Abstract
Objective
A rare mutation in low density lipoprotein receptor-related protein 6 gene (LRP6) was identified as the primary molecular defect underlying monogenic form of coronary artery disease. We hypothesised that common variants in LRP6 could predispose subjects to elevated LDL-cholesterol (LDL-C).
Methods and Results
12 common (minor allele frequency ≥0.1) single nucleotide polymorphisms in LRP6 were genotyped in 703 individuals from 213 Polish pedigrees (Silesian Cardiovascular Study families). The family-based analysis revealed that the minor allele of rs10845493 clustered with elevated LDL-C in offspring more frequently than expected by chance (p=0.0053). The quantitative analysis restricted to subjects free of lipid-lowering treatment confirmed the association between rs10845493 and age-, sex- and BMI-adjusted circulating levels of LDL-C in families as well as 2 additional populations - 218 unrelated subjects from Silesian Cardiovascular Study replication panel and 1138 individuals from Young Men Cardiovascular Association cohort (p=0.0268, p=0.0476 and p=0.0472, respectively). In the inverse variance weighted meta-analysis of the 3 populations each extra minor allele copy of rs10845493 was associated with 0.14 mmol/L increase in age-, sex- and BMI-adjusted LDL-C (SE=0.05, p=0.0038).
Conclusions
Common polymorphism in the gene underlying monogenic form of coronary artery disease impacts on risk of LDL-C elevation.
Keywords: gene, genetics, LDL-cholesterol, lipids, association
Low-density lipoprotein cholesterol (LDL-C) is the most powerful independent predictor of death from cardiovascular disease (1). The association between circulating concentrations of LDL-C and death from coronary artery disease (CAD) is continuous across a wide spectrum of cardiovascular risk (2). Similarly, there is a linear relationship between LDL-C lowering and the attenuation in risk of death from cardiovascular disease - each 1% reduction in LDL-C plasma levels correlates with a 1% decrease in CAD mortality (3). LDL-C shows >50% heritability (4) and correlates with familial predisposition to CAD even in young, apparently healthy subjects (5–6). The recent genome-wide association scans (GWAs) and the subsequent replication studies have identified several common polymorphic variants with very significant impact on LDL-C levels (7–8). However, the collective contribution of the variants identified through GWAs to the overall interindividual variation in LDL-C is modest and does not fully explain its entire heritability (8). Clearly, additional strategies are needed to identify the remaining genetic contributors to human hyperlipidemia. One of the potential approaches to uncover these loci is a systematic analysis of common alleles in genes responsible for rare, monogenic syndromes of human cardiovascular disorders. Using this conceptual strategy in relation to genes underlying monogenic forms of hypertension and hypotension we have recently revealed that common alleles within the locus responsible for type 2 Bartter’s syndrome (KCNJ1) are associated with blood pressure in the general population (9). Low density lipoprotein receptor-related protein 6 gene (LRP6) - responsible for monogenic form of CAD (10) is an excellent model locus for investigations on LDL-C. Indeed, rare missense mutation in LRP6 was implicated as the primary molecular driver of LDL-C elevation and subsequent premature CAD in a pedigree of Asian origin (10). Compared to unaffected members of the family, carriers of the rare mutant alleles in LRP6 exhibited prominent metabolic disturbances (among which LDL-C elevation was the most significant) and died of CAD at an early age (10). Furthermore, at least two other disorders in which LDL-C plays a significant role (Alzheimer’s disease and age-related macular degeneration) were linked to common genetic variants in LRP6 (11–12). Therefore, we sought to determine whether common genetic variation within LRP6 might be associated with to LDL-C.
Methods
Subjects
Silesian Cardiovascular Study – families and replication panel
Silesian Cardiovascular Study (SCS) is an investigation designed to examine genetic background of risk factors underlying cardiovascular disorders. All subjects (Polish Caucasian) were recruited in 3 reference centres for cardiovascular diseases in the south of Poland. The recruitment was conducted through probands - index patients with high cardiovascular risk, defined as i) co-existence of CAD and hypertension or ii) one cardiovascular disease (CAD or hypertension) accompanied by at least 2 cardiovascular risk factors (smoking, waist circumference >102 cm /men/ or >88 cm /women/, clinically documented history of hyperglycemia, hyperlipidemia or lipid-lowering medication, parental history of CAD or hypertension) or iii) clustering of at least 3 cardiovascular risk factors.
