Introduction
At its core, the practice of medicine is an information intensive endeavor. Most of what physicians do involves the collection, review and management of information. Examples of such activities include obtaining and recording patient information, consulting colleagues, reading the scientific literature, planning diagnostic procedures, devising strategies for patient care, interpreting tests, and conducting research. The ever-increasing biomedical knowledge-base that must be considered in order to deliver optimal patient care only adds to the challenges facing medicine today.
Successfully addressing these challenges in order to deliver the best healthcare possible requires not only the existence of valid and generalizable datasets derived from systematic basic, clinical and epidemiologic research efforts, but also the ability to apply the knowledge derived from these research efforts at the point-of-care. It is easy to understand, therefore, why the field of biomedical informatics, a field that is the concerned with collecting, managing and optimally using information in healthcare and biomedicine, is critical to the current and future practice of medicine and the study of healthcare outcomes that result from such practice 1, 2.
Biomedical informatics approaches and related health information technology (Health IT) platforms are key to enabling knowledge-driven healthcare and practice improvement initiatives based on a solid research foundation. Similar biomedical informatics approaches and resources are also critical to advancing outcomes research. Indeed, the emergence of such technologies such as electronic health records (EHRs), clinical data repositories, and research-specific data management systems are already transforming the way we practice medicine and conduct research. This transformation is being further advanced by federally directed funding and research infrastructure development efforts 3, 4.
In the sections that follow, we provide an overview of how Biomedical Informatics and Health IT processes and tools can impact the conduct of research and the delivery of evidence-based healthcare from the authors’ perspective. Given the current state of development in this area, we introduce a conceptual framework by which clinicians, researchers, and the healthcare community at-large can optimally leverage biomedical informatics processes and tools to facilitate outcomes research and drive improvements in healthcare outcomes. We conclude by proposing a course of action intended to support greater collaboration between the clinical, research, and biomedical informatics communities to achieve the goals set forth.
A Framework for Knowledge-driven Outcomes Research
Outcomes research as a cyclical activity
Outcomes research, “seeks to understand the end results of particular health care practices and interventions,” on individual patients and populations 5. This often includes the evaluation of economic impacts linked to health outcomes, such as cost effectiveness, cost utility, and comparative effectiveness. Such research involves a range of data collection and aggregation methods including drawing upon primary studies as well as collecting data de novo for such research 6. The conduct of outcomes research therefore can be thought of as involving an information-cycle that includes collecting data about healthcare practices and patient/population level outcomes, analyzing those data, and the reporting on the findings.
When considering the types of information relevant to the conduct of outcomes research and the practice of evidence-based medicine, two major types are relevant: (1) patient-specific information – or information generated during the care of patients such as numerical values, free text, imaging information etc. as might be found in an EHR; and (2) knowledge-based information – or information derived from the scientific medical literature that is based on biomedical and healthcare research. As the collection, storage, retrieval, and optimal use of these types of information constitute the material focus of biomedical informatics, it becomes clear why the intersection of biomedical informatics and outcomes research is critical to the advancement of medical science and practice.
Indeed, the efficient conduct of outcomes research requires access to robust clinical and population-level datasets as well as existing research and knowledge datasets in order to evaluate relevant metrics such as procedural complications, days of hospitalization, health status, and mortality. Moreover, the results of research can be used to develop system-level interventions designed to facilitate the improvement and/or optimization of healthcare policies and practice, often by leveraging clinical informatics platforms such as EHRs and clinical decision support systems.
Biomedical Informatics Systems and Their Roles in Research
Biomedical informatics can be seen as key a key enabler of the outcomes research cycle. One way to explore Biomedical Informatics’ role is in terms of the resources and platforms developed and used for contemporary clinical practice and research practice. Examples of such systems including EHR platforms, clinical data warehouses, and research information management platforms, all of which are becoming increasingly available in the healthcare environment.
At each stage in the research process, general purpose or research-specific IT systems may be of utility. Payne et al. provided a model for clinical research in general, and as a subset of clinical research, the same can be said of the applicability of this model for outcomes research 3. Examples of general and clinical information systems that are able to support the conduct of clinical research include:
Literature search tools such as PubMed can be used to conduct background research necessary for hypothesis development and study preparation 7–9.
Electronic health records (EHRs) can be utilized to collect clinical data on research participants in a structured form that can reduce redundant data entry and identify patients who are eligible for interventions 10–13.
Data mining tools can be used to identify particular cohorts of potential subjects for studies or conduct retrospective analyses from existing databases 14–16.
Decision support systems can be used to alert providers at the point-of-care that an individual may be eligible for a clinical trial 14, 17, 18.
Computerized physician order entry (CPOE) systems, which collect data describing the therapies delivered to research participants, can be used in both participant tracking and study analyses 11, 19, 20.
