Abstract
Propranolol, a beta adrenergic blocker prevents the blood alcohol (BAL) cycle in rats fed ethanol intragastrically at a constant rate by preventing the cyclic changes in the metabolic rate caused by fluctuating levels of norepinephrine released into the blood. The change in the rate of metabolism changes the rate of alcohol elimination in the blood which causes the BAL to cycle. Microarray analysis of the livers from the rats fed ethanol and propranolol showed similar changes in clusters of functionally related gene expressions. The controls and the trough of the cycle differed dramatically from the cluster pattern seen in the rats at the peaks of the blood alcohol cycle. The changes in gene expression induced by ethanol were similar when propranolol was fed without ethanol especially with the changes in the kinases and phosphatases, Toll-like receptor signaling and cytokine-cytokine receptor interaction were also changed. The changes in gene expression caused by ethanol and propranolol feeding are alike probably because both drugs induce β adrenergic receptor desensitization.
Keywords: Epinephrine, Catecholamine, Adrenergic receptors, Alcoholic liver disease
INTRODUCTION
When rats are fed ethanol intragastrically at a constant rate 24 h/day their blood and urinary alcohol levels equilibrate and cycle up and down over 8-10 days (1). The integrity of the hypothalamic stalk is required for the cycle to occur. When the stalk is cut the cycle does not develop (2). This result indicated that the hypothalamic-pituitary axis provides the mechanism for the cycle. It was shown that propylthiouracil prevented the cycle (2) indicating that thyroid hormone was required for the cycle to develop. Propylthiouracil also caused the reduction in the ethanol elimination rate. Thyroxin given to rats, increased the ethanol elimination rate and eliminated the cycle. It caused central necrosis of the liver as a consequence of the increased metabolic rate as reflected by the cycling of body temperature. The necrosis of the liver was due to an increase in O2 demand by hepatocytes when the metabolic rate increased (3). Use of adrenergic β and α blockers (propranolol and phenoxybenzamine) prevented the cycle by blocking the β and α receptors, indicating that catecholamines were involved in the mechanism of the cycle (4). Catecholamines were markedly elevated in the blood of the rats at the peaks of the cycle (4). When caffeine and ephedrine were fed with ethanol, the ethanol elimination rate was increased, the cycle was prevented and central necrosis and fibrosis developed similar to when thyroid was fed with ethanol (5). Rotenone feeding prevented the cycle (6) indicating that NADH donation of electrons to the mitochondrial electron transport chain was involved in the cycle. NADH increases and NAD+ decreases in the liver at the peak of the cycle (7). Reduction of NADH levels by the mitochondria is required for the cycle to occur (6). The essential nature of electron transport and ATP synthesis by the mitochondria as a cause of the cycle was established by uncoupling of oxidative phosphorylation by feeding dinitrophenol (DNP) with ethanol intragastrically. DNP feeding blocked the cycle and prevented alcohol induced elevation of serum ALT, probably by preventing the hypoxia of the liver which develops at the peaks (8,9).
The pathology of the liver in experimental alcoholic liver disease in the rat is worsened by the cycle (7). The reason for this is not clear. To investigate the effect of inhibiting the cycle by feeding propranolol we subjected the fast frozen liver tissue from the study previously published (4) to microarray analysis in order to determine the effect on gene expression in response to propranolol and alcohol.
MATERIAL AND METHODS
Animals
The animal feeding experiments were performed using the intragastric tube feeding model as previously described (2). In this model, ethanol is fed at a constant rate 24 h/day (13 g/kg body weight/day) through a permanent intragastric tube for one month. Controls were pair-fed the diet supplemented with dextrose isocaloric to the ethanol fed animals. The research protocol was approved by the Research and Education Institute Animal Care Committee in accordance with the guidelines for animal care as described by the National Academy of Sciences (1996).
