Abstract
α-Defensin biosynthesis requires the proteolytic conversion of inactive precursors to microbicidal forms. In mouse Paneth cell pro-α-defensin proCrp4(20–92), anionic amino acids positioned near the proregion N-terminus inhibit proCrp4 activity by an apparent charge neutralization mechanism. Because most pro-α-defensins contain proregions of highly conserved chain length, we tested whether decreasing the distance between the inhibitory acidic residues of the proregion and the α-defensin component of the precursor would alter proCrp4 inhibition. Accordingly, two proCrp4 deletion variants, (Δ44–53)-proCrp4 and (Δ44–58)-proCrp4, truncated in a manner corresponding to deletions between MMP-7 cleavage sites, were prepared and assayed for bactericidal peptide activity. Consistent with the properties of full-length proCrp4(20–92), (Δ44–53)-proCrp4 and (Δ44–58)-proCrp4 were processed effectively by MMP-7, lacked bactericidal activity at high peptide levels and over a 3 h exposure period, and failed to induce permeabilization of live E. coli in vitro. Thus, bringing the inhibitory proregion domain into greater proximity with the Crp4 component of the precursor did not alter the activity of this pro-α-defensin. Therefore, the conserved distance that separates inhibitory acidic proregion residues from the Crp4 peptide is not critical to maintaining proCrp4(20–92) in an inactive state.
Keywords: Antimicrobial peptide, precursor convertase, microbicide, Paneth cell, innate immunity
1. Introduction
Mammalian defensins are effectors of innate immunity that function in neutrophils and at varied epithelial surfaces, including the skin, oral cavity, airway, and the gastrointestinal tract [19,26]. The mammalian defensins comprise three subfamilies, α-, β-, and θ-defensins, with broad spectrum microbicidal activities [12] that generally are mediated by membrane disruptive mechanisms facilitated by net electropositive charge and amphipathicity [6,12,17]. Specifically, the α-defensins are abundant peptide constituents of neutrophil azurophil granules that mediate non-oxidative killing following microbial phagocytosis, and in dense core granules of Paneth cells at the base of small intestinal crypts which are released into the lumen of the small bowel [14,19]. Regardless of their sites of expression, α-defensins are synthesized as inactive precursors that must undergo proteolytic conversion to give rise to processed, mature peptides with bactericidal activities [4,5].
Events associated with pro-α-defensin activation involve proteolytic cleavage of the proregion by lineage-specific proteinases, which may occur during granulogenesis in the regulated pathway or after secretion in certain instances [22]. After co-translational removal of canonical signal sequences, the inactive ~ 8.5 kDa pro-α-defensins consist of an acidic prosegment of ~ 4 kDa that is N-terminal of the 3.5-4.4 kDa α-defensin component of the precursor [4,5]. Most myeloid α-defensin precursors are fully processed in the azurophil granules of neutrophils, and in vitro studies implicate the granule serine proteinases elastase, cathepsin G, and proteinase-3 as the activating convertases [2,8]. Human Paneth cells accumulate unprocessed proforms, e.g., pro-HD5, which are activated by anionic and meso-trypsins after secretion [7]. In contrast, mouse Paneth cell α-defensins, termed cryptdins (Crps), are activated by matrix metalloproteinase-7 (MMP-7) intracellularly and prior to secretion [24]. For example, 8.4 kDa mouse pro-cryptdin-4 (proCrp4(20–92)) lacks in vitro bactericidal and membrane disruptive peptide activities [16] until specific prosegment cleavage events mediated by MMP-7 at Ser43↓Ile44, Ala53↓Leu54, and Ser58↓Leu59 relieve the inhibitory effects of the proregion on the defensin component of the precursor by severing their covalent association [20,23]. Thus, the proregion maintains proCrp4, as well as additional pro-α-defensins [4,7,8], in an inactive state.
Acidic amino acids that cluster near the N terminus of the proCrp4(20–43) proregion mediate inhibition of proCrp4 [3]. Site-directed mutagenesis experiments have shown that Arg residues in Crp4 contribute essential electropositive charge that is required for Crp4 bactericidal peptide activity, although the effect of mutation was independent of the actual Arg residue positions in the peptide primary structure [21]. In nearly all pro-α-defensins, the electropositive charge of the α-defensin component is balanced or neutralized by anionic amino acid residues in the prosegment. For example, in mouse proCrp4, cleavage at Ser43↓Ile44 catalyzed by MMP-7 removes 24 amino acid residues from prosegment N-terminus, including nine acidic amino acids [23]. That proteolytic event is sufficient to activate and enable full bactericidal and membrane disruptive behavior of Crp4 [3,23], because all MMP-7 processing intermediates, Crp4(44–92), Crp4(54–92) and Crp4(59–92), are as bactericidal as the fully-processed Crp4 molecule [23]. Furthermore, charge neutralizing mutagenesis of proregion Asp and Glu residues eliminated prosegment inhibition of proCrp4 [23], and replacement and deletional mutagenesis studies showed that those acidic amino acids positioned nearest the proCrp(20–92) N terminus, between residue positions 20–28, predominate in mediating inhibition of proCrp4 membrane disruptive mechanisms [3].
The role of these N-terminal electronegative residue positions and the highly conserved chain length of α-defensin proregions suggested that the peptide bond distance between these inhibitory elements and the Crp4 moiety of the proform may provide an optimal distance for maintaining proCrp4 in an inactive state. To test this hypothesis, we prepared proCrp4 variants that retain the inhibitory acidic amino acids but have proregion deletions of 10 and 15 amino acids that correspond to proregion fragments produced by MMP-7 proteolysis. The truncated proCrp4 molecules with inhibitory anionic amino acids placed closer to the mature peptide were completely lacking in bactericidal peptide activity, evidence that conserved prosegment chain length is not a critical determinant of its inhibitory activity.
