Abstract
The ionotropic glutamate receptors are localized in the pre- and postsynaptic membrane of neurons in the brain. Activation by the principal excitatory neurotransmitter glutamate allows the ligand binding domain to change conformation, communicating opening of the channel for ion conduction. The free energy of the GluR2 S1S2 ligand binding domain (S1S2) closure transition was computed using a combination of thermodynamic integration and umbrella sampling modeling methods. A path that involves lowering the charge on E705 was chosen to clarify the role of this binding site residue. A continuum electrostatic approach in S1S2 is used to show E705, located in the ligand binding cleft, stabilizes the closed conformation of S1S2. In the closed conformation, in the absence of a ligand, S1S2 is somewhat more closed than reported from X-ray structures. A semi-open conformation has been identified which is characterized by disruption of a single cross-cleft interaction and differs only slightly in energy from the fully closed S1S2. The fully open S1S2 conformation exhibits a wide energy well and shares structural similarity to the apo S1S2 crystal structure. Hybrid continuum electrostatics/MD calculations along the chosen closure transition pathway reveal solvation energies, as well as electrostatic interaction energies between two lobes of the protein increase the relative energetic difference between the open and the closed conformational states. By analyzing the role of several cross-cleft contacts as well as other binding site residues we demonstrate how S1S2 interactions facilitate formation of the closed conformation of the ligand binding domain.
Keywords: Glutamate receptor, GluR2, molecular dynamics simulations, umbrella sampling, thermodynamic integration, electrostatic interaction, solvation energy, potential of mean force, free energy, conformational transition
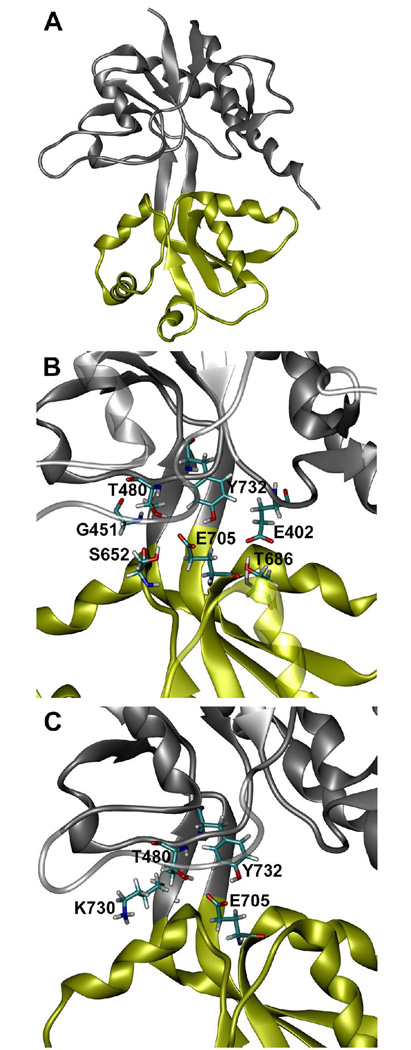
Ionotropic glutamate receptors (iGluR) are a family of ligand-gated ion channels that facilitate excitatory response in neuronal cells. Due to their importance in brain functioning and as potential drug targets, GluRs and particularly, the AMPA-type glutamate receptor of type 2 (GluR2), which is a subject of this paper, received considerable attention of both experimental (1, 2) and theoretical investigators (3–9). In the full receptor, binding of agonists and partial agonists results in an opening of a cation selective ion channel located in a transmembrane domain of the receptor. This process is described as gating of the channel (10). While a three dimensional structure of the full receptor is still unknown, the ligand-binding domain construct (further termed S1S2) forming a monomeric soluble protein, has been crystallized in its apo form (11) and with a variety of bound ligands (11–14). The ligand-binding domain of the GluR2 receptor is formed by two polypeptide segments S1 and S2 in the shape of a two-lobe protein (11) (shown in Figrue 1A). S1S2 can be logically divided into two separate lobes, D1 and D2. The lobes were defined as follows (12): D1 residues 393–498 and 728–775, D2 residues 499–727. The ligand binding site is located in the cleft at the lobe interface. Binding of agonists and partial agonists initiates a cleft closing conformational rearrangement of S1S2, as evident from crystal structures and supported by the time-resolved experiments (15–17). Recently computed free energy profiles of the S1S2 conformational space with and without glutamate (Glu) agonist are also consistent with this mechanism (8). In these calculations performed using molecular dynamics (MD) with enhanced sampling (umbrella sampling technique) an open cleft state of the apo-S1S2 has lower free energy than the closed cleft state, while the opposite is true for the glutamate bound S1S2. A degree of the inter-lobe cleft closure is believed to correlate strongly with an efficacy of the channel (11, 18). However, recent fluorescence resonance energy transfer (FRET) based assays of the isolated S1S2 ligand binding domain revealed smaller cleft closure upon binding various ligands than observed in crystals structures, suggesting other mechanisms contributing to channel activation (17). Thus it is important to develop a detailed atomistic level understanding of the mechanism and energetics of the S1S2 cleft closure transition and how it is influenced by a presence or an absence of a ligand.
Figure 1.
Structure of the GluR2 S1S2 ligand binding core. (A) Ribbon representation of GluR2 S1S2 in a closed conformation. D1 is shown in grey and D2 is shown in yellow. (B) Enlarged view of (A) which shows the orientation of: E705, Y732, and T480 located in the ligand binding pocket and T686, E402, S652, and G451 which interact across the S1S2 domain cleft in the closed conformation (see text). (C) Enlarged view of (A) which shows the orientation of E705, Y732, T480, and K730 in the closed conformation. Licorice representation and CPK coloring are used for the side chains shown.
Specific interactions of ligands with amino acid residues exposed in the binding cleft have been studied using a variety of experimental techniques (19–22) as well as by molecular modeling by us (5, 6) and others (4, 7). Speranskiy and Kurnikova (6) demonstrated that electrostatic potential complementarity between the positively charged protein and a negatively charged ligand is a determining feature in agonist binding of this protein. However, a role of the negatively charged E705 residue exposed in the ligand-binding pocket has not yet been understood. Crucial importance of E705 for the ligand binding ability of S1S2 has been demonstrated in a number of experimental studies (11, 14, 19, 23, 24). While most mutants with a neutral residue substituted at this site nearly abandon ligand binding (23), the direct interaction of E705 with the glutamate ligand (in the closed cleft ligand-bound state) is weak and even somewhat disruptive (6) due to the electrostatic repulsion between the negatively charged glutamate ligand and the negatively charged E705. This has also been noted earlier by Mayer and coworkers (18). One goal of the present study is to clarify the role of E705 in the ligand binding process.
In recent work (9) we used MD simulations, rigidity theory, and computational mutations to investigate major factors that control cleft closing transition of the S1S2 protein in the absence of the ligand. When E705 has been mutated to a neutral residue or its charge has been lowered, the protein underwent a rapid cleft opening transition in free unconstrained MD simulations. Based on this observation we hypothesized that the role of E705 is to facilitate the S1S2 cleft closure via direct electrostatic attraction of the opposite lobe, rather than to affect ligand orientation in the binding cleft as it was presumed earlier. In this paper we report on a careful free energy calculation of the cleft S1S2 closure transition along a path that involves lowering the charge on the E705 residue. To compute the cleft closing energy we applied a combination of thermodynamic integration (TI) (25) and umbrella sampling (US) (26) free energy modeling methods using a previously proposed protocol (27). Thermodynamically, the resulting energy difference between two stable states of the protein (open and closed conformations) is independent from a transition path. Choosing a path that involves mutation of the E705 allowed us to pinpoint the role of E705. Furthermore, using a hybrid continuum electrostatics/MD methodology to estimate relative solvation energies; as well as electrostatic interaction energies between two lobes of the protein in different conformations, we demonstrate how interactions of E705 facilitate formation of the closed conformation of the cleft. We also analyzed roles of the cleft facing K730 residue as well as two pairs of residues: S652-G451 and E402-T686, previously observed to form cross-cleft contacts (9).
