Abstract
The membrane protein folding problem can be articulated by two central questions. How is protein topology established by selective peptide transport to opposite sides of the cellular membrane? And how are transmembrane segments inserted, integrated and folded within the lipid bilayer? In eukaryotes, this process usually takes place in the endoplasmic reticulum (ER) coincident with protein synthesis, and is facilitated by the translating Ribosome and the Sec61 Translocon Complex (RTC). At its core, the RTC forms a dynamic pathway through which the elongating nascent polypeptide moves as it is delivered into cytosolic, lumenal and lipid compartments. This perspective will focus on emerging evidence that the RTC functions as a protein folding machine that restricts conformational space by establishing transmembrane topology and yet provides a permissive environment that enables nascent transmembrane domains to efficiently progress down their folding energy landscape.
The process of polytopic (multispanning) membrane protein folding can be viewed as a series of sequential but potentially overlapping steps that include: i) formation, orientation and integration of transmembrane helices in the lipid bilayer, ii) helical packing within the membrane, iii) localization and folding of cytosolic and extracytoplasmic domains, and for many proteins iv) quaternary organization into functional oligomers. Structural intermediates that populate the folding pathway are therefore conceptually (although not physically) analogous to “molten globules” where collapse of TM domains during helical packing would presumably replace lipid contacts with more energetically favorable helix-helix interactions. However, the folding environment, physical forces, and energetics that give rise to membrane protein structure differ strikingly from the aqueous environment of globular protein folding1,2. In addition, membrane protein folding in eukaryotes is primarily compartmentalized in the endoplasmic reticulum (ER) and facilitated by a complex set of folding machinery3,4. Understanding how membrane proteins fold in cells therefore requires detailed knowledge of how cellular factors impact the folding landscape within the hydrophobic environment of the lipid bilayer.
Cellular basis of membrane protein folding
Nearly three decades ago Günter Blobel proposed that polytopic proteins might be generated by internally encoded topogenic determinants that interact with cytosolic and membrane-bound receptors and a multisubunit protein “translocator” to cotranslationally insert the nascent polypeptide to the ER membrane5. Several aspects of this prescient prediction are particularly noteworthy. Experimental evidence for the underlying signal sequence hypothesis was just emerging, and few receptors had yet been identified. The existence of the putative ER protein translocator was entirely speculative and stimulated a heated debate that lasted for nearly two decades. And skeptics argued that the biological complexity of polytopic proteins was too great to be accommodated by a singular protein machine.
Over the ensuing years, biochemical, genetic, and structural studies confirmed both the presence and identities of proteins involved in virtually all aspects of targeting to, translocation across and integration into the ER and other organellar membranes3,4,6,7. Applying these principles to polytopic proteins, however, has proven challenging, in part due to technical constraints, but perhaps more importantly due to the paucity of solved membrane protein structures with which to test specific hypotheses. Improved methodologies for interrogating nascent polypeptides and expansion of the membrane protein structure database have provided new insights into thermodynamics of protein folding and the molecular structure of cellular machineries that guide the folding process. These topics have been the subject of several recent excellent reviews7–10, and will therefore not be covered in detail here. Rather, this perspective will focus primarily on our emerging understanding of how ER translocation machinery interacts with, and is in turn controlled by, the nascent polypeptide as it facilitates specific topogenesis and folding events.
Membrane protein folding is an extension of translocation
From a mechanistic standpoint it is instructive to consider early steps of polytopic protein folding as an extension of protein secretion, many principles of which are relatively well established3,6,10–12. Protein secretion is initiated when an ER signal sequence emerges from the ribosome, binds the cytosolic signal recognition particle (SRP), and targets the ribosome-nascent chain complex (RNC) to the ER membrane13. GTP hydrolysis by SRP and its receptor releases the signal sequence and transfers the RNC to a large protein conducting channel formed by the Sec61αβγ heterotrimer14 and numerous associated proteins that include: TRanslocation Associated Membrane protein (TRAM)15, the TRanslocation Associated Protein complex (TRAP)16 oligosaccharyl transferase17, signal peptidase complex, and others18. While the precise stoichiometry and structure of actively engaged Sec61 complexes remain unknown, for the purposes of this perspective, I will refer to the fully assembled and functional protein complex as the ER translocon13,19 to distinguish it from the actual channel core which is formed by one or more copies of Sec61αβγ20–22.
As the RNC docks onto the ER membrane, the exit tunnel of the large ribosomal subunit is aligned with the axial translocon pore21,23, and the signal sequence engages a binding site within Sec61α. Biochemical and fluorescence quenching studies have shown that this establishes a tight association between the ribosome and translocon 24,25 that shields the nascent chain from the cytosol26. The signal sequence also opens the lumenal gate of the translocon to create a continuous aqueous translocation pathway from the ribosome exit tunnel through the translocon pore and into the ER lumen26,27. Most secretory and transmembrane proteins move through this pathway coincident with peptide elongation, although the efficiency of translocation is dependent upon the nature of the signal sequence, the passenger domain, and potentially, the presence of regulatory translocon factors28,29.
Bitopic membrane proteins require at least two additional steps that are mediated by so-called stop transfer sequences. Peptide movement into the ER lumen must be terminated. And the hydrophobic transmembrane (TM) segment must be transferred laterally from the proteinaceous environment of the translocon into the surrounding lipid bilayer, a step commonly referred to as membrane integration30. Again, analysis of fluorescent probes incorporated into nascent integration intermediates indicate that these events are orchestrated by dynamic regulation of the ribosome translocon complex (RTC). Synthesis and compaction of the TM within the ribosome exit tunnel31 alters the translocation pathway by closing the lumenal gate of the translocon via ATP-dependent interactions with BiP32,33 and subsequently relaxing the ribosome-translocon junction. Thus instead of continuing to move into the ER lumen, the elongating nascent polypeptide is redirected beneath the base of the ribosome and into the cytosol33,34.