Experiment 1
8 Wistar male rats, weighing about 275 grams (Harley-Sprague Dawley, Hollister, CA) were fed ethanol continuously at 13 g/kg/day for 4 weeks. Four pair-fed control rats were fed the diet with dextrose solution that was isocaloric to ethanol. The diet was fed to provide 271 kcal/kg in both groups. The diet used was modified to include a salt and vitamin mix (Dyets Bethlehem, PA) as described by the American Institute of Nutrition Manual Mix and Vitamin Mix for Optimum Growth of Rats (10). The source of protein was lactalbumin. The diet was supplemented with 500 mg of choline and 1 g of methionine per liter of diet. The dietary calories derived were 33.3% from fat, 25.9% from protein, 6.7% from dextrose, and 34% from ethanol.
Urinary alcohol levels (UALs) (24 h) were measured daily to determine the peaks and troughs of the UAL cycle. Urine was collected under toluene to prevent evaporation. UALs were measured using the QED Saliva Alcohol Test Kit A150 (STC Technologies, Bethlehem, PA). Blood used for blood alcohol levels (BAL) was collected initially by tail vein and terminally from the heart. The BALs were measured enzymatically on a clinical analyzer. The collection of blood was timed to coincide with the peak and trough of the UALs.
Microarray Analysis
RNA extraction of quick frozen liver tissue stored at −80°C was subjected to microarray analysis. Microarray analysis on livers of chronic ethanol fed rats from previously reported studies were compared with the microarray analysis on livers of rats fed ethanol acutely.
RNA extraction
Approximately 50 to 100 μg of frozen liver samples and 1000 μl of Trizol® (Invitrogen™, Carlsbad, CA) were added to a Bio 101 bead tube containing Lysing Matrix D (MP Biomedicals, LLC, Irvine, CA). Each tissue was homogenized by processing the tubes in the FastPrep centrifuge (MP Biomedicals LLC, Irvine, CA) for 30s and incubating them on ice for 1 min. Total RNA was extracted following the Trizol® protocol using chloroform, heavy phase lock gel tubes (Eppendorf, Westbury, NY), and isopropanol. RNA quality was confirmed using Agilent’s 2100 BioAnalyzer (Santa Clara, CA).
About 5 μg of total RNA was used for preparing biotin-labeled cRNA. Fifteen micrograms of labeled and fragmented cRNA were subsequently hybridized to Rat Genome 230 2.0 Array (Affymetrix, Santa Clara, CA). RNA isolation, labeling, and data analysis were performed at the Microarray Core at Los Angeles Biomedical Research Institute. Hybridization, washing, staining, and scanning of the chips were performed at the Microarray Core at Cedars-Sinai Medical Center.
cDNA Microarray
Sample preparation and loading: Equal amount of RNA (5 μg) from each sample were used for Affymetrix GeneChip analysis. RNA was converted to cDNA using GeneChip® One-Cycle cDNA
Synthesis Kit (Affymetrix) and then converted to biotinylated cRNA using GeneChip® IVT Labeling Kit. The quality of labeled RNA was confirmed with the Affymetrix Test 3 Array before placement on chip.
Hybridization and Staining
The hybridization cocktail was prepared, which includes controls at the fragmented cRNA. The samples were hybridized in the array at 45°C for 17 hours using GeneChip Hybridization Oven 640. Immediately following hybridization, the array underwent an automated washing and staining protocol (R-Phycoerythin Streptavidin conjugated, Molecular Probes, Eugene, OR) on the GeneChip Fluidics Station 400. The arrays were then scanned with a GeneChip Scanner 3000 (Affymetrix). The sample quality was checked first, by running the cRNA on the Bioanalyzer and then was checked a second time by using the Test 3 (Affymetrix) array prior to hybridization of the samples on to the Rat Genome Array.
Microarray Data Analysis
Data preparation, analysis, and integration were performed using Affymetrix’s GeneChip Operating Software (GCOS). The software was used to perform image processing, evaluation of data quality, normalization, transformation, and filtering, so that data were ready for further analysis. Wilcoxon’s signed rank test was used in comparison analysis to derive biologically significant results from the raw probe intensities on expression arrays. For comparison analysis, each probe set on the experiment array was compared with its counterpart on the control array to calculate the change in p-value that was used to generate the difference all of increase (I; p<0.04), marginal increase (MI; p < 0.04 to p < 0.06), decrease (D; p > 0.997), marginal decrease (MD; p > 0.992 to p > 0.997), or no change (NC; p > 0.06 to p < 0.997). Comparison analyses were used to generate a signal log ratio for each probe prior to experimental array to the corresponding probe pair on the control array. This strategy canceled out differences resulting from different probe binding coefficients. Signal log ratio was computed by using a one-step Tukey’s biweight method by taking a mean of the log ratio of probe pair intensities across the 2 arrays.