In addition to the preceding general-purpose and clinical IT systems, research-specific IT systems have been developed that include:
Simulation and visualization tools can streamline the pre-clinical research process (e.g., disease models) and assist in the analysis of complex data sets 21, 22.
Protocol authoring tools can allow geographically distributed authors to collaborate on complex protocol documents 23–27.
Participant screening tools can assist in the identification and registration of research participants 14, 17, 28.
Research-specific web portals provide researchers with a single point of access to research-specific documents and information for collaboration 29–31.
Electronic data collection or capture tools (EDC) can be used to collect research-specific data in a structured form, and reduce the need for redundant and potentially error-prone paper-based data collection techniques 11, 32–34.
Research-specific decision support systems provide study-specific guidance to researchers, as for tracking the participants’ status and protocol compliance 11, 27.
Numerous reports have concluded that the use of such tools and platforms can lead to increased data and research quality 7, 11, 32, 35. Furthermore, the use of informatics platforms across multi-site studies has been shown to increase the efficiency and efficacy of such team-science endeavors 32, 34, 36–38. The ability to use IT in support of clinical research relies on the ability to collect, store and analyze data. Examples of information systems that are becoming increasingly common include Electronic Data Capture (EDC) systems as well as integrated clinical trials management systems (CTMS) that target multiple aspects of the clinical trials process, including EDC, financial management, data quality assurance and research participant tracking 39, 40. Indeed, the use of such IT for clinical research is growing rapidly, and it is projected that nearly 50% of all studies will soon utilize information management technologies 41.
Informatics Methods for Data Exchange, Integration, and Utilization
A common problem for the outcomes researcher is the aggregation of data from varied and disparate information resources. Resources that bring together clinical data from otherwise inaccessible and non-integrated medical record, laboratory, hospital-based and other healthcare enterprise systems throughout a given region in order to facilitate more efficient and effective care processes are therefore becoming an important part of the research IT solution set.
The growing availability of such integrative datasets via the development of regional health information exchange (HIE) organizations, is one area of particular relevance to this discussion. While HIEs are in various stages of development across the country and some have struggled to establish themselves, there are early examples of successful HIE models that have enabled the use of population level data for public benefit. Examples include the use of HIEs to significantly improve reporting of common diseases over standard means, allow surveillance for disease outbreaks, and facilitate population level epidemiology studies that would otherwise have been difficult if not impossible 42.
Fundamental to the success and utility of such efforts, whether at the regional or even at the individual institutional level where multiple data sources are common, are the informatics underpinnings that enable the aggregation and integration of data from disparate sources into data repositories. One key element of such Informatics methods underpinning the exchange of data is transactional standards. A commonly employed example is Health Level 7 (HL7) version 2, a health data messaging interchange standard employed to transfer information between systems and which resources like HIEs often employ to exchange information between source and destination systems 43. It is worth noting however, that due to the fact that such standards traditionally define the mechanism of exchanging data but do not define the semantic annotations necessary to ensure a shared understanding of the meaning of such data across platforms or organizations, their adoption alone is not sufficient to enable the seamless exchange of complex data sets. Work is ongoing to address this issue in new versions of such standards such as HL7 version 3 44.
Another key element is the methodologies and approaches used to maintain integrative data repositories of data in resources such as data warehouses. A data warehouse (DW) is a type of database or data repository that is designed to have certain characteristics that enable its use for purposes such as research including the following definitional factors 45–47:
Subject-oriented: data elements being collected and managed via the DW correspond to real world entities, and often have a set of hierarchical and/or semantic interrelationships;
Time-variant: as data and data sources change over time, the natural history of such information in the DW is stored and can be retrieved by end-users;
Non-volatile: no data is deleted or expunged from the DW, allowing it to serve as an authoritative, longitudinal repository of targeted data types; and
Integrated: data stored in the DW is consistent in its scope and comprehensiveness, and appropriate linkages between data sets that have hierarchical, semantic, geographic or temporal interdependencies are maintained regardless of the specific data modeling or management approach utilized in the implementation of the warehouse.
There are many potential uses for a DW 48, 49, but it is the ability to perform longitudinal or episodic queries based upon one or more criteria of interest in support of research activities (e.g., study planning, retrospective data analyses) 45, and the delivery of task or role-specific datamarts to support context-specific access to data sets by other applications or direct query and analysis by authorized end-users that make these resources so useful in the biomedical research domain.
Indeed, within the biomedical domain, numerous reports provide contexts in which data warehouses have been utilized, including:
The retrieval of patient cohorts for either clinical trial feasibility analyses or active participant recruitment 50.
The application of data mining and statistical analysis tools to large-scale data extracts in order to identify or test hypotheses concerning relationships between demographic, phenotypic and bio-molecular parameters 51.