An important and currently unresolved issue in membrane protein folding is how and when TMs are released from Sec61 once translocation is terminated. One view, supported by lipid crosslinking studies is that TMs continuously sample the hydrophobic membrane environment and move into the bilayer by passive thermodynamic partitioning through a lateral cleft in the Sec61α subunit4,20,35,36. Consistent with this, the ability of TMs in bitopic proteins (and to a lesser extent polytopic proteins) to terminate translocation and adopt a membrane-spanning topology appears to be primarily determined by overall hydrophobicity of the TM37–39. Alternatively, TM release from the translocon can be delayed and mechanistically triggered at specific stages of synthesis during or at the end of translation. This is presumably accomplished by a conformational change in the RTC that provides the TM access to bulk lipid40, although the precise mechanism remains unknown. In support of this latter model, chemical and photocrosslinking studies have shown that many and possibly most native TMs appear to reside within the translocon for prolonged periods of time and progress through different proteinaceous environments prior to integration30,40–44. A remarkable finding is that some TMs can be actively retained within the translocon via specific polar interactions even after peptidyl tRNA bond cleavage and released into the membrane in an ATP-dependent manner30. This later finding is quite surprising as there are no known eukaryotic translocon components that contain ATPases activity.
It should be also noted that while the basic principles of translocation are well established, many details are not universally accepted. For example, the cytosolic inaccessibility of nascent chains in functionally engaged translocons as determined by fluorescence collisional quenching has been challenged by cryoEM studies of detergent solubilized RTC complexes lacking substrate. Results of the latter show a constitutive 12–17 Å gap between the ribosome base and translocon that would be sufficient to expose the translocating nascent chain to the cytosolic environment4,21,22. Studies of functional translocons have also implicated a large translocon pore that is gated at its lumenal end by the action of BiP45, whereas the crystal structure of an archaebacterial Sec61 homolog suggests that gating of a small pore is accomplished by displacement of short helical plug20. While further studies are clearly needed to resolve these and other issues, this review will focus primarily on data obtained using functionally intact translocons that contain a translocating substrate and then discuss potential ways in which these results might impact consideration of alternative structural models.
The cotranslational topogenesis model
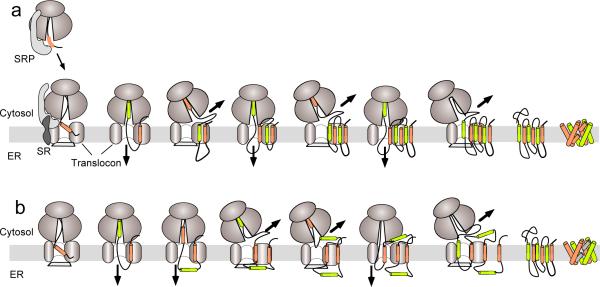
How then does the RTC direct polytopic protein biogenesis when multiple TMs are presented in rapid succession? The simplest biogenesis mechanism would involve a series of iterative cotranslational translation initiation and termination events similar to those used by secretory and bitopic proteins (Fig. 1a). Such a model predicts that the first TM of a polytopic protein would function as a signal (or signal anchor) sequence to target the RNC to the membrane, gate open the translocon and initiate translocation into the ER lumen. The second TM segment would function as a stop transfer sequence to terminate translocation, close the lumenal gate, relax the ribosome junction, and direct the downstream peptide loop into the cytosol. The third TM would then reestablish the ribosome translocon junction, reopen the translocon pore, reinitiate translocation, and so forth. Alternate gating of the translocon channel and ribosome junction by sequential topogenic determinants would therefore direct peptide loops into their proper cellular compartment and establish topology of each TM as the nascent chain emerged from the ribosome11,12.
Figure 1.
Models of polytopic protein biogenesis. (a) Cotranslational biogenesis is initiated as signal recognition particle (SRP) interacts with a signal sequence, binds its receptor (SR) at the ER membrane, and transfers the ribosome nascent chain complex to the Sec61 translocon. Signal sequences (tan cylinders) stimulate ribosome binding and open the translocon pore to initiate peptide movement into the ER lumen. Stop transfer sequences (green cylinders) terminate translocation and redirect the elongating nascent chain beneath the ribosome and into the cytosol. Each sequential TM therefore alters the direction of nascent chain movement through the RTC (bold arrow) to establish transmembrane topology from N- to C-terminus one helix at a time. (b) During AQP1 biogenesis, TM2 fails to terminate translocation, TM3 is initially inserted into the translocon in a type I topology, and TM4 transiently resides on the cytosolic face of the membrane. This sequence of events generates a four-spanning intermediate that is converted to a six spanning topology during or after synthesis of TMs 5–6.
The cotranslational model imposes three major requirements. First, the direction of nascent chain movement and hence TM topology, is determined by the functional state of the translocation pathway. For example, when the ribosome is tightly bound and translocon gate is open, the nascent peptide loop can only translocate into the ER lumen. If, however, the translocon is closed and the ribosome junction is relaxed, then the growing polypeptide must move into the cytosol. Second, the nature of the translocation pathway is strictly controlled by topogenic information encoded within the nascent polypeptide. In order to achieve the correct topology, each TM must be recognized by the RTC as either a signal (anchor) sequence or a stop transfer sequence and reset the translocation pathway for proper delivery of the next hydrophilic peptide loop. Third, the translocation pathway must be highly dynamic and precisely coordinated to change the direction of peptide movement every few seconds as TM segments are synthesized in rapid succession.
While substantial effort has been directed at testing whether the cotranslational model accurately describes native polytopic protein biogenesis, few studies have directly measured RTC gating. Instead, functional properties of topogenic determinants have been inferred by their ability to direct translocation and integration of heterologous reporter domains. A common method involves analysis of truncated fusion proteins in which a passive translocation reporter is ligated C-terminal to each TM to provide a simple readout for topology of sequential loops46. While this approach identified signal anchor and stop transfer activities in some native polytopic proteins, many topogenic determinants were either arranged in unexpected patterns or exhibited unusual topogenic properties47–52. The C-terminal reporter approach therefore failed to confirm a unified biogenesis mechanism and often resulted in ambiguous and/or conflicting topologies. While initially confusing from a topological standpoint, these results provided important insight into folding mechanisms that often went unappreciated. A well-studied example involved two homologous aquaporin water channels, AQP1 and AQP4, where minor differences in primary sequence dramatically change the folding pathway but have no effect on the final folded structure11.