Once the absolute, pair-wise comparison, and replicate comparison data files were created in GCOS, genes were identified with signal intensity differences using BULLFROG v12.3 TG (Lockhart and Lockhart) and GeneSpring (Silicon Genetics). In the BULLFROG analysis, the pair-wise comparisons were used to find consistent differences among all of the samples compared. The criteria used are the following: a change call of increase/marginal increase or decrease/marginal decrease, a fold change >1.5, and a present call in at least one of the arrays. BULLFROG was also used to determine the amount of variability between replicates. In GeneSpring the probes were first normalized using “Per Gene: Normalize to median.” Next, transcripts were determined to be differentially expressed based on the following criteria: a Change Call of Increase, Marginal Increase, Decrease, or Marginal Decrease with a Change in p value <0.0006 or >0.994, a Signal Log Ratio <)0.06 or >0.6, a Present Call for the probe set in either or both experimental conditions, and a minimum signal intensity of 50 in a probe in either or both of the experimental files.
After generating a list of differentially expressed genes, down stream analysis was performed. The filtered transcripts were clustered in GeneSpring using the heatmap and K-means clustering to find similar patterns of gene expression.
The lists of transcripts were also uploaded into GenMapp (Gene Micro Array Pathway Profiler, Gladstone Institutes, University of California at San Francisco, CA). This software clusters the transcripts based on biological function
RESULTS
Heat Map
The clusters were the same when the controls were compared to the ethanol-fed rats which were sacrificed when their urinary and blood alcohol levels were at the troughs (~100 mg %) (Fig 1).
Figure 1.
Heatmaps showing the changes in gene expression of clusters of genes having related function (n=2-3). Green indicates decreased expression and red indicates increasing gene expression. Note that the heatmap at the peak differs from all other group.
However, when the rats were sacrificed at the peak blood and urinary alcohol levels, the expression of a large number of genes were up regulated (Red clusters). A sizable number of genes were down regulated compared to the controls (green). On the other hand, when propranolol was fed with ethanol or propranolol alone the clusters of functionally related up regulated genes were largely the same as those of the controls and troughs. The down regulated gene clusters remained down regulated compared to the controls and troughs. The effect of propranolol on gene expression of the ethanol fed rats could be due largely to propranolol per se, since the heat maps of both were largely the same compared to the controls and troughs. However, the effect of propranolol on the gene expression of ethanol fed rats may partly be due to the fact that the urinary ethanol levels were lower than that seen at the peaks (~200 mg% on propranolol vs. ~ 500 mg% at the peaks).
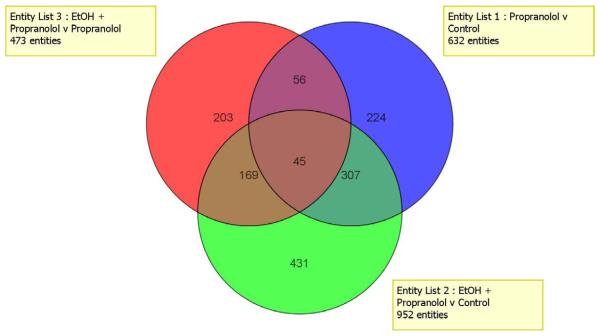
The Venn diagram (Fig 2) shows 952 changes in gene expression when rats fed ethanol and propranolol were compared to controls.
Figure 2.
Venn diagram comparing the changes in gene expression that were changed by different diet treatment. Very few changes in gene expression were the same by all the different treatment (45 genes).
The changes in the expression of 431 genes were unique to ethanol plus propranolol. When rats fed ethanol+propranolol were compared with rats fed propranolol alone, there were changes in the expression of 473 genes, 203 were unique to ethanol plus propranolol vs. propranolol which indicates that ethanol altered the livers response to propranolol feeding. The number of changes in genes expression in response to propranolol feeding alone, was 632 compared with the controls, 224 were unique to propranolol. The overlap of the genes, which were changed by the three diet regimens (577), indicates the complexity of the effects of the dietary ingredient interactions was on the expression of the genes involved.
