Abstract
Holoprosencephaly (HPE) is the most common structural malformation of the developing forebrain in humans and is typically characterized by different degrees of hemispheric separation that are often accompanied by similarly variable degrees of craniofacial and midline anomalies. HPE is a classic example of a complex genetic trait with “pseudo”-autosomal dominant transmission showing incomplete penetrance and variable expressivity. Clinical suspicion of HPE is typically based upon compatible craniofacial findings, the presence of developmental delay or seizures, or specific endocrinological abnormalities, and is then followed up by confirmation with brain imaging. Once a clinical diagnosis is made, a thorough genetic evaluation is necessary. This usually includes analysis of chromosomes by high-resolution karyotyping, clinical assessment to rule-out well recognized syndromes that are associated with HPE (e.g. Pallister-Hall syndrome, Smith-Lemli-Opitz syndrome and others), and molecular studies of the most common HPE associated genes (e.g. SHH, ZIC2 and SIX3). In this review, we provide current step-by-step recommendations that are medically indicated for the genetic evaluation of patients with newly diagnosed HPE. Moreover, we provide a brief review of several available methods used in molecular diagnostics of HPE and describe the advantages and limitations of both currently available and future tests as they relate to high throughput screening, cost, and the results that they may provide.
Keywords: holoprosencephaly, HPE, disease genes, multi-factorial inheritance, molecular diagnostics
Introduction
Holoprosencephaly (HPE) is the most common disorder of the developing forebrain in humans, occurring with a frequency of 1:250 conceptuses [Matsunaga and Shiota, 1977] and 1:10-16,000 live births [Roach et al., 1975]. The HPE phenotypic spectrum results from failure of the forebrain to cleave into two hemispheres. Different degrees of hemispheric separation, ranging from the classically described alobar form, to semilobar, lobar and middle-interhemispheric variant (MIHV) describe the anatomically distinguishable forms of HPE. The mildest end of the spectrum includes subtle midline brain anomalies. These phenotypes are often accompanied by a broad spectrum of craniofacial differences, ranging from the most severe form with cyclopia (one eye) or synophthalmia (two fused eyes) with a proboscis (nose-like appendage), to less severe forms with hypotelorism, mid-face hypoplasia or a single maxillary central incisor (SCI) [Cohen, 2006; Dubourg et al., 2007; Muenke and Beachy, 2000; reviewed in Solomon et al., this issue]. The occurrence and manifestations of HPE are influenced by both genetic causes and environmental risk factors. In cases where a specific gene is known to be causative, it is inherited as a complex trait with incomplete penetrance and variable expressivity. The basis of these phenotypic differences is largely unknown but likely reflects measured and unmeasured genetic and environmental components [Solomon et al., 2009].
Cytogenetic Alterations and Mutations of Developmental Genes are the Most Common Known Causes of HPE
It is estimated that the cause of HPE is due to cytogenetic anomalies in 30-50% of individuals, to well recognized syndromes (e.g. Smith-Lemli-Opitz syndrome (SLOS)) in ∼25%, to either environmental causes and/or unknown genetic alterations in ∼10-15%; and to mutations in established HPE gene(s) in ∼5-10% [Bullen et al., 2001; Ong et al., 2007; Dubourg et al., 2007; Roessler et al., 2009a]. Additional risk factors that may act alone or in concert with genetic alterations include the use of retinoids, statins, or alcohol during pregnancy, alterations in the biosynthesis of cholesterol, and pre-existing or gestational diabetes [Cohen and Shiota, 2002].
Mutations in at least 12 genes have been detected in patients with HPE; however, there is significant variability in the observed mutation rate of each gene (see below). The most common HPE genes were identified as mutational targets within loci defined by chromosomal rearrangements [Dubourg et al., 2004; Muenke and Beachy, 2000]. Among the best characterized HPE genes are SHH [Roessler et al., 1996], ZIC2 [Brown et al., 1998], SIX3 [Wallis et al., 1999], TGIF [Gripp et al., 2000], GLI2 [Roessler et al., 2003], PATCHED-1 [Ming et al., 2002], DISP1 [Roessler et al., 2009c]. Most CLIA-certified laboratories, both commercial fee-for service and those associated with National Institutes of Health (NIH) or similar centers, only screen the first four genes (the named HPE loci 2-5) for mutations on a routine basis. Microdeletions and microduplications have been suggested to play important roles given that some of these alterations occur in the vicinity of known HPE genes [Bendavid et al., 2009].
Currently, there is still a large proportion of individuals with non-syndromic and non-chromosomal HPE (∼20-10% of all HPE patients) in whom no specific genetic cause can be identified [Wallis and Muenke, 2000]. The general consensus regarding the etiology of HPE is that the molecular interactions and pathways are complex [Monuki, 2007], consistent with the theory that a large number of loci or genetic factors are yet to be identified and fully understood. The primary goal of this review is to describe our current recommendations for molecular genetics testing of patients with newly diagnosed HPE, the types of strategies for evaluation that are currently used, what tests are likely to be of use in the future, the advantages and limitations of these technologies, and the importance and benefits of the participation of the patients and their families in research studies.
