Abstract
The human immune system has developed an elaborate network of cascades for dealing with microbial intruders. Owing to its ability to rapidly recognize and eliminate microorganisms, the complement system is an essential and efficient component of this machinery. However, many pathogenic organisms have found ways to escape the attack of complement through a range of different mechanisms. Recent discoveries in this field have provided important insights into these processes on a molecular level. These vital developments could augment our knowledge of the pathology and treatment of infectious and inflammatory diseases.
Evading the manifold attacks of the immune system is a key determinant for the survival of pathogens within their hosts. It is not surprising, therefore, that the coexistence and co-evolution of humans and microorganisms has produced a multitude of microbial mechanisms for attenuating or escaping these attacks. As a first line of defence against pathogenic intruders, and a mediator between the innate and adaptive immune responses, the complement system is a particular focus of these evasion strategies. Although this carefully regulated cascade of enzymes, protein complexes and receptors ensures the rapid recognition and elimination of foreign structures, it also offers many sites of interference that can disrupt this balanced network of protein interactions. A detailed understanding of the individual processes and the underlying interactions on a molecular level is essential for describing the mechanisms of infectious diseases and the development of new therapies. Recent discoveries of complement-targeting proteins, the availability of complete microbial genome sequences and advances in experimental methods have propelled this area of research, and provide fascinating insights into complement attack and evasion. Many pathogens seem to have developed parallel routes for escaping complement, and several evasion principles are shared not only among members of the same genus but even among diverse organisms, such as bacteria, viruses, fungi and parasites.
In this Review, we will provide a comprehensive over-view and update of the exciting recent developments in this field. After a short introduction that will discuss the diverse role of the complement system in defence, disease and infection, the emphasis will be on the functional and structural aspects of the evasion strategies of human pathogens. Rather than separating them by organism, we classify distinct and common mechanisms for all pathogens based on their mode of action. In light of recent findings, the unique evasion strategies of Staphylococcus aureus will be analysed in more detail. Finally, the potential impact of these developments on prospective antimicrobial and complement-specific therapeutics will be discussed.
The human complement system
The complement system is a central component of the innate immune response and fulfils numerous functions, including the recognition of foreign cells, communication with and activation of adaptive immunity and the removal of cellular debris (reviewed in REFS 1–5). Complement consists of a well-balanced network of circulating and cell-surface-bound proteins, which serve as substrates, enzymes or modulators of a hierarchical series of extracellular proteolytic cascades. There are three established mechanisms of complement activation; these are known as the classical, lectin and alternative pathways (FIG. 1a). The initial steps that trigger these activation processes differ considerably. The classical pathway is stimulated by the recognition of antigen–antibody complexes on foreign-cell surfaces by the hexameric complement component C1q. Structurally similar pattern-recognition receptors, mannose-binding lectin (MBL) and ficolins, bind to carbohydrate ligands on microbial intruders and initiate the lectin pathway. Conversely, the alternative pathway is stimulated by the spontaneous hydrolysis of native C3 or the presence of foreign surface structures (FIG. 1a). Recent findings suggest that additional processes, such as the C2-bypass6 and extrinsic protease7 pathways or properdin-mediated direct convertase assembly on microbial surfaces8, can also initiate complement activation.
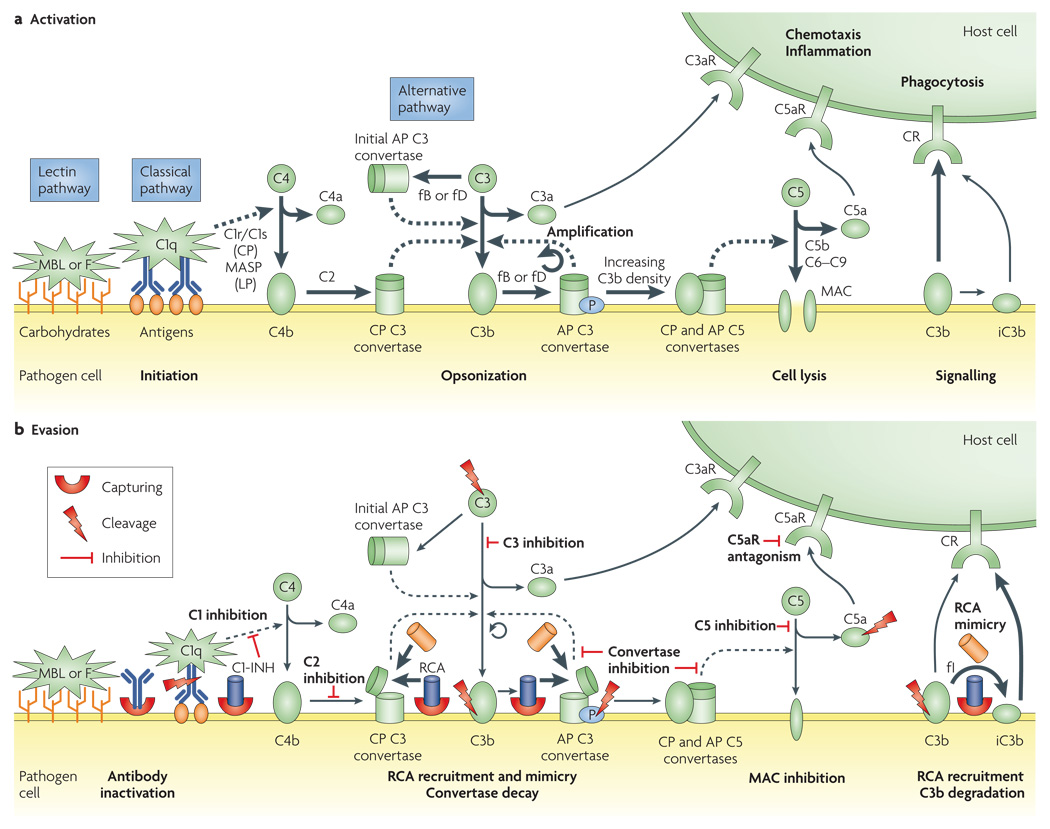
Figure 1. Activation and evasion of complement.
a | After activation of the complement system by antibody complexes (classical pathway (CP)), terminal mannose (lectin pathway (LP)) or by spontaneous and induced C3 hydrolysis (alternative pathway (AP)), the C3 convertases cleave C3 to its active fragments C3a and C3b. Covalent binding of C3b (opsonization) amplifies the cascade and mediates phagocytosis and adaptive immune responses by binding to complement receptors (CRs). Accumulation of deposited C3b also leads to the assembly of C5 convertases that activate C5 to C5a and C5b. Whereas C5b initiates the formation of the lytic membrane-attack complex (MAC), the anaphylatoxins C3a and C5a induce pro-inflammatory and chemotactic responses by binding to their receptors (C3aR and C5aR). On pathogenic surfaces, properdin (P) induces and stabilizes the AP C3 convertase, which leads to enhanced complement activity. b | Microorganisms have developed many ways to evade complement actions. Suppression of CP activation can be achieved by trapping endogenous C1 inhibitor (C1-INH) to the surface or by inactivating antibodies through the capture of their Fc regions. Whereas the recruitment of soluble regulators by capturing host proteins is a common strategy to impair downstream complement actions, certain viruses also produce structural mimics of these regulators. In addition, some microbial proteins have similar activities to CD59 in preventing MAC formation. Direct inhibition of C3, the C3 and C5 convertases, C5 or the C5a receptor (C5aR) is a prominent strategy of Staphylococcus aureus. Finally, a set of different microbial proteases can degrade many of the crucial components of the complement system. These proteases act directly or by capturing and activating a human protease. An extended list of complement evasion proteins can be found in Supplementary information S1 (table). Increased and decreased activity is represented by thick and thin arrows, respectively. F, ficolin; fB, factor B; fD, factor D; fI, factor I, MASP, MBL-associated serine protease; MBL, mannose-binding lectin; RCA, regulators of complement activation.
All of the complement cascades culminate in the central cleavage of C3 and the generation of its active fragments C3a and C3b (FIG. 1a). Opsonization of foreign surfaces by covalently attached C3b fulfils three major functions: cell clearance by phagocytosis; amplification of complement activation by the formation of a surface-bound C3 convertase; and assembly of the C5 convertases. Cleavage of C5 induces the formation of a multiprotein pore complex (the membrane-attack complex (MAC)), which leads to cell lysis. Both the covalent attachment of C3b and the stabilization of the C3 convertase by the complement regulator properdin are greatly encouraged by hydroxyl-rich pathogen surfaces. A series of complement receptors mediate the recognition of opsonized cells by leukocytes, which results in phagocytosis and the stimulation of the adaptive immune system (for example, by B and T cells). Finally, the anaphylatoxins C3a and C5a are released during complement activation and trigger a range of chemotactic and pro-inflammatory responses (for example, the recruitment of inflammatory cells and an increase in microvasculature permeability). In this way, the complement cascade also supports and promotes the function of downstream mechanisms of the immune response.