Aquaporin 4 as a paradigm for cotranslational biogenesis
Aquaporins constitute a large family of proteins that facilitate passive water and glycerol transport across biological membranes53–55. High resolution structures have demonstrated a conserved topological fold in which six membrane spanning helices and two half helices are arranged in the membrane in an inverted two-fold pseudo symmetry56,57. Although each monomer contains an intact pore, mammalian AQPs are believed to function exclusively as homotetramers that fold and assemble in the endoplasmic reticulum (ER) prior to transport to the cell surface11,58–60. Their small size, known structure, and high degree of homology make AQPs ideal candidates to investigate membrane protein folding mechanisms.
Topological analysis of truncated AQP4 fusion proteins confirmed the presence of alternating signal (TMs 1, 3 and 5) and stop transfer (TMs 2, 4 and 6) sequences and provided some of the first evidence that an entire native polytopic protein could be generated via a cotranslational topogenesis mechanism. Specifically, each AQP4 TM was found to act independently to establish the expected six-spanning topology from N- to C- terminus, one TM helix at a time (Fig. 1a)61. Photocrosslinking studies using a series of truncated integration intermediates, which provide static snapshots of the nascent chain environment, also demonstrated that each TM enters a binding site within Sec61 as it exits the ribosome, remains in this location until entry of the next TM, and then progresses through the translocon in a unique and highly ordered manner41. Some TMs (i.e. TM2, and TM4) exhibit a brief well defined interval of crosslinking, while others (i.e. TM1 and TM3) exhibit several phases of crosslinking at different stages of synthesis. As a result of this behavior, multiple TMs were found accumulate within or in close proximity to Sec61α prior to their release into the lipid bilayer.
AQP1, an exception to the cotranslational rule
Surprisingly, AQP1-TMs control the translocation pathway in a very different manner than their AQP4 counterparts (Fig. 1b)50. For example, AQP1-TM2 does not efficiently terminate translocation and therefore transiently passes through the translocon into the ER lumen as it exits the ribosome. Because TM2 does not close the translocation pathway, TM3 encounters an open translocon where it terminates translocation and directs its C-terminal flanking residues into the cytosol. This behavior results in a mixture of topologies in which most nascent AQP1 polypeptides cotranslationally span the membrane only four times11,50. However, the initial four-spanning structure actually represents a folding intermediate that is converted into a mature six-spanning topology during and/or after the completion of synthesis62. AQP1 maturation therefore involves a 180° rotation of TM3 from a type I to a type II topology, which transfers the TM2-3 peptide loop from the ER lumen to cytosol, the TM3-4 loop from the cytosol to the ER lumen, and inserts TM2 and TM4 into the plane of the membrane. These events represent a striking exception to the cotranslational model and raise two obvious questions. Why would two highly homologous proteins with similar structure and function use such different folding pathways? And how is AQP1 converted from a four-spanning to a six-spanning topology?
AQP1 folding illustrates the potential conflict between structural determinants that direct early topogenesis events within the RTC, and the role of the same residues during subsequent stages of protein maturation. A principle difference between AQP1 and AQP4 topogenesis is that AQP1-TM2 fails to terminate translocation as it exits the ribosome. This divergent behavior is caused by two residues in TM2, Asn49 and Lys51 in AQP1 versus Met48 and Leu50 in AQP463. Exchanging these residues converts AQP1-TM2 to an efficient stop transfer sequence but completely disrupts water channel function. Hydrophilic residues are therefore needed to generate a functional channel, but their presence forces AQP1 to deviate from a cotranslational biogenesis mechanism. Such behavior appears to be a relatively common feature of eukaryotic polytopic proteins where rules of cotranslational topogenesis do not apply64,65.
What then is the advantage for polytopic proteins that utilize a non-cotranslational folding pathway? For AQP1, Asn49 and Lys51 both interact with a partially buried aspartic acid residue, Asp185, near the C-terminus of TM5. Asn49 forms an intramolecular H-bond that is needed for proper helical packing of the monomer, while Lys51 forms a nonessential ionic bond that stabilizes the AQP1 tetramer60. Because proteins emerge from the ribosome vectorally from N- to C-terminus, TM2 must enter the translocon before TM5 is synthesized. At early stages of synthesis, the unpaired hydrophilic residues therefore prevent TM2 from terminating translocation. This could be caused by a reduction in hydrophobicity that interferes with translocon interactions needed to terminate translocation38, or alternatively, a delay in helix formation within the ribosome exit tunnel, which would potentially be needed to relax the ribosome junction and allow TM2 to properly span the membrane31,33. Current studies are underway to distinguish these possibilities. Importantly, the same polar residues that dictate these early events of topogenesis are also needed to stabilize a polar residue (Asp185) in a distal C-terminal TM segment. Failure of Asp185 to interact productively with Asn49 in TM2 leads to sequestration of AQP1 in a large complex or aggregate, a finding consistent with the strong energetic potential of Asp residues to stimulate helix-helix interactions in hydrophobic environments66,67. AQP1 biogenesis therefore illustrates the complex ways in which subtle variations in primary sequence can alter multiple steps along the folding pathway: i) cotranslational translocon interactions that direct early topogenesis, ii) tertiary folding and helical packing within the lipid bilayer, and iii) quaternary stabilization of AQP1 tetramers.
Functional implications of the AQP1 folding pathway
In contrast to initial predictions5, delivery of the nascent chain into its proper cellular compartment is not necessarily a constitutive process. TMs that lack strong topogenic properties may allow the nascent chain to enter both the cytosol and ER lumen as they exit the ribosome, an observation that has also been reported for other native and engineered substrates51,68,69. It is currently unknown whether the RTC provides access to both compartments simultaneously, or whether access is achieved stochastically by alternate translocon conformations. Such findings contrast with the view of a rigid, cytosolically inaccessible translocation pathway and suggests that protein movement through the RTC can exhibit substantial variation depending upon topogenic information present in the nascent polypeptide. These aspects of polytopic protein topogenesis are consistent with recent observations that the translocation efficiency of secretory proteins can also be regulated by specific properties of the signal sequence, the availability of translocon associated proteins29, and/or presence of small molecule inhibitors70. Protein secretion and membrane protein folding may therefore share similar molecular mechanisms of translocon control. Further work is needed to understand the molecular basis for these observations, particularly the manner in which the ribosome junction and translocon channel gating are controlled during different states of translocation. An obvious advantage of this non-constitutive translocation behavior is that it enables polytopic proteins to acquire variations in primary sequence (i.e. polar residues) that would not be permitted by a strict cotranslational topogenesis mechanism.