Current Evaluation Strategy
The clinical diagnosis of HPE is confirmed by a combination of physical examination, family history, and brain imaging (MRI, CT, or ultrasound, etc.). Once HPE is clinically confirmed, parental samples should also be obtained in order to allow for a better interpretation of results in the setting of a positive cytogenetic or molecular finding in the proband.
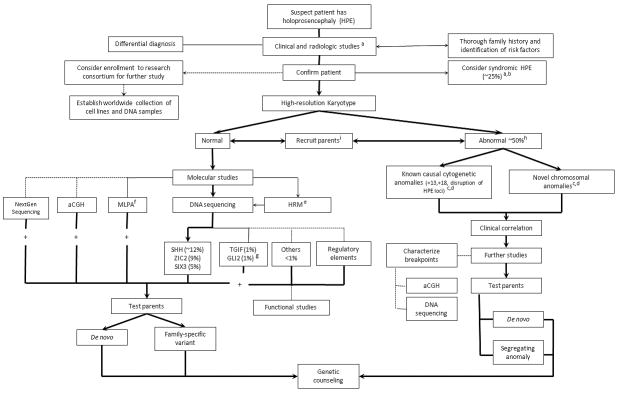
As shown in Figure 1, we propose a general strategy for the genetic evaluation of a patient newly diagnosed with HPE. Holoprosencephaly is usually diagnosed clinically based upon specific phenotypic features (described above) [Cohen, 2006; Dubourg et al., 2007; Orioli and Castilla, 2007], which typically must then be confirmed with brain imaging in order to fully characterize the anomaly [Hahn and Plawner, 2004]. A comprehensive evaluation of a patient with HPE should typically begin with cytogenetic studies, including a high-resolution karyotype with a minimum of 550 band resolution, given that ∼30-50% of patients will have a chromosomal anomaly, including deletions, duplications, but also balanced translocations and inversions that are not detected by array comparative genomic hybridization (array CGH) [Cohen, 2006; Orioli and Castilla, 2007; Bullen et al., 2001; Ong et al., 2007]. In selected patients, medically indicated studies should then be done to rule out syndromes that might cause HPE (e.g. elevated 7-dehydro-cholesterol levels in SLOS). Finally, in all nonsyndromic patients found to have normal chromosomes, molecular analysis should be performed for the most common genes implicated in HPE: SHH, ZIC2 and SIX3 [Dubourg et al., 2004; Wallis and Muenke, 2000].
Figure 1. Algorithm for the genetic study of new holoprosencephaly patients.
Bold lines refer to medically indicated tests; thin lines are optional tests depending on a specific clinical indication or the capabilities of the diagnostic laboratory; dotted lines refer to tests available in research labs that will contribute to a better understanding of HPE. For further details, see the following references: a: [Hahn et al., this issue; [Hahn and Plawner, 2004] b: [Dubourg et al., 2007] c: [Roessler and Muenke, this issue] d: [Bendavid et al., this issue] e: Refers to High-Resolution DNA Melting (HRM) f: Multiplex Ligation-dependent Probe Amplification (MLPA) g: gene specific phenotype, h: [Bullen et al., 2001; Ong et al., 2007]. i: recruit parental samples for better test interpretation in case of a positive result. +: positive diagnostic test results.
Parental samples should be obtained at the initial evaluation of a proband as the study of these individuals can be critical for the interpretation of the proband's test results and for future genetic counseling, whether or not a cytogenetic or molecular diagnosis is immediately established. The strongest predictors of the pathogenicity of new alterations relates to whether the changes are de novo gross cytogenetic, microdeletions/duplications, or mutations [reviewed in Roessler and Muenke in this issue].
For newly diagnosed patients who have an abnormal karyotype, the cytogenetic findings should be correlated with the clinical phenotype and the underlying mechanism involved. For example, well recognized trisomies involving chromosomes 13 and 18 or rearrangements that disrupt one of the major genes implicated in HPE, such as SHH or ZIC2, would be expected to contribute to the etiology of HPE [Dubourg et al., 2007]. Other chromosomal rearrangements can also occur [see Roessler and Muenke, this issue]; however currently there is little proof of the pathogenicity for the majority of them. New technologies, controlled population genetics, and functional studies should allow us to further expand our knowledge. Again, parental studies are important to define whether the anomaly is segregating through the family or if it is a de novo event. In the case of chromosomal rearrangement, more in-depth molecular analysis of the chromosomal breakpoints, using DNA sequencing or array-CGH, can be important given that the vicinity of the breakpoints produces unstable DNA with deletions and duplications frequently occurring beyond the particular locus. These additional studies will allow better characterization of the genetic alterations and phenotypic correlations, which may be helpful for genetic counseling purposes.