This paper is organized as follows: Models and Methods section describes simulated proteins and protocols of the MD simulations. It also briefly describes the TI and US methods and their implementation in this work, as well as the hybrid method to estimate electrostatic free energy differences. The results of the simulations and their analysis are presented and discussed in Results and Discussion sections.
MODELS AND METHODS
Molecular Dynamics (MD) Simulations and simulated systems
Crystal structures of the S1S2 ligand binding protein in the apo and glutamate bound states (PDB ID: 1FTO and 1FTJ chain A (11)) were used as initial structures for modeling. Missing hydrogen atoms and counter ions were added to the x-ray resolved structures; these were then solvated in water and equilibrated as described previously (6). The overall size of the simulated system was 65×70×65 Å. All MD simulations were performed using the AMBER 8 package (28) with the Cornel et al. force-field (29). The TIP3P water model was used. Temperature was controlled using the Berendsen thermostat (30). Equilibrium production runs were performed at constant volume and constant temperature at 300K. Bonds to hydrogen atoms were constrained via the SHAKE algorithm (31). The time step for integration was 2 fs.
MD Thermodynamic Integration (TI)
Five independent TI simulations were performed in this work. TI (25) is used to compute a free energy difference between two systems by running a set of individual equilibrium MD simulations in which an initial system is gradually transformed into a final system. A coupling parameter λ is used to interpolate between the initial and the final structures. When λ = 0 the simulated system corresponds to the initial system and when λ = 1 it corresponds to the final system. In all TI simulations reported in this work the initial and the final systems differed only by the value of the E705 residue electric charge. Its native charge is −1e (where e indicates a charge of a proton). In the perturbed system the charge of E705 was −0.75e. In four TI simulations the initial structure with λ=0 corresponded to the native E705 charge. One TI calculation was performed in a backward direction, where the charge on the E705 residue was restored from −0.75e (λ=0) to its native charge −1e (λ=1). Each transformation used nine λ windows. The mutant was constructed using the Amber Leap program (28). To preserve the protein conformation during a TI simulation the distance between S652 and G451 was constrained with harmonic potentials of 1–6 kcal/mol/Å2 depending on the simulated system. The entire simulation time included in a single TI calculation varied from 21 to 27 ns, of which 3–4 ns trajectories were simulated in λ windows 1 through 6, and 1 ns trajectories were simulated in windows 7–9. Longer trajectories in windows 1–6 are needed to improve statistical error of the entire simulation. Upon completion of all TI simulations the weighted histogram analysis method (WHAM) (32) was used to eliminate energy contributions due to restraining potentials.
MD Umbrella Sampling (US)
US is a well established simulation technique (26) to evaluate a potential of mean force (PMF) along a path between two different conformations of the system. A PMF is the free energy of a system along a chosen coordinate or a set of coordinates; hence PMF is a path dependent function. However, a free energy difference between any two conformations is path independent and therefore it can be computed using any path that connects the two end conformations. While theoretically, any path can be chosen for an US simulation, in practice, numerical errors which must come from insufficient sampling, suppression of the natural fluctuations of the system due to too strong biasing potential, or due to overcoming high energy barriers and deep wells may accumulate, and bias the results of the US simulation. Here we utilized US simulations to convert a protein with the modified electric charge on the E705 residue (E705−0.75) from its closed to the open form. In US the simulated protein system is constrained within a narrow region of a reaction coordinate by applying a quadratic biasing potential also called umbrella, hence the name of the method. In this work a one dimensional umbrella potentials were used as follows,
| (1) |
where k is the biasing potential constant and r→0 is a position vector along the reaction coordinate r→. The coordinate r→ in this study was taken to be the distance between the Cα of S652 and the Cα of G451. For each umbrella potential, characterized by its values of k and r→0, an equilibrium MD simulation is performed to sample configurational space of a simulated protein in the vicinity of r→0. In this work fifteen umbrella windows were applied to cover the range of interest of r→ from 5 to 14 Å. The distances between neighboring umbrella potential centers r→0 were 1 Å or 0.5 Å. Values of the biasing potential constants k ranged from 1 to 6 kcal/mol/Å2. Simulation time varied from 1.2 to 3 ns per window to ensure good convergence of the results and to lower statistical errors. WHAM was used to remove effects of biasing umbrella potentials and compute the final PMF.
Continuum Electrostatics Calculations
The electrostatic free energy E of a protein can be computed as
| (2) |
where qi is a partial electric charge of a protein atom i, and φ (r→i) is the value of the electrostatic potential at the position of charge qi, r→i is a position vector of atom i. The summation in Eq. 2 is over all the charges qi in the system. The electrostatic potential φ (r→i) is found by solving the corresponding Poisson equation subject to Dirichlet boundary conditions:
| (3) |
where ε(r→) is the dielectric constant of a medium, which is a piece –wise constant in this work, δ is the three dimensional Dirac delta function and r→i is the position of charge qi. In all electrostatic calculations partial charges of the protein atoms corresponded to the Cornell et al. force field (29). An internal dielectric constant for a protein was 2, a solvent dielectric constant was 81, and a grid of 271×271×271 Å3 at 3.0 grids/Å was used. The value of the electrostatic potential at the computational grid boundaries was set to equal an approximate Columbic potential of the protein charges in a homogeneous dielectric of 81. Eq. 2 and Eq. 3 were solved using the Poisson equation solver of the HARLEM (33) package.
Hybrid Continuum Electrostatics/Molecular Dynamics Calculations of Electrostatic Free Energy
Hybrid continuum electrostatic/molecular mechanics approaches for computing free energy of proteins and free energy of protein ligand interactions has previously been described and implemented with success (6, 34). This method utilizes the continuum electrostatic theory outlined above applied to protein structures generated from molecular dynamics, extracted from the fully explicit solvent simulations and embedded within a continuum dielectric solvent model. Each new system as a snapshot from a molecular dynamics trajectory, now a single protein structure in implicit solvent, is used to calculate an electrostatic free energy (Eq. 2).
In this work the electrostatic free energy of interaction between lobes D1 and D2 of the S1S2 ligand binding domain Δ GElec Inter was computed as:
| (4) |
where ES1S 2 is the electrostatic free energy as defined in Eq. 2 of the S1S2 protein embedded in the solvent, ED1 is the electrostatic free energy of D1 of a protein alone embedded in the solvent, and ED2 is the energy of D2 of a protein alone embedded in the solvent. The ensemble average indicated by <…> is over a molecular dynamics trajectory and accounts for protein flexibility. The relative interaction free energies ΔΔGInteraction are then calculated according to:
| (5) |
where are the electrostatic interaction energies described by Eq. 4 for the native protein, and for a single charge substitution with all other charges native in the S1S2 protein respectively.