General implications for membrane protein folding
Another remarkable feature of AQP1 biogenesis is that there must exist a mechanism in the ER for reorienting TMs and peptide loops that are initially (cotranslationally) directed into the wrong cellular compartment. While the precise mechanism that drives AQP1 reorientation remains unknown, the folded monomer is likely stabilized in part by alignment of TM2 and TM5 in the membrane and formation of the partially buried H-bond between Asn49 -Asp185 in the relatively apolar core of the protein60. The nascent chain must therefore retain sufficient conformational flexibility during early stages of folding to allow for “topological editing” while synthesis of downstream TMs provides additional folding information. This behavior raises a fundamental question as to the extent to which conformational flexibility is allowed, and where and how it is achieved in the context of the RTC and/or the ER membrane.
Movement of charged residues and/or hydrophilic peptide loops directly across the core of the lipid bilayer imposes a significant energy barrier that would likely limit the transfer rate and hence slow kinetics of AQP1 folding from a 4- to a 6-spanning topology2. The magnitude of such a barrier is difficult to estimate for polytopic proteins because transfer of a given peptide region would take place in an environment formed by both lipids and other TMs within the protein. Studies of prokaryotic transporters indicate that peptide loops and even helical bundles may be spontaneously transferred across the membrane simply by changes in phospholipid composition71. Relatively large peptide domains in tail-anchored proteins have also been shown to translocate unassisted across the ER membrane72. Thus it is possible that AQP1 topological maturation might take place after the nascent chain has been released into the bulk lipid of the ER (Fig. 2b).
Figure 2.
Mechanism of TM integration. (a) During membrane protein biogenesis, TMs are laterally transferred from the proteinaceous environment of the translocon into the lipid bilayer. This may occur in a sequential fashion as each TM is synthesized, in pairs, or in groups. (b)The timing of TM integration will determine, in part, whether helical packing takes place primarily in a lipid or proteinaceous environment. Two potential folding models are illustrated for AQP1. If TMs are sequentially released from the translocon into bulk lipid, then topological maturation of TMs 2–4 will involve spontaneous TM insertion, rotation, and movement of two hydrophilic loops across the lipid bilayer. Alternatively, retention of TMs within or in close proximity to translocon proteins (bottom panel) could potentially reduce energy barriers for peptide transfer and topological maturation.
An alternate scenario is that adjacent translocon proteins might contribute to the folding environment and thereby facilitate peptide reorientation during early stages of folding. This intriguing possibility is supported by numerous crosslinking and membrane extraction studies demonstrating that multiple TMs can accumulate in close proximity to translocon proteins and be released into the bilayer in pairs or groups (Fig. 2a, b)42–44,49,73,74. In the case of AQP1, the close proximity of adjacent translocon proteins could potentially reduce the free energy barrier imposed by TM3 reorientation and thereby provide the nascent polypeptide access to an increased conformational space needed to establish proper helical packing. Once formed, the mature six-spanning structure would presumably be stabilized by proper helix-helix contacts and by formation of the H-bond between TM2 and TM5. The net outcome would be to improve the efficiency with which the nascent chain could progress down its folding energy landscape to a lower free energy state than was achieved in the original, cotranslational four-spanning topology. Currently, however, the precise mechanism of TM reorientation, the temporal sequence of helical packing, and the role of lipids and/or translocon or other ER proteins in carrying out these processes remain unknown for AQP1 or any other eukaryotic polytopic protein of similar complexity.
Crosslinking results suggest two possible mechanisms by which the translocon might facilitate helical packing. TMs could potentially be released from the translocation channel but remain associated with accessory proteins at the translocon periphery as has been postulated for TRAM in eukaryotes and the chaperone-like protein YidC in prokaryotes4,36,40,43. Indeed, TRAM association with nascent TMs been correlated with the presence of charged residues36. Depending upon the architecture of individual subunits, it is also possible that TMs might initially exit from the Sec61 channel to another location within the interior of the translocon that may contain either a large hole 23,75–77 or central depression that has been proposed to be filled with intercalated lipid 21,78. Such an arrangement is appealing because the translocon interior could shield the nascent chain from bulk lipids and yet provide a relatively hydrophobic environment that would permit transient hydrophobic as well as polar interactions. The nature of such interactions would depend upon primary sequence of the substrate TMs and available translocon proteins and lipids in the immediate vicinity, each of which could be potentially tailored for substrate folding.
Is the ER translocon a membrane protein chaperone?
Cellular chaperones are typically defined by their ability to assist the folding and assembly of proteins in a catalytic and non-consumptive manner. The ER translocon was originally viewed as a channel with the capacity to facilitate translocation of secretory proteins from the cytosol to the ER lumen. It is now accepted that the translocon also establishes topology of bitopic and polytopic membrane proteins by recognizing specific sequence determinants and delivering peptide regions into their proper folding compartment. Because localization is critical for folding, this criteria alone would fulfill the requirements for a membrane protein chaperone. Recent studies now raise the more profound possibility that in addition to substrate localization, the translocon may provide a specialized environment that increases conformational flexibility needed for tertiary folding of helical transmembrane domains. If this is the case, then the chaperone functions of the translocon might be conceptually analogous to the folding chamber formed by Hsp60 protein family in which GroEL and the TRIC chaperonin sequester small globular proteins from the bulk cytosol as they acquire their proper tertiary structure79. Recent observations that the translocon can actively retain certain TMs and release them in an ATP-dependent manner further resembles these chaperone functions and suggests that substrate release into the bilayer might also be an active and regulated process30,80.