For the patients with a normal karyotype, DNA sequencing analysis should be performed for the most commonly identified genes associated with HPE. In general, mutations in SHH are present in ∼12% of these patients [Roessler et al., 2009a], ZIC2 in ∼9% [Roessler et al., 2009b], and SIX3 in ∼5% [Lacbawan et al., 2009]. Given the high detection rate of likely pathologic mutations, we consider these genes to be essential for a first line medical assessment. Other genes have been described to be implicated in HPE, such as TGIF (altered in ∼1% of patients) [Gripp et al., 2000; Wallis and Muenke, 2000], GLI2 (∼1%) [Roessler et al., 2003], PATCHED-1 [Ming et al., 2002], DISP1 [Roessler et al., 2009c], FOXH1, NODAL [Roessler et al., 2009d], and others. However, at the present time, we recommend that these latter genes with low mutation frequency rates among HPE patients be tested only in selected cases, or that they be referred to specialized testing centers with the requisite expertise.
One example of a specialized situation that calls for testing of GLI2 is when specific abnormalities occur in the development of the pituitary gland, in the context of variable brain and craniofacial anomalies consistent with the broad spectrum of HPE [Roessler et al., 2003; Roessler et al., 2005]. Likewise, other genes have also been shown to be associated with characteristic brain and craniofacial abnormalities [Solomon et al., this issue; Muenke Lab, unpublished data]. In these special cases where a specific phenotype is present, molecular analysis of the associated locus is considered medically indicated.
From the current molecular diagnostic perspective, exonic mutational analysis via bi-directional DNA sequencing remains the gold standard. Both fee-for-service and free-of-charge CLIA-certified testing based in research laboratories (e.g., at the National Institutes of Health in the United States) are available and give comparable results, which can be used for the clinical management of patients and for genetic counseling for the family. When a novel mutation is identified in a proband, such as a single nucleotide change, insertion, duplication, deletion or a frame-shift mutation, parental samples should subsequently be tested to assess whether the mutation is segregating in the family (familial HPE) [Solomon et al., 2009] or a de novo variant. In general, de novo mutations are more likely to be pathogenic based on functional studies [Domené et al., 2008]. However, a large proportion of patients have unique mutations that are family-specific which can make it very difficult to predict the functional consequences [Roessler et al., 2009a]. Parental studies should also be conducted even if the identified mutation has previously been associated with HPE in order to provide the family with accurate recurrence risk information.
In order to better identify which genetic variants are truly pathogenic, the identified variants must be correlated with their predicted or experimentally determined residual function. Computerized prediction algorithms may be used; however these algorithms may be inconclusive, and therefore, highly specialized functional studies based on animal models, cellular models and conservation analyses among vertebrate species are typically required. Functional consequences of changes in SIX3 [Domené et al., 2008], SHH [Roessler et al., 2009a], ZIC2 [Roessler et al., 2009b], and TGIF [El-Jaick et al., 2007], have been well illustrated [reviewed in Roessler and Muenke, this issue].
Not all variants among the HPE genes are obvious loss-of-function. Although nucleotide changes occurring in very conserved regions of the genome are more likely to cause defects through loss–of-function, further analyses are frequently necessary to determine their precise effects [Kryukov et al., 2007]. Importantly, there is also increasing evidence that gene regulatory elements and non-coding portions of HPE genes can play an important role in disease causation and would be missed by most traditional diagnostic strategies [Jeong et al., 2008].
Local genetic counseling, facilitated by the expertise of tertiary care centers (such as The Carter Centers for Brain Research in Holoprosencephaly and Related Malformations, including the National Institutes of Health (NIH)) and patient groups (“Families for HoPE,”), should be offered to families whether results of the genetic tests are negative or positive. This counseling should be performed based upon state-of-the-art evidence to help to interpret the results and their limitations. When there is inconclusive evidence about the effect of a given variant, it should be made clear to the family that the effect is uncertain.
Finally, we recommend clinicians to assess the willingness of the parents to participate in research in HPE, which is beneficial to the final and broad goal of larger knowledge and better medical management for their children. Moreover, research gives the opportunity for patients to be evaluated by an expert multidisciplinary team, which can advise parents and local physicians in the appropriate treatment of underlying disorders.
