Abstract
Interleukin-15 (IL-15) is a skeletal muscle-derived cytokine with favorable effects on muscle mass and body composition. Modulation of IL-15 levels has been suggested as a treatment for sarcopenia and age-associated increases in adiposity. However, it is unclear whether IL-15 levels change during aging, as measurement of IL-15 at physiological concentrations in mice has been technically difficult, and translational regulation of IL-15 is complex. Moreover, the IL-15 receptor alpha (IL-15Rα) can comprise part of a membrane-associated receptor complex, or appear as a soluble form which stabilizes IL-15 and facilitates IL-15 secretion. Here, we report measurement of physiological levels of murine IL-15, and determine that muscle and serum IL-15 levels decline progressively with age. However, expression of IL-15 mRNA and membrane-associated subunits of the IL-15 receptor did not change with age in muscle. Expression of soluble IL-15Rα (sIL-15Rα) mRNA declined 5-fold with age, and serum IL-15 levels correlated highly with muscle sIL-15 mRNA expression, suggesting declines in sIL-15Rα expression lead to decreased circulating IL-15 levels during aging. These findings complement studies which described several single-nucleotide polymorphisms in the human IL-15Rα gene which impact muscularity and adiposity, and provide a technical basis for further investigation of IL-15 and the sIL-15Rα in determining body composition in aging mice, as a model for humans.
Keywords: Aging, Cytokines, IL-15, Skeletal Muscle, Sarcopenia, IL-15 receptors, Soluble IL-15 alpha receptor, Body composition
1. Introduction
Aging is typically characterized by reduced skeletal muscle mass (sarcopenia) and increased adiposity (Baumgartner, 2000). Age-related changes in body composition result in frailty, insulin resistance, and cardiovascular disease (Baumgartner, 2000; Lebrun et al., 2006; Goodpaster et al., 2006). Interleukin-15 (IL-15) is a cytokine which is highly expressed in skeletal muscle tissue (Grabstein et al., 1994). IL-15 inhibits muscle protein degradation (Carbó et al., 2000; Quinn et al., 2002) and also inhibits fat deposition (Carbó et al., 2001; Alvarez et al., 2002; Nielsen et al., 2008; Quinn et al., 2009). These observations suggest IL-15 supplementation may inhibit age-associated changes in body composition. However, due to technical difficulties in measuring IL-15 protein at physiological levels, it is unclear if IL-15 levels decrease with age in well-characterized mouse models of aging.
Conflicting reports exist concerning changes in rodent muscle IL-15 mRNA and protein expression during aging (Marzetti et al., 2009; Pistilli et al., 2007). Marzetti et al. (2009) found that while muscle IL-15 mRNA expression did not change with age, muscle IL-15 protein content decreased with age in ad lib-fed, but not calorie-restricted, rats, suggesting IL-15 signaling could play a role in preservation of muscle mass during aging. This result underscores previous reports that there is often little correlation between IL-15 mRNA and protein levels (Fehniger and Caligiuri, 2001; Budagian et al., 2006). The lack of correlation of IL-15 mRNA and protein levels is partly due to transcriptional and translational blocks which are present in the IL-15 gene sequence (Fehniger and Caligiuri, 2001; Budagian et al., 2006). Additionally, two mRNA isoforms, which encode identical mature IL-15 proteins, are transcribed from a single IL-15 gene. Short signal peptide- (SSP-) and long signal peptide- (LSP-) IL-15 mRNA isoforms direct expression of intracellular and secreted IL-15 protein species, respectively (Tagaya et al., 1997; Fehniger and Caligiuri, 2001; Budagian et al., 2006). Although Northern blot surveys of IL-15 isoform expression in multiple tissues suggested skeletal muscle preferentially expresses LSP-IL-15 mRNA (Tagaya et al., 1997), this has not been studied in detail using newer, more sensitive technologies, and has not been assessed in different physiological states.
The biochemistry of IL-15 signaling is also complex. IL-15 signaling is transduced through a heterodimeric receptor comprising the beta and gamma subunits of the interleukin-2 receptor (IL-2Rβ and IL-2Rγ), or through a heterotrimeric receptor comprising membrane-associated IL-15 receptor alpha (mIL-15Rα) plus IL-2Rβ and IL-2Rγ (Budagian et al., 2006). RNA splicing also gives rise to a soluble IL-15Rα variant (sIL-15Rα), which facilitates IL-15 secretion, stabilizes IL-15, and increases IL-15 bioactivity (Mortier et al., 2006; Rubinstein et al., 2006; Bulanova et al., 2007; Bergamaschi et al., 2008). Measurement of physiologic levels of murine IL-15 using commercially-available assays has not been reported previously, and no studies of IL-15 and aging or body composition have taken into account the complexities of IL-15 expression and its receptor system (Gangemi et al., 2005; Pistilli et al., 2007; Pistilli and Alway, 2008; Marzetti et al., 2009; Nielsen et al., 2008).
We hypothesized that circulating IL-15 levels decline during normal aging in mice, along with decreased expression of the secreted LSP-IL-15 mRNA isoform in skeletal muscle tissue. To resolve technical problems involved in measurement of the components of the murine IL-15 axis, sensitive bead-based dual-laser technology was utilized to determine serum and muscle levels of IL-15, and branched-DNA (bDNA) RNA signal amplification (Andras et al., 2001; Flagella et al., 2006) was used to assess the expression of the closely homologous LSP- and SSP-IL-15 mRNA isoforms in adult and aging mice. This technology was also used to determine expression of sIL-15Rα and mIL-15Rα mRNA, as well as IL-2Rβ and IL-2Rγ mRNA expression in aging mouse muscle tissue. The results presented here point to age-related changes in sIL-15Rα expression, rather than changes in LSP-IL-15 mRNA expression, as a key regulatory point determining systemic IL-15 levels, with implications for modulation of body composition during aging.