To further analyze the effects of the 3 dietary regimens on gene expression, KEGG functional pathways were assessed (Fig 3-5).
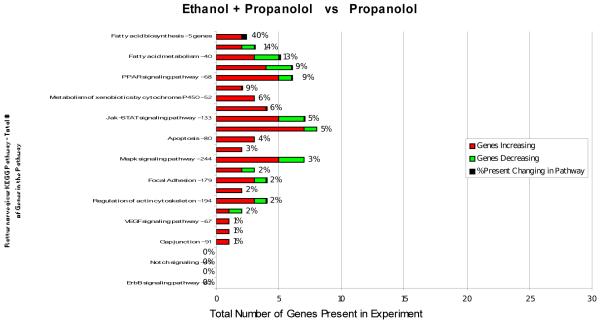
Figure 3.
KEGG functional pathways that were changed by feeding ethanol + propranolol compared with propranolol feeding alone are shown. The affected pathways were fatty acid biosynthesis and fatty acid metabolism.
Figure 5.
KEGG functional pathways that were changed by feeding propranolol compared with controls are shown. The Jak-STAT and Mapk signaling pathways were most affected.
When rats fed ethanol and propranolol were compared with rats fed propranolol only, the expression of genes in most of the functional pathways was affected. Fatty acid synthesis genes were up regulated by ethanol plus propranolol (40%). Genes involved in fat metabolism were mostly up regulated by ethanol + propranolol. Jak-STAT signaling, cytokine-cytokine receptor interaction and MAPK signaling pathways were more up regulated than down regulated.
When rats fed ethanol plus propranolol were compared with controls, gene expression was more up regulated, especially adipocytokine signaling, cytokine-cytokine receptor interaction, Jak-STAT and MAPK signaling, TGFβ signaling, and Toll-like receptor signaling (Fig 4). The effect of propranolol alone, compared with controls, was similar to ethanol plus propranolol compared with controls, although there were more down regulated genes by propranolol feeding (Fig 5). This is interpreted to mean that the up regulation of adipocytokine signaling, Jak-STAT signaling and MAPK signaling seen with ethanol plus propranolol was in part the effect of propranolol feeding rather than ethanol feeding. This would not be the case in explaining the up regulation of cytokine-cytokine receptor interaction and Toll-like receptor signaling seen when ethanol plus propranolol was fed (Fig 4).
Figure 4.
KEGG functional pathways that were changed by feeding ethanol+propranolol compared with control rats are shown. The most pathways affected were the cytokine-cytokine receptor interaction, Jak-STAT signaling pathway and the Mapk signaling pathway.
The changes in the expression of the individual genes are listed in Table I and II where rats fed ethanol and propranolol were compared to controls. Table I lists the kinases and phosphatases genes which were changed. Most of the kinases were up regulated by feeding ethanol and propranolol compared with controls. However, most phosphatases were down regulated. The combined net effect would favor phosphorylation of proteins over dephosphorylation of the proteins that regulate the Erk, p38, MEK and JNK signaling pathways. Table II lists the changes in gene expression in the Toll-like receptor signaling pathway and the cytokine-cytokine receptor interaction. It is clear that the genes which involve cytokine-cytokine receptor interaction are up regulated by ethanol plus propranolol feeding. The genes in the Toll-like receptor signaling pathway were also partially up regulated by ethanol plus propranolol feeding.
Table 1.
Effect of Ethanol plus propranolol vs. propranolol on gene expression of kinases and phosphatases
| Kinases | Phosphatases | ||
|---|---|---|---|
| Gene symbol | Fold change | Gene symbol | Fold change |
| Mapk4 | 7.9±2 | Ppp 2r1b | 2.4±1.1 |
| Jak3 | 4.4±1.7 | Ppap 2c | 1.8±1.2 |
| Prkcc | 2.7±1.4 | Inppl | 1.7±1.0 |
| Dapk1 | 2.6±1.6 | Ppp2r2d | 1.6±1.3 |
| Snf1lk | 2.4±1.2 | Ppp2r1a | −1.6±1.5 |
| Snfl1lk2 | 1.9±1.2 | Ppp1r2 | −1.7±1.2 |
| Cerk | 1.8±1.2 | Ppp1r3c | −1.7±1.3 |
| P4ka | 1.6±1.2 | Ppp3r1 | −1.7±1.4 |
| Gyk | −2.0±1.2 | Pppap2b | −1.9±1.2 |
| Ppp2r5e | −2.4±1.5 | ||
| Ppp1r3b | −2.5±1.8 | ||
Table II.