The extent to which eukaryotic membrane protein folding takes place in the bilayer, the translocon, or both, will obviously depend on the composition, stoichiometry and precise architecture of ribosome-bound, functional, ER translocons (Fig. 2c). Unfortunately, none of these parameters are currently known with precision, and hence any model of membrane protein folding is inherently speculative. However, the crystal structure of a Sec61 homolog from Methanococcus janashsii (SecYEβ) together with cryoEM studies of solubilized ribosome bound translocons has provided some important clues20,21. SecYEβ is a cuboidal structure roughly 40 Å in size that contains a constricted ~8A diameter central pore and a lateral opening through which protein translocation and integration, respectively, are predicted to occur. Based on this structure, several models have been proposed to explain how the eukaryotic homolog Sec61αβγ might facilitate cotranslational membrane protein topogenesis4,7,81. As this topic has been covered extensively elsewhere10, the goal here has been to provide a different viewpoint of the membrane protein folding problem based on novel folding behaviors and requirements of native substrates. Such an approach raises several questions that have been less well appreciated in structurally based models. For example, the predicted pore formed by Sec61αβγ is clearly too small to accommodate multiple TMs, rotation of a single TM, or topological maturation and folding of large protein domains. Thus these events must either occur outside of the Sec61 channel in the bilayer, or within the confines of a larger, fully assembled translocon. In this regard, Sec61 has been shown to form ring-like oligomers with a large central pore-like region 23,75,77. However, higher resolution studies have suggested alternate oligomeric arrangements21,78 and most recently the possibility that a single Sec61αβγ heterotrimer might reside beneath the ribosome22 (Fig. 3). Unfortunately, these structures lack translocon associated subunits such as TRAM, oligosaccharyltransferase, signal peptidase complex, and others. The specific arrangement of Sec61αβγ heterotrimers and/or additional associated proteins may therefore play critical roles in TM retention, rotation, and topological maturation observed for AQP1 and potentially other native substrates (Fig. 3). Solving this difficult problem will undoubtedly require complementary techniques that consolidate structural as well as functional information from fully assembled RTCs that are actively engaged with substrate. A general model derived from such studies that adequately explains membrane protein folding at a molecular level will solve one of the long-standing and persistent questions in modern biology.
Figure 3.
Potential arrangement of Sec61αβγ heterotrimers (gray cylinders) in the assembled translocon (purple disc) and implications for cotranslational folding. Cryo-EM of empty, solubilized mammalian ER RTCs suggest Sec61 may be present beneath the ribosome in a single copy22 (a) or a back-to-back tetramer configuration21,82 (b). Both models propose that only one Sec61 protein is used for translocation which provides TM helices (orange and green cylinders) with a single lateral exit site to the translocon periphery. Alternative orientations include a front-to-front Sec61 dimer configuration observed in CryoEM structures of the E. coli SecYEG complex83 (c), a large central pore derived from fluorescence quenching experiments of functionally intact ER translocons84 and supported by early low resolution EM studies75,76,77 (d), and a related but hypothetical oligomeric front-to-front configuration11 in which TMs could initially exit Sec61 into the translocon interior (e). These latter models provide a potential means to accommodate multiple helices during translocation85 and prior to nascent chain movement between subunits into the bilayer.
Acknowledgments
I would like to thank Dr. Brian Conti and other members of the Skach Lab for helpful comments on this manuscript and the US National Institutes of Health (R01GM53457 and R01 DK51818) for supporting our research in this area.
The abbreviations used are
- ER
endoplasmic reticulum
- RNC
ribosome nascent chain complex
- RTC
ribosome-translocon complex
- TM
transmembrane segment
References
- 1.Popot J, Engelman D. Helical membrane protein folding, stability and evolution. Ann. Rev. Biochem. 2000;69:881–922. doi: 10.1146/annurev.biochem.69.1.881. [DOI] [PubMed] [Google Scholar]
- 2.Bowie J. Solving the membrane protein folding problem. Nature. 2005;438:581–589. doi: 10.1038/nature04395. [DOI] [PubMed] [Google Scholar]
- 3.Johnson A, van Waes M. The Translocon: A dynamic gateway at the ER membrane. Ann. Rev. Cell Dev. Biol. 1999;15:799–842. doi: 10.1146/annurev.cellbio.15.1.799. [DOI] [PubMed] [Google Scholar]
- 4.Rapoport T, Goder V, Heinrich S, Matlack K. Membrane-protein integration and the role of the translocation channel. Trends Cell Biol. 2004;14:568–575. doi: 10.1016/j.tcb.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 5.Blobel G. Intracellular protein topogenesis. Proc. Natl. Acad. Sci. USA. 1980;77:1496–1500. doi: 10.1073/pnas.77.3.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schnell D, Hebert D. Protein translocons: multifunctional mediators of protein translocation across membranes. Cell. 2003;112:491–505. doi: 10.1016/s0092-8674(03)00110-7. [DOI] [PubMed] [Google Scholar]
- 7.White S, von Heijne G. The machinery of membrane protein assembly. Curr. Opin. Struct. Biol. 2004;14:397–404. doi: 10.1016/j.sbi.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 8.Johnson A. Fluorescence approaches for determining protein conformations, interactions and mechanisms at membranes. Traffic. 2005;6:107801092. doi: 10.1111/j.1600-0854.2005.00340.x. [DOI] [PubMed] [Google Scholar]