2. Materials and methods
2.1. Aging mice
Animal procedures were carried out following protocols approved by the VA Puget Sound Health Care System (VAPSHCS) Institutional Animal Care and Use Committee (IACUC), and complied with NIH guidelines. Normally-aging C57BL/6 × C3H F1 hybrid (B6C3) male mice were obtained from the NIA Aging Rodent Colony under contract to Charles River Laboratories (Boston, MA), and were maintained at VAPSHCS in specific pathogen-free conditions for 2-4 weeks. At the ages indicated in the text (12, 18, 24 and 28 months), animals were weighed on an electronic scale, and body composition was determined under deep pentobarbitol anesthesia (75 mg/kg, IP) using a Lunar PIXImus dual-energy X-ray absorptiometry (DEXA) scanner (GE Healthcare, Madison, WI). Body weight determined from the scale corresponded extremely well to the total of lean mass, fat mass, and bone mineral content determined by DEXA (not shown); the latter is reported as body weight in Results. Immediately following DEXA scans, blood was collected by cardiac puncture for serum IL-15 determination, followed by a second dose of pentobarbitol (150 mg/kg, IP) for euthanasia and subsequent tissue collection for IL-15 protein and mRNA analyses. To account for increased mortality and tumor development in the older age groups, initial group sizes were larger in the older age groups, as follows: 6 at 12 months; 8 each at 18 and 24 months; and 9 at 28 months of age. Because tumors can secrete cytokines which would confound our analyses, and because cancer-associated cachexia can affect body composition, mice that exhibited macroscopically-visible tumors upon post-mortem dissection (2 at 24 months and 3 at 28 months of age) were removed from the analysis. Final group sizes were 6 animals each at 12, 24, and 28 months, and 8 at 18 months of age.
2.2. Assay of serum and muscle IL-15 protein levels
Blood was allowed to clot at RT for 30 min, centrifuged at 1,000 × g for 10 min, and serum removed and stored frozen at −20°C in aliquots. Quadriceps muscle tissue samples were frozen and stored at −80°C. Muscle samples were weighed and placed in T-PER Tissue Protein Extraction Reagent (Pierce, Rockford, IL) at 100 mg tissue/ml reagent. “Halt” protease inhibitor cocktail (Pierce) and ETDA (5 mM) were added. Tissue was homogenized on ice, and debris and excess lipid removed by centrifugation at 4°C. Total protein in an aliquot of the tissue sample was determined by the BCA Protein Assay (Pierce) and the remainder of the sample was normalized to 3 mg/ml protein. Serum and muscle IL-15 concentrations were determined using a BioRad (Hercules, CA) mouse IL-15 bead-based quantitative immunoassay performed in a BioPlex 200 dual-laser fluorescent microsphere detection instrument (BioRad). All samples were run in triplicate and the mean of each determination calculated as the value for that sample; standard curves were run concurrently for all assays. The assay was sensitive to 6.6 pg/ml recombinant murine IL-15 standard. Intra- and inter-assay CVs were 6% and 3%, respectively, and cross-reactivity with other cytokines was negligible. Results are reported as the concentration of IL-15 (in pg) per mg muscle tissue protein or the concentration of IL-15 per ml serum. In validation experiments (not shown), this assay was compared to a similar LINCOplex bead-based immunoassay performed in the same instrument and utilized previously in publications from this laboratory describing serum and muscle IL-15 concentrations in IL-15 overexpressing transgenic mice (Quinn et al., 2009). Although both assays could detect similar concentrations of recombinant murine IL-15, when normal (non-transgenic) young adult mouse serum was tested using both assays, the BioRad assay used in this study yielded values in the range of 10-300 pg/ml, while the LINCOplex assay of the same samples yielded values below the manufacturer's reported detection limit of 9.3 pg/ml in all samples (Quinn et al., 2009). The basis of this difference in the assays is unknown, but may be related to the epitopes detected by the respective monoclonal capture and detection antibodies, and could reflect differential abilities of these antibodies to detect IL-15 complexed to sIL-15Rα.
2.3. Assay of mRNA expression
Quadriceps muscle samples were submerged in RNAlater (Qiagen, Valencia, CA), equilibrated at 4°C for 24 h, and stored at −20°C. Samples were homogenized without RNA isolation, and processed according to directions supplied by Panomics (Fremont, CA) for analysis of mRNA expression using the Panomics QuantiGene Plex 2.0 system. This method combines bDNA RNA signal amplification and microspheres with unique fluorescent signatures to enable quantitation of multiple mRNA targets simultaneously in the same sample, without introducing the amplification inaccuracies inherent in RT-PCR, and allows for discrimination of highly homologous messages (Andras et al., 2001; Flagella et al., 2006). Specific oligonucleotide capture and extender probe sets (3 per target), designed to anneal exclusively to each mRNA of interest and each housekeeping mRNA, were designed by Panomics to unique sequences within each message sequence. The probe sets for total IL-15 were located in sequences common to both SSP- and LSP-IL-15, and for total IL-15Rα were located in sequences found in both sIL-15Rα and mIL-15Rα. The probe sets which distinguished total IL-15 from SSP-IL-15, and total IL-15Rα from mIL-15Rα, were located in sequences uniquely found in the short signal peptide of IL-15 and in the membrane-spanning sequence of the IL-15Rα, respectively. Probe sets were located in the following regions of each mRNA species: total IL-15 (NM_008357), bps 624-1117; SSP-IL-15 (NM_00357), bps 400-859; total IL-15Rα (NM_003858), bps 139-568; mIL-15Rα (NM_008358), bps 229-768; IL-2 receptor beta (IL-2Rβ; NM_008368), bps 152-597; IL-2 receptor gamma (IL-2Rγ; NM_013563), bps 67-634. Housekeeping genes were: hypoxanthine phosphoribosyltransferase 1 (HPRT1; NM_013556), beta-actin (Actb; NM_007393), and polymerase (RNA) II (DNA directed) polypeptide A (Polr2a; NM_007393). Probes were combined into a 7-plex and a 5-plex assay; the 7-plex contained reagents to detect total IL-15, total IL-15Rα, IL-2Rβ, IL-2RγC, and the 3 housekeeping genes (HPRT1, Polr2a, and Actb, representing low, medium, and high expressers, respectively), while the 5-plex contained reagents to detect SSP-IL-15, mIL-15Rα, and the 3 housekeeping genes. Specific mRNA transcripts were captured to specific fluorescent beads by hybridization to capture probe-extender probe interactions. Signal from each hybridized unit was amplified by attachment of biotinylated label probes at multiple binding sites on the complexes, which in turn bound to streptavidin-conjugated R-phycoerythrin (SAPE) to produce fluorescence. The fluorescent signals associated with individual capture beads were read using a dual-laser Bio-Plex 200 (BioRad), with the bead signature designating RNA target and the SAPE signal designating abundance. For each well, total fluorescence from each individual bead type (corresponding to individual mRNA species) minus background fluorescence for that bead type was normalized to the geometric mean of the fluorescence of the 3 housekeeping genes also in that well. The normalized signals for individual mRNAs from triplicate wells were averaged to yield a single value for each mRNA species being measured. Values for LSP-IL-15 mRNA and sIL-15Rα were derived by subtraction of SSP-IL15 mRNA from total IL-15 mRNA, and by subtraction of mIL-15Rα from total IL-15Rα, respectively.