Effect of ethanol plus propranolol vs. control on gene expression of TOLL-like receptor signaling and cytokine-cytokine receptor interaction.
| Toll-like receptor signaling | Cytokine-cytokine receptor interaction | ||
|---|---|---|---|
| Gene symbol | Fold change | Gene symbol | Fold change |
| Lbp | 4.6±2.2 | CXCl1 | 30.9±3 |
| Trafd | 1.7±1.2 | Tnfrsf21 | 6.3±3.6 |
| Cd14 | 1.6±1.3 | Ilrn | 7.3±1.5 |
| Fas | 4.4±1.4 | ||
| Irf1 | 2.5±1.7 | ||
| Infrsf11b | 1.9±1.2 | ||
| Ifitm3 | 1.9±1.2 | ||
| Il13ra1 | 1.9±1.5 | ||
The expression of several genes that are involved in ethanol metabolism were changed by ethanol plus propranolol feeding compared to controls as follows: Aldh1a1 3.25±1.9 fold change, Aldh 3b1 2.8±1.2 fold change (up regulation) Aldh1b 1-5, 0±2.6 fold change, and Aldh 1a7-6.6±2.4 (down regulation).
DISCUSSION
Tissue sensitivity in the adrenergic receptors changes after ethanol ingestion when the receptors become desensitized. After ethanol withdrawal, the receptors become supersensitive (11-14). This may be the mechanism how propranolol and phenoxybenzamine, the alpha and beta receptor blockers, alter the livers response to ethanol feeding (14). In the present study, propranolol prevented the blood alcohol cycle observed in the liver. Propranolol would, like ethanol, cause a densitization of the β adrenergic receptors in the liver which would block the cycle by interfering with the change in the metabolic rate which is driven by the changes in norepinephrine blood levels which causes the cyclic change in the rate of ethanol elimination (4). As a consequence, propranolol would prevent the peak alcohol levels from occurring. In this way the changes in the expression of genes would be reflected by the effect of a constant unchanging blood alcohol level of ~ 200 mg%. These changes are the result of the inhibition of the adrenergic receptor responses imposed both by ethanol and propranolol in a synergistic manner. The changes included the up regulation of many kinases and the down regulation of many phosphatases as shown here and previously reported when ethanol was fed alone (15,16).
The up regulation of the Toll-like receptor signaling seen in Table II could be due to ethanol induced leakage of LPS from the gut. LPS activates the Toll-like receptors signaling pathway. The changes in cytokine-cytokine receptor interaction may explain the inflammatory changes seen in the livers of the rats fed ethanol plus propranolol (4) (Table II).
Acknowledgement
The authors thank Adriana Flores for typing the manuscript. This study was supported by Grants NIH/NIAAA 8116 and Alcohol Center Grant 011999 morphology core.