The identification of trends or phenomena surrounding events of interest such as infections 52, adverse events or complications associated with clinical interventions 47, and evidence-based guideline compliance 53.
The support of business intelligence applications that enable operations research or optimization, including the provision of real-time performance indicators and dashboards 45, 54.
The execution of complex, integrative reports for oversight, regulatory and financial monitoring purposes 45, 54.
Along with all of these benefits, there are also challenges associated with the use of DWs, including 45, 54: 1) overcoming regulatory and data privacy and confidentiality concerns; 2) ensuring the provenance and quality of data being included in the DW; 3) implementing and supporting sufficiently robust and timely interfaces between production systems and DWspecific extraction, transformation, and load (ETL) processes; and 4) providing timely access to limited or de-identified data sets for retrospective research or research planning purposes. However, despite such potential limitations and challenges, the use of data warehouse platforms in biomedical settings has been and continues to be associated within major increases in productivity and efficiency surrounding many application scenarios.
Another factor motivating the development of resources that enable connectivity and information exchange within and between major research centers is the increased NIH funding in recent years for research informatics infrastructure. Examples include the Cancer Biomedical Informatics Grid (caBIG), Informatics for Integrating Biology and the Bedside (i2b2), and Clinical and Translational Science Award (CTSA) initiatives 40, 55, 56. These efforts have led to the development of additional resources and approaches to advancing information integration and accessibility to advance all kinds of research.
As previously mentioned, patient or population level data is one key type of information needed to advance evidence-based medicine. The other is knowledge-based information such as that contained in the scientific literature. Here, too, informatics resources such as Medline/Pubmed, the Cochrane database, and other data repositories of medical knowledge including guideline repositories have become critical components of the evidence-driven medicine solution. In addition to relying upon manual review of such resources on an ad hoc basis in the course of hypothesis generation or to answer clinical questions, these repositories have the potential to influence care directly through biomedical informatics approaches.
One such example involves the combination of knowledge-based information with patient-level data in order to drive healthcare support via rules-engines, often as part of Electronic Health Records. Such Clinical Decision Support Systems (CDSS) assesses individual rules and determines their applicability in a specific case or situation 57. At the most basic level, such a rules engine is comprised of two components, a knowledge-base (consisting of rules represented in a computational format), and an inference or execution engine which can reason about incoming data based upon the contents of the knowledge-base in order to generate some form of output such as an instruction set or alert. An additional critical component of a rules engine is the knowledge engineering facility, which provides the capability to curate the contents of the knowledge-base on an automated, semi-automated, or manual basis.
Biomedical Informaticians have contributed various models for representing clinical decision support rules throughout the years. Examples such as the Arden syntax which can be used to write what are called Medical Logic Modules (MLM’s), form the basis of clinical alerting systems 58. Work has also been done on frameworks and systems to represent the knowledge of clinical guidelines. Examples such as the Guideline Interchange Format (GLIF), EON, and EON’s successor, SAGE, allow for computation on guidelines for the purposes of point-of-care decision support, and have been utilized in multiple studies 59, 60. Indeed, products of the EON project have been deployed in a production-level advisory system for the management of hypertensive patients in VA hospitals, named ATHENA 61.
Developing an Integrative Model for Informatics-supported Outcomes Research
As the preceding descriptions make evident, the biomedical informatics community has generated a broad variety of techniques, platforms, and theoretical models capable of supporting the myriad of information management and analysis activities essential to the conduct of outcomes research and ultimately the practice of evidence-based medicine. However, such physical and methodological resources are often underutilized, usually due to a combination of human-factors, socio-technical issues and technology-based barriers 3, 4. In analogous research domains such as clinical research and bio-molecular translational science, the development of conceptual frameworks that illustrate the interrelationships of critical components of the socio-technical system has been shown to help in advancing the development of informatics solutions to similar issues 19, 62. Therefore we propose the basis for such a model to advance the integration of biomedical informatics and outcomes research. The proposed model builds upon the AHRQ description of outcomes research, which states that, “Outcomes research seeks to understand the end results of particular health care practices and interventions… For clinicians and patients, outcomes research provides evidence about benefits, risks, and results of treatments so they can make more informed decisions” 5.
Building upon this description, we first acknowledge the relevant methodological approaches and operational or research products, as follows:
Healthcare policies and practices, that serve to influence behaviors and clinical outcomes;
Direct and surrogate measurements of the preceding behaviors and outcomes, at multiple levels of dimensionality and granularity, including patient data generated during routine clinical care, population based data sets such as those associated with public health interventions and studies, and research data generated during targeted clinical studies;
Synthesized results of analytical operations applied to the preceding data sets in order to elucidate the benefits, risks, and results of the policies and practices enumerated in item 1) above; and
Informatics platforms and components, such as clinical decision support systems (CDSS), guideline delivery systems; and evidence dissemination applications (e.g., literature databases, guideline repositories, etc.), all of which are informed by or leverage the synthesized results associated with item 3) above.