Solvation free energies ΔGSolvation of the ligand binding domain were calculated using the same continuum theory according to:
| (6) |
where the are the electrostatic energy of the S1S2 ligand binding domain in the solvent dielectric and in vacuum respectively. The electrostatic free energy of the protein is then computed as an ensemble average of thus obtained single structure energies.
In this work electrostatic energies were calculated at 10 picosecond intervals and averaged using the molecular dynamics trajectories of approximately 2 ns in length (water and ions stripped out) generated in this study (see Table 1).
Table 1.
Summary of calculated Free Energy Differences
| Protein | Conformation | Method | ΔG, kcal/mol |
|---|---|---|---|
| E705 → E705−0.75 | closed | TI | 32.9 |
| E705−0.75 → E705−0.75 | closed → open | US | −3 |
| E705−0.75 → E705 | open | TI | −34.5 |
| E705 → E705−0.75* | semi-open | TI | 33.6 |
| E705 → E705−0.75** | semi-open | TI | 33.9 |
| WT total | open → closed | 4.3±0.6 | |
CA652 - CA451 distance is 8 Å
CA652 -_CA451 distance is 9 Å
RESULTS
D1-D2 electrostatic interaction energies reveal E705 importance for stabilization of the closed S1S2 conformation
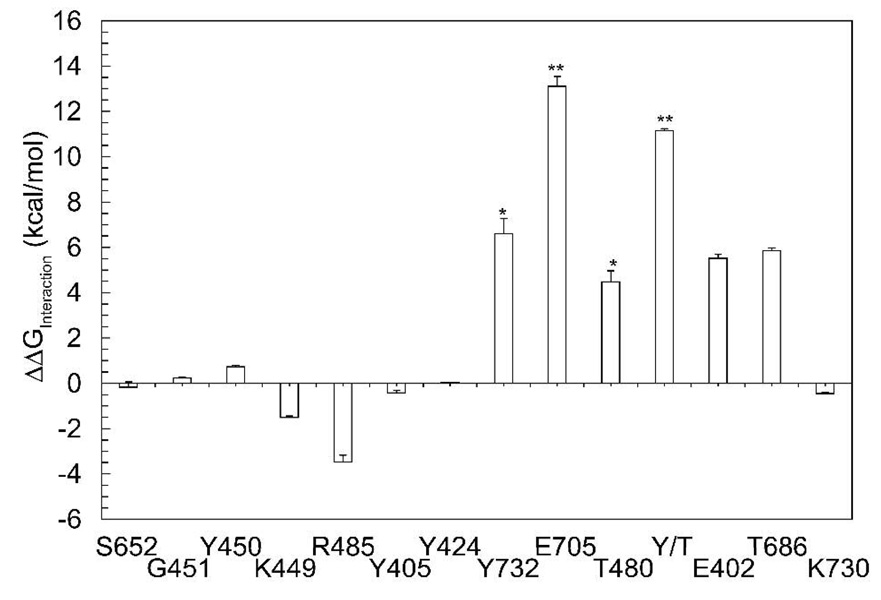
The overall charge on the S1S2 domain is a positive 5e. Individually the domains contribute an overall +1e from D1 and +4e from D2. This native charge distribution creates a repulsive electrostatic interaction between the two lobes of the S1S2 domain. To test the hypothesis that shielding direct electrostatic interactions between the D1 and D2 lobes is important for stabilizing the closed conformation of the S1S2 domain, we computed electrostatic interaction free energies between lobes of the wild type protein and a protein with charges on a single residue set to zero calculated by Eq. 5. The native S1S2 closed conformation and a single residue charge substituted S1S2 closed conformation from equilibrium MD were the two proteins used in each calculation. By substituting the charge on a single residue and calculating the relative electrostatic interaction energy with respect to the native charge distribution, key interactions important to a particular conformation of the S1S2 domain can also be identified. Charges on each residue were substituted according to the following rules, the charged residues were neutralized, N-H or C-O backbone dipoles of neutral residues were eliminated, or O-H side-chain dipoles of polar residues were eliminated. Relative electrostatic interaction free energies between lobes D1 and D2 as charge is substituted per residue are shown in Figure 2, namely for the charge substituted residues: G451, Y450, K449, R485, Y405, Y424, Y732, T480, and E402 located in D1; S652, T686, E705, and K730 located in D2 (Figure 1B and C). In Figure 2 a more positive energy indicates more unfavorable relative interaction between D1 and D2 upon removal of a charge from an indicated residue. Negative energy in Figure 2 indicates more favorable interaction between the lobes upon charge substitution. As seen in Figure 2 removal of charge from numerous residues on D1, the lobe not containing E705 produces little effect on the interaction between the two lobes. Neutralization of residue E705 (E7050) produces a large increase in electrostatic repulsion (13 kcal/mol) between the D1 and D2 domains of S1S2, as indicated by the (**) in Figure 2. Figure 2 also reveals that Y732 and T480 (marked with *) upon neutralization significantly increase repulsion of D1 and D2, destabilizing the closed conformation of S1S2. When substituted together, they produce a destabilization of closed S1S2 of similar magnitude (11 kcal/mol) to the E705 substitution indicated in Figure 2 by Y/T and the (**). These results combined with the observation that an E705 charge substitution along with an S652A modification induces fast cleft opening during equilibrium MD simulations (9), point to this residue as playing a major role in stabilizing the closed conformation of the S1S2 ligand binding domain.
Figure 2.
Electrostatic interaction energy between D1 and D2 lobes of GluR2. Relative interaction free energy between D1 and D2 are shown for the closed conformation with respect to the wild-type as calculated by Eq. 5. Each bar corresponds to a single charge substitution according to the type of interaction (see text). T480 and Y732 form a pair (*) which when removed simultaneously compensate for the effects of E7050 (**).