Past, Present and Future Methods and their Implications
In Table I, we present the advantages and disadvantages of several methods that have been used and that are being proposed for the molecular study of HPE. In the past, SSCP and DHPLC have been used as effective screening methods [Brown et al., 1998; Gripp et al., 2000; Dubourg et al., 2004; Roessler et al., 1996; Wallis et al., 1999]. SSCP was initially the best way to pre-screen individuals for variants for a given DNA product, however it was not an ideal technique given that some materials required special handling and training and constituted a potential hazard for the laboratory environment, and that the sensitivity of the test was low [Orita et al., 1989]. DHLPC was presented as an alternative method, as it had improved sensitivity, provided higher throughput options than SSCP, and the preparation, run and analysis of the experiments were relatively short [O'Donovan et al., 1998]. Nevertheless, due to its good sensitivity, DHLPC is still used in many diagnostic laboratories around the world.
Table I.
| Use | Method | Advantages | Disadvantages |
|---|---|---|---|
| Past | SSCP (Single strand conformational polymorphism) |
|
|
| DHPLC (Denaturing High-Performance liquid Chromatography) |
|
|
|
| Present | Automated capillary DNA sequencing |
|
|
| HRM (High-Resolution DNA Melting) |
|
|
|
| MLPA (Multiplex Ligation dependent Probe Amplification) |
|
|
|
| aCGH (Array comparative genomic hybridization) |
|
|
|
| Future | Next-Generation Sequencing |
|
|
Capillary electrophoresis DNA sequencing is the current gold standard for mutational screening of HPE genes, with its primary advantage being close to 100% sensitivity and specificity. However, data analysis is labor intensive and its interpretation may be challenging due to presence of variants of unknown significance. Furthermore, allelic drop-out and failure to detect deletions/duplications that are larger than the sequence being interrogated may occur. Although DNA sequencing is more readily available than other technologies, and it can be used with equal success on both medically-indicated and research-only genetic tests, there is still a strong need for newer methods given the extensive heterogeneity of causative HPE genes.
Multiplex ligation-dependent probe amplification (MLPA) (MRC-Holland, Amsterdam) is a relatively new molecular method to detect the occurrence of micro deletions/duplications in genes. There is a panel commercially available (SALSA MLPA kit P187 Holoprosencephaly – MCR-Holland, Amsterdam, Netherlands) with probes spanning the 8 HPE genes [Bendavid et al., 2009]. Among the limitations of this method are that it is available in only a few laboratories, a follow-up test is necessary to validate presence of dosage differences (e.g. qPCR and Fluoresce in Situ Hybridization - FISH), and it is unable to detect single nucleotide mutations or smaller deletions or duplications. There is sufficient evidence in the literature of an overwhelming number of single nucleotide mutations or small deletions/duplications causing truncated proteins. For example, in a recent study on patients with HPE and alterations in SHH, there were 125 different mutations tabulated, none of which are detected by MLPA [Roessler et al., 2009a]. Hence, copy number variations and hypothetical promoter or enhancer variations are likely to be among the least common types of variations that are likely to be detected.
High-resolution DNA melting (HRM) strategies have recently been proposed to pre-screen samples for mutations [Reed et al., 2007]. In our experience, amplicons from many individuals can be simultaneously screened from genomic DNA in roughly two hours, followed by direct sequencing of a targeted subset of presumed variants. This method promises considerable savings in terms of money and time in the identification of variants. Some of its greatest advantages are the high sensitivity and specificity (over 95% for heterozygous variants) [Wittwer, 2009], as well as its high throughput nature (up to 384 samples to be screened per run, in a Roche LightCycler 480 II instrument (Roche Appplied Science, Indianapolis, IN and Idaho Technology Inc., Salt Lake City, UT)). HRM loses efficacy when screening GC-rich amplicons due to the difficulty in denaturing them. However, denaturing solutions, such as Roche GC-RICH solution (Roche applied science, Indianapolis, IN), can be added to enhance the melting process to increase the specificity [Tindall et al., 2009]. Moreover, current protocols recommend that amplicons be limited in size, up to 400 base pairs [Wittwer, 2009].
Next-Generation (NextGen) sequencing strategies and array-CGH (aCGH) offer the promise of great amounts of information [Bejjani and Shaffer, 2006; Mardis, 2008] although it is not yet clear which of the many new methods will emerge as the most useful. With both of these new strategies, the interpretation of results should be made carefully, since miscalling of normal variants as mutations presents the risk of misinterpretation. Currently, there is not enough evidence from well-controlled studies to unambiguously differentiate disease-causing alterations from incidental copy number variants except for the ones involved in the known HPE genes [Bendavid et al., 2009]. Further research should help to mitigate these obstacles. Since these techniques are so new and rapidly changing, most of the technologies have not been FDA approved for routine clinical use, but are nevertheless currently used by commercial diagnostic laboratories, such as GeneDx (Gaithersburg, MD, USA) for the diagnostic studies of several diseases. Array-CGH may not be appropriate for use on a routine basis until there is a better understanding of the implications of copy number variants (CNVs) in the pathogenesis of HPE. As with all detection methods, presumptive positive results should be followed up by family studies, since occurrence of novel events are more likely to be pathogenic.