213 families (703 subjects) along with 435 biologically unrelated individuals (an independent sample for replication analysis - replication panel) were recruited into this study. Each subject underwent thorough clinical, anthropometric and biochemical phenotyping (under fasting conditions) according to the previously described protocols and methods (5, 13–15). Total cholesterol (TC), HDL-cholesterol (HDL-C) and triglycerides were measured using colorimetric-enzymatic methods on a Cobas Bio-Autoanalyzer, as reported before (5). In eligible subjects (those with triglycerides <4.5 mmol/L) LDL-C levels were calculated based on the Friedewald formula.
Given that almost 20% of subjects in our pedigrees were on lipid-lowering medication, we used categorised LDL-C as the primary phenotype of interest in the family-based association analyses. We classified subjects as affected if they were on pharmacological lipid-lowering medication or if their fasting LDL-C was ≥ the lower threshold of borderline-high elevation, defined as:
for adults (subjects aged >19 years) - 3.37 mmol/L (as per NCEP ATP III Classification) (1);
for adolescents - age- and gender-specific LDL-C cut-off points (according to the newly developed clinical classification of elevated lipoprotein levels that is linked to the adult NCEP ATP III values) (16)
Moreover, phenotypic information from subjects who were not on lipid-lowering treatment was used to examine whether the findings from the analysis based on the categorised phenotype translate into significant genetic effects on circulating concentrations of LDL-C (continuous trait).
General clinical characteristics of all recruited SCS subjects are presented in Table 1. Further details of phenotyping were included in the Supplement material.
Table 1.
Clinical characteristics of subjects from Silesian Cardiovascular Study.
| Phenotype | Families - fathers |
Families - mothers |
Families - sons |
Families - daughters |
Replication panel |
|---|---|---|---|---|---|
| N | 166 | 194 | 164 | 179 | 435 |
| Age (years) |
55.0 (49.0 – 61.0) | 53.0 (48.0 – 61.5) | 28.0 (21.5 – 37.0) | 30.0 (22.0 – 40.0) | 55.0 (48.0 – 66.0) |
| Sex (M/F) | 166/0 | 0/194 | 164/0 | 0/179 | 294/141 |
| BMI (kg/m2) | 27.1 (25.0 – 30.1) | 27.7 (24.4 – 30.4) | 25.1 (22.5 – 27.8) | 22.7 (20.1 – 26.4) | 27.1 (24.5 – 29.8) |
| SBP (mmHg) | 130.0 (118.5 – 145.8) | 137.5 (122.8 – 149.4) | 128.0 (118.0 – 138.5) | 121.5 (113.8 – 132.9) | 125.5 (116.0 – 141.5) |
| DBP (mmHg) | 77.0 (69.0 – 86.0) | 77.5 (71.0 – 84.0) | 71.5 (65.0 – 78.0) | 70.5 (64.3 – 78.0) | 73.0 (65.5 – 80.7) |
| Hypertension (%) |
129 (77.7) | 131 (67.5) | 52 (31.7) | 49 (27.4) | 316 (72.6) |
| Antihypertensive treatment (%) |
106 (63.9) | 95 (49.0) | 20 (12.2) | 30 (16.8) | 270 (62.1) |
| CAD (%) | 109 (65.7) | 65 (33.5) | 15 (9.1) | 11 (6.1) | 299 (68.7) |
| TC (mmol/L) | 5.1 (4.3 – 6.2) | 5.5 (4.8 – 6.5) | 5.1 (4.1 – 6.0) | 5.0 (4.3 – 5.8) | 5.0 (4.2 – 5.9) |
| ↑ TC (%) | 125 (75.3) | 137 (70.6) | 84 (51.2) | 77 (43.0) | 286 (65.7) |
| LDL-C(mmol/L) | 3.5 (2.7 – 4.4) | 3.7 (3.1 – 4.7) | 3.2 (2.5 – 4.3) | 3.3 (2.7 – 4.0) | 3.2 (2.5 – 4.1) |
| ↑ LDL-C (%) | 129 (77.7) | 139 (71.6) | 83 (50.6) | 77 (43.0) | 291 (66.7) |
| Triglycerides (mmol/L) |
1.6 (1.2 – 2.2) | 1.3 (0.9 – 1.8) | 1.2 (0.8 – 1.8) | 1.0 (0.8 – 1.4) | 1.6 (1.2 – 2.3) |
| ↑ Triglycerides (%) |
111 (66.9) | 82 (42.3) | 53 (32.3) | 31 (17.3) | 272 (62.5) |
| HDL-C (mmol/L) | 0.9 (0.7 – 1.1) | 1.1 (0.9 – 1.3) | 1.0 (0.8 – 1.2) | 1.2 (1.0 – 1.4) | 0.9 (0.7 – 1.1) |
| ↓ HDL-C (%) | 126 (75.9) | 153 (78.9) | 88 (53.7) | 103 (57.5) | 355 (81.6) |
| Lipid-lowering medication (%) |
77 (46.4) | 44 (22.7) | 12 (7.3) | 3 (1.7) | 203 (46.7) |
Data are medians and 25%–75% inter-quartile ranges or counts and percentages, BMI – body mass index, SBP – systolic blood pressure, DBP – diastolic blood pressure, CAD – coronary artery disease, TC – total cholesterol, LDL-C – LDL-cholesterol, HDL-C – HDL-cholesterol, ↑ TC, ↑ LDL-C, ↑ Triglycerides, ↓ HDL-C were defined as serum levels of respective lipid fractions over or below thresholds defined in the Methods and Supplement material or taking lipid-lowering medication
Young Men Cardiovascular Association Study (YMCA)
A demographically and clinically homogenous population of 1157 young (mean age - 19 years), apparently healthy men from the YMCA cohort (5) was used to quantify effects of LRP6 alleles on circulating concentrations of LDL-C. Recruited from randomly selected secondary schools of Silesia (Southern Poland), the YMCA subjects had no history of overt cardiovascular disease and were not treated with lipid-lowering medication (5). The details of phenotyping and clinical characteristics of those subjects were described elsewhere (5).