Excessive complement activation on self tissue has severe effects and can lead to the development of various diseases2,9,10. In addition to a location- and time-based restriction to immediate sites of activation, a finely tuned set of soluble and membrane-bound regulators ensure that any action of complement on host cells is either prevented or actively inhibited. These structurally similar regulators of complement activation (RCA) comprise complement receptor 1 (CR1), factor H, factor H-like protein-1 (FHL-1), C4-binding protein (C4BP), decay-accelerating factor (DAF) and membrane cofactor protein (MCP). RCA either destabilize the C3 convertase (decay-acceleration activity; carried out by CR1, factor H, FHL-1, C4BP and DAF) or promote the factor-I-mediated degradation of C3b to iC3b (cofactor activity; carried out by CR1, factor H, C4BP, FHL-1 and MCP). CR1 also acts as a cofactor for an additional factor-I cleavage of iC3b to C3d. A structurally unrelated regulator protein, CD59, prevents formation of the MAC. Finally, the glycoprotein C1-inhibitor controls the activity of several enzymes that are involved in the first steps of the classical and lectin pathways (C1r, C1s and MBL-associated serine protease).
This arsenal of proteases, active components and regulators is not only essential for the efficient elimination of microbial intruders, but also modulates the functions of the adaptive immune system11. Complement also seems to be able to communicate with other important cascades and networks in the human body. Such connections have, for example, been shown for the coagulation system7. Furthermore, recent data suggest that crosstalk between complement and Toll-like receptors might have synergistic effects in the immune response to infections12,13. Despite all of its benefits, however, the many proteins that are involved in this powerful network also provide pathogenic organisms with numerous locations where anti-complement strategies can be executed.
Mechanisms of complement evasion
The ability to escape the elaborate machinery of the human immune system is a key determinant in the virulence of pathogens. In fact, immune-evasion strategies are often focused on the complement system, which is a centre-piece of innate immunity and is generally regarded as the first line of defence against pathogenic microorganisms. Our knowledge of how these escape mechanisms function on a molecular level has increased remarkably in recent years. Identification of the individual pathogenic proteins and their human targets has been one crucial step in this task and the list of complement-targeting proteins is constantly growing. Despite this plethora of complement-binding proteins (Supplementary information S1 (table)), their mechanisms of action can be condensed to a few successful strategies: the recruitment or mimicking of complement regulators; the modulation or inhibition of complement proteins by direct interactions; and inactivation by enzymatic degradation (FIG. 1b; TABLE 1). In addition to these strategies, many microorganisms also possess passive evasive features — a prominent example is the cell wall of Gram-positive bacteria, which prevents lysis by the MAC14. The following sections focus on the active complement-evasion strategies and illustrate their common and unique features.
Table 1.
Examples of complement evasion proteins and their targets on host cells*
| Complement evasion protein | Host target |
|---|---|
| Antibody depletion | |
| Staphylococcal protein A (SpA) | IgG |
| Complement inhibition | |
| Extracellular fibrinogen-binding protein (Efb) | C3 and C3b-containing convertases |
| Staphylococcal superantigen-like protein-7 (SSL-7) | C5 |
| Staphylococcus complement inhibitor (SCIN) | C3 convertases |
| Complement C2 receptor trispanning protein (CRIT) | C2 |
| Chemotaxis inhibitory protein of Staphylococcus aureus (CHIPS) |
C5a receptor (C5aR) |
| Regulators of complement activation (RCA) recruitment | |
| Complement-regulator-acquiring protein (CRASP) |
Factor H, factor H-like protein-1 (FHL-1) and C4-binding protein (C4BP) |
| M protein family | Factor H, FHL-1 and C4BP |
| RCA mimicry | |
| Variola virus complement-control protein (VCP) | C3b and C3 convertases |
| Smallpox protein of complement enzymes (SPICE) | C3b and C3 convertases |
| Proteolytic degradation | |
| Staphylokinase | C3b and IgG (by activation of plasmin) |
| Pseudomonas elastase (PaE) | C3 |
| 56 kDa protease | C5a |
See Supplementary information S1 (table) for an extended list of microbial evasion proteins.
Making use of the host’s arsenal — acquiring regulators
The presence of RCA on cell surfaces is a key determinant for complement function on self and non-self cells. Although most pathogens do not express RCA on their surfaces, they have found ways to adapt by stably binding the RCA that circulate in human plasma15 (FIG. 1b; TABLE 1). In fact, of all the mechanisms of complement evasion that are used by various pathogens, trapping RCA is by far the most widely disseminated strategy for avoiding the complement response. RCA recruitment is common in bacteria16 (for example, Escherichia coli, Borrelia burgdorferi and streptococci), but has also been described for viruses17 (for example, HIV-1), fungi18,19 (for example, Candida albicans) and parasites20 (for example, Echinococcus spp.) (Supplementary information S1 (table)). The recruitment strategy has important advantages: RCA are the natural regulators of complement and are evolutionarily tuned to fulfil their functions; they are produced by the host and are available in relatively high concentrations; and they share common structural features, called short consensus repeat (SCR) domains, that allow the same pathogen-derived binding protein to recruit different host RCA.
Although they are less potent than the cell-surface-bound CR1, the availability of C4BP, factor H and FHL-1 as soluble proteins means that these proteins are also primary targets. All three regulators show decay acceleration and cofactor activity. Owing to their size and multivalency, the captured regulators do not usually lose their overall activity. In many cases, the recruitment function can be attributed to a family of similar proteins in a bacterial genus. Typical examples are the complement-regulator-acquiring proteins in Borrelia spp., porins in Neisseria spp. and the M protein family in Streptococcus spp.16 The recruitment of regulators is not restricted to soluble RCA, however. The in vivo transfer of membrane-bound regulators (MCP, DAF and CD59) between human cells has been reported21, and the acquisition of CD59 by this mechanism has been suggested for E. coli22 and Helicobacter pylori23. Finally, some viruses camouflage themselves with RCA-containing host membrane during budding or stimulate RCA expression on the host cell24.
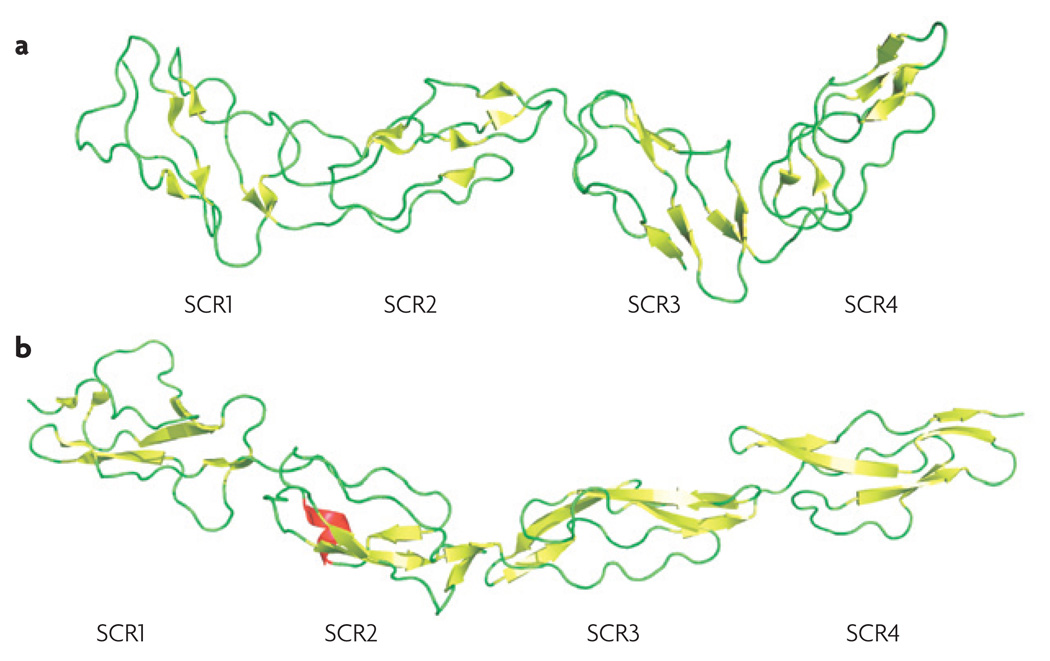
Certain viruses have found ways to produce soluble proteins that closely mimic the structure and function of host regulators (FIG. 1b; TABLE 1). The orthopoxviruses variola (smallpox) and vaccinia express complement- inhibitory proteins25,26 that consist of four SCR domains. These modules are the common building blocks and functional entities of all human RCA, and are also found in complement receptors (for example, CR2) and factors (for example, factor B). Structural analysis of the complete vaccinia virus complement-control protein (VCP)27 (FIG. 2a) clearly demonstrated its similarity to human RCA, such as DAF (FIG. 2b). Despite their small size compared with endogenous factor H (20 SCRs) and CR1 (30 SCRs), both VCP and the smallpox inhibitor of complement enzymes (SPICE) show decay acceleration as well as cofactor activity. Although these proteins vary in only 11 residues, the resulting changes in the electrostatic surface potential seem to have dramatic effects on their regulatory potency; SPICE had a 1000-fold higher inhibitory activity for the alternative pathway than VCP28. Similar RCA-like proteins have also been described for monkeypox29 and cowpox30, two viruses that can infect humans. Interestingly, the monkeypox protein MOPICE (monkeypox inhibitor of complement proteins) contains only three SCR units and lacks decay-accelerating activity. Although a membrane-bound VCP homologue has been reported in vaccinia virus (B5R)31, no complement-based functions could be detected for this protein (J.D.L., unpublished observations). Recent discoveries show that the same class of molecule is also produced in herpes viruses. A type 1 membrane protein that has four SCR repeats and dual complement-regulation activity has been isolated from Kaposi’s sarcoma-associated herpesvirus (HHV-8) and has been named Kaposica32.