New technologies, such as HRM, aCGH and NextGen Sequencing, will allow for the generation of large amounts of data with sensitivities and specificities over 90%, the ability to detect CNVs that were not previously identifiable, and for the routine screening of more genes and regulatory elements to be both cheaper and faster. However, the generation of such overwhelming amounts of data by itself does not always translate into a better understanding of a disorder. Consequently, the application of these tests in a clinical context is presently limited [Bejjani and Shaffer, 2006].
Benefits of Research in Patients with Holoprosencephaly
The current knowledge of holoprosencephaly is the product of nearly four decades of research in several specialized centers. Despite that, our knowledge is still very incomplete, and many important questions remain. To better understand this complex disorder, patients and their families should continue to be encouraged to freely enroll in these studies, whether they have positive or negative genetic testing results.
The participation of a diverse set of parent-child trios, extended families and well-controlled case-control studies will allow for future work to address new genetic associations, modifier screens, and other methods aimed at better understanding how genetic interactions, genetic variations and environmental co-factors may influence the variable penetrance and expressivity of HPE traits, even when a mutation is present in a well-characterized gene. Hence, the participation of parents in the molecular evaluation of their children can have both direct and indirect benefits for HPE research.
Additionally, there are a large number of genomic variants in which the biological effects are unclear. Advances in technology and continued cooperation with research centers can often result in the development of functional tests for novel sequence variants in known and newly identified HPE genes in order to clarify the nature of such alterations and their implications in a clinical context.
Our experiences over the past decade have proven that the value of cooperation amongst multiple international testing centers goes beyond the simple ability to share methodologies and testing strategies, but that the sharing of patient data and test samples enhances the likelihood of identifying additional HPE genes and the understanding of their associated phenotypic manifestations.
One of the benefits of enrolling in a research study is that as new genes are identified, patients in whom mutations were not previously identified can be tested for the new genes. If families consent, they are notified of these novel results, giving them the opportunity to be counseled based on new state-of-the-art evidence.
In the long term, research will allow a more integral understanding of HPE, in which children and parents will directly benefit through better counseling and individualized clinical management, focusing on specific issues arising from a genetic variant and its interactions.
Call for a Holoprosencephaly Consortium
We recommend the formation of a worldwide consortium where research data, DNA samples and cell lines would be shared between the largest possible number of active investigators involved on HPE research in order to accomplish an integration of knowledge that would contribute to a thorough understanding of the clinical and genetic aspects of this disease. While no such formal organization yet exists, the rationale for such an effort is clear. The extensive genetic heterogeneity of HPE and the unresolved issues underlying its characteristic variable expressivity compel researchers in this field to cooperate with one another and to enlist the cooperation of primary care providers, patient groups and families in this effort. Some of the obvious future challenges of this proposed group will be to collect cases for large-scale studies (e.g. to establish routine functional studies based on animal or cellular models, perform family-based association studies, and case-control association studies) to dissect the genomic variants that impact on HPE incidence and severity. Large datasets increase the statistical power of such studies and enhance the certitude of the interpretations. Such an approach, in combination with the technologies mentioned above, should allow, in the future, the expansion of a more comprehensive genetic testing strategy of patients with the HPE phenotypic spectrum and their relatives.
Finally, all of these considerations contribute to difficulties in counseling families with HPE [see Odent, this issue]. The extreme heterogeneity and diverse manifestations of HPE presents considerable challenges to medical geneticists and counselors. No single algorithm is presently sufficient to explain all cases of HPE. However, we hope that by providing a guideline for the busy clinician, we can inspire the clinical genetics community to engage in fostering important research in this area. The sharing of cases and case materials should maximize the ability of clinicians to provide meaningful results to their patients for the present, as new technologies offer the future promise of an even greater understanding of this complex set of malformations.
Summary
In summary, our current recommendations for medically indicated genetic testing of families with HPE is a tiered approach with cytogenetic studies as the first layer of the algorithm (see review by Bendavid et al., this issue), since cytogenetic abnormalities make up the most common causes of HPE. Molecular testing of SHH, SIX3 and ZIC2 is the second layer of evaluation, since they explain at least 20% of non-syndromic and non-chromosomal HPE. Other genes identified in HPE should be tested as complementary studies in special cases, given their low frequency (∼1% or less). These steps should take place in the context of a discussion about whether to pursue commercial lab testing and/or enrollment in a research study.
Acknowledgments
We would like to thank the patients, families, and clinicians from around the world for their continued support of research investigations into the genetic basis of HPE and its clinical manifestations. Likewise, we would like to acknowledge Emily Kauvar (a Howard Hughes Medical Institute (HHMI) scholar) for critically reviewing this manuscript. This work was supported by the Division of Intramural Research of the National Human Genome Research Institute, the National Institutes of Health.