Monocyte Profiling Study
As a part of the Cardiogenics Monocyte Profiling Study 161 patients (mean age - 55.4 years; 89.4% men) with a history of myocardial infarction (MI) before the age of 65 years were recruited in Leicester. Fasting blood samples were collected from these subjects between 3 and 36 months following their MI, and all subjects were ≤66 years at the time of sampling. All patients were non-smokers, had normal full blood count, C-reactive protein <10 mg/L and plasma blood glucose <7.0 mmol/L.
The investigations were approved by the local Bioethical Committees and each subject gave a written informed consent to participate.
Methods
Genotyping
Genetic markers
Eleven common (minor allele frequency ≥0.1 in CEU) tagging single nucleotide polymorphisms (SNPs) (rs11609634, rs12313200, rs207524, rs7980903, rs11054738, rs2417085, rs10492120, rs10743980, rs11054704, rs10845493, rs17302049) within LRP6 were selected from the HapMap under the threshold of r2≥0.8. In addition, a common, potentially functional non-synonymous polymorphism (rs2302685) was added to the array of genotyped SNPs at the pre-experimental stage (17). The selected set of tags provides approximately 96% genetic coverage for LRP6 locus (captures 96% of the ungenotyped common LRP6 alleles in the reference CEU panel) (18).
DNA analysis
DNA was extracted from peripheral leukocytes. Genotyping in SCS was conducted using the iPLEX™ assays on Sequenom MassARRAY® system (Sequenom Inc., San Diego, USA), according to manufacturer's protocols. Analysis of genotypes was carried out using Typer software (version 3.4). Additional genotyping of 2 polymorphisms (rs12313200 and rs11054704) that failed on the MassARRAY platform in SCS along with typing of rs10845493 in the YMCA cohort and Monocyte Profiling Study was conducted using commercially available TaqMan assays on ABI PRISM 7900HT Sequence Detection System (Applied Biosystems).
Monocyte isolation and RNA analysis
Blood was collected into EDTA vacutainer tubes. Within 20 minutes of collection monocytes were isolated from whole blood by positive selection with CD14 magnetic beads using an AutoMACS system (Miltenyi Biotech). Cell purity was confirmed by flow cytometry and in all samples >90% of the cells were CD14+ve monocytes. RNA was extracted immediately, using Trizol, followed by clean-up using RNeasy columns (Qiagen) and DNase. Purified RNA was amplified and labelled using the Illumina TotalPrep RNA Amplification Kit (Applera, Austin, Texas, US). Biotinylated cRNA was applied to Illumina-HG 6v3 whole-genome expression microarrays and LRP6 expression was determined from normalised data from the ILMN 1732912 probe.
Statistical analysis
Deviation from Hardy-Weinberg equilibrium was evaluated on the data from biologically unrelated individuals (parental generation in SCS families, subjects from the replication panel, the YMCA and Monocyte Profiling Study) using the chi2-test and the threshold of p<0.01 (13).