Figure 2. Molecular mimicry of human complement regulators by viruses.
Certain orthopox viruses and herpes viruses express proteins that closely mimic the structure and function of human regulators of complement activation. This structural similarity is illustrated for vaccinia virus complement-control protein (VCP; Protein Data Bank (PDB) code: 1G40 (REF. 27)) (a) and human decay-accelerating factor (DAF; PDB code: 1OJV102) (b), both of which have four short consensus repeat (SCR) domains. These SCRs (or complement control protein modules) are common structural motifs in complement components (for example, complement receptors and regulators) and other proteins (such as selectins).
The structural correspondence between these regulators and host RCA (FIG. 2) might reflect their ancient human origins, as the hijacking and implementation of sequences that encode beneficial host proteins has long been recognized as a hallmark of viral biology and evolution. However, viruses might not be the only microorganisms that follow this strategy. For example, complement-regulatory proteins have been described for various Ixodes spp. ticks. Both the Ixodes scapularis anti-complement protein (ISAC) and its counterpart from Ixodes ricinus (IRAC) exert decay-acceleration activity on the C3 convertases. However, any structural similarities between these proteins and host RCA has yet to be confirmed33,34.
Cutting through complement: pathogenic proteases
The degradation of complement components into smaller, non-functional fragments is the primary task of complement-active proteases. This class of evasion proteins has been reported almost exclusively in bacteria and targets a wide range of substrates. For example, the degradation of immunoglobulins (Igs) and C1q (for example, by Pseudomonas elastase (PaE) and alkaline protease (PaAP)) prevents activation by the classical pathway35, whereas cleavage of C5a (by the 56 kDa protease from Serratia marcescens36 or the C5a peptidases from streptococci37) efficiently disables C5a-mediated pro-inflammatory and chemotactic signalling. Streptococcal pyrogenic exotoxin B was found to degrade the complement regulator proper-din, which subsequently loses its ability to stabilize the convertases on pathogenic surfaces38. Inactivation of C3 by cleavage into non-functional fragments has been described for Pseudomonas spp. proteases (PaE and PaAP) and Porphyromonas spp. proteases (PrtH)35. A rare example of non-bacterial proteases that have complement activity has been observed in the parasitic worm Schistosoma mansoni, which degrades iC3b using a 28-kDa protease, resulting in impaired binding to CR3 (REF. 39).
The acquisition and activation of host proteases is a more recently described alternative to the enzymatic attenuation of complement. For example, the conversion of endogenous plasminogen into plasmin on bacterial surfaces can lead to the degradation of immunoglobulin G (IgG) and C3 fragments. Although a range of bacteria express plasminogen activators and/or receptors40, this mechanism is best described for S. aureus (discussed below)41.
Direct interventions — microbial complement inhibitors
A series of highly specific protein–protein interactions is the driving force behind the sophisticated activation and regulatory mechanisms of the complement system. Any interruption of the individual binding events can break this delicate balance and destabilize complement efficiency. It is surprising, therefore, that only a few direct complement inhibitors have been identified so far; most of these have been discovered in S. aureus and their molecular mechanisms are more thoroughly discussed in the following section. The CD59-like protein from B. burgdorferi is one of the few examples of a direct complement inhibitor in other bacteria. This membrane-bound protein, which has affinity for C9 and C8b, is able to inhibit MAC formation. Similarly, streptococcal inhibitor of complement (SIC) has been found to prevent MAC formation by interfering with the C5b–C7 and C5b–C8 complexes. These functions of SIC seem to be paradoxical, as streptococci are already resistant to MAC-mediated cytolysis owing to their cell-wall structure14,42. This seemingly makes SIC ineffective, or at least redundant, in this respect. Although additional non-complement functionalities have been attributed to SIC, the reason for its MAC-inhibitory potential remains unsolved.
In herpes viruses, anti-complement activities have been attributed to several glycoproteins. The most versatile examples are the transmembrane gC1 and gC2 from herpes simplex virus type 1 and 2, respectively. Both glycoproteins were found to bind C3b and specifically accelerate the decay of the alternative pathway C3 convertase43. Although gC2 has a tenfold stronger affinity for C3b44, only gC1 can also inhibit the interaction of C3b with C5 and properdin45. Other glycoproteins of the herpes virus family have Fc-receptor properties and can deplete antibody recognition and the activation of the classical pathway43,46. Ig-inactivating proteins have also been identified in bacteria, such as S. aureus and group G streptococci35.
Certain human parasites also express direct complement inhibitors. The complement C2 receptor trispanning protein has been detected in species of Schistosoma and Trypanosoma. This protein disrupts the interaction between C2 and C4 and, therefore, prevents formation of the classical pathway C3 convertase47. Furthermore, the Schistosoma spp. protein paramyosin prevents MAC formation by binding C8 and C9 (REF. 48), and has other immunomodulatory activities because of its binding affinity for C1q and IgG49. Finally, soft ticks of the genus Ornithodoros express at least one protein that keeps complement activation in check during their blood meal. The 17 kDa Ornithodoros moubata complement inhibitor has been shown to directly bind to C5 and prevent its cleavage, probably by inhibiting substrate binding to the C5 convertase50,51.
Finding the right mixture of activation and inhibition
Although complement inhibition and regulation is the predominant mechanism that is used to evade host immune defence by most pathogens, there is increasing evidence that some microorganisms actively induce complement activity for their own purposes. One of the most intriguing examples of fine-tuning antipodal actions that is used to ‘outwit’ host immune defence is that accomplished by HIV-1 — although this virus takes advantage of opsonization to enter human cells through complement receptors (BOX 1), the terminal actions, such as MAC formation and phago-cytosis, must be avoided. Strikingly, the same glycoproteins that activate complement through the classical and lectin pathways (gp41 and gp120, respectively) are also responsible for recruiting the regulators factor H and CD59 to the surface of the virus. In addition to inhibition of MAC formation by CD59, the degradation of C3b by factor I and factor H reduces amplification. However, the presence of iC3b still allows HIV-1 to hijack complement receptors to enter immune cells (BOX 1).
Hitchhiking with complement to enter host cells
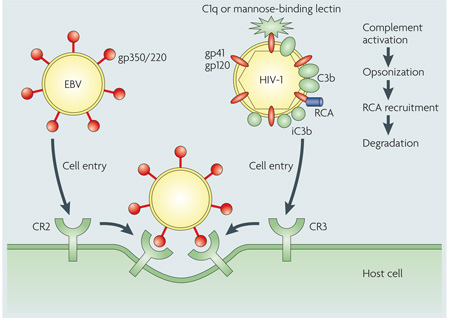
Many human pathogens increase their virulence by avoiding confrontation with complement components or inactivating them through inhibition and degradation. However, a small group of pathogens, predominantly viruses, follow a completely different strategy by using complement receptors (CR) as an entry port into host cells. The general principle of binding to human target receptors using surface proteins, with subsequent internalization, is common to most viruses89, yet only a few target complement proteins3,17. For example, Epstein–Barr virus (EBV) expresses the glycoprotein gp350/220, which specifically interacts with CR2 on B cells and immature T cells (see the figure, left), ultimately causing infectious mononucleosis (Pfeiffer’s disease)90,91. In addition, surface-bound complement regulators serve as targets for the measles virus (haemagglutinin binds to membrane cofactor protein (MCP))92, human herpesvirus 6 (glycoprotein H binds to MCP)93 and some enteroviruses (coxsackievirus and echovirus bind to decay-accelerating factor (DAF))17. Similarly, surface-bound complement regulators, such as DAF and MCP, are recognized by other microbial proteins (for example, the Dr adhesin of Escherichia coli94) and have important roles in the infectivity and colonization of these pathogens.
Even without expressing complement-specific surface proteins, pathogens have found intriguing ways to use the hitchhiking principle — they voluntarily activate complement to become opsonized and take advantage of the deposited C3 fragments to interact with complement receptors on human cells. This formidable strategy has been observed in viruses (for example, HIV-1) (see the figure, right)3,95 and Mycobacterium tuberculosis, both of which use CR3 (REF. 96). However, the destructive potential of complement requires that the pathogen keeps complement activation in check by other mechanisms, such as the recruitment of host regulators of complement activity (RCA). Specific blockage of these activation and attachment steps could be a promising approach in the development of antimicrobial therapies.
The diversity of the complement-targeting proteins (proteases, inhibitors and regulators) suggests that certain bacteria might have developed a similarly elaborate network of balanced complement activation and inhibition. In this respect, S. aureus produces a nearly unparalleled ensemble of immune-evasive molecules that target the complement system.
S. aureus — a master of complement evasion
S. aureus is a prototypic opportunistic pathogen and a leading cause of nosocomial and community-acquired infections52. Perhaps more than any other bacterial pathogen, S. aureus has evolved the ability to adapt to, and persist within, the diverse physiological microenvironments of its host (for example, skin and bones). Recent studies on this versatile bacterium have uncovered a formidable arsenal of virulence-facilitating proteins and structures that contribute to its pathogenesis.
Whereas the impact of S. aureus cell-surface-retained adhesins on host invasion and colonization has been firmly established53,54, the identities and functions of this bacterium’s immune-evading molecules have emerged only recently (reviewed in REFS 35,55–57). Several studies have revealed new mechanisms of immune evasion and modulation, and S. aureus has developed into a model system to study the fascinating interplay of host–pathogen interactions. Some strains synthesize capsular polysaccharides58 that have been shown to impede antibody recognition59,60 and interfere with opsonization and C3b recognition by its receptors61. As the peptidoglycan-rich structure of its Gram-positive cell wall renders the bacterium resistant to MAC formation42,55, most of the active evasion processes of S. aureus target the initial, amplifying and pro-inflammatory steps of complement activation.