- 9.von Heijne G. Membrane-protein topology. Nature Rev. Mol. Cell Biol. 2006;7:909–918. doi: 10.1038/nrm2063. [DOI] [PubMed] [Google Scholar]
- 10.Osborne AR, Rapoport TA, van den Berg B. Protein translocation by the Sec61/SecY channel. Annu. Rev. Cell. Dev. Biol. 2005;21:529–550. doi: 10.1146/annurev.cellbio.21.012704.133214. [DOI] [PubMed] [Google Scholar]
- 11.Pitonzo D, Skach W. Molecular mechanisms of aquaporin biogenesis by the endoplasmic reticulum Sec61 translocon. Bioch. Biophys. Acta. 2006;1758:976–988. doi: 10.1016/j.bbamem.2006.04.021. [DOI] [PubMed] [Google Scholar]
- 12.Sadlish H, Skach W. Biogenesis of CFTR and other polytopic membrane proteins; new roles for the ribosome-translocon complex. J. Mem. Biol. 2004;202:115–126. doi: 10.1007/s00232-004-0715-6. [DOI] [PubMed] [Google Scholar]
- 13.Walter P, Lingappa VR. Mechanisms of protein translocation across the endoplasmic reticulum membrane. Annu. Rev. Cell Biol. 1986;2:499–516. doi: 10.1146/annurev.cb.02.110186.002435. [DOI] [PubMed] [Google Scholar]
- 14.Song W, Raden D, Mandon E, Gilmore R. Role of Sec61alpha in the regulated transfer of the ribosome-nascent chain complex from the signal recognition particle to the translocation channel. Cell. 2000;100:333–343. doi: 10.1016/s0092-8674(00)80669-8. [DOI] [PubMed] [Google Scholar]
- 15.Görlich D, Hartmann E, Prehn S, Rapoport T. A protein of the endoplasmic reticulum involved in early polypeptide translocation. Nature. 1992;357:47–52. doi: 10.1038/357047a0. [DOI] [PubMed] [Google Scholar]
- 16.Wiedmann M, Kurzchalia T, Hartmann E, Rapoport T. A signal sequence receptor in the endoplasmic reticulum membrane. Nature. 1987;328:830–833. doi: 10.1038/328830a0. [DOI] [PubMed] [Google Scholar]
- 17.Kelleher D, Gilmore R. An evolving view of the eukaryotic oligosaccharyltransferase. Glycobiology. 2006;16:47R–62R. doi: 10.1093/glycob/cwj066. [DOI] [PubMed] [Google Scholar]
- 18.Shibatani T, David L, McCormack A, Frueh K, Skach W. Proteomic analysis of mammalian oligosaccharyltransferase reveals multiple subcomplexes that contain Sec61, TRAP and two potential new subunits. Biochemistry. 2005;44:5982–5992. doi: 10.1021/bi047328f. [DOI] [PubMed] [Google Scholar]
- 19.Snapp E, Reinhart G, Bogert B, Lippincott-Schwartz J, Hegde R. The organization of engaged and quiescent translocons in the endoplasmic reticulum of mammalian cells. J. Cell. Biol. 2004;164:997–1007. doi: 10.1083/jcb.200312079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van den Berg B, et al. X-ray structure of a protein-conducting channel. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- 21.Ménétret J-F, et al. Architecture of the ribosome-channel complex derived from native membranes. J. Mol. Biol. 2005;348:445–457. doi: 10.1016/j.jmb.2005.02.053. [DOI] [PubMed] [Google Scholar]
- 22.Ménétret J, et al. Single copies of Sec61 and TRAP associate with a nontranslating mammalian ribosome. Structure. 2008;16:1126–1137. doi: 10.1016/j.str.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beckmann R, et al. Architecture of the protein-conducting channel associated with the translating 80S ribosome. Cell. 2001;107:361–72. doi: 10.1016/s0092-8674(01)00541-4. [DOI] [PubMed] [Google Scholar]
- 24.Belin D, Bost S, Vassalli J-D, Strub K. A two step recognition of signal sequences determines the translocation efficiency of proteins. EMBO J. 1996;15:468–478. [PMC free article] [PubMed] [Google Scholar]
- 25.Jungnickel B, Rapoport T. A posttargeting signal sequence recognition event in the endoplasmic reticulum membrane. Cell. 1995;82:261–270. doi: 10.1016/0092-8674(95)90313-5. [DOI] [PubMed] [Google Scholar]
- 26.Crowley K, Liao S, Worrell V, Reinhart G, Johnson A. Secretory proteins move through the endoplasmic reticulum membrane via an aqueous, gated pore. Cell. 1994;78:461–471. doi: 10.1016/0092-8674(94)90424-3. [DOI] [PubMed] [Google Scholar]
- 27.Crowley K, Reinhart G, Johnson A. The signal sequence moves through a ribosomal tunnel into a noncytoplasmic aqueous environment at the ER membrane early in translocation. Cell. 1993;73:1101–1115. doi: 10.1016/0092-8674(93)90640-c. [DOI] [PubMed] [Google Scholar]
- 28.Kim S, Mitra D, Salerno J, Hegde R. Signal Sequences control gating of the protein translocation channel in a substrate-specific manner. Dev. Cell. 2002;2:207–217. doi: 10.1016/s1534-5807(01)00120-4. [DOI] [PubMed] [Google Scholar]
- 29.Hegde R, Kang S-W. The concept of translocational regulation. J. Cell Biol. 2008;182:225–232. doi: 10.1083/jcb.200804157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pitonzo D, Yang Z, Matsumura Y, Johnson A, Skach W. Sequence-specific retention and regulated integration of a nascent membrane protein by the ER Sec61 Translocon. Mol. Biol. Cell. 2009;20:685–698. doi: 10.1091/mbc.E08-09-0902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Woolhead C, McCormick P, Johnson A. Nascent membrane and secretory proteins differ in FRET-detected folding far inside the ribosome and in their exposure to ribosomal proteins. Cell. 2004;116:725–736. doi: 10.1016/s0092-8674(04)00169-2. [DOI] [PubMed] [Google Scholar]
- 32.Alder N, Shen Y, Brodsky J, Hendershot L, Johnson A. The molecular mechanisms underlying BiP-mediated gating of the Sec61 translocon of the endoplasmic reticulum. J. Cell Biol. 2005;168:389–99. doi: 10.1083/jcb.200409174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haigh N, Johnson A. A new role for BiP: closing the aqueous translocon pore during protein integration into the ER membrane. J. Cell Biol. 2002;156:261–270. doi: 10.1083/jcb.200110074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liao S, Lin J, Do H, Johnson A. Both lumenal and cytosolic gating of the aqueous translocon pore are regulated from inside the ribosome during membrane protein integration. Cell. 1997;90:31–42. doi: 10.1016/s0092-8674(00)80311-6. [DOI] [PubMed] [Google Scholar]