2. Materials and Methods
2.1 Mutagenesis of proCrp4 proregion deletion variants
Amino acids between MMP-7 cleavage sites at Ile44 and Ala53, inclusively, were deleted from the proregion of proCrp4 by a series of mutagenizing PCR reactions to prepare (Δ44–53)-proCrp4 using a previously described approach [17,20]. PCR site-directed mutagenesis was performed using GeneAMP PCR Core Reagents (Applied Biosystems, Foster City, CA). The cDNA template for the first and second reaction was a pET-28a expression construct for native proCrp4 as previously reported [20]. For the first reaction, the forward primer Δ44–53-PC4-f1 (5′-CAGGC TGTGT CTCTT CATGA AAAA-3′) was paired with reverse primer Crp4-R (5′-TATAT GTCGA CTCAG CGGCG TGGCA-3′. For the second reaction, the forward primer EcoR1-Met-PC4-f (5′-GCGCG AATTC ATGGA TCCTA TCCAA AACAC A-3′) and reverse primer Δ44–53-PC4-R1 (5′-TTTTT CATGA AGAGA CACAG CCTG-3′) were used. Each amplification reaction mixture was incubated at 94°C for 5 min followed by 5 cycles of 94°C for 30 s, 38°C for 30 s, and 72°C for 30 s followed by 25 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s and finally an extension reaction for 7 min at 72°C. A third reaction generated the full-length mutagenized product by using 0.01% of amplification products from the first and second PCR reactions as templates and using a forward cloning primer complementary to the 5′-ends of each template strand [10,23]. Forward primer EcoR1-Met-PC4-f was paired with reverse primer Crp4-R in amplification reactions that were incubated at 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s for 25 cycles, followed by a 7 min extension reaction at 72°C to prepare the final proCrp4 variant coding sequence.
A longer deletion variant corresponding to residues between the Ile44 and Ser58 MMP-7 cleavage sites, termed (Δ44–58)-proCrp4, also was prepared to investigate the effect of prosegment length on proCrp4 inhibition. Amino acids Ile44 to Ser58 of the proCrp4 prosegment were deleted inclusively by iterative mutagenizing PCR reactions as described above. For the first reaction, the forward primer Δ44–58-PC4-f1 (5′-CAGGC TGTGT CTTTG AGAGG TTTG-3′) was paired with the reverse primer Crp4-R. For the second reaction, the forward primer EcoR1-Met-PC4-f was paired with the reverse primer Δ44–58-PC4-R1 (5′-CAAAC CTCTC AAAGA CACAG CCTG -3′). Each amplification reaction and the third PCR reaction was carried out as described in the previous section to give the final proCrp4 variant PCR product. ProCrp4 variant PCR products were cloned in pCR-2.1 TOPO, the mutagenesis verified by DNA sequencing, excised with SalI and EcoRI, subcloned into pET28a plasmid DNA (Novagen, Inc.), and transformed into E. coli BL21(DE3)-pLysS cells (Stratagene, La Jolla, CA) for recombinant expression [17,20].
2.2 Induction and Purification of Recombinant Peptides
ProCrp4 variants were purified using methods previously described [16,17,20]. The recombinant peptides were expressed in E. coli BL21(DE3)-pLysS as N-terminal His6-tagged fusion proteins cloned in-frame between the EcoRI and SalI sites of the pET-28a expression vector polylinker (Novagen, Inc., Madison, WI). Met residues were introduced at the peptide N-termini providing a unique CNBr cleavage site so as to release the peptide from the His6 tag fusion partner [17,20]. E. coli cells were grown at 37°C in Terrific Broth medium consisting of 12 g of Bacto Tryptone (Becton Dickenson Microbiological Systems, Inc., Sparks, MD), 24 g of Bacto Yeast Extract (Becton Dickenson), 4 ml of glycerol, 900 ml of H2O, 100 ml of sterile phosphate buffer consisting of 0.17 M KH2PO4, 0.72 M K2HPO4, and 70 μg/ml kanamycin. Recombinant peptide expression was induced in mid-log phase cells by addition of isopropyl-βD-1-thiogalacto-pyranoside at a final concentration of 0.1 mM for 4 h at 37°C [17]. The peptides were purified to homogeneity by HPLC.
Induced bacterial cells were lysed by sonication in 6 M guanidine-HCl in 100 mM Tris-Cl (pH 8.1), and the soluble protein fraction was clarified by centrifugation [16,17,20]. His-tagged fusion peptides were purified using nickel-nitrilotriacetic acid (Qiagen, Valencia, CA) resin affinity chromatography [20]. The peptides were eluted with 1 M imidazole, 6 M guanidine-HCl, 100 mM Tris-HCl and then dialyzed in 5% acetic acid. To cleave the His tag fusion partner, the fusion peptides were lyophilized and reacted with10 mg/ml cyanogen bromide (CNBr) in 80 % formic acid for 18 h at 25°C, diluted with 10 vol of water and lyophilized [23]. After CNBr cleavage, the mutagenized peptides and cleavage products were resuspended in 5% acetic acid and then purified to homogeneity by C18 reverse-phase high performance liquid chromatography (RP-HPLC, data not shown). Homogeneity was assessed by acid urea (AU)-PAGE analyses as described [13,18]. Peptide concentrations were quantified by UV absorption at 280 nm based on their extinction coefficients. The molecular masses of the purified peptides were determined using MALDI-TOF-MS (Voyager-DE MALDI-TOF, PE-Biosystems, Foster City, CA) at the Mass Spectroscopy Facility, Department of Chemistry, University of California, Irvine, CA, and using a Microflex LRF System spectrometer (Bruker Daltonics, Billerica, MA).
2.3 In Vitro MMP- 7 mediated proteolysis of proCrp4 deletion variants
To test proCrp4 deletion variants for proteolytic conversion by MMP-7, samples consisting of 10 μg peptide were incubated with or without 0.5 molar equivalent of MMP-7 in 1 mM HEPES, 15 mM NaCl, and 0.5 mM CaCl2, pH 7.4 for 18 h at 37 °C [1]. Complete peptide digests were resolved by AU-PAGE and stained with Coomassie Blue. After digestion, the molecular masses of the products obtained were determined by MALDI-TOF-MS using a Microflex LRF System spectrometer (Bruker Daltonics).
