Abstract
LepA is a translational GTPase highly conserved in bacterial lineages. While it has been shown that LepA can catalyze reverse ribosomal translocation in vitro, the role of LepA in the cell remains unclear. Here, we show that deletion of the lepA gene (ΔlepA) in E. coli causes hypersensitivity to potassium tellurite and penicillin G, but has no appreciable effect on growth under many other conditions. ΔlepA does not increase miscoding or frameshifting errors under normal or stress conditions, indicating that LepA does not contribute to the fidelity of translation. Overexpression of LepA interferes with tmRNA-mediated peptide tagging and A-site mRNA cleavage, suggesting that LepA is a bona fide translation factor that can act on stalled ribosomes with a vacant A site in vivo. Together these results lead us to hypothesize that LepA is involved in co-translational folding of proteins that are otherwise vulnerable to tellurite oxidation.
Keywords: ribosome, translation, reverse translocation, EF4, BipA
Introduction
Protein synthesis relies on several related GTPases (translational GTPases) that have in common two tandem structural domains, designated I and II [1, 2]. In bacteria, four translational GTPases—initiation factor IF2, elongation factors EF-Tu and EF-G, and release factor RF3— carry out well-established roles in protein synthesis. With the exception of RF3, these factors are essential for cell viability and conserved in all three domains of life.
A number of other translational GTPases have been identified in bacteria whose physiological roles are less clear. LepA (or EF4) is an EF-G paralog encoded upstream of the leader peptidase gene (lepB) in E. coli [3, 4]. Although LepA is widely conserved among bacteria and bacterium-derived organelles, the lepA gene can be deleted from the chromosome of E. coli without an obvious effect in growth rate [5]. LepA bears considerable similarity to EF-G but localizes to the membrane [6]. Sequence and structural data show that the protein domains of LepA are homologous to domains I, II, III, and V of EF-G [7]. LepA lacks regions corresponding to domain IV and sub-domain G', but has a unique C-terminal domain which is unlike any other known proteins. In 2006, Nierhaus and coworkers showed that LepA can catalyze reverse translocation in vitro (i.e., movement of tRNAs from the P and E sites to the A and P sites, respectively) [8]. Cryo-EM reconstructions revealed that LepA binds the ribosome in the same orientation as EF-G, but the lack of domain IV allows LepA to bind simultaneously with A-site tRNA [9]. The C-terminal domain of LepA has a positively-charged surface that interacts extensively with A-site tRNA in the complex, and this interaction may bias tRNA-mRNA movement in the reverse direction [7, 9]. It was also shown by the Nierhaus group that addition of purified LepA can increase the fraction of active Green Fluorescent Protein (GFP) synthesized in a coupled in vitro transcription/translation system [8]. The authors proposed that the increase in GFP activity resulted from an increase in the translation fidelity. According to their model, by promoting reverse translocation, LepA can rescue “defective” posttranslocation complexes that are particularly prone to miscoding. A separate study indicated that LepA of mitochondria (Guf1) plays an important role in the biogenesis of functional cytochrome oxidase in S. cerevisiae [10]. Under suboptimal growth conditions, the absence of LepA/Guf1 decreased the activity of cytochrome oxidase to 10% of the control strain without a substantial decrease in the protein level. These authors suggested that this loss of activity could be due to errors in protein synthesis, consistent with the Nierhaus model, or to a defect in co-translational protein folding and assembly. But the question of whether LepA contributes to translational fidelity in vivo was not experimentally addressed in either study.
BipA is another paralog of EF-G conserved in most bacterial lineages [1, 11]. BipA has the same domain architecture as LepA but a distinct C-terminal domain. BipA exhibits ribosome-stimulated GTPase activity, binds 70S ribosomes in the presence GDPNP, and binds 30S subunits in the presence of the ppGpp [12, 13]. The ΔbipA phenotype is pleiotropic, with defects in cell motility, expression of the K5 capsule system, growth at low temperatures, and resistance to certain antimicrobial peptides [11, 14-17]. While it is clear that BipA influences several processes and the expression of various genes in the cell, the mechanism by which it acts remains unknown. At least two phenotypes of ΔbipA (cold sensitivity and altered capsule synthesis) are suppressed by mutations in rluC, which is responsible for pseudouridylation at positions 955, 2504, and 2580 of 23S rRNA [18]. Base substitutions at these three positions of 23S rRNA are sufficient for suppression, suggesting that the physiological effects of BipA depend on its interaction with the ribosome.
In this study, we find that ΔlepA causes hypersensitivity to potassium tellurite but not to other oxidizing agents, suggesting that ΔlepA increases the accessibility of thiol groups in the cell. Neither ΔlepA nor ΔbipA has an appreciable effect on missense, nonsense, or frameshift suppression, indicating that the accuracy of translation depends on neither LepA nor BipA. Both of these factors can inhibit tmRNA-tagging in vivo, providing evidence that they act as elongation factors, but only LepA inhibits A-site mRNA cleavage. Based on these results, potential functions of LepA in regulating protein biogenesis are discussed.
Materials and methods
Strains, plasmids, and growth conditions
ΔlepA and ΔbipA strains (JW2553 and JW5771, respectively) and their parental strain BW25113 (rrnB3 ΔlacZ4787 hsdR514 Δ[araBAD]567 Δ[rhaBAD]568 rph-1) were obtained from the Keio collection (National Institute of Genetics, Japan) [19]. In each case, the gene was replaced by an in-frame kanamycin resistance gene. For complementation experiments, a ~2.9-kb HindIII fragment of the genomic region encompassing lepA, but not lepB, was cloned into pSC101-derived low-copy-number vector pWSK29 [20] to generate pLEPA. Plasmids for dual-luciferase assays were described previously [21, 22]. Potassium tellurite, penicillin G, methyl viologen (paraquat), and phenazine methosulfate were purchased from Sigma. For growth rate measurements, overnight cultures were diluted by 500-fold into 50 ml of LB medium supplemented with chemicals as indicated. The flasks (250 mL) were shaken at 200 rpm at 37°C, and 1 ml samples were collected for optical density measurements.