Genotypes of the SCS families were tested for Mendelian inconsistencies using family-based association test (FBAT) software (http://www.biostat.harvard.edu/~fbat/fbat.htm). Associations between LRP6 SNPs and categorised LDL-C (primary hypothesis) as well as other lipid traits (secondary analyses) in families were examined using FBAT (19) under null hypothesis of no linkage and no association. The tests were executed under additive model of inheritance (13) with an equal-weight offset option that allows for extraction of information from both affected and unaffected offspring (20). To correct for multiple testing in single locus family-based association analysis we used a spectral decomposition of linkage disequilibrium (LD) matrices generated for each pair of genotyped markers as proposed by Nyholt (21) and Li (22). In brief, the calculation was based on estimation of the effective number of independent genotyped markers (after dissection of their linkage disequilibria) along with the corresponding threshold of statistical significance required to keep type I error rate at 5%. In the current analysis, the number of non-redundant markers and the corrected experiment-wide significance threshold were estimated at 6 and p=0.0085, respectively. The 6 non-redundant SNPs (rs12313200, rs2075241, rs7980903, rs2417085, rs10845493 and rs17302049) identified as the major contributors to >90% variance in SCS data (through inspection of principal component coefficients for varimax-rotated matrix) were included in haplotype analysis. This multi-marker testing was executed under additive model of inheritance using family-based haplotype analysis (23) combined with multiple (10000) permutations.
To provide a quantitative estimate of the effect of LRP6 alleles on age-, sex- and BMI-adjusted LDL-C (as a continuous trait) in SCS families we analysed data from subjects who were not on lipid-lowering medication using additive model of inheritance and within-only option of family-based association test for quantitative traits (QFAM available in PLINK) (24, 25).
Associations between LRP6 SNP and circulating levels of LDL-C amongst biologically unrelated subjects from SCS replication panel and YMCA cohort were assessed by linear regression after adjustment for age, BMI and sex (where appropriate). The regression models were fitted in PLINK under additive model of inheritance. Additional exploratory analyses were also conducted under dominant and recessive model of inheritance. β-coefficients [along with standard errors (SEs)] were used as robust estimates of genetic effects.
Inverse variance weighted averages of β-coefficients and SE from all populations (SCS families, SCS replication panel and YMCA cohort) were then combined together in fixed effects meta-analysis. A summary effect size and overall p-value were calculated for the combined sample under additive model of inheritance using METAL software (http://www.sph.umich.edu/csg/abecasis/metal/index.html). The between-study heterogeneity was evaluated using chi2-test.
All analyses on LDL-C as a quantitative trait were carried out using data from subjects who were not on lipid-lowering treatment.
Baseline descriptive analysis of other phenotypes across LRP6 genotypes was based on Student’s t-test, Mann-Whitney test, Kruskal-Wallis test, Fisher’s exact test and chi2 test for trend dependent on the type of variable (qualitative or quantitative) and distribution.
PBAT-based (26) calculations showed that for the majority of common alleles (0.1–0.5) and under additive model of inheritance the family-based study had from moderate to good power (0.4–1.0) to detect SNP–elevated LDL-C associations of larger magnitude (allelic odds ratio range of 1.6–1.9) but only modest power (0.1–0.4) to detect smaller effect sizes (allelic odds ratio ≤1.3).
Results
LDL-C and genetic variation within LRP6 – family-based association analysis
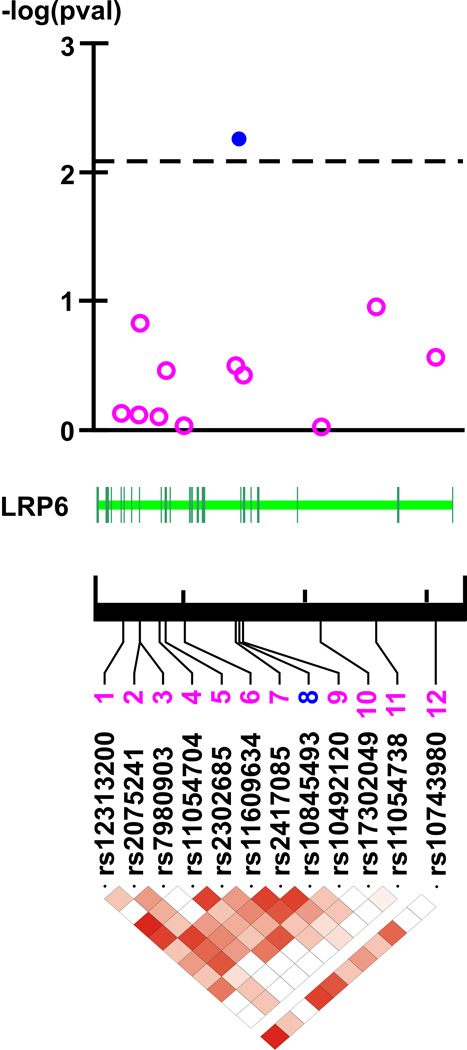
None of the genotyped LRP6 SNPs violated the HWE (Supplementary Table I). Of 12 LRP6 bi-allelic polymorphisms, one (rs10845493) was associated with LDL-C (Figure and extended description of the Figure legend in Supplement material). Specifically, under additive model of inheritance the minor allele of rs10845493 clustered with elevated LDL-C in offspring more frequently than expected by chance (p=0.0053, Figure; Supplementary Table I). This association retained its statistical significance even after using a threshold reflecting the correction for multiple testing (p=0.0085, Figure and extended description of the Figure legend in the Supplement material).