Prevention of complement initiation
There are several mechanisms through which S. aureus impairs initiation of the classical pathway. Among these, the structure and function of staphylococcal protein A (SpA) (FIG. 3a) has been characterized most extensively. This surface-bound protein recognizes the Fc domain of Igs with high affinity62,63, which results in inverted tagging and block-age of the C1q (and Fcγ receptor) binding sites. It has been shown that SpA can also bind to the C1q receptor gC1qR/p33, which is highly expressed on the surface of activated platelets and endothelial cells64. However, this mechanism might be more closely associated with cell adhesion, through bridging to fibrinogen, than immune evasion65. Combined with the recent discovery that SpA can bind to von Willebrand factor and tumour-necrosis factor receptor 1 (REFS 66,67), these findings could reflect a far more versatile role for SpA than was initially realized. Interestingly, a second, less-characterized protein that has Ig-binding properties, called Sbi (S. aureus IgG-binding protein), has been described68. Whether Sbi shares the same physiological versatility as SpA remains to be established.
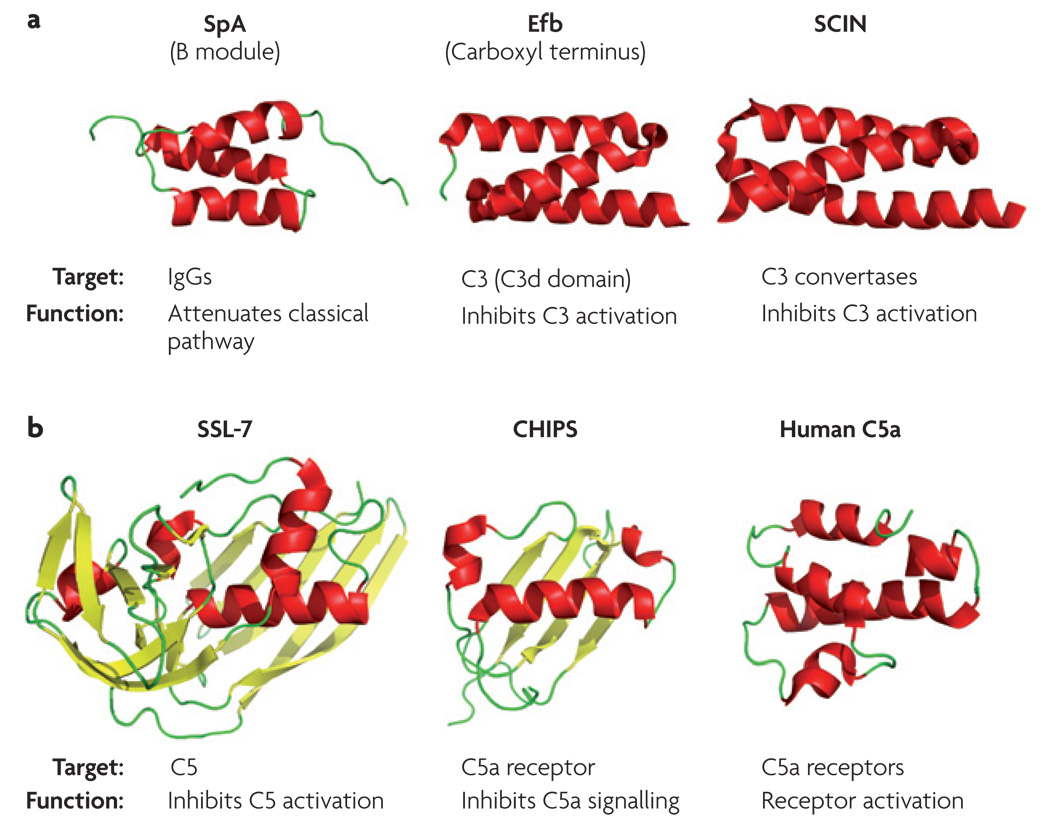
Figure 3. Complement-targeting proteins in Staphylococcus aureus.
A range of S. aureus proteins that interact with targets from the complement system have been structurally analysed. a | The immunoglobulin G (IgG)-binding module of staphylococcal protein A (SpA; Protein Data Bank (PDB) code: 1BDD63), the C3-binding domain of the extracellular fibrinogen-binding protein (Efb; PDB code: 2GOM74) and staphylococcal complement inhibitor (SCIN; PDB code: 2QFF78) share similar structural motifs, but show differential functionality. b | Both staphylococcal superantigen-like protein-7 (SSL-7; PDB code: 1V1O103) and the chemotaxis inhibitory protein of S. aureus (CHIPS; PDB code: 1XEE104) attenuate complement on the level of C5, but bind to unrelated targets (C5 protein and C5a receptor, respectively). The solution structure of human C5a (obtained by nuclear magnetic resonance at pH 5.2; PDB code: 1KJS105) is shown as a comparison. All protein-structure representations are coloured according to their secondary structure (α-helices in red and β-sheets in yellow).
Distinct use of proteolytic enzymes
Even for proteases, S. aureus has developed an indirect but highly efficient way of counteracting complement. Although staphylokinase is not an enzyme itself, the formation of a complex with plasminogen converts this endogenous protein into the active serine protease plasmin. The broad substrate spectrum of plasmin includes C3 and human IgG. Even more strikingly, plasmin was recently shown to bind to the surface of S. aureus and remove deposited IgG, C3b and iC3b from its surface, thereby reversing the effect of opsonization41. Plasmin cleavage of IgG at its hinge region might also attenuate classical-pathway activation through C1q. In addition to plasmin activation, staphylokinase directly abolishes the bactericidal properties of α-defensins, which makes it an important colonization factor69. Although the anti-opsonization activities of surface-activated plasmin have so far only been described for S. aureus, the presence of plasminogen activators and receptors on various other bacteria40 suggests that this strategy could be more widely prevalent.
Suppression of complement amplification
Surprisingly, no RCA-recruiting proteins have been reported for S. aureus so far. However, recent reports have described the factor-I-mediated degradation of C3b to iC3b that is independent of factor H on the surface of certain S. aureus strains. The overall result of this process is the decreased efficiency of bacterial killing by phagocytic neutrophils70,71. Although these findings are suggestive of a bacterially derived cofactor protein, current evidence suggests that S. aureus has evolved different pathways for counteracting the downstream actions of complement. By targeting the central components C3 and C5, most of the opsonization, amplification and signalling steps can be efficiently suppressed. In fact, there are several unique S. aureus proteins that inhibit the conversion of native C3 to C3b. Extracellular fibrinogen-binding protein (Efb) was the first C3b-binding protein to be identified in S. aureus and was shown to inhibit opsonophagocytosis by granulocytes72. Biochemical and structural studies revealed that Efb binds to the thioester-containing C3d domain of C3 through its three-helix bundle carboxy-terminal domain (Efb-C)73,74 (FIG. 3a). Recently, an Efb-homologous protein (Ehp) from S. aureus was reported75 that shares a high level of structural similarity with Efb-C. Surprisingly, Ehp can bind two molecules of C3d and is a more potent inhibitor of C3 conversion through the alternative pathway75. Both proteins produce their effects by inducing conformational changes in C3 that render this central component unable to participate in downstream activation events74,75. A recent study indicates that Efb and Ehp (designated Ecb in this case76) can also bind to the C3b-containing convertases and, therefore, prevent the cleavage of both C3 (amplification) and C5 (generation of C5a)76. This convertase-centred inhibition is probably a manifestation of a conformational change in C3b that has occurred upon the binding of Efb74.
S. aureus also expresses another group of small, helical proteins that block complement activation. These staphylococcal complement inhibitors (SCINs) (FIG. 3a) seem to stabilize the C3 convertases in a non-functional state, thereby efficiently blocking all three pathways77. In contrast to Efb and Ehp, SCINs bind only to the assembled C3 convertases76–78 and not to C3 or its fragments. Both the Efb and SCIN families share a high degree of structural similarity with the IgG-binding module of SpA74,78 (FIG. 3a). However, a closer inspection of these structures reveals that the spatial arrangement of the helices is distinct. SCINs are more similar to SpA than Efb or Ehp, which may partly explain the differential binding specificities of these proteins.
Inhibition on the level of C5
In addition to the interference at the stage of C3, S. aureus also produces two proteins that disrupt the downstream inflammatory responses that are carried out by C5 and its activation products. Staphylococcal superantigen-like protein-7 (SSL-7) (FIG. 3b) has recently been shown to bind native C5 with high affinity, resulting in the inhibition of complement-mediated haemolysis and killing of E. coli in an in vitro setting79. SSL-7 also interacts with IgA in a C5-independent manner79. Separately, the chemotaxis inhibitory protein of S. aureus (CHIPS) (FIG. 3b) was found to impair neutrophil and monocyte responses to C5a signalling, primarily by interacting with and antagonizing the C5a receptor80–82. Although CHIPS does not share substantial homology in either sequence or secondary structure with the natural ligand C5a (FIG. 3b), the similar overall shape and size of these molecules might explain their competition for the same receptor. Interestingly, and despite their structurally and functionally unrelated targets (native C5 and C5a receptor, respectively), the structures of SSL-7 and CHIPS share some similarities55 (FIG. 3b). In this respect, the prominent fold that is observed in both proteins is related to the β-grasp domains that are found in several bacterial superantigens and superantigen-like proteins82,83, and are representative of a large family of bacterial immune-evasion proteins.