Derivatives of E. coli strain X90 (F’ lacIq lac’ pro’/ara Δ[lac-pro] nalA argE[am] rifr thi-1) were used for A-site mRNA cleavage and tmRNA tagging studies [23]. Plasmid pFLAG-mYbeL(PP) expresses the C-terminal 49 residues of ybeL(PP) fused to an N-terminal FLAG epitope under control of the T7 promoter. Plasmid pHis6-YbeL(PP) expresses an N-terminal His6-tagged version of YbeL(PP) under control of the T7 promoter. For overexpression, the lepA and bipA genes were cloned into pCH450, a pACYC184-derived vector containing the PBAD promoter [24].
RT-PCR
Overnight cultures of control and ΔlepA strains were diluted by 500-fold into 20 ml of LB medium. The flasks were shaken at 200 rpm at 37°C until OD550 was 0.3. Cells (~109 cfu) were harvested and total RNA was isolated using RiboPure-Bacteria Kit (Ambion, TX). Complementary DNA strands were synthesized by M-MLV reverse transcriptase (USB, OH) with gene-specific primers, and the gene fragments were amplified using Taq DNA polymerase (New England Biolabs, MA).
Translation fidelity in vivo
BW25113 (control), JW2553 (ΔlepA), and JW5771 (ΔbipA) cells transformed with Rluc-Fluc reporter plasmids [21, 22] were cultured to mid-log phase in 1 ml of LB with or without additives as indicated, the activities of Rluc and Fluc were measured, and the frequency of either missense suppression, stop-codon readthrough, or frameshifting were determined as described [21, 22]. The frequency of the prfB programmed frameshifting was measured by using a prfB’-lacZ reporter as described [21, 25].
Analysis of protein and mRNA
Overnight cultures were diluted into 50 ml of LB medium supplemented with 150 μg/ml ampicillin and 25 μg/ml tetracycline and cells were incubated at 37 °C with aeration. LepA and BipA expression was induced with 0.4% L-arabinose, and uninduced control cultures were supplemented with 0.4% glucose. SDS-PAGE analysis indicated that LepA and BipA were overexpressed to similar levels under these conditions. At OD600 of 0.7 - 0.9, IPTG was added to a final concentration of 1.5 mM to induce the synthesis of YbeL. After incubation for 90 min, cultures were poured onto an equal volume of ice, cells collected by centrifugation, and cell pellets frozen at -80 °C. His6-tagged proteins were purified by Ni2+-affinity chromatography and analyzed by SDS-PAGE as described previously [26]. Total RNA was isolated and analyzed as described [27].
Results
ΔlepA confers hypersensitivity to potassium tellurite
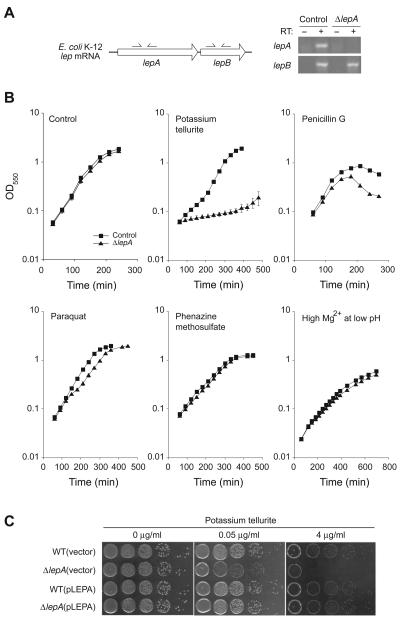
Only a few phenotypes have been attributed to null mutations of lepA, including lethality at low pH in Helicobacter pylori [28] and heat/cold sensitivity in yeast [10], despite wide conservation of the gene [8]. To further investigate the role of LepA, E. coli strains BW25113 (control) and JW2553 (ΔlepA) were analyzed by high-throughput Phenotype Microarrays (Biolog, Inc., Hayward, CA), testing ~1,200 independent growth conditions [29]. Strain JW2553 contains an in-frame kanamycin resistance gene in place of lepA [19]. As expected, RT-PCR analysis of this strain revealed a product corresponding to lepB but not lepA (Fig. 1A). The phenotypic microarray indicated that ΔlepA causes a growth defect in only a few conditions. These included growth on Glu-Asp as the sole carbon source, growth in the presence of potassium tellurite, and growth in the presence of penicillin G. We further investigated the effects of potassium tellurite and penicillin G by independently monitoring growth of the ΔlepA mutant in the presence of 0.05 μg/ml potassium tellurite or 2 μg/ml penicillin G (Fig. 1B). While the ΔlepA mutant grew as well as the control strain in LB medium, its growth was clearly inhibited in the presence of potassium tellurite. In addition, the ΔlepA mutant was somewhat more sensitive to penicillin G. To verify that the tellurite hypersensitivity was due to the absence of lepA, we transformed ΔlepA cells with plasmid pLEPA, carrying a genomic fragment with lepA and its promoter but no flanking genes. Growth of the complemented strain was indistinguishable from a control strain (Fig. 1C), indicating that ΔlepA was solely responsible for the tellurite sensitivity phenotype.
Figure 1. Phenotypes of a ΔlepA strain.