Reorganization energy of the protein using MD simulations
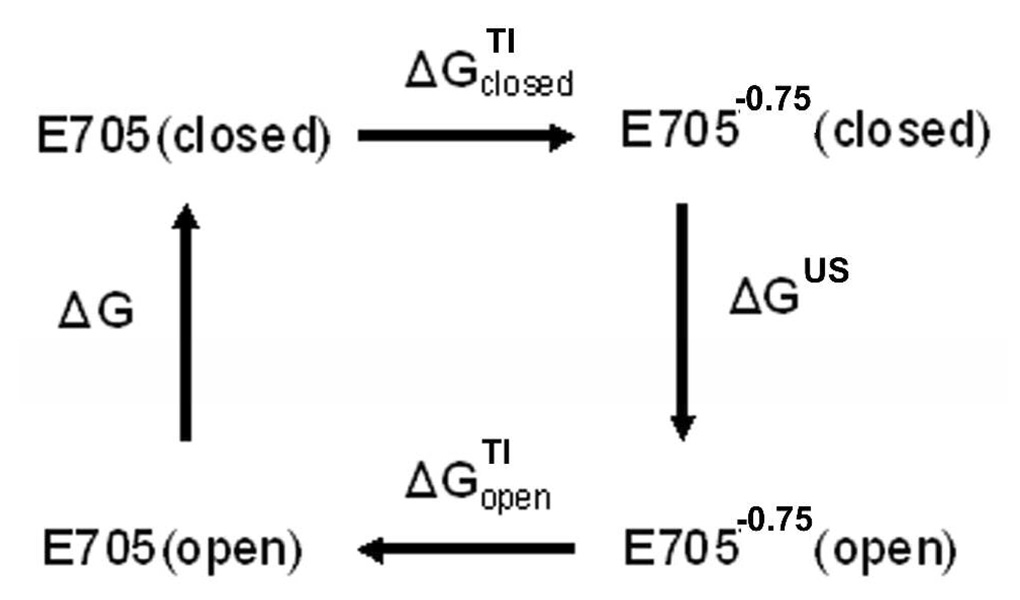
The free energy cost ΔG for converting the S1S2 conformation from open to closed in the absence of a ligand has been calculated using a combination of the enhanced sampling techniques TI and US (see the Methods section for detailed description of TI and US methods). Previously we have successfully utilized this approach to compute the free energy of reorganization of a large flexible protein system (27). A conformational transition path utilized in this work has been obtained in unconstrained MD simulations (9). While ΔG is independent on a path between two end conformations, to achieve a good accuracy of computed ΔG a chosen pathway should avoid high energy barriers and deep wells, also it is desirable to avoid steep gradients of the energy profile. One such “minimally invasive” path for the cleft opening transition involves creating an artificial E705−0.75 mutant to destabilize the closed conformation while this mutant did not spontaneously undergo transition from the closed to the open state in unrestrained MD; it does so under influence of a weak harmonic one-dimensional biasing force. The free energy difference between the open and closed cleft conformations ΔG of the wild type protein was computed in this work using the thermodynamic cycle shown in Figure 3. The sum of all free energy differences in Figure 3 is zero; hence, ΔG can be calculated as
| (7) |
in both Figure 3 and Eq. 7 subscripts “closed” or “open” indicate a corresponding conformational form of the protein and superscripts TI and US indicate a computational method used. ΔGclosedTI and ΔGopenTI are the free energy differences computed using the TI method while the protein is constrained in one conformation (“closed” or “open” respectively). The results of all enhanced sampling MD simulations are summarized in Table 1.
Figure 3.
The thermodynamic cycle utilized in this work to determine the free energy difference ΔG between the closed and open structures of GluR2 S1S2. See details in text.
The free energy change associated with modifying the charge of E705 from −1e (which is its value in the native state) to −0.75e in the closed conformation of S1S2 was computed using the TI method (listed in Table 1 as “E705−0.75 closed”). For this TI set of simulations, the 4 ns MD trajectories were generated for the first six λ windows and the 2 ns trajectories were generated for windows 7 – 9. The distance between the Cα atom of residue 652 and the Cα atom of residue 451 was constrained at 12 Å with the harmonic biasing potential where k=2 kcal/mol/Å2. computed using the 2 ns fragments of 4 ns trajectories is 33.1 kcal/mol with statistical error 0.6 kcal/mol. When full 4 ns trajectories were used for the free energy calculation the resulting is 32.9 kcal/mol (Table 1), indicating that reasonable convergence has been achieved in the simulations. It is useful to note that while an error of 0.6 kcal/mol constitutes a relative error of less then 2% fo , it translates into significantly larger relative error for the total ΔG of the conformational transition as discussed below. Starting at the final point of the E705−0.75 closed TI simulation (λ=1) additional 2 ns of an unconstrained MD has been performed to check for stability of the structure. No conformational changes were observed beyond typical fluctuations of atoms about their average positions, indicating that the mutated protein remained in the closed cleft conformation.
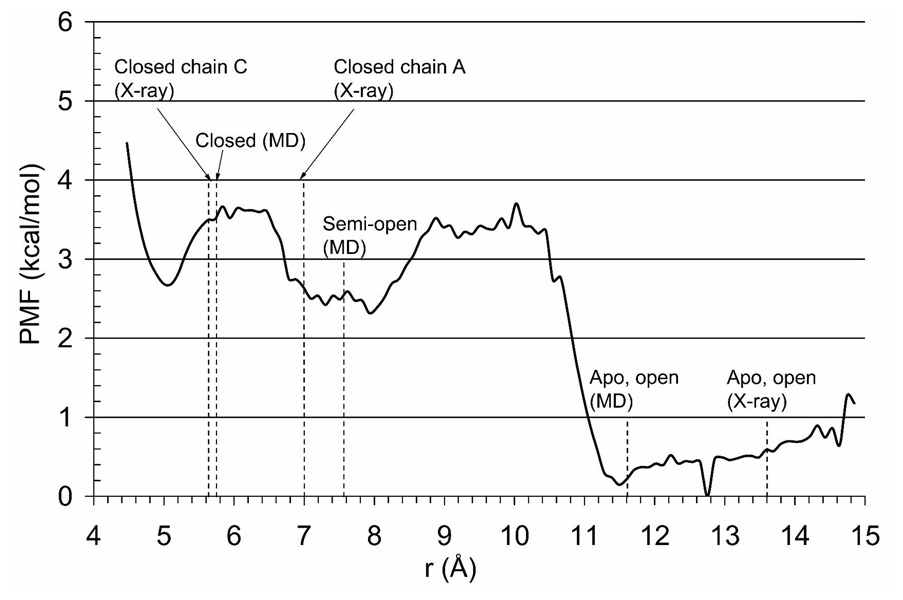
Next, the potential of mean force (PMF) of the conformational reorganization of S1S2 E705−0.75 from the closed to open structure was computed using umbrella sampling as described in the Methods section. The PMF profile is shown in Figure 4. The resulting ΔGUS is ca. −3.3 kcal/mol (Table 1) as measured between the glutamate bound state (chain C) and the apo state.
Figure 4.
PMF profile obtained using the US simulations of the E705−0.75 mutant. The reaction coordinate r is taken to be the distance between the Cα atoms of the residues S652 and G451. Values for the closed and apo-open conformations of the protein found from X-ray structures are indicated with dotted lines. The values of the closed, semi-open conformation (average chain A), and apo-open (average chain A) from averaged MD simulations are also indicated.
To complete the thermodynamic cycle (Figure 3) the open S1S2 E705−0.75 protein obtained at the end of the US simulations was restrained in the open conformation and the second TI run was performed restoring the partial charge of E705 to its native state. The protocol of this TI simulation was the same as described above. The free energy change along this transformation is −34.5 kcal/mol (Table 1).
Two additional charge modifying TI calculations were performed with the protein constrained at intermediate conformations along the reaction coordinate. Starting conformations for these calculations were taken from a pool of configurations generated in the US simulations. In these simulations the distance between the Cα atom of S652 and the Cα atom of G451 was constrained with a harmonic potential with the strength of k = 4 kcal/mol/Å2 and k = 6 kcal/mol/Å2 centered at the distance of 8 Å and 9 Å respectively. 3 ns MD simulations were performed for each λ window of each TI set, and corresponding free energy changes were calculated using the last 1.5 ns fragments of trajectories. The results of these two TI simulations are listed in Table 1. These calculations serve as a validation and confirm that there is no significant difference in destabilization upon charge substitution for intermediate conformations of the protein.