Therapeutic potential
Our increasing knowledge of individual microbial agents, their human targets and their mechanisms of action could not only facilitate our understanding of microbial infection but also pave the way towards new therapeutics. Currently, our arsenal of antibiotic drugs is becoming alarmingly ineffective against many severe infectious diseases and the pipeline of novel therapeutic concepts is thin. In the past, drug-screening initiatives focused mainly on bacterial metabolism targets rather than immune evasion84. Nevertheless, recent discoveries of complement-attenuating proteins have revealed some promising, albeit indirect, targets for therapeutic intervention. One potential limitation of this approach is that microorganisms often have multiple, sometimes redundant, evasion mechanisms15, and not all of these are equally important in pathogenesis. For example, the acquisition of factor H is not essential for the mammalian infection of B. burgdorferi85. Although confirmation of individual proteins as virulence factors by in vivo mutational studies should be a first and crucial step for selecting drug targets, it is clear that classifying promising targets by basic research will provide the foundation of any such development efforts. To this end, governmental initiatives, such as that implemented by the United States Food and Drug Administration (FDA) — for example, the Antibiotic Safety and Innovation amendment to the Revitalization Act and the Orphan Drug Act — may provide new incentives for antibiotic drug development in which evasion strategies are targeted86.
Whereas the development of new antibiotics might represent a speculative endeavour, the path towards the development of new therapeutics for complement-mediated diseases seems clearer. Increased activation or insufficient regulation of this attack system on human cells is known to induce an ever-growing list of autoimmune, inflammatory and ischaemic conditions9. In many cases, the mechanisms that are involved have been revealed and numerous attractive pharmacological targets within the complement network have been identified2. Nonetheless, the number of anti-complement therapeutics on the market remains surprisingly limited10. In this respect, many microbial proteins are adept at inhibiting the complement response, and these molecules provide a broad structural and functional frame-work for complement-modulating agents. Although the therapeutic administration of soluble forms of human RCA has long been considered to be an ideal method for supporting the natural regulation of complement, their size (for example, soluble CR1) or restricted activity (for example, soluble DAF and MCP) has limited this approach. In this context, the development of modified, non-immunogenic viral RCA mimics based on SPICE and VCP could overcome some of these limitations, as they have high decay-acceleration and cofactor activity but are comparatively small. Indeed, VCP has been tested in an in vitro model of Alzheimer’s disease and animal models of head trauma87,88. Furthermore, the structural information on these highly potent mimics27,28 could serve as a template for designing modified host RCA that have enhanced functional properties.
The versatile arsenal of direct complement inhibitors that is provided by S. aureus could be another promising source of drug templates. As potent inhibitors of the alternative pathway, proteins such as Efb, Ehp and SCIN have clinical potential for pathological conditions in which the excessive or uncontrolled generation of C3b seems to result in disease (for example, age-related macular degeneration). Owing to its broad specificity for all C3 convertases, SCIN could also be effective for conditions in which activation of the classical pathway is involved (for example, systemic lupus erythematosus). Finally, impairing the signalling function of the C5a receptor using CHIPS could serve as a new approach for a range of inflammatory disorders. Although all these proteins are highly soluble and can be expressed in high quantities, their potential immunogenicity could severely limit their direct use as therapeutics, as has recently been shown for CHIPS82. However, the availability of robust structural data for these proteins (in some cases, even as co-crystals with their human target (BOX 2)) should facilitate the rational design of smaller, synthetic compounds that mimic the potent functions of these microbial proteins.
Building structure into complement evasion
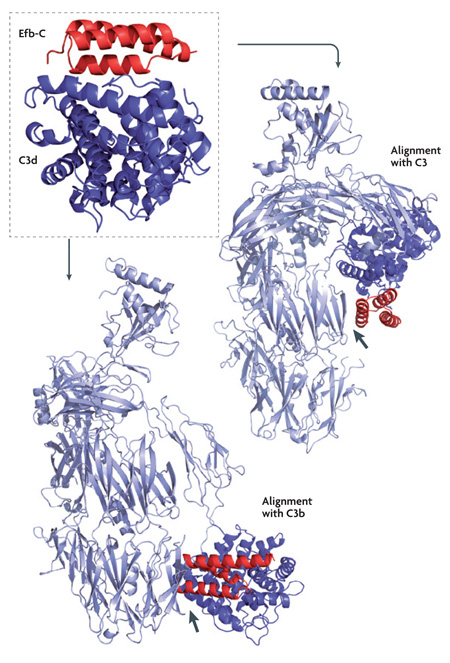
The recent increase in structural information for microbial evasion proteins and individual complement components (reviewed in REFS 97,98) has vitally enriched our comprehension of immune-escape mechanisms. However, for a detailed understanding of the underlying processes, and the development of therapeutic interventions, knowledge of the co-crystal structures between pathogenic proteins and their complement targets would be highly beneficial. Except for the early examples of staphylococcal protein A (SpA) and streptococcal protein G (SpG), which bind to the Fc portion of immunoglobulin G (IgG)99,100, no such structural information has been available. Fortunately, the recent release of the complex structures between C3d and two staphylococcal inhibitors — the carboxyl terminus of extracellular fibrinogen-binding protein (Efb-C)74 (Protein Data Bank (PDB) code: 2GOX) and Efb-homologous protein (Ehp)75 (PDB code: 2NOJ) — have provided a first and important step in this direction. On the one hand, a detailed characterization of the binding interface between bacterial and host proteins can identify key residues and essential thermodynamic properties for the interaction. For example, electrostatic contributions have not only been found to be highly important in the case of Efb and Ehp, in which the mutation of two charged residues to alanine completely abolished C3d binding and complement inhibition74,75, but also for vaccinia virus complement control protein (VCP) and smallpox inhibitor of complement enzymes (SPICE)28. This could indicate a general principle of complement-targeted interaction that needs to be observed and validated in the future. On the other hand, the combination of structural information with biophysical and biochemical methods provides a fascinating insight into the molecular mechanism of C3 inhibition by Efb and Ehp. Whereas direct binding studies indicate preferential binding to native C3, rather than its active fragment C3b, biochemical data suggest a conformational change is induced upon complex formation. Structural alignment of the Efb-C–C3d co-crystal with the C3d domains of native C3 and C3b supported this hypothesis; whereas Efb-C might contact additional sites in C3, its docking to C3b led to steric hindrance (see the figure). This example shows how detailed structural information, combined with biochemical and interaction data101, can further propel this area of research.
Perspectives
The coexistence of humans and microorganisms has generated a fascinating interplay of mechanisms that maintain the balance between infection, defence and evasion. For many infectious diseases, however, this balance is changed in favour of the pathogenic intruders. Alongside many other factors, such as cell adhesion and entry or colonization efficiency, avoiding the attacks of the immune system is a key determinant of microbial survival inside an often hostile host environment. The diverse and elaborate evasion strategies that are discussed here illustrate the importance of the human complement system as a target for immune escape.
The new structural insights into complement evasion, combined with our increased knowledge of complement functions in microbial defence and human disease, provide an exciting platform for long-awaited progress in research. In this context, combinations of various techniques and more systemic approaches are an important premise for coping with the complex nature of host–pathogen interactions. The continued availability of sequenced microbial genomes can only supplement our list of known complement-targeting proteins. With an eye to the future, continued studies on such mechanisms of immune evasion and complement inhibition should not only further our under-standing of these host–pathogen interactions, but also provide vital insights into better treatment strategies for many types of human diseases.
DATABASES
Entrez Genome: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=genome
cowpox | HHV-8 | HIV-1 | monkeypox | vaccinia | variola
Entrez Genome Project: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=genomeprj
Borrelia burgdorferi | Candida albicans | Escherichia coli | Helicobacter pylori | Schistosoma mansoni | Serratia marcescens | Staphylococcus aureus
Protein Data Bank: http://www.rcsb.org/pdb/home/home.do
human C5a | CHIPS | DAF | Efb | SCIN | SpA | SSL-7 | VCP
FURTHER INFORMATION
John D. Lambris’s homepage: http://www.lambris.net
SUPPLEMENTARY INFORMATION
See online article: S1 (table)
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
Supplementary Material
Acknowledgements
This work was supported by National Institutes of Health grants GM-069736, GM-62134, AI-30040, AI-072106 and AI-068730, and by the research incentive funds of the School of Biological Sciences at the University of Missouri-Kansas City.
Glossary
- Pattern-recognition receptor
A highly diverse group of soluble and surface-bound proteins that can detect specific molecular surface structures. These receptors are important for discriminating between self and non-self cells (for example, microorganisms) and are, therefore, found in various pathways of the immune system. Prominent examples in the complement system are C1q and mannose-binding lectin.
- Anaphylatoxin
A small protein fragment (approximately 10 kDa) that is generated during the activation of complement components. C3a and C5a trigger a range of inflammatory and immune-stimulating responses by binding to their receptors (C3aR and C5aR) on various effector cells. The chemotactic activity of C5a is 100-fold higher than that of C3a; no such activities or receptors have so far been described for C4a.