Figure 1. LRP6 and LDL-C in Silesian Cardiovascular Study – family-based association analysis.
Top panel. -log transformed p-values from the association analysis.
Middle panel. Structure of LRP6 gene.
Bottom panel. The linkage disequilibrium (LD) map (r2-coefficients-based) of LRP6 (dark red corresponds to r2=1 – maximal LD and white to r2=0 – no LD)
None of 15 six-allelic LRP6 haplotypes identified in SCS families were associated with LDL-C (Supplementary Table II). Using a smaller sliding window of 5, 4, 3 and 2 consecutive alleles did not have any effect on the results of the haplotype analysis (data not shown).
Given that LDL-C is a by-product of TC, HDL-C and triglycerides we also examined the associations of these lipid fractions with LRP6 SNPs in the secondary analyses. LRP6 SNPs were not associated with either HDL-C or triglycerides while rs10845493 showed a consistent significant association with TC but of lesser magnitude than LDL-C (data not shown).
Estimation of rs10845493 effect on circulating concentrations of LDL-C in families – family-based association tests for quantitative traits (QFAM)
The quantitative analysis restricted to subjects with no history of lipid-lowering treatment confirmed the association between rs10845493 and age-, sex- and BMI-adjusted circulating levels of LDL-C in families - the effect of each extra minor allele copy of rs10845493 on age-, sex- and BMI-adjusted LDL-C was estimated at approximately 0.4 mmol/L (SE=0.20, p=0.0268).
Estimation of rs10845493 effect on circulating concentrations of LDL-C in unrelated subjects from SCS replication panel and YMCA cohort
After exclusion of subjects on lipid-lowering medication and those with missing genotypes/phenotypes 218 individuals from the SCS panel and 1138 men from YMCA cohorts were included in the quantitative analysis. Distribution of rs10845493 genotypes was in agreement with Hardy-Weinberg equilibrium in both populations (p=0.253 and p=0.765, respectively). Minor allele frequency of rs10845493 was calculated at 12.4% and 14.5% in the replication panel and the YMCA, respectively. Mean LDL-C levels stratified on rs10845493 genotypes are presented in Supplementary Table III (SCS replication panel) and IV (YMCA cohort). Other clinical characteristics of both populations (after stratification based on rs10845493 genotypes) are included in Supplementary Tables V – VI.
Under additive model of inheritance and after adjustment for age, sex and BMI each minor allele copy of rs10845493 was associated with 0.31 mmol/L increase in LDL-C (SE=0.15, p=0.0476) in SCS replication panel. The association between rs10845493 and circulating concentrations of LDL-C was also apparent amongst YMCA individuals - under the same model of inheritance each minor allele copy of rs10845493 increased age- and BMI-adjusted LDL-C by 0.10 mmol/L (SE=0.05, p=0.0472).
Secondary exploratory analyses showed that neither dominant nor recessive model of inheritance offered a significant advantage over the additive one in examination of association between rs10845493 and LDL-C in SCS replication and YMCA cohort (Supplementary Tables VII and VIII).
rs10845493 and LDL-C in SCS families, SCS replication panel and YMCA cohort - inversed variance fixed effects meta-analysis
In the combined meta-analysis of the data from 1658 informative subjects each minor allele copy of rs10845493 was associated with 0.14 mmol/L increase in LDL-C after adjustment for age, BMI and sex (where appropriate) (SE=0.05, p=0.0038, Table 2). There was no evidence of heterogeneity in effect sizes across 3 populations included in the meta-analysis (p=0.1941)
Table 2.
rs10845493 SNP and circulating levels of LDL-C – inverse variance weighted fixed effects meta-analysis.
| Sample | No of informative subjects |
Allele - reference |
β- coefficient |
Standard error (SEs) |
P-value |
|---|---|---|---|---|---|
| SCS families |
302* | T | 0.40 | 0.20 | 0.0268 |
| SCS replication panel |
218** | T | 0.31 | 0.15 | 0.0476 |
| YMCA cohort |
1138** | T | 0.10 | 0.05 | 0.0472 |
| Combined | 1658 | T | 0.14 | 0.05 | 0.0038 |
SNP – single nucleotide polymorphism, SCS – Silesian Cardiovascular Study, YMCA – Young Men Cardiovascular Association Study; individual β-coefficients, SEs and p-values were obtained from QFAM-family-based association tests for quantitative traits (SCS families) or by linear regression (SCS replication panel and YMCA); all p-values were obtained after adjustment of LDL-C for age, sex and BMI (SCS) or age and BMI (YMCA men)
number of subjects free of lipid-lowering treatment that contributed information in QFAM-family-based association tests for quantitative traits
number of subjects with available genotype and phenotype information who were not on lipid-lowering medication.