(A) RT-PCR of the lep transcript in ΔlepA cells. Genomic organization of the lep operon is shown in the left panel. Total RNA was extracted from control or ΔlepA cells in mid-log phase and amplified by gene-specific primers (indicated by thin arrows on each gene). (B) Growth curves of a wild-type and ΔlepA strain in LB medium at 37°C in the absence (control) or presence of 0.05 μg/ml potassium tellurite, 20 μg/ml penicillin G, 40 μg/ml paraquat, 15 μg/ml phenazine methosulfate, or 100 mM MgCl2 (as indicated). In the latter case, the pH of media was adjusted to 5 with HCl prior to inoculation. All data points in the figure include error bars; many of them are smaller than the symbols. (C) Complementation of phenotypes of ΔlepA cells by plasmid pLEPA harboring the lepA gene. Overnight culture of wild-type and ΔlepA strains carrying either empty vector or pLEPA were diluted in LB medium by a factor of 102, 104, 105, 106, and 107. A 10 μl aliquot of each dilution was spotted on LB plates containing 0.05 or 0.4 μg/ml potassium tellurite, and the plates were incubated at 37°C until colonies became visible.
Tellurite toxicity is thought to be due to oxidization of reduced thiol groups in the cell or generation of superoxide radicals [30, 31]. To determine if the ΔlepA mutant is susceptible to oxidative stress in general, we tested the effects of other superoxide radical generators. Paraquat (40 μg/ml) and phenazine methosulfate (15 μg/ml) had little or no effect on growth of the ΔlepA mutant compared to the control (Fig. 1B). In line with these observations, data from Phenotype Microarrays showed no significance difference in the growth rates of the ΔlepA and control strains in the presence of a variety of superoxide radical generators, such as menadione and plumbagin, as well as paraquat (data not shown).
Previous work suggested that LepA is critical for growth in media containing high concentrations of salts (e.g., 100 mM MgCl2 or 200 mM KCl) [8, 32]. However, we observed no growth defect of strain JW2553 (ΔlepA) in neutral or acidic (pH 5) LB media supplemented with 100 mM MgCl2 (Fig. 1B, data not shown). The reason for this discrepancy is unclear, but differences in the nature of the null mutations might have contributed to these results.
LepA has no apparent role in translation fidelity in vivo
Based on in vitro experiments, it was proposed that LepA functions to increase the fidelity of protein synthesis [8]. To test this idea, we first measured the frequency of miscoding in control, ΔlepA, and ΔbipA strains using a dual-luciferase reporter system developed previously [21, 22]. This system employs a gene fusion of Rluc (encoding Renilla luciferase) and Fluc (encoding firefly luciferase) in which codon 529 of Fluc is changed. Codon 529 corresponds to an essential active-site Lys, and hence the specific activity of Fluc relative to that of Rluc reflects the level of miscoding of the codon with Lys-tRNALys. Suppression of missense and nonsense mutations was unaffected by ΔlepA or ΔbipA (Tables 1-3), arguing against a role for either factor in decoding fidelity. To test if stress conditions reveal any effect of ΔlepA on translation fidelity that was otherwise masked, we supplemented the medium with potassium tellurite (0.05 μg/ml) or MgCl2 (100 mM) and HCl (to pH 5) and measured the miscoding frequency. Again, neither ΔlepA nor ΔbipA increased the frequency of the decoding errors (Table 1). Addition of streptomycin, a well-known miscoding agent, significantly increased the miscoding frequency for each of the constructs, confirming that decoding errors can be readily detected in this experimental system.
Table 1.
Effects of ΔlepA and ΔbipA on miscoding
| Frequency (× 10−4)b |
|||||
|---|---|---|---|---|---|
| Medium | Plasmid | Codona | Control | ΔlepA | ΔbipA |
| LB | pEK5 | UUU | 1.4 ± 0.2 | 1.3 ± 0.1 | 1.7 ± 0.2 |
| pEK6 | AAC | 1.7 ± 0.2 | 1.1 ± 0.1 | 1.3 ± 0.2 | |
| pEK7 | AAU | 6.0 ± 0.6 | 4.9 ± 0.4 | 5.5 ± 0.2 | |
| pEK13 | AGA | 16 ± 1 | 15 ± 1 | 13 ± 1 | |
| pEK24 | AGG | 13 ± 2 | 13 ± 2 | 10 ± 1 | |
| pEK15 | UAG | 29 ± 5 | 31 ± 3 | 22 ± 2 | |
| LB | pEK5 | UUU | 2.2 ± 0.2 | 2.4 ± 0.5 | 2.8 ± 0.3 |
| + 0.05 μg/ml | pEK6 | AAC | 2.2 ± 1 | 1.5 ± 0.3 | 2.3 ± 0.3 |
| K2TeO3 | pEK7 | AAU | 11 ± 1 | 12 ± 3 | 14 ± 2 |
| pEK13 | AGA | 20 ± 2 | 19 ± 3 | 23 ± 2 | |
| pEK24 | AGG | 15 ± 3 | 15 ± 2 | 21 ± 3 | |
| pEK15 | UAG | 30 ± 3 | 31 ± 6 | 30 ± 3 | |
| LB, low pHc | pEK5 | UUU | 2.3 ± 0.3 | 1.9 ± 0.4 | 2.2 ± 0.2 |
| + 0.1 M | pEK6 | AAC | 2.2 ± 0.6 | 1.8 ± 0.2 | 3.2 ± 0.7 |
| MgCl2 | pEK7 | AAU | 18 ± 3 | 12 ± 1 | 17 ± 1 |
| pEK13 | AGA | 31 ± 4 | 24 ± 3 | 29 ± 1 | |
| pEK24 | AGG | 12 ± 2 | 8.3 ± 1.2 | 12 ± 2 | |
| pEK15 | UAG | 8.7 ± 1.3 | 4.8 ± 0.5 | 8.3 ± 2.0 | |
| LB | pEK5 | UUU | 4.8 ± 1.5 | 3.4 ± 0.6 | 4.1 ± 0.9 |
| + 2 μg/ml | pEK6 | AAC | 15 ± 4 | 15 ± 2 | 12 ± 3 |
| streptomycin | pEK7 | AAU | 110 ± 30 | 99 ± 13 | 82 ± 17 |
| pEK13 | AGA | 49 ± 11 | 45 ± 5 | 51 ± 14 | |
| pEK24 | AGG | 43 ± 10 | 39 ± 5 | 40 ± 9 | |
| pEK15 | UAG | 120 ± 30 | 76 ± 7 | 78 ± 16 | |
Codon corresponding to the amino acid 529 of firefly luciferase. Differences from a Lys codon (AAA or AAG) are underlined.