Spanning the preceding four methodological and operational/research products are a set of enabling biomedical informatics practice areas, which are concerned with the design, application, and evaluation of tools and techniques concerned with:
Data capture and storage, as exemplified by electronic health records, personal health records, data warehouses, and clinical trial management systems, which collectively allow for the population of data sets based upon the outcomes/impact of healthcare practices and policies;
Data integration and exchange, as exemplified by mechanisms used to aggregate disparate and heterogeneous data sets that are generated via data capture and storage platforms, in order to enable a full spectrum of hypothesis discovery and testing activities; and
Knowledge representation and reasoning, as exemplified by inferencing applications used to deliver appropriate clinical decision support or practice guidelines based upon the best available evidence and patient or context-specific variables.
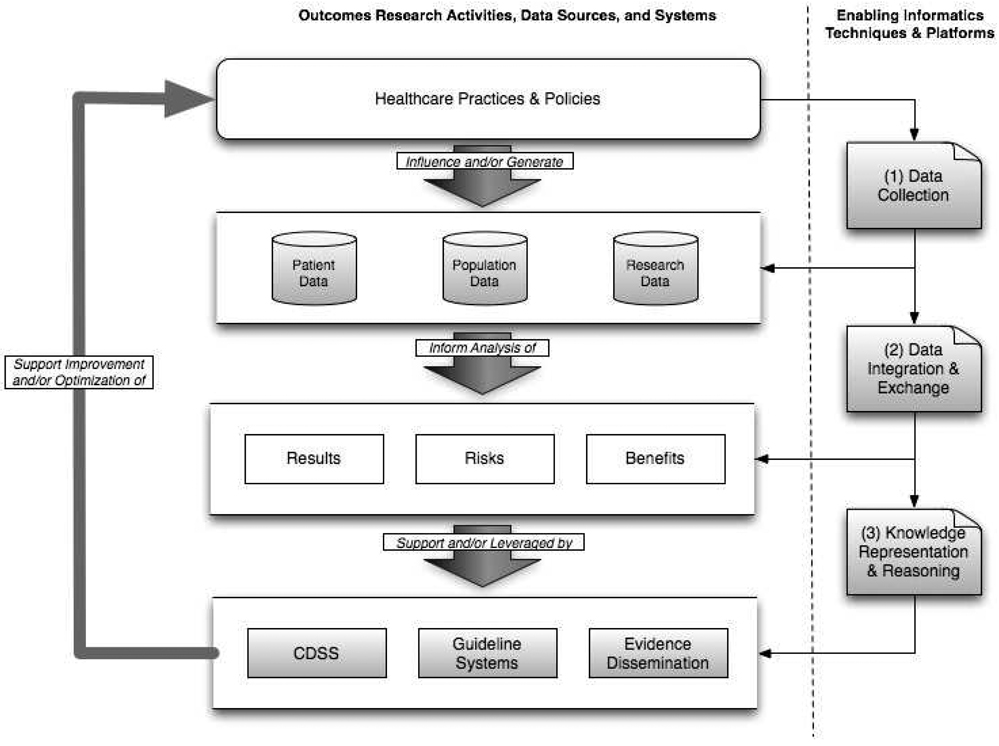
Taken together, the combination of the four methodological and research/operational products introduced earlier, and the corresponding and enabling informatics practice areas described above, create a framework to facilitate and support the improvement or optimization of healthcare practice and policies, based upon the systems-level understanding of outcomes, risks, and benefits that they afford researchers, practitioners, and policy-makers (Figure 1). It is important to note that this model includes an iterative feedback cycle to ensure that experiences with interventions inform the creation and optimization of healthcare practices and policies including those related to the development of clinical and research informatics solutions.
Figure 1.
Overview of conceptual model for informatics-enabled outcomes research, illustrating research activities, data sources, systems, and enabling informatics techniques and platforms.
Proposed Courses of Action
As reviewed in the preceding sections, the conduct of outcomes research and the corresponding provision of evidence-based medicine is both a complex and information-intensive endeavor. Central to such activities is a systems-level approach to information and knowledge generation, collection, analysis, and dissemination. We believe that the development of a community-accepted conceptual model that advances a collective understanding of the complex interplay between the constituent operational, research, and informatics components is both necessary and advantageous to those working in these domains. In the preceding section, we outlined the basis for such a model in the hopes of catalyzing the further discussion and research required to formalize such a construct. However, to realize the benefit of such a model, and the processes by which it will be fully conceived and validated, a number of important factors must be addressed by our clinical and research communities, namely:
Efforts must be undertaken at the local, state, and national level to promote and support the development of formal and informal collaboratories that span the clinical, outcomes research, and biomedical informatics communities. Currently, the formation of such collaborative-research-alliances is limited by insufficient or inadequately distributed resources, policy-based barriers to the conduct of team-science activities (including tenure and promotion criteria in academic settings that often do not reward interdisciplinary science), and the absence of appropriate venues for the training and career development of multi-disciplinary outcomes researchers who span the preceding practice and scholarly domains.