Continuum electrostatics calculations point to E705 as stabilizing the closed conformation of S1S2
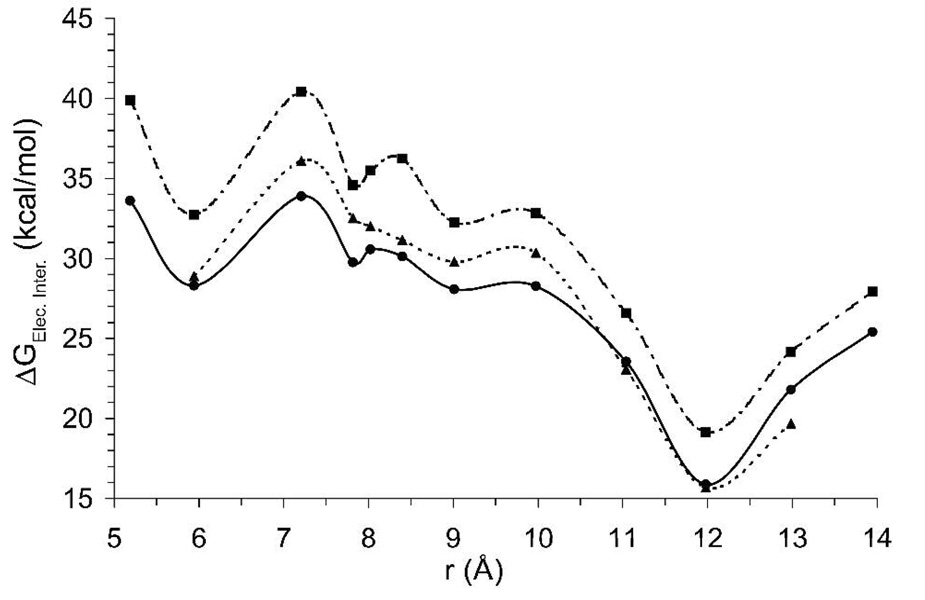
The electrostatic interaction free energy ΔGElec.Inter between the D1 and D2 lobes was calculated for the protein with the wild type charge distribution (further referred to as E705) as well as for a modified protein with neutral E705 (further referred to as E7050). These calculations were performed as defined by Eq. 5 and described in the Methods section. Figure 5 shows how the electrostatic free energy of the inter-lobe interaction varies over the total reorganization pathway determined in the previous section (Figure 4). Electrostatic disruption of E705 in the S1S2 domain results in an increase of unfavorable interaction between the D1 and D2 lobes with respect to the wild type protein, indicated by a more positive shift in the electrostatic interaction curve around an inter-lobe distance of 6 Å, defined in this study as a closed S1S2 conformation. In fact, this calculation shows that all the states along this conformational pathway have been destabilized by the introduction of E7050.ΔGElec.Inter. was also calculated for a charge substituted S652, where the dipole on the side chain hydroxyl group was removed, to eliminate the hydrogen bound with G451 across the cleft. The S652 substitution increases unfavorable interaction between the D1 and D2 lobes with respect to the wild type protein although not to the extent as seen with the E7050 protein.
Figure 5.
D1, D2 lobe electrostatic interaction energy. The electrostatic interaction energy between the D1 and D2 lobes of the ligand binding domain for the wild-type (solid), E7050 charge substitution (dashed), and S652 charge substitution (grey) are shown as a function of the sane reaction coordinate as in Figure 4. Each point represents the ensemble average interaction energy from 2 ns of umbrella sampling trajectories.
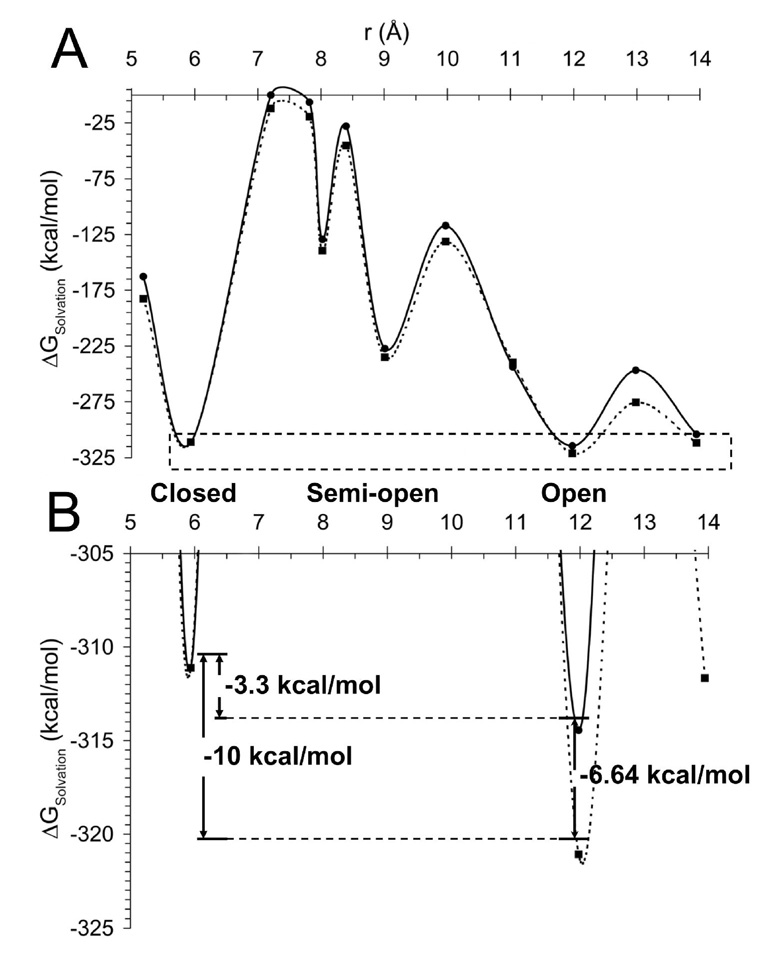
In the presence of the E7050substitution, the solvation free energy of the S1S2 domain as calculated from Eq. 4 becomes more favorable over the entire closed to open reaction coordinate. Figure 6B emphasizes the effect of the E7050 substitution on the relative difference between the closed and open conformations of S1S2. The charge substitution significantly increases the relative solvation energy difference between the open and closed conformational states of S1S2 where −3.3 kcal/mol separate the wild-type E705 S1S2 states and −10 kcal/mol separate the E7050 substituted S1S2 states.
Figure 6.
S1S2 Ligand binding domain solvation energy. (A) The ligand binding domain solvation energy is shown for the wild-type (solid) and E7050 charge substitution (dashed) referenced to the fully closed conformation of S1S2 as a function of the smae reaction coordinate as in Figure 4. Each point represents the ensemble average interaction energy from 2 ns of umbrella sampling trajectories. (B) Solvation energy differences between open and closed conformations of the native and charge substituted proteins shown more clearly by a focused plot of the dashed region shown in (A).
DISCUSSION
Based on x-ray crystallography, it has been proposed that in the absence of a ligand, the open form of the S1S2 protein is energetically preferable to the closed conformation (11). A recent theoretical study (8) corroborated this fact by calculating the PMF landscape of the cleft closure conformational transition of this protein using two dimensional US simulations. However, in general computing free energy landscapes of proteins capable of complex conformational transitions is not easy and has been rarely done in practice. Several factors can limit accuracy of the simulations employing sampling enhancing methods, such as US. These include using strong biasing potentials needed along a path with large energy gradients which may influence underlying dynamics of the protein, as well as the existence of an a priori unknown and, therefore, unaccounted for slow modes which may not be properly sampled in the simulations. One example of such possible mode is water entering the ligand binding cleft. Currently there is no rigorous method for assessing accuracy of computed PMFs. However, confidence in the results can be significantly improved by performing simulations along several independently chosen paths. The reorganization energies should remain path independent for transition between same end conformational states. In this paper a path chosen to compute the PMF for the cleft opening conformational transition of the S1S2 protein differs drastically from a path chosen in (8). We have found the free energy difference between the open and closed conformations is similar for both paths, i.e. ca. 4 kcal/mol (Table 1) which increases the confidence in the results.