2.4. Statistical procedures
Analyses comprised 6-8 animals per age group, using SigmaStat for Windows Version 3.00 (SPSS/Systat, Chicago, IL). The age effects on each factor were determined by Kruskal-Wallis one-way ANOVA on age ranks, with pairwise post-hoc comparisons performed by Dunn's method. Correlations between parameters for individual mice were determined to suggest mechanistic relationships among variables, and were determined by Spearman rank order correlation. Significance was accepted at p ≤ 0.05.
3. Results
3.1. Body composition
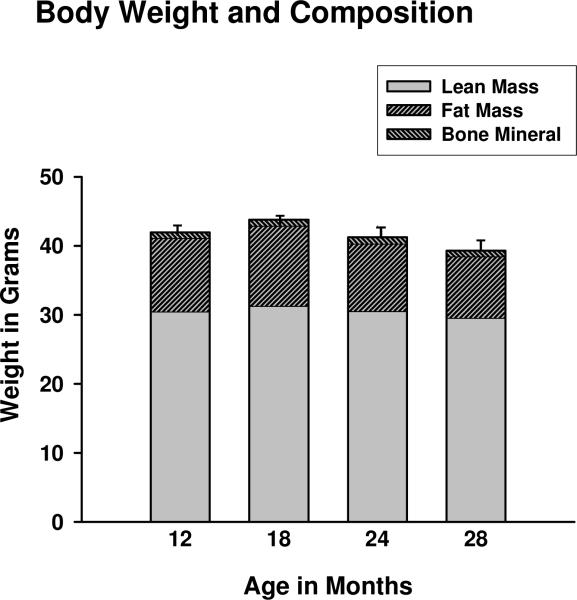
Body weight and body composition were assessed cross-sectionally in B6C3 male mice at 12, 18, 24, and 28 months of age using DEXA. Mice with tumors visible upon postmortem dissection, present in the 2 oldest age groups, were not included in the analysis. Trends towards lower lean body mass (which includes, but is not limited to, skeletal muscle) and total body weight were observed at 24 and 28 months (Fig. 1). However, significant differences in body weight, lean body mass, fat mass, and bone mineral content were not detected among age groups. The lack of change in body composition is typical of cross-sectional, as opposed to longitudinal, studies of aging humans and rodents (Lesser et al., 1973; Bunout et al., 2007), and indicates the hybrid mice studied here were undergoing healthy aging.
Figure 1.
Mean total body weight, lean body mass, fat mass, and bone mineral content in male B6C3 mice at 12, 18, 24 and 28 months of age. Error bars represent SEM for total body weight. None of these parameters differed significantly among age groups; n = 6-8 animals per age group.
3.2. IL-15 protein and mRNA
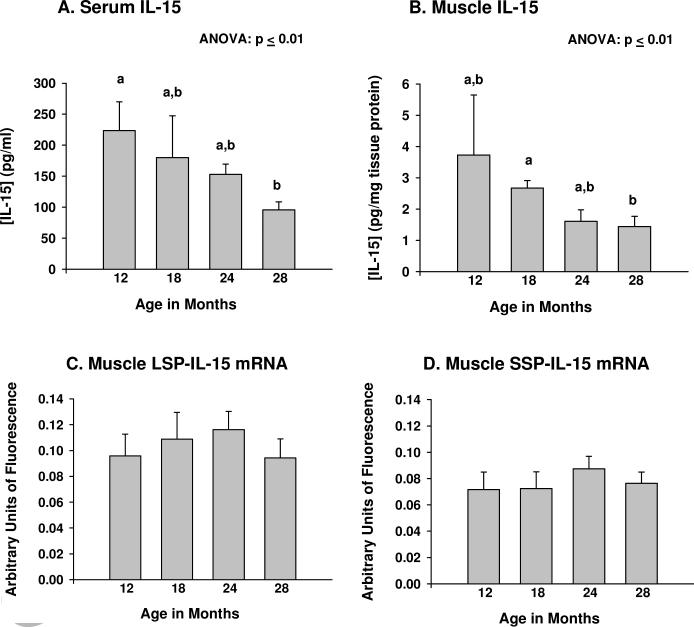
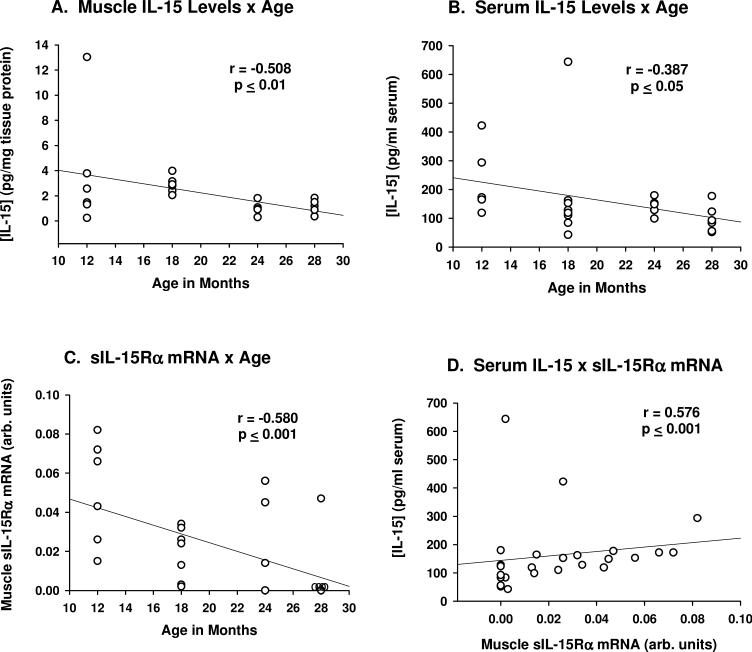
Significant (p ≤ 0.01, ANOVA) age-related declines in both serum and muscle IL-15 protein levels were observed (Fig. 2A,B). Post-hoc analyses indicated serum IL-15 levels at 12 months of age differed significantly from those at 28 months of age (p ≤ 0.05); muscle IL-15 levels differed significantly (p ≤ 0.05) between 18 and 28 months of age. Mouse quadriceps muscle tissue expressed both LSP- and SSP-IL-15 mRNA isoforms, with LSP-IL-15 mRNA being slightly more abundant (Fig. 2C, D). However, LSP- and SSP-IL-15 mRNA expression did not change with age (Fig. 2C, D). These findings indicate that the age-related decreases in IL-15 protein expression in muscle and serum are not due to decreases in muscle IL-15 mRNA expression. For individual mice, muscle and serum IL-15 protein levels correlated negatively with age (Fig. 3A,B).