2.4 Bactericidal Peptide Assays
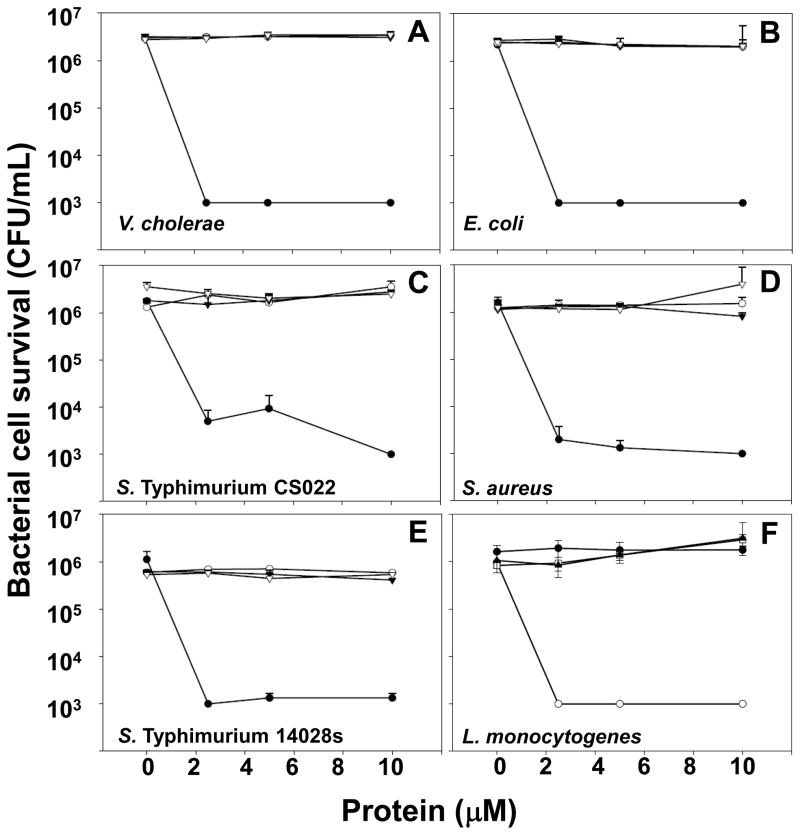
To test whether decreasing prosegment chain length relieved proCrp4 inhibition, the activities of purified recombinant peptides were tested in vitro against E. coli ML35, Vibrio cholerae, L. monocytogenes, Staphylococcus aureus, and Salmonella enterica serovar Typhimurium (S. Typhimurium) strains as described previously [17,23]. Samples consisting of exponentially growing bacterial cells were collected by centrifugation, washed, and resuspended in 10 mM PIPES (pH 7.4) supplemented with 0.01 vol (1% v/v) trypticase soy broth (PIPES-TSB). In triplicate experiments, bacteria (5 × 106 CFU/ml) were incubated with 0–10 μM of each peptide in 50 μl of PIPES-TSB. After 60 min at 37°C, 20 μl samples of each incubation mixture were diluted 1:1000 with 10 mM PIPES (pH 7.4), and 50 μl samples of the diluted mixtures were plated on trypticase soy agar plates using a Spiral Biotech Autoplate 4000 (Spiral Biotech Inc., Bethesda, MD). Surviving bacteria were quantitated as CFU/ml on plates after incubation at 37°C for 18 h, and data were analyzed and plotted using Sigma Plot (Systat Software, Inc., San Jose, CA) (Figs. 6A–F).
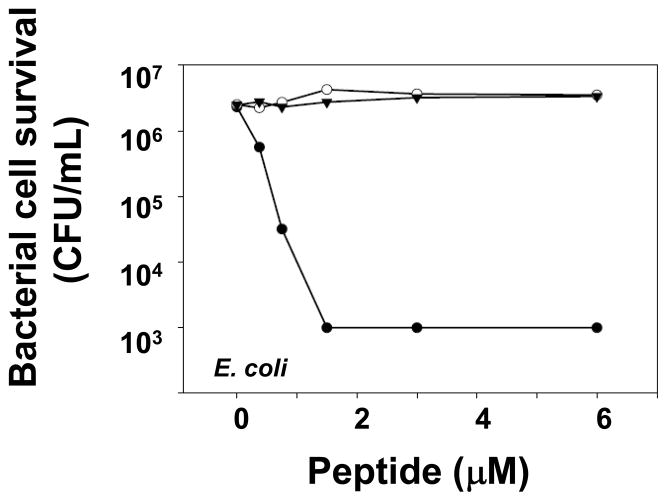
Fig. 6. Bactericidal activity of truncated prosegment variants is fully inhibited.
Exponentially-growing bacteria (~ 5 × 106 CFU/mL) Vibrio cholerae (A), Escherichia coli (B), Salmonella Typhimurium CS022 (C), Staphylococcus aureus (D), Salmonella Typhimurium 14028S (E), and Listeria monocytogenes (F) were exposed to peptides at 37 °C in 50 μl 10 mM PIPES buffer supplemented with 1% TSB for 1 h. Assays were performed in triplicate, and error bars denote standard deviation. Following peptide exposure, the bacteria were plated on TSB-agar, incubated overnight at 37 °C (see Materials and Methods, Section 2.4) [17], and surviving bacteria were counted as CFU at each peptide concentration. Values at or below 1 × 103 CFU/ml signify absence of colonies after overnight growth, the detection limit of the assay. Symbols: Crp4 (-●-), proCrp4(20–92) (-○-), (Δ44–53)-proCrp4(-▼-), (Δ44–58)-proCrp4 (-▽-). For all species tested, exposure to native proCrp4 and truncated variants has no effect on bacterial cell survival.
2.5 Peptide mediated permeabilization of live E. coli
To investigate the effect of decreased prosegment length on inducing permeabilization of live Escherichia coli cells, truncated proCrp4 peptides were tested in vitro against E. coli ML35. Exponentially growing E. coli ML35 cells were washed, and resuspended in 10 mM PIPES-TSB as described [17,20]. Bacteria were incubated in triplicate with 0–6 μM of peptide and 2.5 mM 2-nitrophenyl β-D-galactopyranoside (ONPG), a chromogenic substrate. To 10 μl of ONPG and 10 μl of peptide solution, 80 μl of E. coli ML35 cells in PIPES-TSB (~ 5 × 106 CFU/mL) were incubated at 37° C for 2 h in a 96-well plate format. E. coli ML35 cells are β-galactosidase constitutive and permease-negative, so that ONPG diffusion into bacterial cells and ONP production is dependent on peptide-mediated membrane disruption. ONP was measured at 405 nm on a 96-well Spectra-Max plate spectrophotometer (Molecular Devices, Sunnyvale, CA), and data were analyzed using Sigma Plot.