The binding energy of the glutamate ligand with the closed conformation of the protein has been previously computed by Speranskiy and Kurnikova (6) using the MD/PBSA approach; the binding energy was ca. −13 kcal/mol. Combining this value with the protein reorganization energy computed in this work (ca. −4 kcal/mol), the total binding energy of the glutamate ligand with S1S2 protein is estimated to be ca. −9 kcal/mol, which compares well with the experimentally measured value of −8.62 kcal/mol (35). This result further validates the model of the S1S2 protein previously validated by predicting the ligand vibrational frequency shifts upon binding with the protein (5, 6) and by comparing this observation with the experimental FTIR spectra (19).
The fact that the closed to semi-open conformational transition has occurred in an unmodified protein in equilibrium MD simulations is indicative that the free energy profile of a slightly modified protein with E705−0.75 differs only slightly from that of a wild type protein. The comparison of ΔGclosedTI and ΔGopenTI (Table 1) reveal that the cost of removing 0.25e from the protein in the open and closed conformation is similar.
The PMF plot in Figure 4 exhibits three energy wells. The protein conformation in the 5 Å well corresponds to the closed state of S1S2 in the absence of the ligand, which is somewhat more closed than the conformation found in the crystal structure with the glutamate ligand bound. The degree of cleft closure is ligand dependent. It has been noted that the ligands, which bind more tightly, e.g. AMPA and glutamate, promote higher degree of the cleft closure (11). The energy well at the 7–9 Å distance corresponds to a semi-open conformation of the protein, which has also been observed in the equilibrium MD simulations of the S652G mutant protein (9). This semi-open conformation is characterized by a disrupted hydrogen bond between S652 and G451 while all other hydrogen bonds remain the same as in the closed structure of the wild type protein. The open cleft conformation of E705−0.75 corresponds to the wide energy well seen in Figure 4, spanning from ca. 11.5 to 14 Å. The average structure within this energy well obtained in the US simulations is similar to the apo-open crystal structure (9). In unconstrained MD simulations of the wild-type protein in the presence of the glutamate ligand, a spontaneous transition between the fully closed and the semi-open conformations has been observed (9). This observation is corroborated by the present study. In Figure 4 the energies of the fully closed and the semi-open conformations are approximately the same and separated by only a small barrier of about 1 kcal/mol. Such a barrier can be easily overcome by a protein in solution at room temperature and thus, it seems reasonable to suggest that both conformations exist at equilibrium in similar proportion. Indeed in a recent NMR experiment (21) disruption of this bond has been observed. Our findings are also in agreement with crystallographic evidence revealing that in the presence of glutamate ligand, this bond may or may not be observed (due to the flip-flop transition of the backbone), as well as the NMR evidence revealing that the flip-flop transition of the backbone, which may disrupt this H-bond, occurs on the millisecond time-scale (21, 36).
In an effort to better understand the effect of removing charge from a single residue and the subsequent response of the protein structure, we turn to continuum electrostatics. A continuum electrostatics approach is used here for assessing the effect of a large disruption of the system such as a charge substitution. We first examine the E402-T686 hydrogen bond that has been previously identified for its role in stabilizing the closed conformation of the S1S2 domain (11, 20, 21, 23, 36). Experimental T686 mutations decrease agonist affinity and increase recovery from desensitization which may indicate a destabilized closed conformation of the ligand binding domain (20). Theoretical calculations of the free energy barrier between an intermediate state and the closed state of S1S2 was observed to be reduced for a mutant T686A/S (8). By evaluating effects of charge substitution or charge removal of this known interaction, namely the electrostatic cost of removing a hydrogen bond between E402 and T686 in the closed conformation, we can asses the utility of a continuum electrostatics approach for such interactions. The results from relative electrostatic interaction free energies of the closed conformation of S1S2 presented in Figure 2 are consistent with the idea that charge substitution for both T686 and E402 produce an unfavorable D1–D2 interaction of equal magnitude suggesting a direct electrostatic interaction between these two residues contributing to the stabilization of the S1S2 domain closed conformation. In Addition to increase in electrostatic repulsion between the D1 and D2 lobes in the closed cleft conformation removal of charge from E402 could lead to more flexibility due to loss of protein-protein interactions between lobes. E402 has been described as one such “anchor” of the D2 lobe flexibility (9).
Since we are interested in the role of E705, we can use the same ideas to sort out its role in stabilizing the S1S2 domain. Direct electrostatic interactions between residues of D1 and D2 as evidenced by continuum electrostatics calculations of E7050, K7300, Y7320, and T4800 (Figure 2) suggest that these residues form an electrostatic network (Figure 1C). A key salt bridge between E705 and K730 has been identified in the crystal structure of S1S2 (11, 37). Recently this interaction has been suggested to be involved in part of the mechanism for the S1S2 domain conformational transition (9). We suggest and have further emphasized in Figure 2 that K730 acts to effectively short circuit E705 from Y732 and T480 and does not contribute any significant direct D1 or D2 lobe interactions. In light of an overall positive charge on both D1 and D2, a newly formed E705-K730 salt bridge would result in increased electrostatic repulsion of lobes. Taken together these results reveal a network consisting of E705, T480, Y732, and K730. E705, T480, and Y732 control the stability of closed S1S2. The role of K730 is to interact favorably with E705 thus allowing the transition from the closed to open conformation state of the S1S2 domain.
Electrostatic interaction energies between the D1 and D2 lobes for both the E705 and E7050 proteins over the entire reorganizational pathway further bolsters the idea that E705 plays major role in the stability of the closed S1S2 conformation as plotted in Fig 5. The upward shift of the electrostatic interaction energy curve (Figure 5, dotted curve) represents a destabilized S1S2 state. Compared to the fully closed S1S2 conformation, E705 is required for the fully closed state to be optimally stable.
This stability can also be captured through the solvation energy of S1S2 over the same reorganizational pathway which allows a quantitative measure of the stability of each conformational state facilitated by the solvent. In the E7050 substituted proteins, the open conformation of the S1S2 domain is stabilized by −6.64 kcal/mol as shown in Figure 6B. More interestingly, the stability of the open conformation compared to the closed conformation is significantly increased by the E7050 charge substitution. Shown more clearly in Figure 6B, the relative difference in solvation energy between closed and open conformations for the E7050 protein is −10 kcal/mol, whereas the solvation energy between closed and open conformations for the wild-type protein are a modest −3 kcal/mol. The overall result is suggestive that relative stability has shifted towards the open conformation, implying once more that E705 is vital in stabilizing the closed conformation of S1S2. From this analysis it is clear that the presence of a negative charge exposed in the binding cleft indeed serves to regulate stabilization of the closed conformation, is absolutely essential for the cleft to close, and participates in ligand orientation despite its minor destabilization of the ligand/protein interaction (6).
In Summary, the quantitative agreement of free energies computed in our studies with experimentally measured values significantly increases confidence in the quality of our models. Namely, computing the free energy of the glutamate ligand binding with the protein at −9 kcal/mol, compares well with the experimental value of −8.62 kcal/mol (35).