- Toll-like receptor
A pattern-recognition receptor that recognizes a range of surface structures on pathogens (for example, proteoglycans and lipopolysaccharides). Toll-like receptors are expressed on most immune cells and their activation and signalling induces numerous inflammatory, innate and adaptive immune responses.
- Short consensus repeat
The structural building block of many complement regulators and receptors (for example, factor H and CR1). These β-sheet-rich domains, which are composed of approximately 60 residues, are also found in viral complement-evasion factors and other proteins (for example, selectins, clotting factor XIII B and GABA receptors).
- α-defensin
A cationic, cyclic, cysteine-rich peptide of 15–20 amino acids that belongs to a family of antimicrobial peptides. Whereas α-defensins are mainly expressed in mammalian neutrophils, other members of this family have been described in various species within mammals, insects and plants. Although these peptides are thought to primarily disrupt microbial cell walls, they might also act as immunomodulators.
- β-grasp domain
A structural fold that consists of anti-parallel β-strands that ‘grasp’ a single α-helix. Initially described as the central structural element of ubiquitin, this fold was later identified in several other proteins. Despite its small size, it has a wide range of functions and is present in enzymatic, binding and signalling proteins. β-grasp domains are central elements in the structures of a number of bacterial immune-evasion proteins.
Footnotes
Competing interest statement
The authors declare competing financial interests: see web version for details.
References
- 1.Lambris JD, Sahu A, Wetsel RA. In: The Human Complement System in Health and Disease. Volanakis JE, Frank MM, editors. New York: Marcel Dekker; 1998. pp. 83–118. [Google Scholar]
- 2.Lambris JD, Holers VM, editors. Therapeutic Interventions in the Complement System. Totowa: Humana; 2000. [Google Scholar]
- 3.Walport MJ. Complement. First of two parts. N. Engl. J. Med. 2001;344:1058–1066. doi: 10.1056/NEJM200104053441406. [DOI] [PubMed] [Google Scholar]
- 4. Walport MJ. Complement. Second of two parts. N. Engl. J. Med. 2001;344:1140–1144. doi: 10.1056/NEJM200104123441506. References 3 and 4 provide a brief overview of the complement system and its ambiguous involvement in infections and other diseases.
- 5.Lambris JD, editor. Current Topics in Complement. New York: Springer; 2006. [Google Scholar]
- 6.Atkinson JP, Frank MM. Bypassing complement: evolutionary lessons and future implications. J. Clin. Invest. 2006;116:1215–1218. doi: 10.1172/JCI28622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Markiewski MM, Nilsson B, Nilsson Ekdahl K, Mollnes TE, Lambris JD. Complement and coagulation: strangers or partners in crime? Trends Immunol. 2007;28:184–192. doi: 10.1016/j.it.2007.02.006. Presents the interesting connections between the complement and the coagulation systems, and discusses their impact on disease and the immune response.
- 8.Spitzer D, Mitchell LM, Atkinson JP, Hourcade DE. Properdin can initiate complement activation by binding specific target surfaces and providing a platform for de novo convertase assembly. J. Immunol. 2007;179:2600–2608. doi: 10.4049/jimmunol.179.4.2600. [DOI] [PubMed] [Google Scholar]
- 9.Volanakis JE, Frank MM, editors. The Human Complement System in Health and Diseases. New York: Marcel Dekker; 1998. [Google Scholar]
- 10. Ricklin D, Lambris JD. Complement-targeted therapeutics. Nature Biotechnol. 2007;25:1265–1275. doi: 10.1038/nbt1342. A current overview of the complement-specific drugs that are on the market and in clinical trials, and a discussion of how endogenous, pathogen-derived or synthetic drugs can be used to develop such therapeutics.
- 11.Morgan BP, Marchbank KJ, Longhi MP, Harris CL, Gallimore AM. Complement: central to innate immunity and bridging to adaptive responses. Immunol. Lett. 2005;97:171–179. doi: 10.1016/j.imlet.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 12.Hawlisch H, et al. C5a negatively regulates toll-like receptor 4-induced immune responses. Immunity. 2005;22:415–426. doi: 10.1016/j.immuni.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 13.Zhang X, et al. Regulation of Toll-like receptor-mediated inflammatory response by complement in vivo. Blood. 2007;110:228–236. doi: 10.1182/blood-2006-12-063636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joiner K, Brown E, Hammer C, Warren K, Frank M. Studies on the mechanism of bacterial resistance to complement-mediated killing. III. C5b-9 deposits stably on rough and type 7 S. pneumoniae without causing bacterial killing. J. Immunol. 1983;130:845–849. [PubMed] [Google Scholar]
- 15. Zipfel PF, Wurzner R, Skerka C. Complement evasion of pathogens: common strategies are shared by diverse organisms. Mol. Immunol. 2007;44:3850–3857. doi: 10.1016/j.molimm.2007.06.149. A supplementary description of the current topics in the field of complement evasion, with the focus on redundancy and multiplicity, the acquisition of host regulators and simultaneous binding to other host proteins.
- 16.Kraiczy P, Würzner R. Complement escape of human pathogenic bacteria by acquisition of complement regulators. Mol. Immunol. 2006;43:31–44. doi: 10.1016/j.molimm.2005.06.016. [DOI] [PubMed] [Google Scholar]
- 17. Bernet J, Mullick J, Singh AK, Sahu A. Viral mimicry of the complement system. J. Biosci. 2003;28:249–264. doi: 10.1007/BF02970145. A broad overview of the use of complement proteins by viruses that discusses immune evasion and cell attachment and entry.
- 18.Meri T, et al. The hyphal and yeast forms of Candida albicans bind the complement regulator C4b-binding protein. Infect. Immun. 2004;72:6633–6641. doi: 10.1128/IAI.72.11.6633-6641.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meri T, et al. The yeast Candida albicans binds complement regulators factor H and FHL-1. Infect. Immun. 2002;70:5185–5192. doi: 10.1128/IAI.70.9.5185-5192.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inal JM. Parasite interaction with host complement: beyond attack regulation. Trends Parasitol. 2004;20:407–412. doi: 10.1016/j.pt.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 21.Kooyman DL, et al. In vivo transfer of GPI-linked complement restriction factors from erythrocytes to the endothelium. Science. 1995;269:89–92. doi: 10.1126/science.7541557. [DOI] [PubMed] [Google Scholar]
- 22.Rautemaa R, Jarvis GA, Marnila P, Meri S. Acquired resistance of Escherichia coli to complement lysis by binding of glycophosphoinositol-anchored protectin (CD59) Infect. Immun. 1998;66:1928–1933. doi: 10.1128/iai.66.5.1928-1933.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rautemaa R, et al. Survival of Helicobacter pylori from complement lysis by binding of GPI-anchored protectin (CD59) Gastroenterology. 2001;120:470–479. doi: 10.1053/gast.2001.21197. [DOI] [PubMed] [Google Scholar]
- 24.Tortorella D, Gewurz BE, Furman MH, Schust DJ, Ploegh HL. Viral subversion of the immune system. Annu. Rev. Immunol. 2000;18:861–926. doi: 10.1146/annurev.immunol.18.1.861. [DOI] [PubMed] [Google Scholar]
- 25.McKenzie R, Kotwal GJ, Moss B, Hammer CH, Frank MM. Regulation of complement activity by vaccinia virus complement-control protein. J. Infect. Dis. 1992;166:1245–1250. doi: 10.1093/infdis/166.6.1245. [DOI] [PubMed] [Google Scholar]
- 26.Rosengard AM, Liu Y, Nie Z, Jimenez R. Variola virus immune evasion design: expression of a highly efficient inhibitor of human complement. Proc. Natl Acad. Sci. USA. 2002;99:8808–8813. doi: 10.1073/pnas.112220499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murthy KH, et al. Crystal structure of a complement control protein that regulates both pathways of complement activation and binds heparan sulfate proteoglycans. Cell. 2001;104:301–311. doi: 10.1016/s0092-8674(01)00214-8. [DOI] [PubMed] [Google Scholar]
- 28.Sfyroera G, Katragadda M, Morikis D, Isaacs SN, Lambris JD. Electrostatic modeling predicts the activities of orthopoxvirus complement control proteins. J. Immunol. 2005;174:2143–2151. doi: 10.4049/jimmunol.174.4.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liszewski MK, et al. Structure and regulatory profile of the monkeypox inhibitor of complement: comparison to homologs in vaccinia and variola and evidence for dimer formation. J. Immunol. 2006;176:3725–3734. doi: 10.4049/jimmunol.176.6.3725. [DOI] [PubMed] [Google Scholar]
- 30.Miller CG, Shchelkunov SN, Kotwal GJ. The cowpox virus-encoded homolog of the vaccinia virus complement control protein is an inflammation modulatory protein. Virology. 1997;229:126–133. doi: 10.1006/viro.1996.8396. [DOI] [PubMed] [Google Scholar]
- 31.Engelstad M, Howard ST, Smith GL. A constitutively expressed vaccinia gene encodes a 42-kDa glycoprotein related to complement control factors that forms part of the extracellular virus envelope. Virology. 1992;188:801–810. doi: 10.1016/0042-6822(92)90535-w. [DOI] [PubMed] [Google Scholar]