2.6 Time Course of Bactericidal Peptide Activity
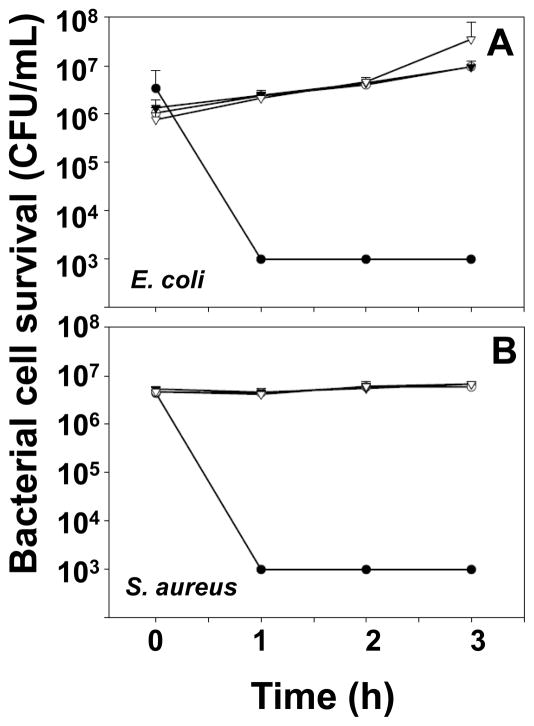
To test whether truncated proCrp4 variants would exhibit bactericidal peptide activities with extended exposure times, the peptides were assayed for activity in vitro against E. coli ML35 and S. aureus for a period of 3 h. Samples consisting of exponentially-growing bacterial cells were collected by centrifugation, washed, and resuspended in 10 mM PIPES (pH 7.4) supplemented with 0.01 vol (1% v/v) trypticase soy broth (PIPES-TSB). In triplicate experiments, bacteria (5 × 106 CFU/ml) were incubated with 3 μM of peptide in 50 μl of PIPES-TSB. After incubation time intervals of 0, 1h, 2h and 3h at 37°C, the samples were processed as described above in Section 2.4 (Figs. 8A & B).
Fig. 8. Truncated prosegment variants remain inhibited during prolonged bacterial exposure.
Exponentially-growing Escherichia coli (A) and Staphylococcus aureus (B), were exposed to peptides in triplicate assays as in Figure 6. After incubation with 3 μM peptides for 0, 1, 2 or 3 h, samples of bacterial-peptide mixtures were plated on TSB-agar and incubated overnight at 37 °C to quantitate surviving bacteria as CFU with error bars denoting standard deviation. Symbols: Crp4 (-●-), proCrp4(20–92) (-○-), (Δ44–53)-proCrp4(-▼-), (Δ44–58)-proCrp4 (-▽-). Native proCrp4 and truncated variants remain inhibited throughout the course of the assay.
3. Results
To date, most pro-α-defensins investigated have electronegative proregions that balance the cationic charge of their covalently linked defensin partners [4,11], and such propeptides lack bactericidal and membrane disruptive activities [4,9,20,25]. For example, charge neutralization or deletion of anionic amino acids located near the proregion N terminus of mouse proCrp4 relieved the inhibition of bactericidal peptide activity [23], apparently by disrupting electrostatic interactions between the proregion and defensin regions of the precursors. This, the conserved proregion chain length in pro-α-defensins, and knowledge that anionic residues near the proCrp4(20–92) N-terminus block Crp4 activity suggested that the physical distance between the inhibitory residue positions and Crp4 may provide an optimal distance required for maintaining proCrp4 in an inactive state.
3.1 Analysis of (Δ44–53)-proCrp4 and (Δ44–58)-proCrp4 peptide homogeneity
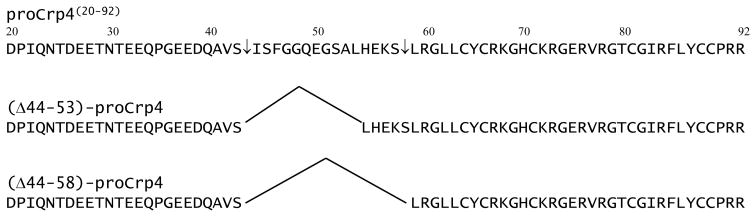
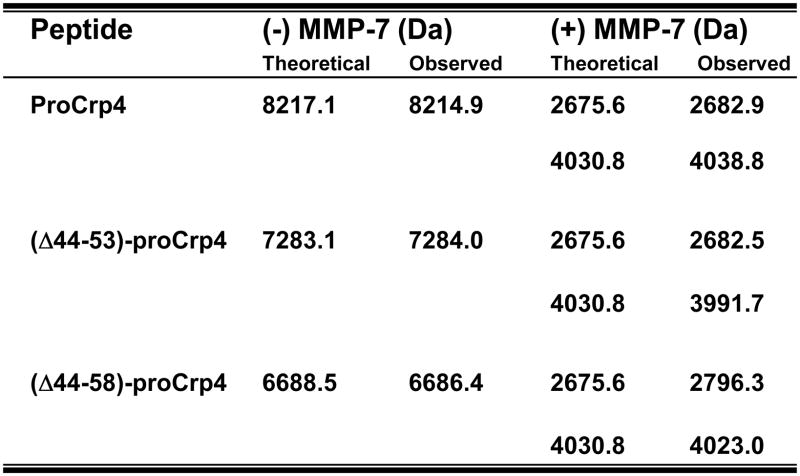
To test this hypothesis, we prepared truncated variants of proCrp4 that retained the inhibitory acidic amino acids found between proCrp4(20–44) but with the distance between those residue positions and Crp4 reduced by 10 and 15 amino acids (Fig. 1). Those deletions were chosen to correspond to proregion fragments resulting from MMP-7 proteolysis. Before the effects of these deletions on bactericidal peptide activity and membrane permeabilization could be assayed, the quality of the peptides was characterized biochemically. Subsequent to their purification by affinity chromatography and C18-HPLC [20], the homogeneity of the (Δ44–53)-proCrp4 and (Δ44–58)-proCrp4 variants was demonstrated by migration in AU-PAGE gels and compared to native Crp4 and proCrp4(20–92) (Fig. 2). Both truncated proCrp4 peptide variants had higher mobilities than full-length, native proCrp4(20–92). Because AU-PAGE resolves peptides primarily on the basis of their net cationic charge-to-size [14], and, as shown below, the Crp4 component of these molecules is folded correctly, the enhanced mobility of proCrp4 deletion mutants is likely due their reduced masses. Only modest changes in overall charge would result from the truncations in that the Δ44–53 deletion would increase charge by +1 due to the deletion of Glu50, and the net charge of (Δ44–58)-proCrp4 would be reduced by −0.5 with the removal of Glu50, His55, Glu56, and Lys57. Peptide homogeneity was further confirmed by mass spectrometry of the purified peptides. The molecular masses of truncated proCrp4 variants were measured using MALDI-TOF MS and corresponded to the theoretical values of their oxidized forms, with experimental masses of 7283.1 A.M.U. for (Δ44–53)-proCrp4 and 6688 A.M.U. for (Δ44–58)-proCrp4, attesting to the quality of the preparations (Fig. 4).