Analysis of correlation between rs10845493 and LRP6 mRNA expression in monocytes
Genotypes of rs10845493 in 161 subjects from Monocyte Profiling Study were in Hardy-Weinberg equilibrium (p=0.993). There were no significant differences in normalised LRP6 mRNA expression in monocytes between subjects with CC genotype and carriers of T-allele of rs10845493 (6.298±0.184 vs. 6.317±0.188, respectively; p=0.5916). CT and TT genotypes of rs10845493 were analyzed together because of the rarity of homozygous combination of the T-allele (3 subjects).
Discussion
Our study provided the first evidence for association between a common genetic variant within LRP6 and predisposition to elevated LDL-C. This association was apparent in 3 independent population samples of subjects irrespective of the type of statistical approach (family-based studies and analysis of biologically unrelated subjects) or the phenotype definition (categorised and continuous trait). The direction of the allelic association was identical in all investigations - the minor allele (T) of rs10845493 promoted LDL-C elevation. The magnitude of the genetic effect of rs10845493 on LDL-C (as shown in the meta-analysis) is clinically modest but corresponds well with anticipated contribution of a single allele to inter-individual variation in a trait regulated by multiple genes and environmental factors.
The most significant association signal between LDL-C and LRP6 was mapped to rs10845493 in intron 7, only 546 base pairs from the exon-intron junction. Our transcriptomic analysis did not lend support to a functional role of rs10845493 in regulation of LRP6 expression. Therefore, we suspect that other statistically similar (in LD with rs10845493) and possibly yet unidentified functional variant(s) of LRP6 drive(s) the detected association. In-depth sequencing analysis of LRP6 followed by genotyping in large population samples will be needed to reveal the causative genetic allele. Given that LRP6 functions as a receptor for cellular uptake of LDL-C particles (27) reduced clearance and subsequent increase in circulating concentrations of LDL-C may be the most likely functional consequence of the identified genetic association. Indeed, LRP6 belongs to a family of ubiquitously expressed multifunctional cell surface receptors (28) integrated into signal transmission within Wnt/β-catenin pathway – a novel regulatory system of lipid and glucose metabolism (29). Future experiments based on cellular models of LDL-C uptake/internalization are warranted to elucidate the precise mechanisms of our findings. However, it would be fair to acknowledge that rs10845493 maps not only to LRP6 but also to an overlapping gene BCL2L14 – a member of BCL2 protein family that has been shown to facilitate apoptosis and tumorigenesis (30). In contrast to the causal role of LRP6 variants in human monogenic hyperlipidemia (10) and LDL-C clearance (27) there is no data (either in vitro or in vivo) in support of a contribution of BCL2L14 to cholesterol regulation. Thus, the existing level of functional evidence strongly favours LRP6 as the more likely driver of the association between the chromosome 12 locus and LDL-C.
We recognize that our association findings are based only on subjects recruited in the south of Poland. Replication analysis in samples from other regions will be needed to provide evidence that our association is not population-specific. Larger samples will be necessary in examination of genetic interactions between LRP6 and other loci that regulate LDL-C metabolism (such as low-density lipoprotein receptor gene) and in detection of more subtle genetic effects.
Nevertheless, within highlighted interpretational limitations our study shows that common genetic variation within LRP6 impacts on the risk of elevated LDL-C. These data also implicate LRP6 as a potential molecular regulator of lipid metabolism and a novel target for pharmacological interventions. Most importantly, the demonstrated associations indicate that common alleles in loci responsible for rare monogenic syndromes may indeed contribute to polygenic forms of cardiovascular disease.
Supplementary Material
Acknowledgment
Sources of funding
This study was supported by NIH Fogarty International Research Collaboration Award (R03 TW007165) (to MT). TB is supported by a British Heart Foundation project grant (PG/06/097) (to MT). NJS holds a British Heart Foundation Chair of Cardiology.
We are very grateful to physicians, research technicians, nurses and students involved in the recruitment of subjects in the Silesian Cardiovascular Study.
Footnotes
Disclosure
None.