- 32.Mullick J, Bernet J, Singh AK, Lambris JD, Sahu A. Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) open reading frame 4 protein (kaposica) is a functional homolog of complement control proteins. J. Virol. 2003;77:3878–3881. doi: 10.1128/JVI.77.6.3878-3881.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Daix V, et al. Ixodes ticks belonging to the Ixodes ricinus complex encode a family of anticomplement proteins. Insect Mol. Biol. 2007;16:155–166. doi: 10.1111/j.1365-2583.2006.00710.x. [DOI] [PubMed] [Google Scholar]
- 34.Valenzuela JG, Charlab R, Mather TN, Ribeiro JM. Purification, cloning, and expression of a novel salivary anticomplement protein from the tick, Ixodes scapularis. J. Biol. Chem. 2000;275:18717–18723. doi: 10.1074/jbc.M001486200. [DOI] [PubMed] [Google Scholar]
- 35.Rooijakkers SH, van Strijp JA. Bacterial complement evasion. Mol. Immunol. 2007;44:23–32. doi: 10.1016/j.molimm.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 36.Oda T, et al. Inactivation of chemotactic activity of C5a by the serratial 56-kilodalton protease. Infect. Immun. 1990;58:1269–1272. doi: 10.1128/iai.58.5.1269-1272.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chmouryguina I, Suvorov A, Ferrieri P, Cleary PP. Conservation of the C5a peptidase genes in group A and B streptococci. Infect. Immun. 1996;64:2387–2390. doi: 10.1128/iai.64.7.2387-2390.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsao N, et al. Streptococcal pyrogenic exotoxin B cleaves properdin and inhibits complement-mediated opsonophagocytosis. Biochem. Biophys. Res. Commun. 2006;339:779–784. doi: 10.1016/j.bbrc.2005.11.078. [DOI] [PubMed] [Google Scholar]
- 39.Ghendler Y, Parizade M, Arnon R, McKerrow JH, Fishelson Z. Schistosoma mansoni: evidence for a 28-kDa membrane-anchored protease on schistosomula. Exp. Parasitol. 1996;83:73–82. doi: 10.1006/expr.1996.0051. [DOI] [PubMed] [Google Scholar]
- 40.Lahteenmaki K, Kuusela P, Korhonen TK. Bacterial plasminogen activators and receptors. FEMS Microbiol. Rev. 2001;25:531–552. doi: 10.1111/j.1574-6976.2001.tb00590.x. [DOI] [PubMed] [Google Scholar]
- 41.Rooijakkers SH, van Wamel WJ, Ruyken M, van Kessel KP, van Strijp JA. Anti-opsonic properties of staphylokinase. Microbes Infect. 2005;7:476–484. doi: 10.1016/j.micinf.2004.12.014. [DOI] [PubMed] [Google Scholar]
- 42.Frank MM. Annihilating host defense. Nature Med. 2001;7:1285–1286. doi: 10.1038/nm1201-1285. [DOI] [PubMed] [Google Scholar]
- 43.Lubinski J, Nagashunmugam T, Friedman HM. Viral interference with antibody and complement. Semin. Cell Dev. Biol. 1998;9:329–337. doi: 10.1006/scdb.1998.0242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rux AH, et al. Kinetic analysis of glycoprotein C of herpes simplex virus types 1 and 2 binding to heparin, heparan sulfate, and complement component C3b. Virology. 2002;294:324–332. doi: 10.1006/viro.2001.1326. [DOI] [PubMed] [Google Scholar]
- 45.Kostavasili I, et al. Mechanism of complement inactivation by glycoprotein C of herpes simplex virus. J. Immunol. 1997;158:1763–1771. [PubMed] [Google Scholar]
- 46.Favoreel HW, Van de Walle GR, Nauwynck HJ, Pensaert MB. Virus complement evasion strategies. J. Gen. Virol. 2003;84:1–15. doi: 10.1099/vir.0.18709-0. [DOI] [PubMed] [Google Scholar]
- 47.Inal JM, Sim RB. A Schistosoma protein, Sh-TOR, is a novel inhibitor of complement which binds human C2. FEBS Lett. 2000;470:131–134. doi: 10.1016/s0014-5793(00)01304-1. [DOI] [PubMed] [Google Scholar]
- 48.Deng J, Gold D, LoVerde PT, Fishelson Z. Inhibition of the complement membrane attack complex by Schistosoma mansoni paramyosin. Infect. Immun. 2003;71:6402–6410. doi: 10.1128/IAI.71.11.6402-6410.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gobert GN, McManus DP. Update on paramyosin in parasitic worms. Parasitol. Int. 2005;54:101–107. doi: 10.1016/j.parint.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 50.Nunn MA, et al. Complement inhibitor of C5 activation from the soft tick Ornithodoros moubata. J. Immunol. 2005;174:2084–2091. doi: 10.4049/jimmunol.174.4.2084. [DOI] [PubMed] [Google Scholar]
- 51.Roversi P, et al. The structure of OMCI, a novel lipocalin inhibitor of the complement system. J. Mol. Biol. 2007;369:784–793. doi: 10.1016/j.jmb.2007.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lowy FD. Staphylococcus aureus infections. N. Engl. J. Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 53.Patti JM, Allen BL, McGavin MJ, Hook M. MSCRAMM-mediated adherence of microorganisms to host tissues. Annu. Rev. Microbiol. 1994;48:585–617. doi: 10.1146/annurev.mi.48.100194.003101. [DOI] [PubMed] [Google Scholar]
- 54.Foster TJ, Hook M. Surface protein adhesins of Staphylococcus aureus. Trends Microbiol. 1998;6:484–488. doi: 10.1016/s0966-842x(98)01400-0. [DOI] [PubMed] [Google Scholar]
- 55. Chavakis T, Preissner KT, Herrmann M. The anti-inflammatory activities of Staphylococcus aureus. Trends Immunol. 2007;28:408–418. doi: 10.1016/j.it.2007.07.002. This comprehensive review discusses how S. aureus uses various target points (including complement) to escape the attack of the immune system.
- 56.Foster TJ. Immune evasion by staphylococci. Nature Rev. Microbiol. 2005;3:948–958. doi: 10.1038/nrmicro1289. [DOI] [PubMed] [Google Scholar]
- 57.Rooijakkers SH, van Kessel KP, van Strijp JA. Staphylococcal innate immune evasion. Trends Microbiol. 2005;13:596–601. doi: 10.1016/j.tim.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 58.O’Riordan K, Lee JC. Staphylococcus aureus capsular polysaccharides. Clin. Microbiol. Rev. 2004;17:218–234. doi: 10.1128/CMR.17.1.218-234.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peterson PK, et al. Dichotomy between opsonization and serum complement activation by encapsulated staphylococci. Infect. Immun. 1978;20:770–775. doi: 10.1128/iai.20.3.770-775.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cunnion KM, Lee JC, Frank MM. Capsule production and growth phase influence binding of complement to Staphylococcus aureus. Infect. Immun. 2001;69:6796–6803. doi: 10.1128/IAI.69.11.6796-6803.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Verbrugh HA, Peterson PK, Nguyen BY, Sisson SP, Kim Y. Opsonization of encapsulated Staphylococcus aureus: the role of specific antibody and complement. J. Immunol. 1982;129:1681–1687. [PubMed] [Google Scholar]
- 62.Cedergren L, Andersson R, Jansson B, Uhlen M, Nilsson B. Mutational analysis of the interaction between staphylococcal protein A and human IgG1. Protein Eng. 1993;6:441–448. doi: 10.1093/protein/6.4.441. [DOI] [PubMed] [Google Scholar]
- 63.Gouda H, et al. Three-dimensional solution structure of the B domain of staphylococcal protein A: comparisons of the solution and crystal structures. Biochemistry. 1992;31:9665–9672. doi: 10.1021/bi00155a020. [DOI] [PubMed] [Google Scholar]
- 64.Nguyen T, Ghebrehiwet B, Peerschke EI. Staphylococcus aureus protein A recognizes platelet gC1qR/p33: a novel mechanism for staphylococcal interactions with platelets. Infect. Immun. 2000;68:2061–2068. doi: 10.1128/iai.68.4.2061-2068.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Peerschke EI, Bayer AS, Ghebrehiwet B, Xiong YQ. gC1qR/p33 blockade reduces Staphylococcus aureus colonization of target tissues in an animal model of infective endocarditis. Infect. Immun. 2006;74:4418–4423. doi: 10.1128/IAI.01794-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hartleib J, et al. Protein A is the von Willebrand factor binding protein on Staphylococcus aureus. Blood. 2000;96:2149–2156. [PubMed] [Google Scholar]
- 67.Gómez MI, et al. Staphylococcus aureus protein A induces airway epithelial inflammatory responses by activating TNFR1. Nature Med. 2004;10:842–848. doi: 10.1038/nm1079. [DOI] [PubMed] [Google Scholar]
- 68.Zhang L, Jacobsson K, Vasi J, Lindberg M, Frykberg L. A second IgG-binding protein in Staphylococcus aureus. Microbiology. 1998;144:985–991. doi: 10.1099/00221287-144-4-985. [DOI] [PubMed] [Google Scholar]
- 69.Jin T, et al. Staphylococcus aureus resists human defensins by production of staphylokinase, a novel bacterial evasion mechanism. J. Immunol. 2004;172:1169–1176. doi: 10.4049/jimmunol.172.2.1169. [DOI] [PubMed] [Google Scholar]
- 70.Cunnion KM, Hair PS, Buescher ES. Cleavage of complement C3b to iC3b on the surface of Staphylococcus aureus is mediated by serum complement factor I. Infect. Immun. 2004;72:2858–2863. doi: 10.1128/IAI.72.5.2858-2863.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cunnion KM, Buescher ES, Hair PS. Serum complement factor I decreases Staphylococcus aureus phagocytosis. J. Lab. Clin. Med. 2005;146:279–286. doi: 10.1016/j.lab.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 72.Lee LY, et al. Inhibition of complement activation by a secreted Staphylococcus aureus protein. J. Infect. Dis. 2004;190:571–579. doi: 10.1086/422259. [DOI] [PubMed] [Google Scholar]
- 73.Lee LY, Liang X, Hook M, Brown EL. Identification and characterization of the C3 binding domain of the Staphylococcus aureus extracellular fibrinogen-binding protein (Efb) J. Biol. Chem. 2004;279:50710–50716. doi: 10.1074/jbc.M408570200. [DOI] [PubMed] [Google Scholar]
- 74. Hammel M, et al. A structural basis for complement inhibition by Staphylococcus aureus. Nature Immunol. 2007;8:430–437. doi: 10.1038/ni1450. Describes the first co-crystal structure between a microbial complement-evasion protein and its host target domain; also elucidates the inhibition mechanism on a molecular level by combining biochemical and biophysical approaches.