Biographies
Dr. Pineda-Alvarez is a Colombian trained medical graduate who is currently a Clinical Molecular Genetics fellow in the Medical Genetics Branch, National Human Genome Research Institute, National Institutes of Health, Bethesda, MD, USA.
Dr. Dubourg is a faculty member in the branch “Génétique des pathologies Liées au Développement”, UMR 6061 CNRS, IGDR, University of Rennes 1, Faculty of Medicine, Rennes, France, and a hospital member in the Laboratory of Molecular Genetics, CHU Pontchaillou, Rennes, France. Her diagnostic and research interests include holoprosencephaly (HPE) and mental retardation.
Professor David is the chief of Holoprosencephaly group in the Branch of “genetics of developmental pathologies”, UMR 6061 CNRS IGDR, Faculty of Medicine, University of Rennes 1, France. She is the chief of the Molecular Diagnosis laboratory, CHU Pontchaillou, Rennes, France. Her research interest mainly concerns HPE, and her clinical research also focuses on iron overload genetic diseases.
Dr. Roessler is a faculty member in the Medical Genetics Branch, National Human Genome Research Institute, National Institutes of Health, Bethesda, MD, USA. His research interests include malformations of the forebrain associated with HPE as well as disturbances in organ sidedness, or laterality.
Dr. Muenke is the Branch Chief of the Medical Genetics Branch. His research interests include HPE, craniofacial malformation syndromes, and Attention Deficit Hyperactivity Disorder (ADHD).
References
- Bejjani BA, Shaffer LG. Application of array-based comparative genomic hybridization to clinical diagnostics. J Mol Diagn. 2006;8:528–533. doi: 10.2353/jmoldx.2006.060029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendavid C, Rochard L, Dubourg C, Seguin J, Gicquel I, Pasquier L, Vigneron J, Laquerriere A, Marcorelles P, Jeanne-Pasquier C, Rouleau C, Jaillard S, Mosser J, Odent S, David V. Array-CGH analysis indicates a high prevalence of genomic rearrangements in holoprosencephaly: an updated map of candidate loci. Hum Mutat. 2009;30:1175–1182. doi: 10.1002/humu.21016. [DOI] [PubMed] [Google Scholar]
- Brown SA, Warburton D, Brown LY, Yu CY, Roeder ER, Stengel-Rutkowski S, Hennekam RC, Muenke M. Holoprosencephaly due to mutations in ZIC2, a homologue of Drosophila odd-paired. Nat Genet. 1998;20:180–183. doi: 10.1038/2484. [DOI] [PubMed] [Google Scholar]
- Bullen PJ, Rankin JM, Robson SC. Investigation of the epidemiology and prenatal diagnosis of holoprosencephaly in the North of England. Am J Obstet Gynecol. 2001;184:1256–1262. doi: 10.1067/mob.2001.111071. [DOI] [PubMed] [Google Scholar]
- Cohen MM., Jr Holoprosencephaly: clinical, anatomic, and molecular dimensions. Birth Defects Res A Clin Mol Teratol. 2006;76(9):658–673. doi: 10.1002/bdra.20295. [DOI] [PubMed] [Google Scholar]
- Cohen MM, Jr, Shiota K. Teratogenesis of holoprosencephaly. Am J Med Genet. 2002;109:1–15. doi: 10.1002/ajmg.10258. [DOI] [PubMed] [Google Scholar]
- Domene S, Roessler E, El-Jaick KB, Snir M, Brown JL, Velez JI, Bale S, Lacbawan F, Muenke M, Feldman B. Mutations in the human SIX3 gene in holoprosencephaly are loss of function. Hum Mol Genet. 2008;17(24):3919–3928. doi: 10.1093/hmg/ddn294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubourg C, Bendavid C, Pasquier L, Henry C, Odent S, David V. Holoprosencephaly. Orphanet J Rare Dis. 2007;2:8. doi: 10.1186/1750-1172-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubourg C, Lazaro L, Pasquier L, Bendavid C, Blayau M, Le Duff F, Durou MR, Odent S, David V. Molecular screening of SHH, ZIC2, SIX3, and TGIF genes in patients with features of holoprosencephaly spectrum: Mutation review and genotype-phenotype correlations. Hum Mutat. 2004;24:43–51. doi: 10.1002/humu.20056. [DOI] [PubMed] [Google Scholar]