References
- 1.Third Report of the National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults (Adult Treatment Panel III) final report. Circulation. 2002;106:3143–3421. [PubMed] [Google Scholar]
- 2.Stamler J, Wentworth D, Neaton JD. Is relationship between serum cholesterol and risk of premature death from coronary heart disease continuous and graded? Findings in 356,222 primary screenees of the Multiple Risk Factor Intervention Trial (MRFIT) JAMA. 1986;256:2823–2828. [PubMed] [Google Scholar]
- 3.Grundy SM, Cleeman JI, Merz CN, Brewer HB, Jr, Clark LT, Hunninghake DB, Pasternak RC, Smith SC, Jr, Stone NJ. Implications of recent clinical trials for the National Cholesterol Education Program Adult Treatment Panel III Guidelines. Circulation. 2004;110:227–239. doi: 10.1161/01.CIR.0000133317.49796.0E. [DOI] [PubMed] [Google Scholar]
- 4.Snieder H, van Doornen LJ, Boomsma DI. Dissecting the genetic architecture of lipids, lipoproteins, and apolipoproteins: lessons from twin studies. Arterioscler Thromb Vasc Biol. 1999;19:2826–2834. doi: 10.1161/01.atv.19.12.2826. [DOI] [PubMed] [Google Scholar]
- 5.Charchar FJ, Tomaszewski M, Lacka B, Zakrzewski J, Zukowska-Szczechowska E, Grzeszczak W, Dominiczak AF. Association of the human Y chromosome with cholesterol levels in the general population. Arterioscler Thromb Vasc Biol. 2004;24:308–312. doi: 10.1161/01.ATV.0000113291.39267.0a. [DOI] [PubMed] [Google Scholar]
- 6.Kark JD, Sinnreich R, Rosenberg IH, Jacques PF, Selhub J. Plasma homocysteine and parental myocardial infarction in young adults in Jerusalem. Circulation. 2002;105:2725–2729. doi: 10.1161/01.cir.0000017360.99531.26. [DOI] [PubMed] [Google Scholar]
- 7.Lusis AJ, Pajukanta P. A treasure trove for lipoprotein biology. Nat Genet. 2008;40:129–130. doi: 10.1038/ng0208-129. [DOI] [PubMed] [Google Scholar]
- 8.Manolio TA. Cohort studies and the genetics of complex disease. Nat Genet. 2009;41:5–6. doi: 10.1038/ng0109-5. [DOI] [PubMed] [Google Scholar]
- 9.Tobin MD, Tomaszewski M, Braund PS, Hajat C, Raleigh SM, Palmer TM, Caulfield M, Burton PR, Samani NJ. Common variants in genes underlying monogenic hypertension and hypotension and blood pressure in the general population. Hypertension. 2008;51:1658–1664. doi: 10.1161/HYPERTENSIONAHA.108.112664. [DOI] [PubMed] [Google Scholar]
- 10.Mani A, Radhakrishnan J, Wang H, Mani MA, Nelson-Williams C, Carew KS, Mane S, Najmabadi H, Wu D, Lifton RP. LRP6 mutation in a family with early coronary disease and metabolic risk factors. Science. 2007;315:1278–1282. doi: 10.1126/science.1136370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haines JL, Schnetz-Boutaud N, Schmidt S, Scott WK, Agarwal A, Postel EA, Olson L, Kenealy SJ, Hauser M, Gilbert JR, Pericak-Vance MA. Functional candidate genes in age-related macular degeneration: significant association with VEGF, VLDLR, and LRP6. Invest Ophthalmol Vis Sci. 2006;47:329–335. doi: 10.1167/iovs.05-0116. [DOI] [PubMed] [Google Scholar]
- 12.De Ferrari GV, Papassotiropoulos A, Biechele T, Wavrant De-Vrieze F, Avila ME, Major MB, Myers A, Saez K, Henriquez JP, Zhao A, Wollmer MA, Nitsch RM, Hock C, Morris CM, Hardy J, Moon RT. Common genetic variation within the low-density lipoprotein receptor-related protein 6 and late-onset Alzheimer's disease. Proc Natl Acad Sci U S A. 2007;104:9434–9439. doi: 10.1073/pnas.0603523104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tomaszewski M, Charchar FJ, Lynch MD, Padmanabhan S, Wang WY, Miller WH, Grzeszczak W, Maric C, Zukowska-Szczechowska E, Dominiczak AF. Fibroblast growth factor 1 gene and hypertension: from the quantitative trait locus to positional analysis. Circulation. 2007;116:1915–1924. doi: 10.1161/CIRCULATIONAHA.107.710293. [DOI] [PubMed] [Google Scholar]