Abbreviation
- DNP
Dinitrophenol
- UALs
urinary alcohol levels
- BALs
blood alcohol levels
- BAC
Blood alcohol cycle
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Tsukamoto H, French SW, Reidelberger RD, Largman C. Cyclic pattern of blood alcohol levels during continuous intragastric ethanol infusion in rats. Alcoholism: Clin_Exp Res. 1985;9:31–37. doi: 10.1111/j.1530-0277.1985.tb05046.x. [DOI] [PubMed] [Google Scholar]
- 2.Li J, Nguyen V, French BA, Parlow AF, Su GL, Fu P, Yuan QX, French SW. Mechanism of the cyclic pattern of urinary ethanol levels in rats fed ethanol. The role of the hypothalamic-pituitary-thyroid axis. Am J Physiol. 2000;279:G118–125. doi: 10.1152/ajpgi.2000.279.1.G118. [DOI] [PubMed] [Google Scholar]
- 3.Li J, French BA, Fu P, French SW. Liver necrosis induced by thyroid hormone administration in rats fed ethanol. Exp Molec Pathol. 2001;71:79–88. doi: 10.1006/exmp.2001.2381. [DOI] [PubMed] [Google Scholar]
- 4.Li J, French BA, Fu P, Bardag-Gorce F, French SW. Catecholamines are involved in the mechanism of the urinary alcohol level cycle in rats fed ethanol intragastrically at a constant rate. Life Sci. 2004;75:3043–3051. doi: 10.1016/j.lfs.2004.05.032. [DOI] [PubMed] [Google Scholar]
- 5.Li J, French BA, Fu P, Bardag-Gorce F, French SW. Mechanism of alcohol cyclic pattern: Role of catecholamines. Am J Physiol Gastrointest Liver Physiol. 2003;285:G442–G448. doi: 10.1152/ajpgi.00093.2003. [DOI] [PubMed] [Google Scholar]
- 6.Li J, Fu P, French BA, French SW. The effect of rotenone on the urinary ethanol cycle in rats fed ethanol intragastrically. Exp Mol Path. 2004;77:210–213. doi: 10.1016/j.yexmp.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 7.Bardag-Gorce F, French BA, Li J, Riley NE, Yuan QX, Reitz R, Cai Y, Wan Y-JY, French SW. The importance of cycling of blood alcohol levels in the pathogenesis of experimental alcoholic liver disease in rats fed ethanol intragastrically. Gastroenterology. 2002;123:325–35. doi: 10.1053/gast.2002.34177. [DOI] [PubMed] [Google Scholar]
- 8.Li J, French BA, Nan L, Fu P, French SW. Uncoupling of oxidative phosphorylation prevents the urinary alcohol level cycling caused by feeding ethanol continuously at a constant rate. Exp Mol Pathol. 2005;78:228–232. doi: 10.1016/j.yexmp.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 9.French SW. The pathogenesis and significance of the urinary alcohol cycle in rats fed ethanol intragastrically. Alcoholism Clin Exp Res. 2005;29:158S–161S. [PubMed] [Google Scholar]
- 10.French SW, Morimoto M, Reitz R, Koop D, Klopfenstein B, Estes K, Clot P, Ingelman-Sundberg M, Albano E. Lipid peroxidation, CYP2E1, and fatty acid metabolism in alcoholic liver disease. J Nutr. 1997;127:9075–9115. doi: 10.1093/jn/127.5.907S. [DOI] [PubMed] [Google Scholar]
- 11.French SW, Palmer DS. Adrenergic supersensitivity during ethanol withdrawal in the rat. Res Commun Chem Path Pharmacol. 1973;6:651. [PubMed] [Google Scholar]
- 12.French SW, Reid PE, Palmer DS, Narod ME, Ramey CW. Adrenergic subsentivity of the rat brain during chronic ethanol ingestion. Res Commun Chem Path Pharmacol. 1974;9:575. [PubMed] [Google Scholar]
- 13.French SW, Palmer DS, Narod ME, Reid PE, Ramey CW. Noradrenergic sensitivity of the cerebral cortex after chronic ethanol ingestion and withdrawal. J Pharmacol Exp Ther. 1975;194:319–326. [PubMed] [Google Scholar]
- 14.French SW, Palmer DS, Narod ME. Noradrenergic subsensitivity of rat liver homogenates during ethanol ingestion. Res Commun Chem Path Pharmacol. 1976;13:283. [PubMed] [Google Scholar]
- 15.French BA, Dedes J, Bardag-Gorce F, Li J, Wilson L, Fu P, Nan L, French SW. Microarray analysis of gene expression of the liver during urinary ethanol cycle in rats fed ethanol intragastrically at a constant rate. Exp Mol Pathol. 2005;79:87–94. doi: 10.1016/j.yexmp.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 16.Bardag-Gorce F, French BA, Dedes J, Li J, French SW. Gene expression patterns of the liver in response to alcohol: in vivo and in vitro models compared. Exp Molec Pathol. 2006;80:241–251. doi: 10.1016/j.yexmp.2005.12.006. [DOI] [PubMed] [Google Scholar]