Fig 1. Recombinant proCrp4 deletion variant peptides prepared by site-directed mutagenesis.
The primary structures of the recombinant peptides prepared and assayed in this study are aligned. Lines shown in (Δ44–58)-proCrp4 and (Δ44–53)-proCrp4 sequences indicate the residue positions deleted in the truncated variants. MMP-7 cleavage sites are indicated by arrows.
Fig 2. Homogeneity of peptides investigated as assessed by AU-PAGE.
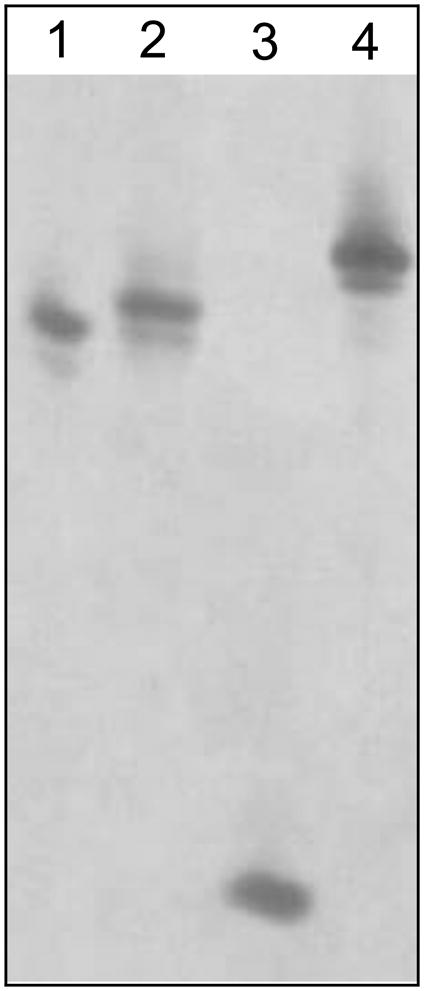
Following their purification, 2 μg samples of recombinant peptides were analyzed by AU-PAGE and stained with Coomassie Blue. Lanes: 1, proCrp4; 2, (Δ44–58)-proCrp4; 3, Crp4; 4, (Δ44–53)-proCrp4. The lower bands in lanes of proCrps denote the peptides lacking the N-terminal aspartate due to hydrolysis of the acid-labile aspartate-proline bonds [20].
Fig. 4. The masses obtained for the major processed products of both truncated and native proCrp4 are similar.
Peptide samples (10 μg) were incubated in the presence (+) or absence (−) of MMP-7 as described in Fig. 3. The digestion products were analyzed by mass spectrometry. Theoretical masses for the recombinant peptides and cleaved products corresponding to MMP-7 cleavage sites (residues 20–43 and 59–92) were identified using the ExPASy ProtParam tool. Observed masses were analyzed by MALDI-TOF-MS.
3.2 In vitro processing of truncated proCrp4 variants by MMP-7
To test whether the deleting the region between Ile44 to Ser58 in the prosegment would induce precursor misfolding and have adverse effects on proteolytic conversion by alteration of MMP-7 processing sites, the (Δ44–53)-proCrp4 and (Δ44–58)-proCrp4 peptides were analyzed after exposure to the activating convertase [1,24]. This assay for co-migration of processing products with native Crp4 is a useful index of peptide folding, because Crp4 and its misfolded peptide variants have distinct mobilities in AU-PAGE, and Crp4 is highly resistant to MMP-7-mediated proteolysis when folded correctly [1,10]. The major processed products generated were clearly evident when the deletion variants were incubated in vitro with MMP-7, as they co-migrated with Crp4 in AU-PAGE (Fig. 3) similar to products obtained on MMP-7 activation of proCrp4 [3,20,23]. As has been noted previously [1], the proregions of mouse proCrps and their MMP-7 mediated cleavage products cannot be visualized by standard peptide staining methods [3], even though cleavage sites were determined by Edman N-terminal sequencing [1]. However, both the proregion and the mature peptide were detected by MALDI-TOF-MS (Fig. 4), and the major MMP-7 conversion products of (Δ44–53)-proCrp4 and (Δ44–58)-proCrp4 were the same as those resulting from native proCrp4 activation by MMP-7 [20], as observed by MALDI-TOF-MS. Thus, reducing the distance between inhibitory proregion acidic amino acids and the Crp4 moiety in proCrp4 did not destabilize the α-defensin peptide fold or induce defective in vitro processing of Crp4 from the truncated precursors.
Fig. 3. In vitro MMP-7 mediated processing of truncated proCrp4 variants.
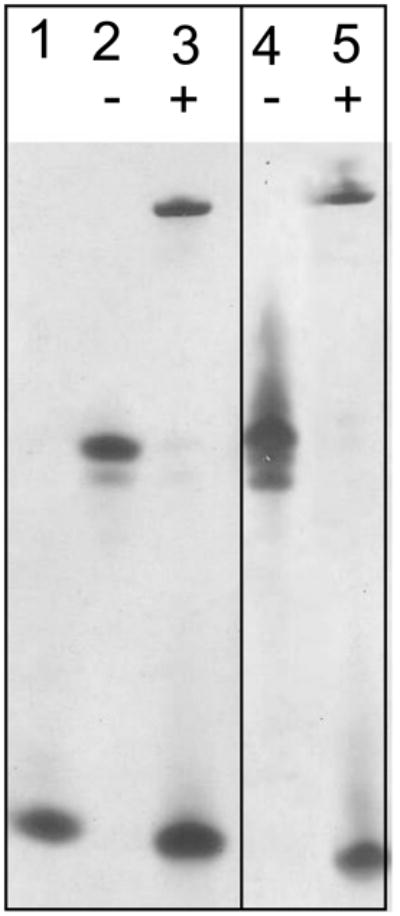
Peptide samples (10 μg) were incubated in the presence (+) or absence (−) of 0.5 molar equivalents of MMP-7 in 1 mM HEPES (pH 7.4), 15 mM NaCl, and 0.5 mM CaCl2 for 18 h at 37°C [24]. The digestion products were resolved by AU-PAGE and visualized by staining with Coomassie blue as in Fig. 2. Lanes: 1, Crp4 control peptide; 2, (Δ44–53)-proCrp4 alone; 3, (Δ44–53)-proCrp4 + MMP-7; 4, (Δ44–58)-proCrp4 alone; 5, (Δ44–58)-proCrp4 + MMP-7.