Significant rationalization of regulatory frameworks is needed to enable the secondary use of clinical data in an efficient, timely, and secure manner, while still ensuring patient privacy and confidentiality. The current regulatory environment is rife with often contradictory and uncoordinated policies and regulations concerning the use of patient data, even in a de-identified format, for secondary research purposes. Such a confusing environment makes it difficult if not impossible to conduct large-scale outcomes research programs without expensive and resource intensive prospective consent processes, even when the use of existing data for such research activities in a retrospective manner is exceptionally low risk in terms of patient privacy and confidentiality. Furthermore, these same regulatory frameworks also contribute to the complexity surrounding contemporary, time and personnel intensive Institutional Review Board (IRB) processes at most if not all academic health centers and can significantly delay research conduct 63.
Greater integration between clinical care and clinical research should be fostered. Currently, organizational and regulatory policies and procedures situate the conduct of clinical research such that it is usually a distinct activity from conventional clinical care. However, in many settings, clinical care and research are often interrelated, even in the traditional sense. When one begins to view clinical care and the related collection and use of healthcare data as part of the research cycle described above, one begins to recognize the value of considering the benefits of integrating research and clinical care in the same environment. Indeed, the differentiation and formal decoupling of these activities only serves to complicate workflows, impede efficient information exchange, and reduce patient access to cutting-edge treatment modalities. Ultimately, to realize the benefits of outcomes research, especially in the modern informatics era, we must embrace an approach to clinical medicine in which all patients are given the opportunity to be involved in some aspect of the research cycle. Without such a model, we will be hard pressed to realize the powerful benefits of our informatics techniques and platforms, and our ability to generate the evidence that outcomes research is intended to enable.
Summary
The conduct of outcomes research is an information intensive endeavor and therefore benefits from the application of biomedical informatics approaches, resources, and platforms. It is our contention that a tighter integration of biomedical informatics, clinical care, healthcare policy, and outcomes research can serve to advance improvements in healthcare research and practice. As the preceding overview of the current state of knowledge in these domains illustrates, such integration will require at the most basic level the development of team-science approaches to outcomes research that include clinicians, researchers, and biomedical informaticians. However, the formation and support of such teams will not be possible without addressing some of the barriers and requirements we have summarized in our discussion. By building upon the recent and ongoing advances in biomedical informatics and addressing the issues raised above, there exists a tremendous opportunity to link research, healthcare delivery and policy in such a way as to have direct and demonstrable impact on the health and quality of life of the public.
Acknowledgments
Funding Sources
Dr. Embi’s contributions were supported in part by grants from the NIH/NLM (R01-LM009533) and NIH/NCRR (UL1-RR026314). Dr. Payne’s contributions were supported in part by grants from the NIH/NCI (P01-CA081534, R01-CA134232) and NIH/NCRR (U54-RR024384).
Footnotes
Disclosures
The authors have no disclosures to report in relation to this manuscript.
References
- 1.Shorliffe EH, Cimino JJ. Biomedical Informatics: Computer Applications in Health Care and Biomedicine. 3rd ed. New York, NY: Springer; 2005. [Google Scholar]
- 2.Hersh WR. Medical informatics: improving health care through information. JAMA. 2002;288:1955–1958. doi: 10.1001/jama.288.16.1955. [DOI] [PubMed] [Google Scholar]
- 3.Payne PR, Johnson SB, Starren JB, Tilson HH, Dowdy D. Breaking the translational barriers: the value of integrating biomedical informatics and translational research. J Investig Med. 2005;53:192–200. doi: 10.2310/6650.2005.00402. [DOI] [PubMed] [Google Scholar]
- 4.Embi PJ, Payne PR. Clinical Research Informatics: Challenges, Opportunities and Definition for an Emerging Domain. J Am Med Inform Assoc. 2009 doi: 10.1197/jamia.M3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. [Accessed March 30, 2009];AHRQ outcomes research fact sheet. Available at: http://www.ahrq.gov/clinic/outfact.htm.
- 6. [Accessed March 30, 2009];National Information Center on Health Services Research and Health Care Technology NICHSR) glossary. Available at: http://www.nlm.nih.gov/nichsr/hta101/ta101014.html.