Figure 2.
IL-15 protein and mRNA expression during aging. (A) Serum IL-15 protein levels, in pg/ml serum; (B) Skeletal muscle IL-15 protein levels, in pg/mg tissue protein; (C) Skeletal muscle long signal peptide (LSP)-IL-15 mRNA expression; (D) Skeletal muscle short signal peptide (SSP)-IL-15 mRNA expression. LSP- and SSP-IL-15 mRNA expression was determined by branched DNA (bDNA) signal amplification; signals were normalized to the geometric mean of 3 housekeeping genes of differing abundance. For serum and muscle IL-15 mRNA levels, ANOVAs on age rank indicated significant differences among age groups at p ≤ 0.01. Bars with different superscripts differ significantly (p ≤ 0.05) based on pairwise post-hoc analyses. N = 6-8 animals per age group.
Figure 3.
Scatter plots indicating correlation of muscle and serum IL-15 protein levels and age (A,B), muscle soluble IL-15R alpha receptor subunit (sIL-15Rα) mRNA expression and age (B), and serum IL-15 levels and muscle sIL-15Rα mRNA expression (C). Correlations were determined by Spearman rank order test.
3.3. IL-15 receptor subunit mRNA expression
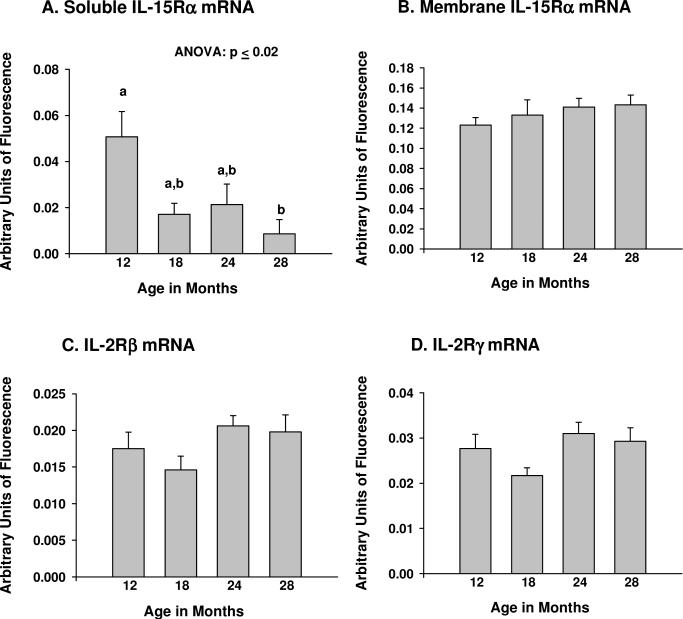
Expression of soluble IL-15Rα (sIL-15Rα) mRNA decreased (p ≤ 0.02, ANOVA) with age (Fig. 4A), while expression of membrane IL-15Rα (mIL-15Rα), IL-2Rβ and IL-2Rγ mRNA did not change (Fig. 4B-D). Post-hoc analyses of sIL-15Rα mRNA expression indicated a significant (p ≤ 0.05) difference between 12 and 28 months of age. For individual mice, expression of sIL-15Rα mRNA in muscle tissue exhibited a strong negative correlation with age (p ≤ 0.001), and was highly correlated (p ≤ 0.001) with serum (but not muscle) IL-15 levels (Fig. 3C,D). These data suggest there may be a relationship between age-related declines in muscle sIL-15Rα levels and serum IL-15 concentrations.
Figure 4.
Expression of mRNA for IL-15 receptor subunits in skeletal muscle tissue during aging. (A) Soluble IL-15 receptor-alpha (sIL-15Rα); (B) Membrane-associated IL-15 receptor-alpha (mIL-15Rα); (C) IL-2 receptor-beta (IL-2Rβ); (D) IL-2 receptor-gamma (IL-2Rγ). Branched DNA (bDNA) signal amplification was used to determine expression of each mRNA species; signals were normalized to the geometric mean of 3 housekeeping genes of differing abundance. For muscle soluble IL-15Rα mRNA expression, ANOVA on age rank indicated significant differences among groups at p ≤ 0.02. Bars with different superscripts differ significantly (p ≤ 0.05) based on pairwise post-hoc analyses. N = 6-8 animals per age group.