One important result presented in this study is understanding the role of the E705 residue, known to play a key role in ligand binding (11, 14, 19, 23, 24). Neutralizing E705 in the closed conformation of the protein produced a small (1 kcal/mol) destabilizing effect on protein-ligand affinity – contrary to the experimentally known facts (6). In the present study we have demonstrated that when the charge on E705 is reduced or neutralized the open conformation of the protein is stabilized more than the closed conformation due to two factors: 1) a more favorable solvation energy; 2) the inter-lobe electrostatic repulsion in the closed conformation increases more than in the open conformation, both effects contributing to stabilization of the open conformation with respect to the closed conformation. These observations led us to conclude that the main role of the E705 residue in the binding cleft is to lower the inter-lobe repulsion upon cleft closure through electrostatic shielding. E705 direct interaction with the ligand via formation of a hydrogen bond does not significantly affect energy of the ligand – protein interaction, which its presence actually destabilizes. Therefore, both, the negatively charged glutamate ligand and the negatively charged glutamate 705 provide electrostatic screening of the lobes D1 and D2 which otherwise strongly repel each other. In the absence of E705 and the ligand, the open conformation is favored.
The final conclusion of this study is that the S652 - G451 hydrogen bond, located at the tip of the binding cleft thought to participate in stabilization of the closed conformation, does not contribute to the energetics of the cleft closure. This conclusion is based on comparing free energies of the fully closed and the semi-open conformations which differ mainly by the presence or absence of this bond. The two conformations have equal free energies separated by a small barrier of less than 1 kcal/mol, which in practice would result in equal population of both conformations and a rapid interconversion between conformations in solution at room temperature. Indeed, in unconstrained MD simulations in the presence of the ligand the hydrogen bond between S652 and G451 can break and reform on the nanosecond time-scale (9). Our findings are in accord with crystallographic evidence which reveals in the presence of glutamate this bond may or may not be observed (due to the flip-flop transition of the backbone), as well as the NMR evidence of the backbone flip-flop transition, which may disrupt this H-bond, occurs on the millisecond time-scale (21).
This study concludes a small series of modeling projects aimed at developing clearer mechanistic understanding of the GluR2 ligand binding domain interaction with the glutamate ligand and conformational reorganization upon ligand binding. A naïve picture of this process is of a clam-shell shaped protein, which upon ligand binding closes the cleft formed by two lobes connected by a hinge. A picture emergent as a result of our modeling studies and analyzing experimental results is significantly more multifaceted and includes interplay between electrostatic interactions and rigidity/flexibility properties of this protein. Upon analyzing the structure using rigidity theory, one lobe was found to be rigid, while the other one deformable (9), which is inconsistent with the hinge closing mechanism. The flexible lobe is slightly deformed in the closed conformation by comparison with the open conformation and is held together by a number of hydrogen bonds formed with the ligand and across the lobe interface. At the same time, a detailed analysis of the electrostatic interactions presented in this paper and elsewhere (6), revealed that i) the ligand- protein electrostatic complementarity is the main factor in the ligand binding with the protein, and the negatively charged glutamate E705 exposed to the otherwise positively charge binding cleft plays little role in direct interaction with the ligand, ii) in the absence of the ligand the open conformation of the protein is favorable mainly due to the inter-lobe electrostatic repulsion and its more favorable water solvation, iii) two glutamate residues that form electrostatic and hydrogen bond interactions directly with the residues on the opposite lobe play important role in stabilizing the closed conformation of the cleft: E402 at the moderate level, and E705, are crucially important because of their location deeper in the cleft. This shielding effect can be transiently weakened once E705 forms a salt-bridge with K730 located in the bottom of the cleft. The third cross-cleft hydrogen bond formed between S652 and G451 plays little role in stabilization of the closed cleft conformation in the presence or the absence of glutamate ligand.
The relative importance of individual residues in the binding cleft may be somewhat ligand-dependent. In the future it would be interesting to analyze the described interactions in the presence of other ligands.
Acknowledgments
Computational support of this study came in part from the Pittsburgh Supercomputing Center (PSC) obtained through the National Science Foundation PACI grant. This research was supported by grants from the National Institutes of Health (R01GMD67962 to MGK), and the American Heart Association (0715286U to MJY).
Abbreviations
- GluR2
ionotropic glutamate receptor type 2
- MD
molecular dynamics
- MD-PBSA
molecular dynamics-Poisson-Boltzmann/surface area
- TI
thermodynamic integration
- FTIR
Fourier transform infrared spectroscopy
- WT
wild-type protein
- PDB
protein data bank
- RMSD
root-mean-squared deviation
- RMSF
root mean square fluctuation
- US
umbrella sampling
- PMF
potential of mean force
- LBD
ligand binding domain.
REFERENCES
- 1.Mayer ML. Glutamate receptors at atomic resolution. Nature. 2006;440:456–462. doi: 10.1038/nature04709. [DOI] [PubMed] [Google Scholar]
- 2.Oswald RE, Ahmed A, Fenwick MK, Loh AP. Structure of glutamate receptors. Curr. Drug Targets. 2007;8:573–582. doi: 10.2174/138945007780618526. [DOI] [PubMed] [Google Scholar]
- 3.Mendieta J, Ramirez G, Gago F. Molecular dynamics simulations of the conformational changes of the glutamate receptor ligand-binding core in the presence of glutamate and kainate. Proteins: Struct., Func., Bioinf. 2001;44:460–469. doi: 10.1002/prot.1111. [DOI] [PubMed] [Google Scholar]
- 4.Arinaminpathy Y, Sansom MSP, Biggin PC. Molecular dynamics simulations of the ligand-binding domain of the ionotropic glutamate receptor GluR2. Biophys. J. 2002;82:676–683. doi: 10.1016/S0006-3495(02)75430-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Speranskiy K, Kurnikova M. Accurate theoretical prediction of vibrational frequencies in an inhomogeneous dynamic environment: A case study of a glutamate molecule in water solution and in a protein-bound form. J. Chem. Phys. 2004;121:1516–1524. doi: 10.1063/1.1752887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Speranskiy K, Kurnikova M. On the binding determinants of the glutamate agonist with the glutamate receptor ligand binding domain. Biochemistry. 2005;44:11508–11517. doi: 10.1021/bi050547w. [DOI] [PubMed] [Google Scholar]