- 7.Briggs B. Clinical trials getting a hand. Health Data Manag. 2002;10:56–60, 62. [PubMed] [Google Scholar]
- 8.Ebbert JO, Dupras DM, Erwin PJ. Searching the medical literature using PubMed: a tutorial. Mayo Clin Proc. 2003;78:87–91. doi: 10.4065/78.1.87. [DOI] [PubMed] [Google Scholar]
- 9.Eysenbach G, Tuische J, Diepgen TL. Evaluation of the usefulness of Internet searches to identify unpublished clinical trials for systematic reviews. Med Inform Internet Med. 2001;26:203–218. doi: 10.1080/14639230110075459. [DOI] [PubMed] [Google Scholar]
- 10.Bates DW, Ebell M, Gotlieb E, Zapp J, Mullins HC. A proposal for electronic medical records in U.S. primary care. J Am Med Inform Assoc. 2003;10:1–10. doi: 10.1197/jamia.M1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marks RG, Conlon M, Ruberg SJ. Paradigm shifts in clinical trials enabled by information technology. Stat Med. 2001;20:2683–2696. doi: 10.1002/sim.736. [DOI] [PubMed] [Google Scholar]
- 12.McDonald CJ, Overhage JM, Tierney WM, Dexter PR, Martin DK, Suico JG, Zafar A, Schadow G, Blevins L, Glazener T, Meeks-Johnson J, Lemmon L, Warvel J, Porterfield B, Cassidy P, Lindbergh D, Belsito A, Tucker M, Williams B, Wodniak C. The Regenstrief Medical Record System: a quarter century experience. Int J Med Inf. 1999;54:225–253. doi: 10.1016/s1386-5056(99)00009-x. [DOI] [PubMed] [Google Scholar]
- 13.Padkin A, Rowan K, Black N. Using high quality clinical databases to complement the results of randomised controlled trials: the case of recombinant human activated protein C. Bmj. 2001;323:923–926. doi: 10.1136/bmj.323.7318.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marks L, Power E. Using technology to address recruitment issues in the clinical trial process. Trends Biotechnol. 2002;20:105–109. doi: 10.1016/s0167-7799(02)01881-4. [DOI] [PubMed] [Google Scholar]
- 15.Gupta R, Tang WH, Young JB. Patterns of beta-blocker utilization in patients with chronic heart failure: experience from a specialized outpatient heart failure clinic. Am Heart J. 2004;147:79–83. doi: 10.1016/j.ahj.2003.07.022. [DOI] [PubMed] [Google Scholar]
- 16.Mullens W, Abrahams Z, Sokos G, Francis GS, Starling RC, Young JB, Taylor DO, Wilson Tang WH. Gender differences in patients admitted with advanced decompensated heart failure. Am J Cardiol. 2008;102:454–458. doi: 10.1016/j.amjcard.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 17.Butte AJ, Weinstein DA, Kohane IS. Enrolling patients into clinical trials faster using RealTime Recuiting. Proc AMIA Symp. 2000:111–115. [PMC free article] [PubMed] [Google Scholar]
- 18.Embi PJ, Jain A, Clark J, Bizjack S, Hornung R, Harris CM. Effect of a clinical trial alert system on physician participation in trial recruitment. Arch Intern Med. 2005;165:2272–2277. doi: 10.1001/archinte.165.19.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sung NS, Crowley WF, Jr, Genel M, Salber P, Sandy L, Sherwood LM, Johnson SB, Catanese V, Tilson H, Getz K, Larson EL, Scheinberg D, Reece EA, Slavkin H, Dobs A, Grebb J, Martinez RA, Korn A, Rimoin D. Central challenges facing the national clinical research enterprise. JAMA. 2003;289:1278–1287. doi: 10.1001/jama.289.10.1278. [DOI] [PubMed] [Google Scholar]
- 20.Teich JM, Merchia PR, Schmiz JL, Kuperman GJ, Spurr CD, Bates DW. Effects of computerized physician order entry on prescribing practices. Arch Intern Med. 2000;160:2741–2747. doi: 10.1001/archinte.160.18.2741. [DOI] [PubMed] [Google Scholar]
- 21.Holford NH, Kimko HC, Monteleone JP, Peck CC. Simulation of clinical trials. Annu Rev Pharmacol Toxicol. 2000;40:209–234. doi: 10.1146/annurev.pharmtox.40.1.209. [DOI] [PubMed] [Google Scholar]
- 22.Kim JH, Kohane IS, Ohno-Machado L. Visualiztion and evaluation of clusters for exploratory analysis of gene expression data. Journal of Biomedical Informatics. 2002;35:25–36. doi: 10.1016/s1532-0464(02)00001-1. [DOI] [PubMed] [Google Scholar]
- 23.Fazi P, Grifoni P, Luzi D, Ricci FL, Vignetti M. Is workflow technology suitable to represent and manage clinical trials? Stud Health Technol Inform. 2000;77:302–306. [PubMed] [Google Scholar]