Miscoding frequency is calculated as relative Fluc activity (i.e. Fluc/Rluc) from the indicated reporter plasmid divided by that from pEK4, with AAA (wild-type) codon as 529, as described [22]. The data represent the quotient ± standard error from at least three independent experiments.
pH of media was adjusted to 5 with HCl prior to inoculation.
Table 3.
Effects of ΔlepA and ΔbipA on +1 programmed frameshifting
| β-galactosidase activity (Miller units)b |
|||||
|---|---|---|---|---|---|
| Medium | Plasmida | Frameshift | Control | ΔlepA | ΔbipA |
| LB | pJC27 | 0 | 8300 ± 200 | 8600 ± 100 | 8300 ± 10 |
| pJC216 | +1 | 5300 ± 100 | 5500 ± 100 | 5700 ± 200 | |
The plasmids contain prfB'-lacZ with the programmed frameshift removed (pJC27) or intact (pJC216) [25].
The data represent mean ± SEM from at least three independent experiments.
Next, we investigated the effects of ΔlepA on spontaneous frameshifting. Constructs in which a single base pair was removed or added within the linker region of Rluc-Fluc were used, so that translation of Fluc requires either −1 or +1 frameshifting, respectively [21]. In all cases, spontaneous frameshifting was unaffected by ΔlepA or ΔbipA in the absence or presence of potassium tellurite.
Finally, the effect of ΔlepA on +1 programmed frameshifting was measured. Here, we used plasmid pJC216, in which the programmed frameshifting site derived from prfB is inserted at the 5’ end of lacZ. The level of [3-galactosidase dependent on frameshifting was ~60% of that observed for the no-frameshift control (pJC27) [25], and neither ΔlepA nor ΔbipA influenced expression from either construct.
Effects of LepA and BipA on tmRNA activity and A-site mRNA cleavage
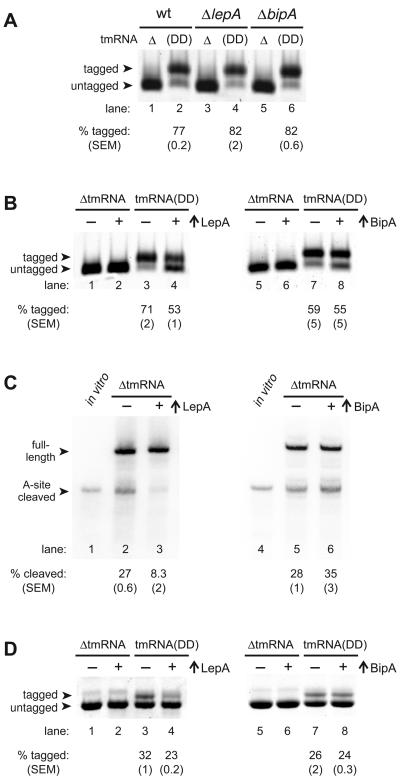
The tmRNA quality control system facilitates the recycling of ribosomes stalled at the 3′-end of truncated messages. tmRNA acts first as a tRNA to bind the A site of the stalled ribosome, and then as an mRNA to add a short peptide tag to the C-terminus of the incompletely synthesized nascent protein [33, 34]. Because EF-Tu delivers alanyl-tmRNA to the A site [35], the putative elongation factors LepA and BipA may compete for the factor-binding site of the paused ribosome. To investigate this, we first tested the effect of ΔlepA on tmRNA activity using a model non-stop mRNA encoding a His6-tagged version of γ phage cI repressor N-terminal domain [23]. Most of the protein produced from this construct was tagged in cells expressing tmRNA(DD), which encodes a protease-resistant peptide tag. In contrast, only untagged cI repressor accumulated in cells lacking tmRNA (AtmRNA). Protein purified from tmRNA(DD) ΔlepA cells showed a modest, but reproducible increase in tmRNA(DD)-tagging compared the control (wt) cells (Fig. 2A, lanes 2 and 4). A similar increase was observed for the strain lacking BipA (Fig. 2A, lane 6). Overexpression of either LepA or BipA from a plasmid in lepA+bipA+ cells reduced the proportion of tmRNA(DD)-tagged cI repressor expressed from a non-stop mRNA (Fig. 2B). This effect was most pronounced in the presence of arabinose, but was also apparent in its absence, particularly in the case of BipA (compare Fig. 2A, lane 2 and Fig. 2B, lane 7). Collectively, these data suggest that LepA and BipA compete with EF-Tu•GTP•alanyl-tmRNA for ribosome binding.
Figure 2. Effects of LepA and BipA on the tmRNA quality control system.