- 75.Hammel M, et al. Characterization of Ehp, a secreted complement inhibitory protein from Staphylococcus aureus. J. Biol. Chem. 2007;282:30051–30061. doi: 10.1074/jbc.M704247200. [DOI] [PubMed] [Google Scholar]
- 76.Jongerius I, et al. Staphylococcal complement evasion by various convertase-blocking molecules. J. Exp. Med. 2007;204:2461–2471. doi: 10.1084/jem.20070818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rooijakkers SHM, et al. Immune evasion by a staphylococcal complement inhibitor that acts on C3 convertases. Nature Immunol. 2005;6:920–927. doi: 10.1038/ni1235. [DOI] [PubMed] [Google Scholar]
- 78. Rooijakkers SHM, et al. Staphylococcal complement inhibitor: structure and active sites. J. Immunol. 2007;179:2989–2998. doi: 10.4049/jimmunol.179.5.2989. The authors describe the crystal structure of a SCIN protein and identify its functional sites by analysing protein chimeras of SCIN and an inactive homologue.
- 79.Langley R, et al. The staphylococcal superantigen-like protein 7 binds IgA and complement C5 and inhibits IgA-FcaRI binding and serum killing of bacteria. J. Immunol. 2005;174:2926–2933. doi: 10.4049/jimmunol.174.5.2926. [DOI] [PubMed] [Google Scholar]
- 80.de Haas CJ, et al. Chemotaxis inhibitory protein of Staphylococcus aureus, a bacterial antiinflammatory agent. J. Exp. Med. 2004;199:687–695. doi: 10.1084/jem.20031636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Postma B, et al. Chemotaxis inhibitory protein of Staphylococcus aureus binds specifically to the C5a and formylated peptide receptor. J. Immunol. 2004;172:6994–7001. doi: 10.4049/jimmunol.172.11.6994. [DOI] [PubMed] [Google Scholar]
- 82.Wright AJ, et al. Characterisation of receptor binding by the chemotaxis inhibitory protein of Staphylococcus aureus and the effects of the host immune response. Mol. Immunol. 2007;44:2507–2517. doi: 10.1016/j.molimm.2006.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Geisbrecht BV, Hamaoka BY, Perman B, Zemla A, Leahy DJ. The crystal structures of EAP domains from Staphylococcus aureus reveal an unexpected homology to bacterial superantigens. J. Biol. Chem. 2005;280:17243–17250. doi: 10.1074/jbc.M412311200. [DOI] [PubMed] [Google Scholar]
- 84. Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nature Rev. Drug Discov. 2007;6:29–40. doi: 10.1038/nrd2201. Although not directly related to complement evasion, this insider view on new screening initiatives for antibiotic drugs by pharmaceutical companies provides noteworthy information on targets and challenges for this endeavour.
- 85.Woodman ME, et al. Borrelia burgdorferi binding of host complement regulator factor H is not required for efficient mammalian infection. Infect. Immun. 2007;75:3131–3139. doi: 10.1128/IAI.01923-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.A step in the right direction. Nature Rev. Drug Discov. 2007;6:419. doi: 10.1038/nrd2352. [DOI] [PubMed] [Google Scholar]
- 87.Daly J, Kotwal GJ. Pro-inflammatory complement activation by the Aβ peptide of Alzheimer’s disease is biologically significant and can be blocked by vaccinia virus complement control protein. Neurobiol. Aging. 1998;19:619–627. doi: 10.1016/s0197-4580(98)00100-6. [DOI] [PubMed] [Google Scholar]
- 88.Pillay NS, Kellaway LA, Kotwal GJ. Vaccinia virus complement control protein significantly improves sensorimotor function recovery after severe head trauma. Brain Res. 2007;1153:158–165. doi: 10.1016/j.brainres.2007.03.056. [DOI] [PubMed] [Google Scholar]
- 89.Marsh M, Helenius A. Virus entry: open sesame. Cell. 2006;124:729–740. doi: 10.1016/j.cell.2006.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nemerow GR, Mold C, Schwend VK, Tollefson V, Cooper NR. Identification of gp350 as the viral glycoprotein mediating attachment of Epstein–Barr virus (EBV) to the EBV/C3d receptor of B cells: sequence homology of gp350 and C3 complement fragment C3d. J. Virol. 1987;61:1416–1420. doi: 10.1128/jvi.61.5.1416-1420.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tsoukas CD, Lambris JD. Expression of EBV/C3d receptors on T cells: biological significance. Immunol. Today. 1993;14:56–59. doi: 10.1016/0167-5699(93)90059-T. [DOI] [PubMed] [Google Scholar]
- 92.Naniche D, et al. Human membrane cofactor protein (CD46) acts as a cellular receptor for measles virus. J. Virol. 1993;67:6025–6032. doi: 10.1128/jvi.67.10.6025-6032.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Santoro F, et al. Interaction of glycoprotein H of human herpesvirus 6 with the cellular receptor CD46. J. Biol. Chem. 2003;278:25964–25969. doi: 10.1074/jbc.M302373200. [DOI] [PubMed] [Google Scholar]
- 94.Nowicki B, Hart A, Coyne KE, Lublin DM, Nowicki S. Short consensus repeat-3 domain of recombinant decay-accelerating factor is recognized by Escherichia coli recombinant Dr adhesin in a model of a cell–cell interaction. J. Exp. Med. 1993;178:2115–2121. doi: 10.1084/jem.178.6.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Datta PK, Rappaport J. HIV and complement: hijacking an immune defense. Biomed. Pharmacother. 2006;60:561–568. doi: 10.1016/j.biopha.2006.07.087. [DOI] [PubMed] [Google Scholar]
- 96.Schorey JS, Carroll MC, Brown EJ. A macrophage invasion mechanism of pathogenic mycobacteria. Science. 1997;277:1091–1093. doi: 10.1126/science.277.5329.1091. [DOI] [PubMed] [Google Scholar]
- 97.Janssen BJ, Gros P. Structural insights into the central complement component C3. Mol. Immunol. 2007;44:3–10. doi: 10.1016/j.molimm.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 98.Lambris JD, Morikis D, editors. Structural Biology of the Complement System. Boca Raton: CRC; 2005. [Google Scholar]
- 99.Deisenhofer J, Jones TA, Huber R, Sjodahl J, Sjoquist J. Crystallization, crystal structure analysis and atomic model of the complex formed by a human Fc fragment and fragment B of protein A from Staphylococcus aureus. Hoppe-Seyler’s Z. Physiol. Chem. 1978;359:975–985. doi: 10.1515/bchm2.1978.359.2.975. [DOI] [PubMed] [Google Scholar]
- 100.Sauer-Eriksson AE, Kleywegt GJ, Uhlen M, Jones TA. Crystal structure of the C2 fragment of streptococcal protein G in complex with the Fc domain of human IgG. Structure. 1995;3:265–278. doi: 10.1016/s0969-2126(01)00157-5. [DOI] [PubMed] [Google Scholar]
- 101.Ricklin D, Lambris JD. Exploring the complement interaction network using surface plasmon resonance. Adv. Exp. Med. Biol. 2007;598:260–278. doi: 10.1007/978-0-387-71767-8_19. [DOI] [PubMed] [Google Scholar]
- 102.Lukacik P, et al. Complement regulation at the molecular level: the structure of decay-accelerating factor. Proc. Natl Acad. Sci. USA. 2004;101:1279–1284. doi: 10.1073/pnas.0307200101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Al-Shangiti AM, et al. Structural relationships and cellular tropism of staphylococcal superantigen-like proteins. Infect. Immun. 2004;72:4261–4270. doi: 10.1128/IAI.72.7.4261-4270.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Haas PJ, et al. The structure of the C5a receptor-blocking domain of chemotaxis inhibitory protein of Staphylococcus aureus is related to a group of immune evasive molecules. J. Mol. Biol. 2005;353:859–872. doi: 10.1016/j.jmb.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 105.Zhang X, Boyar W, Toth MJ, Wennogle L, Gonnella NC. Structural definition of the C5a C terminus by two-dimensional nuclear magnetic resonance spectroscopy. Proteins. 1997;28:261–267. doi: 10.1002/(sici)1097-0134(199706)28:2<261::aid-prot13>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.