- 35.Martoglio B, Hofmann M, Brunner J, Dobberstein B. The protein-conducting channel in the membrane of the endoplasmic reticulum is open laterally toward the lipid bilayer. Cell. 1995;81:207–214. doi: 10.1016/0092-8674(95)90330-5. [DOI] [PubMed] [Google Scholar]
- 36.Heinrich SU, Mothes W, Brunner J, Rapoport T. The Sec61p complex mediates the integration of a membrane protein by allowing lipid partitioning of the transmembrane domain. Cell. 2000;102:233–244. doi: 10.1016/s0092-8674(00)00028-3. [DOI] [PubMed] [Google Scholar]
- 37.Hessa T, et al. Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature. 2005;433:377–381. doi: 10.1038/nature03216. [DOI] [PubMed] [Google Scholar]
- 38.Hessa T, et al. Molecular code for transmembrane-helix recognition by the Sec61 translocon. Nature. 2007;450:1026–1030. doi: 10.1038/nature06387. [DOI] [PubMed] [Google Scholar]
- 39.Enquist K, et al. Membrane-integration characteristics of two ABC Transporters, CFTR and P-glycoprotein. J. Mol. Biol. 2009 doi: 10.1016/j.jmb.2009.02.035. In Press, Epub, PMID: 19236881. [DOI] [PubMed] [Google Scholar]
- 40.Do H, Falcone D, Lin J, Andrews D, Johnson A. The cotranslational integration of membrane proteins into the phospholipid bilayer is a multistep process. Cell. 1996;85:369–378. doi: 10.1016/s0092-8674(00)81115-0. [DOI] [PubMed] [Google Scholar]
- 41.Sadlish H, Pitonzo D, Johnson A, Skach W. Sequential triage of transmembrane segments by Sec61α during biogenesis of a native multispanning membrane protein. Nat. Struct. Mol. Biol. 2005;12:870–878. doi: 10.1038/nsmb994. [DOI] [PubMed] [Google Scholar]
- 42.McCormick P, Miao Y, Shao Y, Lin J, Johnson A. Cotranslational protein integration into the ER membrane is mediated by the binding of nascent chains to translocon proteins. Mol. Cell. 2003;12:329–341. doi: 10.1016/s1097-2765(03)00304-6. [DOI] [PubMed] [Google Scholar]
- 43.Meacock S, Lecomte F, Crawshaw S, High S. Different transmembrane domains associate with distinct endoplasmic reticulum components during membrane integration of a polytopic protein. Mol. Biol. Cell. 2002;13:4114–4129. doi: 10.1091/mbc.E02-04-0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ismail N, Crawshaw S, Cross B, Haagsma A, High S. Specific transmembrane segments are selectively delayed at the ER translocon during opsin biogenesis. Biochem. J. 2008;411:495–506. doi: 10.1042/BJ20071597. [DOI] [PubMed] [Google Scholar]
- 45.Hamman B, Hendershot L, Johnson A. BiP maintains the permeability barrier of the ER membrane by sealing the lumenal end of the translocon pore before and early in translocation. Cell. 1998;92:747–758. doi: 10.1016/s0092-8674(00)81403-8. [DOI] [PubMed] [Google Scholar]
- 46.Skach W. Topology of P-glycoproteins. In: Ambudkar S, Gottesman M, editors. Methods in Enzymology. Academic Press Inc.; San Diego: 1998. pp. 265–277. [DOI] [PubMed] [Google Scholar]
- 47.Xie Y, Langhans-Rajasekaran S, Bellovino D, Morimoto T. Only the first and the last hydrophobic segments in the COOH-terminal third of the Na,KATPase alpha subunit initiate and halt, respectively, membrane translocation of the newly synthesized polypeptide. J. Biol. Chem. 1996;271:2563–2573. doi: 10.1074/jbc.271.5.2563. [DOI] [PubMed] [Google Scholar]
- 48.Skach W, Calayag MC, Lingappa V. Evidence for an alternate model of human P-glycoprotein structure and biogenesis. J Biol. Chem. 1993;268:6903–6908. [PubMed] [Google Scholar]
- 49.Skach W, Lingappa V. Amino terminus assembly of human P-glycoprotein at the endoplasmic reticulum is directed by cooperative actions of two internal sequences. J. Biol. Chem. 1993;268:23552–23561. [PubMed] [Google Scholar]
- 50.Skach W, et al. Biogenesis and transmembrane topology of the CHIP28 water channel in the endoplasmic reticulum. J. Cell Biol. 1994;125:803–815. doi: 10.1083/jcb.125.4.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moss K, Helm A, Lu Y, Bragin A, Skach W. Coupled translocation events generate topologic heterogeneity at the endoplasmic reticulum membrane. Mol. Biol. Cell. 1998;9:2681–2697. doi: 10.1091/mbc.9.9.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Audigier Y, Friedlander M, Blobel G. Multiple topogenic sequences in bovine opsin. Proc. Natl. Acad. Sci. USA. 1987;84:5783–5787. doi: 10.1073/pnas.84.16.5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Agre P, et al. Aquaporin water channels - from atomic structure to clinical medicine. J. Physiol. 2002;542:3–16. doi: 10.1113/jphysiol.2002.020818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.King L, Kozono D, Agre P. From structure to disease, the evolving tale of aquaporin biology. Nat. Rev. Mol. Cell Biol. 2004;5:687–98. doi: 10.1038/nrm1469. [DOI] [PubMed] [Google Scholar]
- 55.Fujiyoshi Y, et al. Structure and function of water channels. Curr. Opin. Struc. Biol. 2002;12:509–15. doi: 10.1016/s0959-440x(02)00355-x. [DOI] [PubMed] [Google Scholar]
- 56.Sui H, Han B-G, Lee J, Walian P, Jap B. Structural basis of water specific transport through the AQP1 water channel. Nature. 2001;414:872–878. doi: 10.1038/414872a. [DOI] [PubMed] [Google Scholar]
- 57.Fu D, et al. Structure of a glycerol-conducting channel and the basis for its selectivity. Science. 2000;290:481–486. doi: 10.1126/science.290.5491.481. [DOI] [PubMed] [Google Scholar]
- 58.Verbavatz JM, et al. Tetrameric assembly of CHIP28 water channels in liposomes and cell membranes. A freeze-fracture study. J. Cell. Biol. 1993;123:605–618. doi: 10.1083/jcb.123.3.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smith BL, Agre P. Erythrocyte Mr 28,000 transmembrane protein exists as a multisubunit oligomer similar to channel proteins. J. Biol. Chem. 1991;266:6407–6415. [PubMed] [Google Scholar]