- 14.Tomaszewski M, Brain NJ, Charchar FJ, Wang WY, Lacka B, Padmanabahn S, Clark JS, Anderson NH, Edwards HV, Zukowska-Szczechowska E, Grzeszczak W, Dominiczak AF. Essential hypertension and β2-adrenergic receptor gene: linkage and association analysis. Hypertension. 2002;40:286–291. doi: 10.1161/01.hyp.0000029105.21202.fe. [DOI] [PubMed] [Google Scholar]
- 15.Tomaszewski M, Charchar FJ, Lacka B, Pesonen U, Wang WY, Zukowska-Szczechowska E, Grzeszczak W, Dominiczak AF. Epistatic interaction between β2-adrenergic receptor and neuropeptide Y genes influences LDL-cholesterol in hypertension. Hypertension. 2004;44:689–694. doi: 10.1161/01.HYP.0000143844.81979.61. [DOI] [PubMed] [Google Scholar]
- 16.Jolliffe CJ, Janssen I. Distribution of lipoproteins by age and gender in adolescents. Circulation. 2006;114:1056–1062. doi: 10.1161/CIRCULATIONAHA.106.620864. [DOI] [PubMed] [Google Scholar]
- 17.Tomaszewski M, Charchar FJ, Samani NJ. Association studies in current cardiovascular genetics - functional variants, tags or both? J Hum Hypertens. 2007;21:425–426. doi: 10.1038/sj.jhh.1002197. [DOI] [PubMed] [Google Scholar]
- 18.de Bakker PI, Yelensky R, Pe'er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 19.Horvath S, Xu X, Laird NM. The family based association test method: strategies for studying general genotype-phenotype associations. Eur J Hum Genet. 2001;9:301–306. doi: 10.1038/sj.ejhg.5200625. [DOI] [PubMed] [Google Scholar]
- 20.Bertram L, Hiltunen M, Parkinson M, Ingelsson M, Lange C, Ramasamy K, Mullin K, Menon R, Sampson AJ, Hsiao MY, Elliott KJ, Velicelebi G, Moscarillo T, Hyman BT, Wagner SL, Becker KD, Blacker D, Tanzi RE. Family-based association between Alzheimer's disease and variants in UBQLN1. N Engl J Med. 2005;352:884–894. doi: 10.1056/NEJMoa042765. [DOI] [PubMed] [Google Scholar]
- 21.Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet. 2004;74:765–769. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li J, Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity. 2005;95:221–227. doi: 10.1038/sj.hdy.6800717. [DOI] [PubMed] [Google Scholar]
- 23.Horvath S, Xu X, Lake SL, Silverman EK, Weiss ST, Laird NM. Family-based tests for associating haplotypes with general phenotype data: application to asthma genetics. Genet Epidemiol. 2004;26:61–69. doi: 10.1002/gepi.10295. [DOI] [PubMed] [Google Scholar]
- 24.Lowe JK, Maller JB, Pe'er I, Neale BM, Salit J, Kenny EE, Shea JL, Burkhardt R, Smith JG, Ji W, Noel M, Foo JN, Blundell ML, Skilling V, Garcia L, Sullivan ML, Lee HE, Labek A, Ferdowsian H, Auerbach SB, Lifton RP, Newton-Cheh C, Breslow JL, Stoffel M, Daly MJ, Altshuler DM, Friedman JM. Genome-wide association studies in an isolated founder population from the Pacific island of Kosrae. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000365. e1000365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lange C, DeMeo D, Silverman EK, Weiss ST, Laird NM. PBAT: tools for family-based association studies. Am J Hum Genet. 2004;74:367–369. doi: 10.1086/381563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu W, Mani S, Davis NR, Sarrafzadegan N, Kavathas PB, Mani A. Mutation in EGFP domain of LDL receptor-related protein 6 impairs cellular LDL clearance. Circ Res. 2008;103:1280–1288. doi: 10.1161/CIRCRESAHA.108.183863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jaeger S, Pietrzik CU. Functional role of lipoprotein receptors in Alzheimer's disease. Curr Alzheimer Res. 2008;5:15–25. doi: 10.2174/156720508783884675. [DOI] [PubMed] [Google Scholar]
- 29.Manolagas SC, Almeida M. Gone with the Wnts: β-catenin, T-cell factor, forkhead box O, and oxidative stress in age-dependent diseases of bone, lipid, and glucose metabolism. Mol Endocrinol. 2007;21:2605–2614. doi: 10.1210/me.2007-0259. [DOI] [PubMed] [Google Scholar]
- 30.Guo B, Godzik A, Reed JC. Bcl-G, a novel pro-apoptotic member of the Bcl-2 family. J Biol Chem. 2001;276:2780–2785. doi: 10.1074/jbc.M005889200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.