(A) Analysis of tmRNA(DD)-mediated tagging of protein expressed from a non-stop mRNA. The N-terminal domain of cI repressor was purified by Ni2+-affinity chromatography and analyzed by SDS-PAGE and Coomassie blue staining. Untagged cI repressor accumulates in ΔtmRNA cells, whereas most of the repressor protein is tagged in tmRNA(DD) cells. The percentage (mean ± SEM) of tmRNA(DD)-tagged protein for each strain is indicated. (B) Effect of LepA and BipA overexpression on tmRNA(DD) tagging. The N-terminal domain of cI repressor was purified from cells containing plasmid-borne PBAD-lepA (left panel) or PBAD-bipA (right panel) grown in the absence (−) or presence (+) of arabinose. The percentage (mean ± SEM) of tmRNA(DD)-tagged protein for each strain is indicated. (C) Northern blot analysis of flag-(m)ybeL(PP) mRNA for A-site mRNA cleavage. RNA was isolated from strains carrying plasmid-borne PBAD-lepA (left panel) or PBAD-bipA (right panel) grown in the absence (−) or presence (+) of arabinose. Two mRNA species corresponding to full-length and A-site cleaved messages were detected (as indicated). An in vitro transcript corresponding to A-site cleaved mRNA was used as a size marker (lanes 1 and 4). (D) Analysis of tmRNA(DD)-mediated tagging of protein expressed from ybeL(PP) mRNA. His6-YbeL(PP) was purified by Ni2+-affinity chromatography, followed by SDS-PAGE analysis and Coomassie blue staining.
Ribosome pausing during translation termination elicits A-site mRNA cleavage in E. coli. The unknown RNase responsible cleaves stop codons in the A site of paused ribosomes, thereby generating non-stop mRNA and facilitating tmRNA recruitment to the ribosome [24]. We examined A-site cleavage using a model mRNA derived from the E. coli ybeL(PP) gene [24] in ΔlepA and ΔbipA strains, and found no significant differences in A-site cleavage activity compared to wild-type control (data not shown). However, the overproduction of LepA reduced the accumulation of A-site cleaved mRNA in AtmRNA cells substantially (Fig. 2C, lane 3). In contrast, overproduction of BipA failed to decrease A-site mRNA cleavage (Fig. 2C, lane 6). Because tmRNA is not recruited to ribosomes paused on full-length messages [36], we predicted that LepA overproduction would also inhibit tmRNA tagging resulting from A-site mRNA cleavage. Indeed, LepA overproduction reduced tmRNA(DD)-mediated tagging of His6-YbeL(PP), whereas BipA overexpression had no effect (Fig. 2D).
Discussion
It was shown previously that addition of purified LepA to an in vitro transcription/translation system increased the percentage of active protein product from ~50% to >90% [8]. The authors attributed this effect to miscoding in the absence of added LepA. They proposed that, by catalyzing reverse translocation, LepA rescues “defective” posttranslocation complexes that are highly prone to decoding errors in the next round of elongation. However, no evidence for “defective” posttranslocation complexes was provided, and the effect of LepA on amino acid misincorporation was not directly measured. Here, we show that the fidelity of translation in vivo is unaffected by ΔlepA (Table 1-3), ruling out the possibility that LepA normally contributes to the accuracy of decoding.
How then does LepA increase the active fraction of translational product in vitro? We consider it most likely that LepA-catalyzed reverse translocation slows down translation elongation and thereby facilitates co-translational protein folding in the in vitro transcription/translation system. Expression of eukaryotic proteins like GFP and luciferase in E. coli often results in production of unfolded proteins [37-40]. The rate of translation elongation is substantially faster in bacteria (~15 amino acids/sec) [41, 42] than in eukaryotes (~5 amino acids/sec) [43, 44], and folding of eukaryotic proteins normally occurs in a co-translational manner [38]. In the heterologous system, fast elongation by bacterial ribosomes may uncouple folding from translation, resulting in misfolded proteins. By slowing elongation, LepA could suppress this effect. The idea that LepA slows elongation is consistent with the observation that LepA decreases the yield of translation product in a dose-dependent manner [8].
While neither LepA nor BipA contributes to the fidelity of translation, both seem to act as bona fide translation factors in vivo. Overexpression of LepA or BipA inhibits tmRNA activity on non-stop messages, suggesting that both GTPases compete with EF-Tu'GTP'alanyl-tmRNA for the factor-binding site of the ribosome. Intriguingly, only LepA was able to inhibit A-site cleavage during ribosome pausing at stop codons. An attractive hypothesis is that this is due to LepA-catalyzed reverse translocation, which should increase occupancy of the A site (by peptidyl-tRNA) and hence protect the A codon from nucleolytic attack. Further experiments will be necessary to test this hypothesis.
Our phenotypic microarray screen identified only a few conditions under which LepA is important for growth, one of which was tellurite stress. Tellurite (TeO32-) is highly toxic to a wide range of bacterial species and has been used as an antimicrobial therapeutic agent [30]. It has been suggested that the tellurite toxicity stems from its strong oxidizing ability toward free thiol groups in the cell [45-47], or the generation of superoxide radicals caused by tellurite reduction, resulting in a redox imbalance in the intracellular environment [48, 49]. Hypersensitivity to tellurite has been observed in a number of mutants, including those of nitrate reductases [50], superoxide dismutase [51], components of the thiol-disulfide redox system [51], enzymes of disulfide bond formation [45], and components of cysteine metabolism [52]. Among these, mutants of dsbA and dsbB, which encode the periplasmic protein thiol:disulfide oxidoreductases, have been reported to exhibit the highest sensitivity to tellurite (MICs of 0.008-0.0015 μg/ml, as opposed to 1-2 μg/ml for a wild-type strain) [45], raising the possibility that the major targets of tellurite are thiol groups of periplasmic and/or membrane proteins. A majority of these proteins are co-translationally translocated through the cytoplasmic membrane by Sec machinery. We speculate that LepA might play a role in co-translational coupling of membrane protein biogenesis, ensuring correct folding of the proteins. Consistent with this idea, LepA localizes in the membrane fraction [6], its gene is co-transcribed with the leader peptidase (lepB) gene in E. coli [3], and its mitochondrial homolog Guf1 is critical for biogenesis of functional cytochrome oxidase [10]. Increased tellurite toxicity due to ΔlepA might be related to oxidation of thiol groups of partially unfolded proteins rather than superoxide radical formation, because other superoxide generating reagents impose relatively small effects on its growth (Fig. 1B). Further experiments will be necessary to determine whether LepA, like Guf1[10], plays a role in membrane protein biogenesis.