3.3 (Δ44–53)-proCrp4 and (Δ44–58)-proCrp4 lack bactericidal peptide activity
To test whether the natural proregion provides a distance between the acidic residues near the proregion N-terminus and the Crp4 moiety that is critical for precursor inhibition, the proCrp4 deletion variants (Δ44–53)-proCrp4 and (Δ44–58)-proCrp4 were tested for in vitro bactericidal peptide activity relative to native Crp4 and proCrp4(20–92). Consistent with previous results showing that (DE/G)-proCrp4(20–60) is not microbicidal, the native proregion component of the precursor, i.e., proCrp1(20–60), was assayed and shown to have no inherent bactericidal activity against E. coli (Fig. 5). If that distance were a determinant of inhibition, (Δ44–53)-proCrp4 and (Δ44–58)-proCrp4 would be predicted to have bactericidal activity. However, against the Gram positive and Gram negative bacterial species tested, e.g., S. Typhimurium CS022, S. aureus and E. coli, truncated proCrp4 variants displayed the same inhibited state as native proCrp4(20–92) (Figs. 6A–F). Although native and truncated precursors lacked activity in these assays at 10 μM peptide concentrations, we cannot exclude the possibility that higher levels of these peptides, e.g., 20 μM or greater, would have microbicidal effects. Furthermore, even against L. monocytogenes and V. cholerae, two species that are highly susceptible to cell killing by Crp4, proCrp4 and the (Δ44–53)-proCrp4 and (Δ44–58)-proCrp4 deletion variants again lacked bactericidal activity, exhibiting the same fully-inhibited property of full-length, native proCrp4(20–92) during 1 h of 10 μM peptide exposure (Figs. 6A and F).
Fig. 5. Mouse pro-α-defensin proregion, proCrp1(20–60) lacks bactericidal activity.
Exponentially-growing Escherichia coli ML35 (~ 5 × 106 CFU/mL) cells were exposed to the peptide as shown at 37 °C in 50 μl 10 mM PIPES buffer supplemented with 1% TSB for 1 h. Following peptide exposure, the bacteria were plated on TSB-agar, incubated overnight at 37 °C (see Materials and Methods, Section 2.4) [17], and surviving bacteria were counted as CFU at each peptide concentration. Values at or below 1 × 103 CFU/ml signify absence of colonies after overnight growth, the detection limit of the assay. Symbols: Crp4 (-●-), proCrp4 (-○-), proCrp1(20–60) (-▼-)
To test whether truncated (Δ44–53)-proCrp4 and (Δ44–58)-proCrp4 are less fully inhibited over longer peptide exposure times, in vitro bactericidal peptide assays were performed on bacteria incubated with peptides for up to 3 h and compared to native Crp4 and proCrp4(20–92). Against E. coli and S. aureus, Crp4 killed all bacteria at each time point. The deletion variants and native proCrp4(20–92) had no bactericidal effects over 3 h. In fact, during the 3 h incubation of E. coli with proCrp4 and truncated variants, bacterial cell numbers increased approximately 10-fold, evidence of cell proliferation over the course of the assay. S. aureus cell survival was unaffected by native or deletion proCrp4 variants, but numbers of S. aureus (CFU) did not increase over the 3 h assay (Figs. 8A and B).
3.4 (Δ44–53)-proCrp4 and (Δ44–58)-proCrp4 lack E. coli cell membrane permeabilization activity
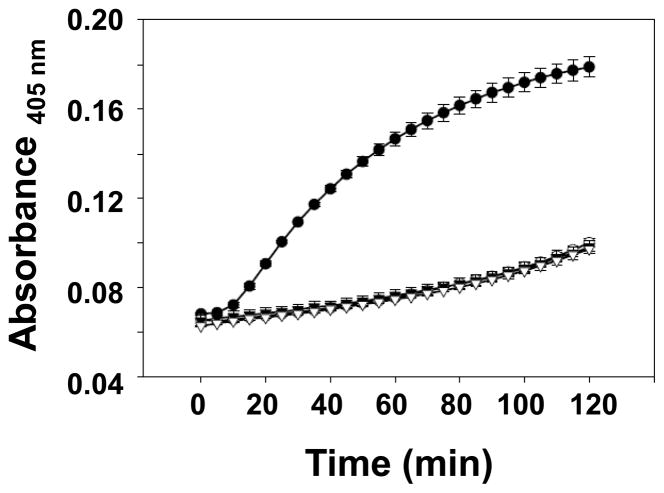
To determine whether truncations of the prosegment in proCrp4 permit sublethal permeabilization of live bacterial cells, (Δ44–53)-proCrp4 and (Δ44–58)-proCrp4 peptides were compared to Crp4 and native proCrp4 in ONPG conversion assays (see “Materials and methods”). In contrast to the robust permeabilizing effects of Crp4, (Δ44–53)-proCrp4 and (Δ44–58)-proCrp4 were unable to induce detectable permeabilization of E. coli ML35 over a 2 h exposure period (Fig. 7). Consistent with their lack of bactericidal activity, the membrane disruptive abilities of the proCrp4 variants were as fully inhibited as the native precursor as judged by the absence of detectable ONP accumulation even at 6 μM peptide concentrations (data not shown).
Fig. 7. Prosegment truncation does not enable sublethal permeabilization of live E. coli ML35 cells.