4. Discussion
This study showed that circulating and quadriceps muscle IL-15 levels decline with age in mice, independent of changes in IL-15 transcription. Additionally, expression of soluble, but not membrane-bound, IL-15Rα mRNA declined with age in muscle tissue and correlated highly with serum IL-15 levels. The sIL-15Rα subunit has been shown to facilitate IL-15 secretion and stability (Bulanova et al., 2007; Bergamaschi et al., 2008), and to potentiate IL-15 bioactivity (Mortier et al., 2006; Rubinstein et al., 2006). Although experimental interventions were not performed in this study, our findings support a model in which decreased sIL-15Rα expression with age reduces IL-15 protein secretion and stability, leading to lower circulating IL-15 levels in aging mice. These, in turn, may contribute to decreased IL-15 signaling and age-related changes in muscle and fat content (Quinn, 2008; Nielsen et al., 2008).
No previous studies have addressed IL-15 mRNA expression during aging in mice. Our findings of no change in either LSP- or SSP-IL-15 mRNA expression with age in the mixed fiber-type quadriceps muscle are similar to those reported in long-lived F344/BN rats by Marzetti et al. (2009), who observed no age-related changes in total IL-15 mRNA expression in the mixed fiber-type gastrocnemius muscle. In contrast, using the same strain of rats, Pistilli et al. (2007) reported significantly increased total IL-15 mRNA expression with age in the slow fiber-type soleus muscle, but reported no significant age-related change in total IL-15 mRNA expression in the predominantly fast plantaris. Therefore, the assay of muscles differing in fiber-type distribution may explain these conflicting results in rodents. However, it is unclear if measurement of IL-15 mRNA levels has functional significance, as previous studies have shown little correlation between IL-15 mRNA and protein levels due to numerous translational and post-translational complexities which impact IL-15 protein expression and stability (Fehniger and Caligiuri, 2001; Budagian et al., 2006; Nielsen et al., 2007).
This study indicated muscle tissue expresses both LSP- and SSP-IL-15 mRNA isoforms, which code for identical mature IL-15 peptides, but differ in intracellular trafficking (Tagaya et al., 1997; Fehniger and Caligiuri, 2001; Budagian et al., 2006). Specifically, the SSP-IL-15 isoform encodes intracellular IL-15, whereas the protein encoded by LSP-IL-15 can be secreted (Tagaya et al., 1997; Fehniger and Caligiuri, 2001; Budagian et al., 2006). Most published studies of IL-15 mRNA expression in skeletal muscle did not distinguish the SSP- and LSP-IL-15 mRNA isoforms (Pistilli et al., 2007; Nielsen et al., 2007; Marzetti et al., 2009). Northern blot surveys of IL-15 isoform expression in multiple tissues suggested skeletal muscle preferentially expresses the secreted LSP-IL-15 mRNA isoform (Tagaya et al., 1997), but this technology is relatively insensitive compared to the bDNA methodology used here (Andras et al., 2001; Flagella et al., 2006). We had originally hypothesized that an isoform switch between LSP- and SSP-IL-15 mRNA could account for declining circulating IL-15 protein levels, but no change in expression of either isoform with age was detected. We therefore determined muscle and serum IL-15 protein levels in adult and aging mice.
This is the first study in which measurement of physiological levels of IL-15 protein in mice using a commercially-available assay has been reported. Previous studies from this laboratory utilized a bead-based immunoassay using dual laser technology from a different manufacturer, which could quantify muscle and serum IL-15 in IL-15-overexpressing transgenic mice, but yielded values for wild-type mouse muscle homogenates and serum which were below the detection limit of the assays (Quinn et al., 2009). Inasmuch as both manufacturer's assays exhibited very similar sensitivities in measurement of recombinant murine IL-15, it is possible that the difference in the monoclonal capture and detection antibodies used could reflect differential reactivity to free IL-15 or IL-15 complexed with sIL-15Rα. Indeed, Bulanova et al. (2007) demonstrated a commercial IL-15 ELISA assay could not detect IL-15 complexed with sIL-15Rα, whereas an in-house two-site ELISA detecting IL-15/sIL-15Rα complexes suggested that most, but not all, IL-15 in mouse serum appears to reside in sIL-15Rα/IL-15 heterodimers, at concentrations similar to those reported in the present study. In a study in which recombinant human IL-15 was infused into young and aging rats using mini-osmotic pumps (Pistilli and Alway, 2008), identical infusion regimes yielded nominal plasma levels of human IL-15 which were more than 10-fold higher in the aging rats. It is possible lower circulating sIL-15Rα levels in the aging rats resulted in an elevation of free IL-15 which was detectable by the anti-human IL-15 ELISA used in that study.
Our findings of age-related decreases in mouse quadriceps muscle IL-15 protein levels are in agreement with those of Marzetti et al. (2009) who found that rat gastrocnemius muscle IL-15 levels declined with age in ad lib-fed, but not calorie-restricted rats. Since ad lib-fed rats underwent sarcopenia, but calorie-restricted rats did not, this suggests increasing or maintaining IL-15 levels in aging muscle might inhibit sarcopenia. This hypothesis is consistent with previous observations indicating that systemic administration of recombinant IL-15 can inhibit muscle protein degradation in vivo and in vitro (Carbó et al., 2000; Quinn et al., 2002), inhibit muscle nuclear apoptosis in young rats (Figueras et al., 2004), maintain muscle mass in cancer cachexia (Carbó et al., 2000) and ameliorate some aspects of muscle degeneration in murine models of muscular dystrophy (Harcourt et al., 2005). However, the only study of artificial enhancement of IL-15 levels in aging rodents did not support this hypothesis. Pistilli and Alway (2008) used osmotic pumps to infuse human recombinant IL-15 into aging rats and observed significant increases, rather than decreases, in muscle apoptotic markers, and also observed decreases in muscle weight in IL-15-infused aging rats. These markers were impacted less in young rats infused with IL-15, suggesting another age-modulated factor inhibits or modifies IL-15 stability, activity, or bioavailability in the aging rodent. Our findings suggest this factor may be sIL-15Rα. Use of a heterologous system (infusion of human recombinant IL-15 into rats) could also be a factor in that study. Rodent and human IL-15 exhibit only 70% homology, and human recombinant IL-15 is 200-fold less active than murine IL-15 in mouse systems in the absence of IL-15Rα (Eisenman et al., 2002). Therefore, the potential for manipulation of the IL-15 signaling axis to ameliorate age-associated muscle wasting remains unresolved; coordinated upregulation of IL-15 and sIL-15Rα should be considered in aging systems.