- El-Jaick KB, Powers SE, Bartholin L, Myers KR, Hahn J, Orioli IM, Ouspenskaia M, Lacbawan F, Roessler E, Wotton D, Muenke M. Functional analysis of mutations in TGIF associated with holoprosencephaly. Mol Genet Metab. 2007;90:97–111. doi: 10.1016/j.ymgme.2006.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gripp KW, Wotton D, Edwards MC, Roessler E, Ades L, Meinecke P, Richieri-Costa A, Zackai EH, Massague J, Muenke M, Elledge SJ. Mutations in TGIF cause holoprosencephaly and link NODAL signalling to human neural axis determination. Nat Genet. 2000;25:205–208. doi: 10.1038/76074. [DOI] [PubMed] [Google Scholar]
- Hahn JS, Plawner LL. Evaluation and management of children with holoprosencephaly. Pediatr Neurol. 2004;31:79–88. doi: 10.1016/j.pediatrneurol.2004.03.006. [DOI] [PubMed] [Google Scholar]
- Jeong Y, Leskow FC, El-Jaick K, Roessler E, Muenke M, Yocum A, Dubourg C, Li X, Geng X, Oliver G, Epstein DJ. Regulation of a remote Shh forebrain enhancer by the Six3 homeoprotein. Nat Genet. 2008;40:1348–53. doi: 10.1038/ng.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryukov GV, Pennacchio LA, Sunyaev SR. Most rare missense alleles are deleterious in humans: implications for complex disease and association studies. Am J Hum Genet. 2007;80:727–739. doi: 10.1086/513473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacbawan F, Solomon BD, Roessler E, El-Jaick K, Domene S, Velez JI, Zhou N, Hadley D, Balog JZ, Long R, Fryer A, Smith W, Omar S, McLean SD, Clarkson K, Lichty A, Clegg NJ, Delgado MR, Levey E, Stashinko E, Potocki L, Vanallen MI, Clayton-Smith J, Donnai D, Bianchi DW, Juliusson PB, Njolstad PR, Brunner HG, Carey JC, Hehr U, Musebeck J, Wieacker PF, Postra A, Hennekam RC, van den Boogaard MJ, van Haeringen A, Paulussen A, Herbergs J, Schrander-Stumpel CT, Janecke AR, Chitayat D, Hahn J, McDonald-McGinn DM, Zackai EH, Dobyns WB, Muenke M. Clinical spectrum of SIX3-associated mutations in holoprosencephaly: correlation between genotype, phenotype and function. J Med Genet. 2009;46:389–398. doi: 10.1136/jmg.2008.063818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis ER. The impact of next-generation sequencing technology on genetics. Trends Genet. 2008;24:133–141. doi: 10.1016/j.tig.2007.12.007. [DOI] [PubMed] [Google Scholar]
- Matsunaga E, Shiota K. Holoprosencephaly in human embryos: epidemiologic studies of 150 cases. Teratology. 1977;16:261–272. doi: 10.1002/tera.1420160304. [DOI] [PubMed] [Google Scholar]
- Ming JE, Kaupas ME, Roessler E, Brunner HG, Golabi M, Tekin M, Stratton RF, Sujansky E, Bale SJ, Muenke M. Mutations in PATCHED-1, the receptor for SONIC HEDGEHOG, are associated with holoprosencephaly. Hum Genet. 2002;110:297–301. doi: 10.1007/s00439-002-0695-5. [DOI] [PubMed] [Google Scholar]
- Monuki ES. The morphogen signaling network in forebrain development and holoprosencephaly. J Neuropathol Exp Neurol. 2007;66:566–575. doi: 10.1097/nen.0b013e3180986e1b. [DOI] [PubMed] [Google Scholar]
- Muenke M, Beachy PA. Genetics of ventral forebrain development and holoprosencephaly. Curr Opin Genet Dev. 2000;10:262–269. doi: 10.1016/s0959-437x(00)00084-8. [DOI] [PubMed] [Google Scholar]
- O'Donovan MC, Oefner PJ, Roberts SC, Austin J, Hoogendoorn B, Guy C, Speight G, Upadhyaya M, Sommer SS, McGuffin P. Blind analysis of denaturing high-performance liquid chromatography as a tool for mutation detection. Genomics. 1998;52:44–49. doi: 10.1006/geno.1998.5411. [DOI] [PubMed] [Google Scholar]
- Ong S, Tonks A, Woodward ER, Wyldes MP, Kilby MD. An epidemiological study of holoprosencephaly from a regional congenital anomaly register: 1995-2004. Prenat Diagn. 2007;27:340–347. doi: 10.1002/pd.1677. [DOI] [PubMed] [Google Scholar]
- Orioli IM, Castilla EE. Clinical epidemiologic study of holoprosencephaly in South America. Am J Med Genet A. 2007;143A:3088–3099. doi: 10.1002/ajmg.a.32104. [DOI] [PubMed] [Google Scholar]
- Orita M, Iwahana H, Kanazawa H, Hayashi K, Sekiya T. Detection of polymorphisms of human DNA by gel electrophoresis as single-strand conformation polymorphisms. Proc Natl Acad Sci U S A. 1989;86:2766–2770. doi: 10.1073/pnas.86.8.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed GH, Kent JO, Wittwer CT. High-resolution DNA melting analysis for simple and efficient molecular diagnostics. Pharmacogenomics. 2007;8(6):597–608. doi: 10.2217/14622416.8.6.597. [DOI] [PubMed] [Google Scholar]