Table 2.
Effects of ΔlepA and ΔbipA on spontaneous frameshifting
| Frequency (× 10−4)b |
|||||
|---|---|---|---|---|---|
| Medium | Plasmida | Frameshift | Control | ΔlepA | ΔbipA |
| LB | pAD2 | −1 | 21 ± 1 | 19 ± 1 | 16 ± 1 |
| pAD4 | −1 | 25 ± 2 | 19 ± 1 | 22 ± 1 | |
| pAD3 | +1 | 290 ± 20 | 380 ± 60 | 300 ± 35 | |
| pAD5 | +1 | 380 ± 80 | 270 ± 50 | 210 ± 4 | |
| LB | pAD2 | −1 | 36 ± 2 | 27 ± 5 | 25 ± 7 |
| + 0.05 μg/ml | pAD4 | −1 | 27 ± 5 | 27 ± 6 | 22 ± 2 |
| K2TeO3 | pAD3 | +1 | 310 ± 40 | 430 ± 50 | 360 ± 30 |
| pAD5 | +1 | 680 ± 130 | 430 ± 10 | 450 ± 30 | |
Acknowledgement
We thank B. Ahmer for vector pWSK29, R. Dalbey for helpful suggestions, and C. H. Yang for experimental supports. S. Popova-Butler and K. Green-Church for proteomic analysis of the ΔlepA mutant. This work was supported by the National Institutes of Health [grant numbers GM072528 (to K.F.) & GM078634 (to C.S.H.)].
List of abbreviations
- GTPase
guanosine 5′-triphosphate hydrolase
- mRNA
messenger RNA
- tRNA
transfer RNA
- cryo-EM
cryo-electron microscopy
- A site
aminoacyl site
- P site
peptidyl site
- E site
exit site
- GDPNP
guanosine 5′-[β,γ-imido]-triphosphate
- ppGpp
guanosine 5′-diphosphate-3′-diphosphate
- RT-PCR
reverse transcription-polymerase chain reaction
- tmRNA
transfer messenger RNA
- HCl
hydrogen chloride
- MgCl2
magnesium chloride
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Margus T, Remm M, Tenson T. Phylogenetic distribution of translational GTPases in bacteria. BMC Genomics. 2007;8:15. doi: 10.1186/1471-2164-8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leipe DD, Wolf YI, Koonin EV, Aravind L. Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol. 2002;317:41–72. doi: 10.1006/jmbi.2001.5378. [DOI] [PubMed] [Google Scholar]
- 3.March PE, Inouye M. Characterization of the lep operon of Escherichia coli. Identification of the promoter and the gene upstream of the signal peptidase I gene. J. Biol. Chem. 1985;260:7206–7213. [PubMed] [Google Scholar]
- 4.Date T, Wickner W. Isolation of the Escherichia coli leader peptidase gene and effects of leader peptidase overproduction in vivo. Proc. Natl. Acad. Sci. USA. 1981;78:6106–6110. doi: 10.1073/pnas.78.10.6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dibb NJ, Wolfe PB. lep operon proximal gene is not required for growth or secretion by Escherichia coli. J. Bacteriol. 1986;166:83–87. doi: 10.1128/jb.166.1.83-87.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.March PE, Inouye M. GTP-binding membrane protein of Escherichia coli with sequence homology to initiation factor 2 and elongation factors Tu and G. Proc. Natl. Acad. Sci. USA. 1985;82:7500–7504. doi: 10.1073/pnas.82.22.7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evans RN, Blaha G, Bailey S, Steitz TA. The structure of LepA, the ribosomal back translocase. Proc. Natl. Acad. Sci. USA. 2008;105:4673–4678. doi: 10.1073/pnas.0801308105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qin Y, Polacek N, Vesper O, Staub E, Einfeldt E, Wilson DN, Nierhaus KH. The highly conserved LepA is a ribosomal elongation factor that back-translocates the ribosome. Cell. 2006;127:721–733. doi: 10.1016/j.cell.2006.09.037. [DOI] [PubMed] [Google Scholar]
- 9.Connell SR, Topf M, Qin Y, Wilson DN, Mielke T, Fucini P, Nierhaus KH, Spahn CM. A new tRNA intermediate revealed on the ribosome during EF4-mediated back-translocation. Nat. Struct. Mol. Biol. 2008;15:910–915. doi: 10.1038/nsmb.1469. [DOI] [PubMed] [Google Scholar]
- 10.Bauerschmitt H, Funes S, Herrmann JM. The membrane-bound GTPase Guf1 promotes mitochondrial protein synthesis under suboptimal conditions. J. Biol. Chem. 2008;283:17139–17146. doi: 10.1074/jbc.M710037200. [DOI] [PubMed] [Google Scholar]
- 11.Farris M, Grant A, Richardson TB, O'Connor CD. BipA: A tyrosine-phosphorylated GTPase that mediates interactions between enteropathogenic Escherichia coli (EPEC) and epithelial cells. Mol. Microbiol. 1998;28:265–279. doi: 10.1046/j.1365-2958.1998.00793.x. [DOI] [PubMed] [Google Scholar]
- 12.deLivron MA, Makanji HS, Lane MC, Robinson VL. A novel domain in translational GTPase BipA mediates interaction with the 70S ribosome and influences GTP hydrolysis. Biochemistry. 2009 doi: 10.1021/bi901026z. [DOI] [PubMed] [Google Scholar]
- 13.deLivron MA, Robinson VL. Salmonella enterica serovar Typhimurium BipA exhibits two distinct ribosome binding modes. J Bacteriol. 2008;190:5944–5952. doi: 10.1128/JB.00763-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grant AJ, Haigh R, Williams P, O'Connor CD. An in vitro transposon system for highly regulated gene expression: Construction of Escherichia coli strains with arabinose-dependent growth at low temperatures. Gene. 2001;280:145–151. doi: 10.1016/s0378-1119(01)00769-7. [DOI] [PubMed] [Google Scholar]