The present study documented progressive decreases in serum IL-15 concentrations in normally-aging mice. Serum IL-15 levels correlated with muscle sIL-15Rα mRNA expression at all ages studied, suggesting there is a mechanistic relationship between these two parameters. Although broadly expressed by many tissue and cell types, the secreted LSP-IL-15 mRNA isoform is much more highly expressed in skeletal muscle tissue compared to other tissues, suggesting muscle may be a major source of circulating IL-15 (Grabstein et al., 1994; Tagaya et al., 1997). Because we observed little correlation between IL-15 mRNA and protein levels, and only skeletal muscle IL-15Rα mRNA expression and muscle and serum IL-15 protein were assessed in this study, no conclusions can be drawn from our data as to the major source(s) of circulating IL-15. Nevertheless, Riechman et al. (2004) observed plasma IL-15 levels increased immediately following a bout of intense resistance exercise in young human subjects, suggesting muscular activity caused release of IL-15 from the tissue into the circulation. The findings reported here indicate further investigation of the expression of sIL-15Rα in skeletal muscle in different physiological states (such as exercise) is warranted.
In human subjects, plasma IL-15 levels are negatively correlated with total and trunk fat mass, and negative correlations between muscle IL-15 mRNA and obesity parameters have been demonstrated (Nielsen et al., 2008). In the present study, no correlations between any aspect of body composition and levels of IL-15 protein, expression of IL-15 mRNA, or expression of mRNA for any IL-15 receptor subunits were observed. This may be due to the cross-sectional nature of the experimental design, which found no significant age-related changes in any parameter constituting body composition. Additionally, little variation in body composition was noted within each age group, as mice were raised in essentially identical laboratory conditions and feeding regimens. The cross-sectional experimental design was necessary in order to harvest muscle tissue for both protein and mRNA analyses. Longitudinal studies of both aging rodents and humans, which unlike cross-sectional studies do not select for healthy (and alive) individuals at each age group, allow for more individual variation and reveal larger age-related changes in body composition (Lesser et al., 1973; Bunout et al., 2007). It is also possible that muscle mass, as opposed to lean body mass, may have correlated with IL-15 or sIL-15Rα expression, but this was not assessed in the present study. The decline of IL-15 protein levels in healthy aging mice observed in this study suggests, but does not prove, that reductions in IL-15 signaling may contribute to, rather than result from, age-related changes in body composition.
We previously observed that transgenic mice with elevated circulating IL-15 levels exhibited lower fat deposition and resistance to diet-induced obesity than control mice (Quinn et al., 2009). Similarly, systemic overexpression of IL-15, via injection of recombinant IL-15 or electrotransfer of IL-15 expression plasmids, inhibited fat deposition in rodents (Carbó et al., 2001; Alvarez et al., 2002; Nielsen et al., 2008). In adipogenic cell cultures, IL-15 inhibited adipocyte differentiation, and independently stimulated lipolysis and secretion of adiponectin by differentiated adipocytes (Ajuwon and Spurlock, 2004; Quinn et al., 2005). These findings suggest declining circulating IL-15 levels may contribute to age-related changes in body composition and possibly decreased insulin sensitivity, which are major determinants of morbidity and mortality in the elderly (Baumgartner, 2000; Lebrun et al., 2006, Facchini et al., 2001). Indeed, Gangemi et al. (2005) observed that while serum IL-15 concentrations exhibited a downward trend with age in unselected human subjects, individuals with unusually long lifespans (95 to more than 100 years) and still living independently had significantly higher circulating IL-15 levels, suggesting elevated IL-15 levels conferred some protection against age-related disability.
The findings reported here complement human genetic studies by three separate laboratories which have identified several single-nucleotide polymorphisms (SNPs) in the IL-15Rα gene which impact muscularity, fat deposition, and insulin sensitivity (Riechman et al., 2004; DiRenzo et al., 2006; Pistilli et al., 2008). Many of these SNPs are located in intron/exon borders, suggesting they may differentially impact IL-15Rα mRNA splicing to produce mIL-15Rα or sIL-15Rα mRNA transcripts. Our findings in this observational study suggest that age-related declines in sIL-15Rα expression lead to declining systemic IL-15 levels, which in turn may affect body composition. Our findings provide a technical and experimental basis for further investigation of IL-15, sIL-15Rα, and their interactions in determining body composition in aging mice, as a model for humans.