- Roach E, Demyer W, Conneally PM, Palmer C, Merritt AD. Holoprosencephaly: birth data, benetic and demographic analyses of 30 families. Birth Defects Orig Artic Ser. 1975;11:294–313. [PubMed] [Google Scholar]
- Roessler E, Belloni E, Gaudenz K, Jay P, Berta P, Scherer SW, Tsui LC, Muenke M. Mutations in the human Sonic Hedgehog gene cause holoprosencephaly. Nat Genet. 1996;14:357–360. doi: 10.1038/ng1196-357. [DOI] [PubMed] [Google Scholar]
- Roessler E, Du YZ, Mullor JL, Casas E, Allen WP, Gillessen-Kaesbach G, Roeder ER, Ming JE, Ruiz i Altaba A, Muenke M. Loss-of-function mutations in the human GLI2 gene are associated with pituitary anomalies and holoprosencephaly-like features. Proc Natl Acad Sci U S A. 2003;100:13424–13429. doi: 10.1073/pnas.2235734100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessler E, El-Jaick KB, Dubourg C, Velez JI, Solomon BD, Pineda-Alvarez DE, Lacbawan F, Zhou N, Ouspenskaia M, Paulussen A, Smeets HJ, Hehr U, Bendavid C, Bale S, Odent S, David V, Muenke M. The mutational spectrum of holoprosencephaly-associated changes within the SHH gene in humans predicts loss-of-function through either key structural alterations of the ligand or its altered synthesis. Hum Mutat. 2009a doi: 10.1002/humu.21090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessler E, Ermilov AN, Grange DK, Wang A, Grachtchouk M, Dlugosz AA, Muenke M. A previously unidentified amino-terminal domain regulates transcriptional activity of wild-type and disease-associated human GLI2. Hum Mol Genet. 2005;14:2181–2188. doi: 10.1093/hmg/ddi222. [DOI] [PubMed] [Google Scholar]
- Roessler E, Lacbawan F, Dubourg C, Paulussen A, Herbergs J, Hehr U, Bendavid C, Zhou N, Ouspenskaia M, Bale S, Odent S, David V, Muenke M. The full spectrum of holoprosencephaly-associated mutations within the ZIC2 gene in humans predicts loss-of-function as the predominant disease mechanism. Hum Mutat. 2009b;30:E541–554. doi: 10.1002/humu.20982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessler E, Ma Y, Ouspenskaia MV, Lacbawan F, Bendavid C, Dubourg C, Beachy PA, Muenke M. Truncating loss-of-function mutations of DISP1 contribute to holoprosencephaly-like microform features in humans. Hum Genet. 2009c;125:393–400. doi: 10.1007/s00439-009-0628-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessler E, Pei W, Ouspenskaia MV, Karkera JD, Velez JI, Banerjee-Basu S, Gibney G, Lupo PJ, Mitchell LE, Towbin JA, Bowers P, Belmont JW, Goldmuntz E, Baxevanis AD, Feldman B, Muenke M. Cumulative ligand activity of NODAL mutations and modifiers are linked to human heart defects and holoprosencephaly. Mol Genet Metab. 2009d;98:225–234. doi: 10.1016/j.ymgme.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon BD, Lacbawan F, Jain M, Domene S, Roessler E, Moore C, Dobyns WB, Muenke M. A novel SIX3 mutation segregates with holoprosencephaly in a large family. Am J Med Genet A. 2009;149A:919–925. doi: 10.1002/ajmg.a.32813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tindall EA, Petersen DC, Woodbridge P, Schipany K, Hayes VM. Assessing high-resolution melt curve analysis for accurate detection of gene variants in complex DNA fragments. Hum Mutat. 2009;30:876–883. doi: 10.1002/humu.20919. [DOI] [PubMed] [Google Scholar]
- Wallis D, Muenke M. Mutations in holoprosencephaly. Hum Mutat. 2000;16:99–108. doi: 10.1002/1098-1004(200008)16:2<99::AID-HUMU2>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Wallis DE, Roessler E, Hehr U, Nanni L, Wiltshire T, Richieri-Costa A, Gillessen-Kaesbach G, Zackai EH, Rommens J, Muenke M. Mutations in the homeodomain of the human SIX3 gene cause holoprosencephaly. Nat Genet. 1999;22:196–198. doi: 10.1038/9718. [DOI] [PubMed] [Google Scholar]
- Wittwer CT. High-resolution DNA melting analysis: advancements and limitations. Hum Mutat. 2009;30:857–859. doi: 10.1002/humu.20951. [DOI] [PubMed] [Google Scholar]