- 15.Grant AJ, Farris M, Alefounder P, Williams PH, Woodward MJ, O'Connor CD. Co-ordination of pathogenicity island expression by the BipA GTPase in enteropathogenic Escherichia coli (EPEC) Mol. Microbiol. 2003;48:507–521. doi: 10.1046/j.1365-2958.2003.t01-1-03447.x. [DOI] [PubMed] [Google Scholar]
- 16.Rowe S, Hodson N, Griffiths G, Roberts IS. Regulation of the Escherichia coli K5 capsule gene cluster: evidence for the roles of H-NS, BipA, and integration host factor in regulation of group 2 capsule gene clusters in pathogenic E. coli. J. Bacteriol. 2000;182:2741–2745. doi: 10.1128/jb.182.10.2741-2745.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barker HC, Kinsella N, Jaspe A, Friedrich T, O'Connor CD. Formate protects stationary-phase Escherichia coli and Salmonella cells from killing by a cationic antimicrobial peptide. Mol. Microbiol. 2000;35:1518–1529. doi: 10.1046/j.1365-2958.2000.01820.x. [DOI] [PubMed] [Google Scholar]
- 18.Krishnan K, Flower AM. Suppression of ΔbipA phenotypes in Escherichia coli by abolishment of pseudouridylation at specific sites on the 23S rRNA. J Bacteriol. 2008;190:7675–7683. doi: 10.1128/JB.00835-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2 doi: 10.1038/msb4100050. 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang RF, Kushner SR. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene. 1991;100:195–199. [PubMed] [Google Scholar]
- 21.Devaraj A, Shoji S, Holbrook ED, Fredrick K. A role for the 30S subunit E site in maintenance of the translational reading frame. RNA. 2009;15:255–265. doi: 10.1261/rna.1320109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kramer EB, Farabaugh PJ. The frequency of translational misreading errors in E. coli is largely determined by tRNA competition. RNA. 2007;13:87–96. doi: 10.1261/rna.294907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roche ED, Sauer RT. SsrA-mediated peptide tagging caused by rare codons and tRNA scarcity. EMBO J. 1999;18:4579–4589. doi: 10.1093/emboj/18.16.4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hayes CS, Sauer RT. Cleavage of the A site mRNA codon during ribosome pausing provides a mechanism for translational quality control. Mol. Cell. 2003;12:903–911. doi: 10.1016/s1097-2765(03)00385-x. [DOI] [PubMed] [Google Scholar]
- 25.Sanders CL, Curran JF. Genetic analysis of the E site during RF2 programmed frameshifting. RNA. 2007;13:1483–1491. doi: 10.1261/rna.638707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garza-Sánchez F, Janssen BD, Hayes CS. Prolyl-tRNA(Pro) in the A-site of SecM-arrested ribosomes inhibits the recruitment of transfer-messenger RNA. J. Biol. Chem. 2006;281:34258–34268. doi: 10.1074/jbc.M608052200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garza-Sánchez F, Gin JG, Hayes CS. Amino acid starvation and colicin D treatment induce A-site mRNA cleavage in Escherichia coli. J. Mol. Biol. 2008;378:505–519. doi: 10.1016/j.jmb.2008.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bijlsma JJ, Lie ALM, Nootenboom IC, Vandenbroucke-Grauls CM, Kusters JG. Identification of loci essential for the growth of Helicobacter pylori under acidic conditions. J. Infect. Dis. 2000;182:1566–1569. doi: 10.1086/315855. [DOI] [PubMed] [Google Scholar]
- 29.Bochner BR, Gadzinski P, Panomitros E. Phenotype microarrays for high-throughput phenotypic testing and assay of gene function. Genome Res. 2001;11:1246–1255. doi: 10.1101/gr.186501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taylor DE. Bacterial tellurite resistance. Trends. Microbiol. 1999;7:111–115. doi: 10.1016/s0966-842x(99)01454-7. [DOI] [PubMed] [Google Scholar]
- 31.Perez JM, Calderon IL, Arenas FA, Fuentes DE, Pradenas GA, Fuentes EL, Sandoval JM, Castro ME, Elias AO, Vasquez CC. Bacterial toxicity of potassium tellurite: unveiling an ancient enigma. PLoS ONE. 2007;2:e211. doi: 10.1371/journal.pone.0000211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qin Y. Fachbereich Biologie. Freie Universität Berlin; Berlin: 2006. The highly conserved LepA is a ribosomal elongation factor that back-translocates the ribosome and is essential for viability at high ionic strength. [DOI] [PubMed] [Google Scholar]