Acknowledgements
Supported by NIH grant #RO1AG024136 from the National Institute on Aging (LSQ), and use of resources and facilities at VA Puget Sound Health Care System.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ajuwon KM, Spurlock ME. Direct regulation of lipolysis by interleukin-15 in primary pig adipocytes. Am. J. Physiol. 2004;287:R608–R611. doi: 10.1152/ajpregu.00192.2004. [DOI] [PubMed] [Google Scholar]
- Alvarez B, Carbo N, Lopez-Soriano J, Drivdahl RH, Busquets S, Lopez-Soriano FJ, Argiles JM, Quinn LS. Effects of interleukin-15 on adipose mass in rodent obesity models: Evidence for direct IL-15 action on adipose tissue. Biochem. Biophys. Acta. 2002;1570:33–37. doi: 10.1016/s0304-4165(02)00148-4. [DOI] [PubMed] [Google Scholar]
- Andras SC, Power JB, Cocking EC, Davey MR. Strategies for signal amplification in nucleic acid detection. Molec. Biotechnol. 2001;19:29–44. doi: 10.1385/MB:19:1:029. [DOI] [PubMed] [Google Scholar]
- Baumgartner RN. Body composition in healthy aging. Ann. NY Acad. Sci. 2000;904:437–448. doi: 10.1111/j.1749-6632.2000.tb06498.x. [DOI] [PubMed] [Google Scholar]
- Bergamasch i C., Rosati M, Jalah R, Valentin A, Kulkarni V, Alicea C, Zhang G-N, Patel V, Felber BK, Pavlakis GN. Intracellular interaction of interleukin-15 with its receptor α during production leads to mutual stabilization and increased bioactivity. J. Biol. Chem. 2008;283:4189–4199. doi: 10.1074/jbc.M705725200. [DOI] [PubMed] [Google Scholar]
- Budagian V, Bulanova E, Paus R, Bulfone-Paus S. IL-15/IL-15 receptor biology: A guided tour through an expanding universe. Cytok. Growth Factor Rev. 2006;17:259–280. doi: 10.1016/j.cytogfr.2006.05.001. [DOI] [PubMed] [Google Scholar]
- Bulanova E, Budagian V, Duitman E, Orinska A, Krause H, Rückert R, Reiling N, Bulfone-Paus S. Soluble IL-15Rα is generated by alternative splicing or proteolytic cleavage and forms functional complexes with IL-15. J. Biol. Chem. 2007;282:13167–13179. doi: 10.1074/jbc.M610036200. [DOI] [PubMed] [Google Scholar]
- Bunout D, de la Maza MP, Barrera G, Leiva L, Gattas V, Hirsch S. Assessment of sarcopenia: longitudinal versus corss sectional body composition. Aging Clin. Exp. Res. 2007;19:295–299. doi: 10.1007/BF03324705. [DOI] [PubMed] [Google Scholar]
- Carbó N, López-Soriano J, Costelli P, Busquets S, Alvarez B, Baccino FM, Quinn LS, López-Soriano FJ, Argilés JM. Interleukin-15 antagonizes muscle protein waste in tumour-bearing rats. Brit. J. Cancer. 2000;83:526–531. doi: 10.1054/bjoc.2000.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbó N, López-Soriano J, Costelli P, Alvarez B, Busquets S, Baccino FM, Quinn LS, López-Soriano FJ, Argilés JM. Interleukin-15 mediates reciprocal regulation of adipose and muscle mass: a potential role in body weight control. Bioch. Biophys. Acta. 2001;1526:17–24. doi: 10.1016/s0304-4165(00)00188-4. [DOI] [PubMed] [Google Scholar]
- Di Renzo L, Bigioni M, Bottini FG, Del Grobbo V, Premrov MG, Cianci R, De Lorenzo A. Normal Weight Obese syndrome: role of single nucleotide polymorphism of IL-15Rα and MTHFR 677→T genes in the relationship between body composition and resting metabolic state. Eur. Rev. Med. Pharm. Sci. 2006;10:235–245. [PubMed] [Google Scholar]
- Eisenman J, Ahdieh M, Beers C, Brasel K, Kennedy MK, Le T, Bonnert TP, Paxton RJ, Park LS. Interleukin-15 interactions with interleukin-15 receptor complexes: Characterization and species specificity. Cytokine. 2002;20:121–129. doi: 10.1006/cyto.2002.1989. [DOI] [PubMed] [Google Scholar]
- Facchini FS, Hua N, Abbasi F, Reaven GM. Insulin resistance as a predictor of age-related diseases. J. Clin. Endo. Metab. 2001;86:3574–3578. doi: 10.1210/jcem.86.8.7763. [DOI] [PubMed] [Google Scholar]
- Fehniger TA, Caligiuri MA. Interleukin-15: biology and relevance to human disease. Blood. 2001;97:14–32. doi: 10.1182/blood.v97.1.14. [DOI] [PubMed] [Google Scholar]
- Figueras M, Busquets S, Carbó N, Barreiro E, Almendro V, Argilés JM, López-Soriano FJ. Interleukin-15 is able to suppress the increased DNA fragmentation associated with muscle wasting in tumour-bearing rats. FEBS Lett. 2004;569:201–206. doi: 10.1016/j.febslet.2004.05.066. [DOI] [PubMed] [Google Scholar]
- Flagella M, Bui S, Zheng Z, Nguyen CT, Zhang A, Pastor L, Ma Y, Yang W, Crawford KL, McMaster GK, Witney F, Luo Y. A multiplex branched DNA assay for parallel quantitative gene expression profiling. Anal Biochem. 2006;352:50–60. doi: 10.1016/j.ab.2006.02.013. [DOI] [PubMed] [Google Scholar]
- Gangemi S, Basile G, Monti D, Merendino RA, Di Pasquale G, Bisigano U, Nicita-Mauro N, Franceschi C. Age-related modifications in circulating IL-15 levels in humans. Mediators Inflamm. 2005;4:245–247. doi: 10.1155/MI.2005.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodpaster BH, Park SW, Harris TB, Kritchevsky SG, Nevit M, Schwartz AV, Simonisck EM, Tylavsky FA, Visser M, Newman AB. The loss of skeletal muscle strength, mass, and quality in older adults: the Health, Aging and Body Composition study. J. Gerontol. 2006;61A:1059–1064. doi: 10.1093/gerona/61.10.1059. [DOI] [PubMed] [Google Scholar]