- 60.Buck T, Wagner J, Grund S, Skach W. A novel tripartite structural motif involved in aquaporin topogenesis, monomer folding and tetramerization. Nat. Struct. Mol. Biol. 2007;14:762–769. doi: 10.1038/nsmb1275. [DOI] [PubMed] [Google Scholar]
- 61.Shi L-B, Skach W, Ma T, Verkman A. Distinct biogenesis mechanisms for water channels MIWC and CHIP28 at the endoplasmic reticulum. Biochemistry. 1995;34:8250–8256. doi: 10.1021/bi00026a006. [DOI] [PubMed] [Google Scholar]
- 62.Lu Y, et al. Reorientation of Aquaporin-1 topology during maturation in the endoplasmic reticulum. Mol. Biol. Cell. 2000;11:2973–2985. doi: 10.1091/mbc.11.9.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Foster W, et al. Identification of sequence determinants that direct different intracellular folding pathways for AQP1 and AQP4. J. Biol. Chem. 2000;275:34157–34165. doi: 10.1074/jbc.M000165200. [DOI] [PubMed] [Google Scholar]
- 64.Tu LW, Wang J, Helm A, Skach W, Deutsch C. Transmembrane Biogenesis of Kv1.3. Biochemistry. 2000;39:824–836. doi: 10.1021/bi991740r. [DOI] [PubMed] [Google Scholar]
- 65.Lu Y, et al. Co- and Posttranslational mechanisms direct CFTR N-terminus transmembrane assembly. J. Biol. Chem. 1998;273:568–576. doi: 10.1074/jbc.273.1.568. [DOI] [PubMed] [Google Scholar]
- 66.Gratkowski H, Lear J, DeGrado W. Polar side chains drive the association of model transmembrane peptides. Proc. Natl. Acad. Sci. USA. 2001;98:880–885. doi: 10.1073/pnas.98.3.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhou F, Cocco M, Russ W, Brunger A, Engelman D. Interhelical hydrogen bonding drives strong interactions in membrane proteins. Nat. Struct. Biol. 2000;7:154–160. doi: 10.1038/72430. [DOI] [PubMed] [Google Scholar]
- 68.Gafvelin G, von Heijne G. Topological “frustration” in multispanning E. coli inner membrane. Cell. 1994;77:401–412. doi: 10.1016/0092-8674(94)90155-4. [DOI] [PubMed] [Google Scholar]
- 69.Goder V, Bieri C, Spiess M. Glycosylation can influence topogenesis of membrane proteins and reveals dynamic reorientation of nascent polypeptides within the translocon. J. Cell Biol. 1999;147:257–266. doi: 10.1083/jcb.147.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Garrison J, Kunkel E, Hegde R, Taunton J. A substrate-specific inhibitor of protein translocation into the endoplasmic reticulum. nature. 2005;436:285–289. doi: 10.1038/nature03821. [DOI] [PubMed] [Google Scholar]
- 71.Bogdanov M, Xie J, Heacock P, Dowhan W. To flip or not to flip: lipid-protein charge interactions are a determinant of final membrane protein topology. J. Cell Biol. 2008;182:925–935. doi: 10.1083/jcb.200803097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brambillasca S, Yabal M, Makarow M, Borgese N. Unassisted translocation of large polypeptide domains across phospholipd bilayers. J. Cell. Biol. 2006;175:767–777. doi: 10.1083/jcb.200608101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lin J, Addison R. A novel integration signal that is composed of two transmembrane segments is required to integrate the Neurospora plasma membrane H+-ATPase into microsomes. J. Biol. Chem. 1995;270:6935–6941. doi: 10.1074/jbc.270.12.6935. [DOI] [PubMed] [Google Scholar]
- 74.Ismail N, Crawshaw S, High S. Active and passive displacement of transmembrane domains during opsin biogenesis at the Sec61 translocon. J. Cell Sci. 2006;119:2826–2836. doi: 10.1242/jcs.03018. [DOI] [PubMed] [Google Scholar]
- 75.Beckmann R, et al. Alignment of conduits for the nascent polypeptide chain in the ribosome-Sec61 complex. Science. 1997;278:2123–2126. doi: 10.1126/science.278.5346.2123. [DOI] [PubMed] [Google Scholar]
- 76.Ménétret J, et al. The structure of ribosome-channel complexes engaged in protein translocation. Mol. Cell. 2000;6:1219–32. doi: 10.1016/s1097-2765(00)00118-0. [DOI] [PubMed] [Google Scholar]
- 77.Hanein D, et al. Oligomeric rings of the Sec61p complex induced by ligands required for protein translocation. Cell. 1996;87:721–732. doi: 10.1016/s0092-8674(00)81391-4. [DOI] [PubMed] [Google Scholar]
- 78.Morgan D, Menetret J, Neuhof A, Rapoport T, Akey C. Structure of the mammalian ribosome-channel complex at 17Å resolution. J. Mol. Biol. 2002;324:871–886. doi: 10.1016/s0022-2836(02)01111-7. [DOI] [PubMed] [Google Scholar]
- 79.Young J, Agashe V, Siegers K, Hartl F. Pathways of chaperone-mediated protein folding in the cytosol. Nat. Rev. Mol. Cell Biol. 2004;5:781–191. doi: 10.1038/nrm1492. [DOI] [PubMed] [Google Scholar]
- 80.Oberdorf J, Pitonzo D, Skach W. An energy-dependent maturation step is required for release of the cystic fibrosis transmembrane conductance regulator from early endoplasmic reticulum biosynthetic machinery. J. Biol. Chem. 2005;280:38193–38202. doi: 10.1074/jbc.M504200200. [DOI] [PubMed] [Google Scholar]
- 81.Clemons W, Menetret J-F, Akey C, Rapoport T. Structural insight into the protein translocation channel. Curr. Opin. Struct. Biol. 2004;14:390–396. doi: 10.1016/j.sbi.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 82.Breyton C, Haase W, Rapoport T, Kuehlbrandt W, Collinson I. Three-dimensional structure of the bacterial protein-translocation complex SecYEG. Nature. 2002;418:662–665. doi: 10.1038/nature00827. [DOI] [PubMed] [Google Scholar]
- 83.Mitra K, et al. Structure of the E. coli protein-conducting channel bound to a translating ribosome. Nature. 2005;438:318–324. doi: 10.1038/nature04133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hamman B, Chen J-C, Johnson E, Johnson A. The aqueous pore through the translocon has a diameter of 40–60Å during cotranslational protein translocation at the ER membrane. Cell. 1997;89:535–544. doi: 10.1016/s0092-8674(00)80235-4. [DOI] [PubMed] [Google Scholar]
- 85.Kida Y, Morimoto F, Sakaguchi M. Two translocating hydrophilic segments of a nascent chain span the ER membrane during multispanning protein topogenesis. J. Cell. Biol. 2007;179:1441–1452. doi: 10.1083/jcb.200707050. [DOI] [PMC free article] [PubMed] [Google Scholar]