- 7.Arinaminpathy Y, Sansom MSP, Biggin PC. Binding site flexibility: Molecular simulation of partial and full agonists within a glutamate receptor. Mol Pharmacol. 2006;69:5–12. doi: 10.1124/mol.105.016691. [DOI] [PubMed] [Google Scholar]
- 8.Lau AY, Roux B. The free energy landscapes governing conformational changes in a glutamate receptor ligand-binding domain. Structure. 2007;15:1203–1214. doi: 10.1016/j.str.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mamonova T, Speranskiy K, Kurnikova M. Interplay between structural rigidity and electrostatic interactions in the ligand binding domain of GluR2. Proteins: Struct., Func., Bioinf. 2008 doi: 10.1002/prot.22090. in press. [DOI] [PubMed] [Google Scholar]
- 10.Meir A, Ginsburg S, Butkevich A, Kachalsky SG, Kaiserman I, Ahdut R, Demirgoren S, Rahamimoff R. Ion channels in presynaptic nerve terminals and control of transmitter release. Physiol. Rev. 1999;79:1019–1088. doi: 10.1152/physrev.1999.79.3.1019. [DOI] [PubMed] [Google Scholar]
- 11.Armstrong N, Gouaux E. Mechanisms for activation and antagonism of an AMPA-Sensitive glutamate receptor: Crystal structures of the GluR2 ligand binding core. Neuron. 2000;28:165–181. doi: 10.1016/s0896-6273(00)00094-5. [DOI] [PubMed] [Google Scholar]
- 12.Armstrong N, Sun Y, Chen GQ, Gouaux E. Structure of a glutamate-receptor ligand-binding core in complex with kainate. Nature. 1998;395:913–917. doi: 10.1038/27692. [DOI] [PubMed] [Google Scholar]
- 13.Jin R, Gouaux E. Probing the function, conformational plasticity, and dimer-dimer contacts of the GluR2 ligand-binding core: studies of 5-substituted willardiines and GluR2 S1S2 in the crystal. Biochemistry. 2003;42:5201–5213. doi: 10.1021/bi020632t. [DOI] [PubMed] [Google Scholar]
- 14.Mayer ML. Crystal structures of the GluR5 and GluR6 ligand binding cores: molecular mechanisms underlying kainate receptor selectivity. Neuron. 2005;45:539–552. doi: 10.1016/j.neuron.2005.01.031. [DOI] [PubMed] [Google Scholar]
- 15.Du M, Reid SA, Jayaraman V. Conformational changes in the ligand-binding domain of a functional ionotropic glutamate receptor. J. Biol. Chem. 2005;280:8633–8636. doi: 10.1074/jbc.C400590200. [DOI] [PubMed] [Google Scholar]
- 16.Li G, Sheng Z, Huang Z, Niu L. Kinetic mechanism of channel opening of the GluRDflip AMPA receptor. Biochemistry. 2005;44:5835–5841. doi: 10.1021/bi047413n. [DOI] [PubMed] [Google Scholar]
- 17.Ramanoudjame G, Du M, Mankiewicz KA, Jayaraman V. Allosteric mechanism in AMPA receptors: a FRET-based investigation of conformational changes. Proc. Natl. Acad. Sci. U. S. A. 2006;103:10473–10478. doi: 10.1073/pnas.0603225103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hogner A, Kastrup JS, Jin R, Liljefors T, Mayer ML, Egebjerg J, Larsen IK, Gouaux E. Structural basis for AMPA receptor activation and ligand selectivity: crystal structures of five agonist complexes with the GluR2 ligand-binding core. J. Mol. Biol. 2002;322:93–109. doi: 10.1016/s0022-2836(02)00650-2. [DOI] [PubMed] [Google Scholar]
- 19.Jayaraman V, Keesey R, Madden DR. Ligand--protein interactions in the glutamate receptor. Biochemistry. 2000;39:8693–8697. doi: 10.1021/bi000892f. [DOI] [PubMed] [Google Scholar]
- 20.Robert A, Armstrong N, Gouaux JE, Howe JR. AMPA receptor binding cleft mutations that alter affinity, efficacy, and recovery from desensitization. J. Neurosci. 2005;25:3752–3762. doi: 10.1523/JNEUROSCI.0188-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahmed AH, Loh AP, Jane DE, Oswald RE. Dynamics of the S1S2 glutamate binding domain of GluR2 measured using F-19 NMR spectroscopy. J. Biol. Chem. 2007;282:12773–12784. doi: 10.1074/jbc.M610077200. [DOI] [PubMed] [Google Scholar]
- 22.Mankiewicz KA, Rambhadran A, Du M, Ramanoudjame G, Jayaraman V. Role of the chemical interactions of the agonist in controlling alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor activation. Biochemistry. 2007;46:1343–1349. doi: 10.1021/bi062270l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abele R, Keinanen K, Madden DR. Agonist-induced isomerization in a glutamate receptor ligand-binding domain - A kinetic and mutagenetic analysis. J. Biol. Chem. 2000;275:21355–21363. doi: 10.1074/jbc.M909883199. [DOI] [PubMed] [Google Scholar]
- 24.Jin R, Horning M, Mayer ML, Gouaux E. Mechanism of activation and selectivity in a ligand-gated ion channel: structural and functional studies of GluR2 and quisqualate. Biochemistry. 2002;41:15635–15643. doi: 10.1021/bi020583k. [DOI] [PubMed] [Google Scholar]
- 25.Leach AR. Molecular modelling: principles and applications. Harlow, England: Pearson Education Limited; 2001. [Google Scholar]
- 26.Torrie GM, Valleau JP. Nonphysical sampling distributions in Monte Carlo free-energy estimation: umbrella sampling. Journal of Computational Physiology. 1977;23:187–199. [Google Scholar]
- 27.Mamonova T, Kurnikova M. Structure and energetics of channel-forming protein-polysaccharide complexes inferred via computational statistical thermodynamics. J. Phys. Chem. B. 2006;110:25091–25100. doi: 10.1021/jp065009n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Case DA, Darden TA, Cheatham TE, III, Simmerling CL, Wang J, Duke RE, Luo R, Merz KM, Wang B, Pearlman DA, Crowley M, Brozell S, Tsui V, Gohlke H, Mongan J, Hornak G, Beroza P, Schafmeister C, Caldwell JW, Ross WS, Kollman PA. AMBER 8. University of California San Francisco; 2004. [Google Scholar]
- 29.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. A 2Nd generation force-field for the simulation of proteins, nucleic-acids, and organic-molecules. J. Am. Chem. Soc. 1995;117:5179–5197. [Google Scholar]
- 30.Berendsen HJC, Postma JPM, Vangunsteren WF, Dinola A, Haak JR. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 31.Ryckaert JPCG, Berendsen HJC. Numerical-integration of cartesian equations of motion of a system with constraints - molecular-dynamics of n-alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 32.Grossfield A. An implementation of WHAM: the weighted histogram analysis method. 2004 http://dasher.wustl.edu/alan/wham.
- 33.Kurnikov IV. HARLEM - molecular modeling program. Pittsburgh: 1996. http://harlemprog.org. [Google Scholar]
- 34.Kollman PA, Massova I, Reyes C, Kuhn B, Huo S, Chong L, Lee M, Lee T, Duan Y, Wang W, Donini O, Cieplak P, Srinivasan J, Case DA, Cheatham TE., 3rd Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc. Chem. Res. 2000;33:889–897. doi: 10.1021/ar000033j. [DOI] [PubMed] [Google Scholar]
- 35.Deming D, Cheng Q, Jayaraman V. Is the isolated ligand binding domain a good model of the domain in the native receptor? J. Biol. Chem. 2003;278:17589–17592. doi: 10.1074/jbc.C300105200. [DOI] [PubMed] [Google Scholar]
- 36.Weston MCGC, Mayer ML, Rosenmund C. Interdomain interactions in AMPA and kainate receptors regulate affinity for glutamate. J. Neurosci. 2006;26:7650–7658. doi: 10.1523/JNEUROSCI.1519-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mayer ML, Olson R, Gouaux E. Mechanisms for ligand binding to GluR0 ion channels: crystal structures of the glutamate and serine complexes and a closed apo state. J. Mol. Biol. 2001;311:815–836. doi: 10.1006/jmbi.2001.4884. [DOI] [PubMed] [Google Scholar]