Exponentially-growing E. coli cells were exposed to 1.5 μM peptide levels in the presence of 2-nitrophenyl β-D-galactopyranoside (ONPG) for 2 h at 37 °C. Hydrolysis of ONPG by the constitutively expressed β-galactosidase of this strain was measured by absorbance at 405 nm. Symbols: Crp4 (-●-), proCrp4 (-○-), (Δ44–53)-proCrp4 (-▼-), and (Δ44–58)-proCrp4 (-▽-). Similar results were obtained in assays of 0.75 to 6 μM peptide levels.
Although the molecular contacts associated with the mechanism or proregion inhibition remain undefined, these collective findings show that the N-terminal anionic amino acid residues still inhibit the Crp4 component of the proforms, even though the spacing between these regions was reduced by approximately 50% along the polypeptide backbone. Given that proregion truncation did not alter inhibition of proCrp4, it appears that pro-α-defensin proregion chain length has not been conserved to position inhibitory residues at a minimum distance critical for blocking precursor function.
4. Discussion
These studies support the conclusion that conservation of pro-α-defensin proregion chain length is not critical to maintenance of the precursor in an inactive state. Specifically, up to fifteen residues in proCrp4 proregion were deleted to test whether an optimal distance exists between inhibitory acidic amino acids in the prosegment and the Crp4 component of the precursor. Although the truncated proforms have markedly decreased chain lengths, the shortened precursors are both processed by MMP-7 and remain biologically inactive. The effects of truncation on proCrp4 inhibition were measured in in vitro bactericidal peptide assays, which showed that proCrp4 and proCrp4 variants with deletions of ten ((Δ44–53)-proCrp4) and fifteen ((Δ44–58)-proCrp4) amino acids also lack activity against several species of bacterial cell targets, three of which are highly susceptible to killing by mouse α-defensins. Thus, as measured along the polypeptide backbone, decreasing the length of the linear proregion is not critical, per se, for proCrp4 inhibition.
The role(s) of individual prosegment residue positions in maintaining pro-α-defensin inhibition may vary as a function of the distribution of electropositive charge or the orientation of peptide amphipathicity along the α-defensin triple-stranded β-sheet structure. For example, proCrp4 and proRMAD-4 contain charge neutralizing anionic amino acids in the prosegment that have the same inhibitory effects as proCrp4(20–92) [8,23]. On the other hand, in human neutrophil pro-α-defensin proHNP1, hydrophobic, not electrostatic, interactions determine proregion-defensin binding and precursor inhibition [27]. Because the proregions of mouse pro-α-defensin primary structures are highly conserved, including sequences between residue positions Ile/Val44 to Ser58, the findings we report may be applicable to all the mouse precursors. Since membrane-disruptive pro-α-defensins could initiate an adverse innate immune response in the ER or the Golgi stack [9], perhaps by triggering an unfolded protein response [15], we speculate that pro-α-defensins that lack bactericidal and membrane disruptive activities maybe under selection.
The inferred interactions between inhibitory anionic amino acids in the N-terminal region of the precursor and the mature defensin peptide that inhibit proCrp4 are independent of proregion truncations. Preliminary solution structures of proCrp4 as determined by NMR showed the unstructured proregion folded loosely over the well-defined β-sheet structure of the Crp4 component of the precursor, apparently in sufficient proximity to inhibit activity (K.J. Rosengren, et al., unpublished). Given the disordered structure of the proCrp4 proregion, it appears that even truncated proregions have sufficient flexibility to position the inhibitory acidic amino acids at an effective distance to block the Crp4 component. Ongoing but preliminary biophysical studies show that anionic residues in the proregion are within 12 A° of cationic Crp4 residue positions in proCrp4 (S.M. Figueredo, et al., unpublished). Perhaps, the lack of an adequate prosegment length may result in inefficient folding in vivo, perhaps resulting in accumulation of misfolded pro-α-defensins that may induce ER stress. Introduction of constructs containing truncated pro-α-defensins into cell lines should provide the means for testing this hypothesis. Because membrane-disruptive interactions of proCrp4 with truncated proregions remain fully inhibited, prosegment chain length may be conserved to facilitate peptide folding or interactions with chaperones in the ER.
Acknowledgments
This work was supported by National Institutes of Health Grants DK044632, AI059346 and the Human Frontiers Science Program. We thank Dr. Michael E. Selsted for useful discussions and Ms. Xiaoqing Qu, Mr. Steven K. Young, and Ms. Valerie Le for excellent technical assistance.
Abbreviations
- AU-PAGE
acid-urea polyacrylamide gel electrophoresis
- Crps
cryptdins
- Crp4
cryptdin-4
- HD5
human defensin 5
- HNPs
human neutrophil alpha-defensins
- proCrps
procryptdins
- proCrp4(20–92)
pro-cryptdin-4(20–92)
- MALDI-TOF MS
matrix-assisted laser desorption ionization time-of-flight mass spectrometry
- MMP-7
matrix metalloproteinase -7
- PIPES
Piperazine-1,4-bis (2-ethanesulfonic acid)
- RMAD
rhesus myeloid alpha defensin
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ayabe T, Satchell DP, Pesendorfer P, Tanabe H, Wilson CL, Hagen SJ, Ouellette AJ. Activation of Paneth cell alpha-defensins in mouse small intestine. J Biol Chem. 2002;277:5219–28. doi: 10.1074/jbc.M109410200. [DOI] [PubMed] [Google Scholar]