- 24.Fazi P, Luzi D, Manco M, Ricci FL, Toffoli G, Vignetti M. WITH: a system to write clinical trials using XML and RDBMS. Proc AMIA Symp. 2002;240:240–244. [PMC free article] [PubMed] [Google Scholar]
- 25.Goodman KW. Using the Web as a research tool. MD Comput. 2000;17:13–14. [PubMed] [Google Scholar]
- 26.Rubin DL, Gennari J, Musen MA. Knowledge representation and tool support for critiquing clinical trial protocols. Proc AMIA Symp. 2000:724–728. [PMC free article] [PubMed] [Google Scholar]
- 27.Tai BC, Seldrup J. A review of software for data management, design and analysis of clinical trials. Ann Acad Med Singapore. 2000;29:576–581. [PubMed] [Google Scholar]
- 28.Lutz S, Henkind SJ. Recruiting for clinical trials on the Web. Healthplan. 2000;41:36–43. [PubMed] [Google Scholar]
- 29.Greaves AW, Payne PRO, Rassenti L, Kipps TJ. CRC Tissue Core Management System (TCMS): Integration of Basic Science and Clinical Data for Translational Research. Paper presented at: AMIA 2003 Symposium; Washington, DC, USA. 2003. [PMC free article] [PubMed] [Google Scholar]
- 30.Payne PRO, Greaves AW, Kipps TJ. CRC Clinical Trials Management System (CTMS): An Integrated Information Management Solution for Collaborative Clinical Research. Paper presented at: AMIA 2003 Symposium; Washington, DC, USA. 2003. [PMC free article] [PubMed] [Google Scholar]
- 31.Westgren M, Kublickas M. To use Internet in collaborative studies and registers. Acta Obstet Gynecol Scand. 2000;79:329–330. [PubMed] [Google Scholar]
- 32.Kuchenbecker J, Dick HB, Schmitz K, Behrens-Baumann W. Use of internet technologies for data acquisition in large clinical trials. Telemed J E Health. 2001;7:73–76. doi: 10.1089/153056201300093976. [DOI] [PubMed] [Google Scholar]
- 33.Merzweiler A, Knaup P, Weber R, Ehlerding H, Haux R, Wiedemann T. Recording clinical data--from a general set of record items to case report forms (CRF) for clinics. Medinfo. 2001;10:653–657. [PubMed] [Google Scholar]
- 34.Wubbelt P, Fernandez G, Heymer J. Clinical trial management and remote data entry on the Internet based on XML case report forms. Stud Health Technol Inform. 2000;77:333–337. [PubMed] [Google Scholar]
- 35.Crerand WJ, Lamb J, Rulon V, Karal B, Mardekian J. Building data quality into clinical trials. J Ahima. 2002;73:44–46. 48–53, 42; quiz 55–46. [PubMed] [Google Scholar]
- 36.Craver JM, Gold RS. Research collaboratories: their potential for health behavior researchers. Am J Health Behav. 2002;26:504–509. doi: 10.5993/ajhb.26.6.11. [DOI] [PubMed] [Google Scholar]
- 37.Dorman K, Saade GR, Smith H, Moise KJ., Jr Use of the World Wide Web in research: randomization in a multicenter clinical trial of treatment for twin-twin transfusion syndrome. Obstet Gynecol. 2000;96:636–639. doi: 10.1016/s0029-7844(00)00978-9. [DOI] [PubMed] [Google Scholar]
- 38.Kearney N, Miller M, Sermeus W, Hoy D, Vanhaecht K. Multicentre research and the WISECARE experience. Workflow Information Systems for European Nursing Care. J Adv Nurs. 2000;32:999–1007. [PubMed] [Google Scholar]
- 39.The Cancer Biomedical Informatics Grid (caBIG): infrastructure and applications for a worldwide research community. Stud Health Technol Inform. 2007;129:330–334. [PubMed] [Google Scholar]
- 40.Zerhouni EA. Clinical research at a crossroads: the NIH roadmap. J Investig Med. 2006;54:171–173. doi: 10.2310/6650.2006.X0016. [DOI] [PubMed] [Google Scholar]
- 41.Forrester. Global Trial Starts Using Web-enabled EDC Capture. Cambridge, MA: Forrester Research, Inc.; 2006. [Google Scholar]
- 42.Shapiro JS. Evaluating public health uses of health information exchange. J Biomed Inform. 2007;40:S46–S49. doi: 10.1016/j.jbi.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Orlova AO, Dunnagan M, Finitzo T, Higgins M, Watkins T, Tien A, Beales S. Electronic health record - public health (EHR-PH) system prototype for interoperability in 21st century healthcare systems. AMIA Annu Symp Proc. 2005:575–579. [PMC free article] [PubMed] [Google Scholar]
- 44.Blobel BG, Engel K, Pharow P. Semantic interoperability--HL7 Version 3 compared to advanced architecture standards. Methods Inf Med. 2006;45:343–353. [PubMed] [Google Scholar]
