequence. EMBO J. 2005;24:3202–3213. doi: 10.1038/sj.emboj.7600788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kida Y., Morimoto F., Sakaguchi M. Two translocating hydrophilic segments of a nascent chain span the ER membrane during multispanning protein topogenesis. J. Cell Biol. 2007;179:1441–1452. doi: 10.1083/jcb.200707050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kida Y., Morimoto F., Sakaguchi M. Signal anchor sequence provides motive force for polypeptide chain translocation through the endoplasmic reticulum membrane. J. Biol. Chem. 2009;284:2861–2866. doi: 10.1074/jbc.M808020200. [DOI] [PubMed] [Google Scholar]
- Kida Y., Sakaguchi M., Fukuda M., Mikoshiba K., Mihara K. Membrane topogenesis of a type I signal-anchor protein, mouse synaptotagmin II, on the endoplasmic reticulum. J. Cell Biol. 2000;150:719–730. doi: 10.1083/jcb.150.4.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura-Someya T., Iwaki S., Yamaguchi A. Site-directed chemical modification of cysteine-scanning mutants as to transmembrane segment II and its flanking regions of the Tn10-encoded metal-tetracycline/H+ antiporter reveals a transmembrane water-filled channel. J. Biol. Chem. 1998;273:32806–32811. doi: 10.1074/jbc.273.49.32806. [DOI] [PubMed] [Google Scholar]
- Koide K., Maegawa S., Ito K., Akiyama Y. Environment of the active site region of RseP, an Escherichia coli regulated intramembrane proteolysis protease, assessed by site-directed cysteine alkylation. J. Biol. Chem. 2007;282:4553–4560. doi: 10.1074/jbc.M607339200. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc. Natl. Acad. Sci. USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwabara N., Inoue H., Tsuboi Y., Nakamura N., Kanazawa H. The fourth transmembrane domain of the Helicobacter pylori Na+/H+ antiporter NhaA faces a water-filled channel required for ion transport. J. Biol. Chem. 2004;279:40567–40575. doi: 10.1074/jbc.M401132200. [DOI] [PubMed] [Google Scholar]
- Martoglio B., Hofmann M. W., Brunner J., Dobberstein B. The protein-conducting channel in the membrane of the endoplasmic reticulum is open laterally toward the lipid bilayer. Cell. 1995;81:207–214. doi: 10.1016/0092-8674(95)90330-5. [DOI] [PubMed] [Google Scholar]
- Meacock S. L., Lecomte F. J., Crawshaw S. G., High S. Different transmembrane domains associate with distinct endoplasmic reticulum components during membrane integration of a polytopic protein. Mol. Biol. Cell. 2002;13:4114–4129. doi: 10.1091/mbc.E02-04-0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menetret J. F., Hegde R. S., Aguiar M., Gygi S. P., Park E., Rapoport T. A., Akey C. W. Single copies of Sec61 and TRAP associate with a nontranslating mammalian ribosome. Structure. 2008;16:1126–1137. doi: 10.1016/j.str.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra K., Schaffitzel C., Shaikh T., Tama F., Jenni S., Brooks C. L., 3rd, Ban N., Frank J. Structure of the E. coli protein-conducting channel bound to a translating ribosome. Nature. 2005;438:318–324. doi: 10.1038/nature04133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori H., Tsukazaki T., Masui R., Kuramitsu S., Yokoyama S., Johnson A. E., Kimura Y., Akiyama Y., Ito K. Fluorescence resonance energy transfer analysis of protein translocase. SecYE from Thermus thermophilus HB8 forms a constitutive oligomer in membranes. J. Biol. Chem. 2003;278:14257–14264. doi: 10.1074/jbc.M300230200. [DOI] [PubMed] [Google Scholar]
- Mothes W., Heinrich S. U., Graf R., Nilsson I., von Heijne G., Brunner J., Rapoport T. A. Molecular mechanism of membrane protein integration into the endoplasmic reticulum. Cell. 1997;89:523–533. doi: 10.1016/s0092-8674(00)80234-2. [DOI] [PubMed] [Google Scholar]
- Mothes W., Prehn S., Rapoport T. A. Systematic probing of the environment of a translocating secretory protein during translocation through the ER membrane. EMBO J. 1994;13:3973–3982. doi: 10.1002/j.1460-2075.1994.tb06713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne A. R., Rapoport T. A. Protein translocation is mediated by oligomers of the SecY complex with one SecY copy forming the channel. Cell. 2007;129:97–110. doi: 10.1016/j.cell.2007.02.036. [DOI] [PubMed] [Google Scholar]
- Osborne A. R., Rapoport T. A., van den Berg B. Protein translocation by the Sec61/SecY channel. Annu. Rev. Cell Dev. Biol. 2005;21:529–550. doi: 10.1146/annurev.cellbio.21.012704.133214. [DOI] [PubMed] [Google Scholar]
- Rapoport T. A. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature. 2007;450:663–669. doi: 10.1038/nature06384. [DOI] [PubMed] [Google Scholar]
- Sadlish H., Pitonzo D., Johnson A. E., Skach W. R. Sequential triage of transmembrane segments by Sec61alpha during biogenesis of a native multispanning membrane protein. Nat. Struct. Mol. Biol. 2005;12:870–878. doi: 10.1038/nsmb994. [DOI] [PubMed] [Google Scholar]
- Sakaguchi M. Eukaryotic protein secretion. Curr. Opin. Biotechnol. 1997;8:595–601. doi: 10.1016/s0958-1669(97)80035-3. [DOI] [PubMed] [Google Scholar]
- Sakaguchi M., Hachiya N., Mihara K., Omura T. Mitochondrial porin can be translocated across both endoplasmic reticulum and mitochondrial membranes. J. Biochem. 1992;112:243–248. doi: 10.1093/oxfordjournals.jbchem.a123884. [DOI] [PubMed] [Google Scholar]
- Schaletzky J., Rapoport T. A. Ribosome binding to and dissociation from translocation sites of the endoplasmic reticulum membrane. Mol. Biol. Cell. 2006;17:3860–3869. doi: 10.1091/mbc.E06-05-0439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon S. M., Blobel G. A protein-conducting channel in the endoplasmic reticulum. Cell. 1991;65:371–380. doi: 10.1016/0092-8674(91)90455-8. [DOI] [PubMed] [Google Scholar]
- Tsukazaki T., et al. Conformational transition of Sec machinery inferred from bacterial SecYE structures. Nature. 2008;455:988–991. doi: 10.1038/nature07421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Berg B., Clemons W. M., Jr, Collinson I., Modis Y., Hartmann E., Harrison S. C., Rapoport T. A. X-ray structure of a protein-conducting channel. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- Walter P., Blobel G. Preparation of microsomal membranes for cotranslational protein translocation. Methods Enzymol. 1983;96:84–93. doi: 10.1016/s0076-6879(83)96010-x. [DOI] [PubMed] [Google Scholar]