- 33.Keiler KC, Waller PR, Sauer RT. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science. 1996;271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- 34.Moore SD, Sauer RT. The tmRNA system for translational surveillance and ribosome rescue. Annu. Rev. Biochem. 2007;76:101–124. doi: 10.1146/annurev.biochem.75.103004.142733. [DOI] [PubMed] [Google Scholar]
- 35.Valle M, Gillet R, Kaur S, Henne A, Ramakrishnan V, Frank J. Visualizing tmRNA entry into a stalled ribosome. Science. 2003;300:127–130. doi: 10.1126/science.1081798. [DOI] [PubMed] [Google Scholar]
- 36.Ivanova N, Pavlov MY, Felden B, Ehrenberg M. Ribosome rescue by tmRNA requires truncated mRNAs. J. Mol. Biol. 2004;338:33–41. doi: 10.1016/j.jmb.2004.02.043. [DOI] [PubMed] [Google Scholar]
- 37.Baneyx F. Recombinant protein expression in Escherichia coli. Curr. Opin. Biotechnol. 1999;10:411–421. doi: 10.1016/s0958-1669(99)00003-8. [DOI] [PubMed] [Google Scholar]
- 38.Agashe VR, Guha S, Chang HC, Genevaux P, Hayer-Hartl M, Stemp M, Georgopoulos C, Hartl FU, Barral JM. Function of trigger factor and DnaK in multidomain protein folding: increase in yield at the expense of folding speed. Cell. 2004;117:199–209. doi: 10.1016/s0092-8674(04)00299-5. [DOI] [PubMed] [Google Scholar]
- 39.Chang HC, Kaiser CM, Hartl FU, Barral JM. De novo folding of GFP fusion proteins: high efficiency in eukaryotes but not in bacteria. J. Mol. Biol. 2005;353:397–409. doi: 10.1016/j.jmb.2005.08.052. [DOI] [PubMed] [Google Scholar]
- 40.Fukuda H, Arai M, Kuwajima K. Folding of green fluorescent protein and the cycle3 mutant. Biochemistry. 2000;39:12025–12032. doi: 10.1021/bi000543l. [DOI] [PubMed] [Google Scholar]
- 41.Andersson DI, Bohman K, Isaksson LA, Kurland CG. Translation rates and misreading characteristics of rpsD mutants in Escherichia coli. Mol. Gen. Genet. 1982;187:467–472. doi: 10.1007/BF00332630. [DOI] [PubMed] [Google Scholar]
- 42.Bohman K, Ruusala T, Jelenc PC, Kurland CG. Kinetic impairment of restrictive streptomycin-resistant ribosomes. Mol. Gen. Genet. 1984;198:90–99. doi: 10.1007/BF00328706. [DOI] [PubMed] [Google Scholar]
- 43.Berridge MV, Lane CD. Translation of Xenopus liver messenger RNA in Xenopus oocytes: vitellogenin synthesis and conversion to yolk platelet proteins. Cell. 1976;8:283–297. doi: 10.1016/0092-8674(76)90012-x. [DOI] [PubMed] [Google Scholar]
- 44.Spirin AS, Ryazanov AG. Regulation of elongation rate. In: Trachsel H, editor. Translation in eukaryotes. CRC Press; Boca Raton: 1991. pp. 325–352. [Google Scholar]
- 45.Turner RJ, Weiner JH, Taylor DE. Tellurite-mediated thiol oxidation in Escherichia coli. Microbiology. 1999;145:2549–2557. doi: 10.1099/00221287-145-9-2549. [DOI] [PubMed] [Google Scholar]
- 46.Turner RJ, Aharonowitz Y, Weiner JH, Taylor DE. Glutathione is a target in tellurite toxicity and is protected by tellurite resistance determinants in Escherichia coli. Can. J. Microbiol. 2001;47:33–40. [PubMed] [Google Scholar]
- 47.Dyllick-Brenzinger M, Liu M, Winstone TL, Taylor DE, Turner RJ. The role of cysteine residues in tellurite resistance mediated by the TehAB determinant. Biochem. Biophys. Res. Commun. 2000;277:394–400. doi: 10.1006/bbrc.2000.3686. [DOI] [PubMed] [Google Scholar]
- 48.Calderon IL, Arenas FA, Perez JM, Fuentes DE, Araya MA, Saavedra CP, Tantalean JC, Pichuantes SE, Youderian PA, Vasquez CC. Catalases are NAD(P)H-dependent tellurite reductases. PLoS ONE. 2006;1:e70. doi: 10.1371/journal.pone.0000070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tremaroli V, Fedi S, Zannoni D. Evidence for a tellurite-dependent generation of reactive oxygen species and absence of a tellurite-mediated adaptive response to oxidative stress in cells of Pseudomonas pseudoalcali genes KF707. Arch. Microbiol. 2007;187:127–135. doi: 10.1007/s00203-006-0179-4. [DOI] [PubMed] [Google Scholar]
- 50.Avazeri C, Turner RJ, Pommier J, Weiner JH, Giordano G, Vermeglio A. Tellurite reductase activity of nitrate reductase is responsible for the basal resistance of Escherichia coli to tellurite. Microbiology. 1997;143:1181–1189. doi: 10.1099/00221287-143-4-1181. [DOI] [PubMed] [Google Scholar]
- 51.Turner RJ, Weiner JH, Taylor DE. The tellurite-resistance determinants tehAtehB and klaAklaBtelB have different biochemical requirements. Microbiology. 1995;141:3133–3140. doi: 10.1099/13500872-141-12-3133. [DOI] [PubMed] [Google Scholar]
- 52.Rojas DM, Vasquez CC. Sensitivity to potassium tellurite of Escherichia coli cells deficient in CSD, CsdB and IscS cysteine desulfurases. Res. Microbiol. 2005;156:465–471. doi: 10.1016/j.resmic.2004.12.010. [DOI] [PubMed] [Google Scholar]