- 45.Dewitt JG, Hampton PM. Development of a data warehouse at an academic health system: knowing a place for the first time. Academic medicine : journal of the Association of American Medical Colleges. 2005;80:1019–1025. doi: 10.1097/00001888-200511000-00009. [DOI] [PubMed] [Google Scholar]
- 46.Goeke RJ, Faley RH. Leveraging the flexibility of your data warehouse. Communications of the ACM. 2007;50 [Google Scholar]
- 47.Sahama TR, Croll PR. A data warehouse architecture for clinical data warehousing. Proceedings of the fifth Australasian symposium on ACSW frontiers; 2007. p. 68. [Google Scholar]
- 48.Lawrence D. All hype? Have business intelligence tools actually changed, or have they just been repackaged and renamed? Healthcare informatics : the business magazine for information and communication systems. 2007;24:16, 18, 20. [PubMed] [Google Scholar]
- 49.Coan T. Business intelligence: using insight to improve the value and performance of your practice. The Journal of medical practice management: MPM. 2007;23:34–36. [PubMed] [Google Scholar]
- 50.Kamal J, Borlawsky T, Ding J, Erdal S, Liu JF, Payne PR. ASAP: A Data-driven Advanced Screening Tool for Active Clinical Protocols. Paper presented at: AMIA Spring Congress; Phoenix, AZ. 2008. [Google Scholar]
- 51.Payne PR, Mendonca EA, Johnson SB, Starren JB. Conceptual knowledge acquisition in biomedicine: A methodological review. J Biomed Inform. 2007;40:582–602. doi: 10.1016/j.jbi.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wisniewski MF, Kieszkowski P, Zagorski BM, Trick WE, Sommers M, Weinstein RA. Development of a clinical data warehouse for hospital infection control. Journal of the American Medical Informatics Association : JAMIA. 2003;10:454–462. doi: 10.1197/jamia.M1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aarts J, Ash J, Berg M. Extending the understanding of computerized physician order entry: Implications for professional collaboration, workflow and quality of care. International journal of medical informatics. 2007;76 Suppl 1:4–13. doi: 10.1016/j.ijmedinf.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 54.Glaser J, Stone J. Effective use of business intelligence. Healthcare financial management : journal of the Healthcare Financial Management Association. 2008;62:68–72. [PubMed] [Google Scholar]
- 55.Kakazu KK, Cheung LW, Lynne W. The Cancer Biomedical Informatics Grid (caBIG): pioneering an expansive network of information and tools for collaborative cancer research. Hawaii Med J. 2004;63:273–275. [PubMed] [Google Scholar]
- 56.Murphy SN, Mendis ME, Berkowitz DA, Kohane I, Chueh HC. Integration of clinical and genetic data in the i2b2 architecture. AMIA Annu Symp Proc. 2006:1040. [PMC free article] [PubMed] [Google Scholar]
- 57.Shortliffe EH, Perreault LE, Wiederhold G, Fagan LM, editors. Medical Informatics: Computer Applications in Health Care and Biomedicine. No. 2. New York: Springer-Verlag; 2001. [Google Scholar]
- 58.Hripcsak G. Arden Syntax for Medical Logic Modules. MD Comput. 1991;8:76, 78. [PubMed] [Google Scholar]
- 59.Sonnenberg FA, Hagerty CG. Computer-interpretable clinical practice guidelines. Where are we and where are we going? Yearb Med Inform. 2006:145–158. [PubMed] [Google Scholar]
- 60.Wang D, Peleg M, Tu SW, Boxwala AA, Greenes RA, Patel VL, Shortliffe EH. Representation primitives, process models and patient data in computer-interpretable clinical practice guidelines: a literature review of guideline representation models. Int J Med Inform. 2002;68:59–70. doi: 10.1016/s1386-5056(02)00065-5. [DOI] [PubMed] [Google Scholar]
- 61.Tu S, Musen MA, Shankar R. Modeling guidelines for integration into clinical workflow. Paper presented at: Medinfo; San Francisco. 2004. [PubMed] [Google Scholar]
- 62.Butte AJ. Medicine. The ultimate model organism. Science. 2008;320:325–327. doi: 10.1126/science.1158343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ness RB. Influence of the HIPAA Privacy Rule on health research. JAMA. 2007;298:2164–2170. doi: 10.1001/jama.298.18.2164. [DOI] [PubMed] [Google Scholar]