- 2.Borregaard N, Sorensen OE, Theilgaard-Monch K. Neutrophil granules: a library of innate immunity proteins. Trends Immunol. 2007;28:340–5. doi: 10.1016/j.it.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 3.Figueredo SM, Weeks CS, Young SK, Ouellette AJ. Anionic amino acids near the pro-alpha-defensin N terminus mediate inhibition of bactericidal activity in mouse pro-cryptdin-4. J Biol Chem. 2009;284:6826–31. doi: 10.1074/jbc.M807024200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ganz T. Defensins: antimicrobial peptides of innate immunity. Nat Rev Immunol. 2003;3:710–20. doi: 10.1038/nri1180. [DOI] [PubMed] [Google Scholar]
- 5.Ganz T. Defensins: antimicrobial peptides of vertebrates. C R Biol. 2004;327:539–49. doi: 10.1016/j.crvi.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 6.Ganz T, Lehrer RI. Defensins. Curr Opin Immunol. 1994;6:584–9. doi: 10.1016/0952-7915(94)90145-7. [DOI] [PubMed] [Google Scholar]
- 7.Ghosh D, Porter E, Shen B, Lee SK, Wilk D, Drazba J, Yadav SP, Crabb JW, Ganz T, Bevins CL. Paneth cell trypsin is the processing enzyme for human defensin-5. Nat Immunol. 2002;3:583–90. doi: 10.1038/ni797. [DOI] [PubMed] [Google Scholar]
- 8.Kamdar K, Maemoto A, Qu X, Young SK, Ouellette AJ. In Vitro Activation of the Rhesus Macaque Myeloid {alpha}-Defensin Precursor proRMAD-4 by Neutrophil Serine Proteinases. J Biol Chem. 2008;283:32361–8. doi: 10.1074/jbc.M805296200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu L, Ganz T. The pro region of human neutrophil defensin contains a motif that is essential for normal subcellular sorting. Blood. 1995;85:1095–103. [PubMed] [Google Scholar]
- 10.Maemoto A, Qu X, Rosengren KJ, Tanabe H, Henschen-Edman A, Craik DJ, Ouellette AJ. Functional analysis of the alpha-defensin disulfide array in mouse cryptdin-4. J Biol Chem. 2004;279:44188–96. doi: 10.1074/jbc.M406154200. [DOI] [PubMed] [Google Scholar]
- 11.Michaelson D, Rayner J, Couto M, Ganz T. Cationic defensins arise from charge-neutralized propeptides: a mechanism for avoiding leukocyte autocytotoxicity? J Leukoc Biol. 1992;51:634–9. doi: 10.1002/jlb.51.6.634. [DOI] [PubMed] [Google Scholar]
- 12.Ouellette AJ. Paneth cell alpha-defensins: peptide mediators of innate immunity in the small intestine. Springer Semin Immunopathol. 2005;27:133–46. doi: 10.1007/s00281-005-0202-x. [DOI] [PubMed] [Google Scholar]
- 13.Ouellette AJ, Hsieh MM, Nosek MT, Cano-Gauci DF, Huttner KM, Buick RN, Selsted ME. Mouse Paneth cell defensins: primary structures and antibacterial activities of numerous cryptdin isoforms. Infect Immun. 1994;62:5040–7. doi: 10.1128/iai.62.11.5040-5047.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rice WG, Ganz T, Kinkade JM, Jr, Selsted ME, Lehrer RI, Parmley RT. Defensin-rich dense granules of human neutrophils. Blood. 1987;70:757–65. [PubMed] [Google Scholar]
- 15.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–29. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 16.Satchell DP, Sheynis T, Kolusheva S, Cummings J, Vanderlick TK, Jelinek R, Selsted ME, Ouellette AJ. Quantitative interactions between cryptdin-4 amino terminal variants and membranes. Peptides. 2003;24:1795–805. doi: 10.1016/j.peptides.2003.08.020. [DOI] [PubMed] [Google Scholar]
- 17.Satchell DP, Sheynis T, Shirafuji Y, Kolusheva S, Ouellette AJ, Jelinek R. Interactions of mouse Paneth cell alpha-defensins and alpha-defensin precursors with membranes. Prosegment inhibition of peptide association with biomimetic membranes. J Biol Chem. 2003;278:13838–46. doi: 10.1074/jbc.M212115200. [DOI] [PubMed] [Google Scholar]
- 18.Selsted ME, Harwig SS. Purification, primary structure, and antimicrobial activities of a guinea pig neutrophil defensin. Infect Immun. 1987;55:2281–6. doi: 10.1128/iai.55.9.2281-2286.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Selsted ME, Ouellette AJ. Mammalian defensins in the antimicrobial immune response. Nat Immunol. 2005;6:551–7. doi: 10.1038/ni1206. [DOI] [PubMed] [Google Scholar]
- 20.Shirafuji Y, Tanabe H, Satchell DP, Henschen-Edman A, Wilson CL, Ouellette AJ. Structural determinants of procryptdin recognition and cleavage by matrix metalloproteinase-7. J Biol Chem. 2003;278:7910–9. doi: 10.1074/jbc.M210600200. [DOI] [PubMed] [Google Scholar]
- 21.Tanabe H, Qu X, Weeks CS, Cummings JE, Kolusheva S, Walsh KB, Jelinek R, Vanderlick TK, Selsted ME, Ouellette AJ. Structure-activity determinants in paneth cell alpha-defensins: loss-of-function in mouse cryptdin-4 by charge-reversal at arginine residue positions. J Biol Chem. 2004;279:11976–83. doi: 10.1074/jbc.M310251200. [DOI] [PubMed] [Google Scholar]
- 22.Valore EV, Ganz T. Posttranslational processing of defensins in immature human myeloid cells. Blood. 1992;79:1538–44. [PubMed] [Google Scholar]
- 23.Weeks CS, Tanabe H, Cummings JE, Crampton SP, Sheynis T, Jelinek R, Vanderlick TK, Cocco MJ, Ouellette AJ. Matrix metalloproteinase-7 activation of mouse Paneth cell pro-alpha-defensins: SER43 down arrow ILE44 proteolysis enables membrane-disruptive activity. J Biol Chem. 2006;281:28932–42. doi: 10.1074/jbc.M602041200. [DOI] [PubMed] [Google Scholar]
- 24.Wilson CL, Ouellette AJ, Satchell DP, Ayabe T, Lopez-Boado YS, Stratman JL, Hultgren SJ, Matrisian LM, Parks WC. Regulation of intestinal alpha-defensin activation by the metalloproteinase matrilysin in innate host defense. Science. 1999;286:113–7. doi: 10.1126/science.286.5437.113. [DOI] [PubMed] [Google Scholar]
- 25.Wu Z, Li X, Ericksen B, de Leeuw E, Zou G, Zeng P, Xie C, Li C, Lubkowski J, Lu WY, Lu W. Impact of pro segments on the folding and function of human neutrophil alpha-defensins. J Mol Biol. 2007;368:537–49. doi: 10.1016/j.jmb.2007.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–95. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 27.Zou G, de Leeuw E, Lubkowski J, Lu W. Molecular Determinants for the Interaction of Human Neutrophil alpha Defensin 1 with its Propeptide. J Mol Biol. 2008;381:1281–91. doi: 10.1016/j.jmb.2008.06.066. [DOI] [PMC free article] [PubMed] [Google Scholar]