- Grabstein KH, Eisenman J, Shanebeck K, Rauch C, Srinivasan S, Fung V, Beers C, Richardson J, Schoenborn MA, Ahdieh M, Johnson L, Alderson MR, Watson JD, Anderson DM, Giri J. Cloning of a T cell growth factor that interacts with the beta chain of the interleukin-2 receptor. Science. 1994;264:965–968. doi: 10.1126/science.8178155. [DOI] [PubMed] [Google Scholar]
- Harcourt LJ, Holmes AG, Gregorevic P, Schertzer JD, Stupka N, Plant DR, Lynch GS. Interleukin-15 administration improves diaphragm muscle pathology and function in dystrophic mdx mice. Am. J. Pathol. 2005;166:1131–1141. doi: 10.1016/S0002-9440(10)62333-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebrun CEI, van der Schouw YT, de Jong FH, Grobbee DE, Lamberts SW. Fat mass rather than muscle strength is the major determinant of physical function and disability in postmenopausal women younger than 75 years of age. Menopause. 2006;13:474–481. doi: 10.1097/01.gme.0000222331.23478.ec. [DOI] [PubMed] [Google Scholar]
- Lesser GT, Deutsch S, Markofsky J. Aging in the rat: longitudinal and cross-sectional studies of body composition. Am. J. Physiol. 1973;225:1472–1478. doi: 10.1152/ajplegacy.1973.225.6.1472. [DOI] [PubMed] [Google Scholar]
- Marzetti E, Carter CS, Wohlgemuth SE, Lees H, Giovannini S, Anderson B, Quinn LS, Leewenburgh C. Modulation of skeletal muscle IL-15 expression and death-receptor apoptotic signaling by aging and calorie restriction. Mech. Aging Devel. 2009;130:272–280. doi: 10.1016/j.mad.2008.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortier E, Quemener A, Vusio P, Lorenzen I, Boublik Y, Grotzinger J, Plet A, Jacques Y. Soluble IL-15Ralpha-sushi as a selective and potent agonist of IL-15 action through IL-15Rbeta/gamma: hyper-agonist IL-15-IL-15Ralpha fusion proteins. J. Biol. Chem. 2006;281:1612–1619. doi: 10.1074/jbc.M508624200. [DOI] [PubMed] [Google Scholar]
- Nielsen AR, Mounier R, Plomgaared P, Mortensen OH, Penkowa M, Speerschneider T, Pilegaard H, Pedersen BK. Expression of interleukin-15 in human skeletal muscle-effect of exercise and muscle fibre type composition. J. Physiol. 2007;584:305–312. doi: 10.1113/jphysiol.2007.139618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen AR, Hojman P, Erikstrup C, Fischer CP, Plomgaard P, Mounier R, Mortensen OH, Broholm C, Taudorf S, Krogh-Madsen R, Lindegaard B, Petersen AMW, Gehl J, Pedersen BK. Association between interleukin-15 and obesity: Interelukin-15 as a potential regulator of fat mass. J. Clin. Endo. Metab. 2008;93:4486–4493. doi: 10.1210/jc.2007-2561. [DOI] [PubMed] [Google Scholar]
- Pistilli EE, Alway SE. Systemic elevation of interleukin-15 in vivo promotes apoptosis in skeletal muscles of young adult and aged rats. Biochem. Biophys. Res. Commun. 2008;373:20–24. doi: 10.1016/j.bbrc.2008.05.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pistilli EE, Siu PM, Alway SE. Interleukin-15 responses to aging and unloading-induced skeletal muscle atrophy. Am. J. Physiol. 2007;292:C1298–C1304. doi: 10.1152/ajpcell.00496.2006. [DOI] [PubMed] [Google Scholar]
- Pistilli EE, Devaney JM, Gordish-Dressman H, Bradbury MK, Seip RL, Thompson PD, Angelopoulos TJ, Clarkson PM, Myna NM, Pescatello LS, Visich PS, Zoeller RF, Gordon PM, Hoffman EP. Interelukin-15 and interleukin-15R alpha SNPs and associations with muscle, bone, and predictors of the metabolic syndrome. Cytokine. 2008;43:45–53. doi: 10.1016/j.cyto.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn LS. Interleukin-15: A muscle-derived cytokine regulating fat-to-lean body composition. J. Anim. Sci. 2008;86:E75–E83. doi: 10.2527/jas.2007-0458. [DOI] [PubMed] [Google Scholar]
- Quinn LS, Anderson BG, Drivdah l R.H., Alvarez B, Argilés JM. Overexpression of interleukin-15 induces skeletal muscle hypertrophy in vitro: Implications for treatment of muscle wasting disorders. Exp. Cell Res. 2002;280:55–63. doi: 10.1006/excr.2002.5624. [DOI] [PubMed] [Google Scholar]
- Quinn LS, Strait-Bodey L, Anderson BG, Argilés JM, Havel PJ. Interleukin-15 stimulates adiponectin secretion by 3T3-L1 adipocytes: Evidence for a skeletal muscle-to-fat signaling pathway. Cell Biol. Int. 2005;29:449–457. doi: 10.1016/j.cellbi.2005.02.005. [DOI] [PubMed] [Google Scholar]
- Quinn LS, Anderson BG, Strait-Bodey L, Stroud AM, Argilés JM. Oversecretion of interleukin-15 from skeletal muscle reduces adiposity. Am. J. Physiol. 2009;296:E191–E202. doi: 10.1152/ajpendo.90506.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechman SE, Balasekaran G, Roth SM, Ferrell RE. Association of interleukin-15 protein and interleukin-15 receptor genetic variation with resistance exercise training responses. J. Appl. Physiol. 2004;97:2214–2219. doi: 10.1152/japplphysiol.00491.2004. [DOI] [PubMed] [Google Scholar]
- Rubinstein MP, Kovar M, Purton JF, Cho J-H, Boyman O, Surh CD, Sprent J. Convertng IL-15 to a superagonist by binding to soluble IL-15Rα. Proc. Natl. Acad. Sci. USA. 2006;103:9166–9171. doi: 10.1073/pnas.0600240103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagaya Y, Kurys G, Thies TA, Losi JM, Azimi N, Hanover JA, Bamford RN, Waldmann TA. Generation of secretable and nonsecretable interleukin 15 isoforms through alternate usage of signal peptides. Proc. Natl. Acad. Sci. USA. 1997;94:14444–14449. doi: 10.1073/pnas.94.26.14444. [DOI] [PMC free article] [PubMed] [Google Scholar]