Abstract
Background
Familial pulmonary arterial hypertension (FPAH) is a rare, autosomal-dominant inherited disease with low penetrance. Mutations in the Bone Morphogenetic Protein Receptor 2 (BMPR2) have been identified in at least 70% of FPAH patients. However, the lifetime penetrance of these BMPR2 mutations is 10-20%, suggesting that genetic and/or environmental modifiers are required for disease expression. Our goal in this study is to identify genetic loci that may influence FPAH expression in BMPR2-mutation-carriers.
Methods
We performed a genome-wide linkage scan in 15 FPAH families segregating for BMPR2 mutations. We used a dense SNP array and a novel multi-scan linkage procedure that provides increased power and precision for the localization of linked loci.
Results
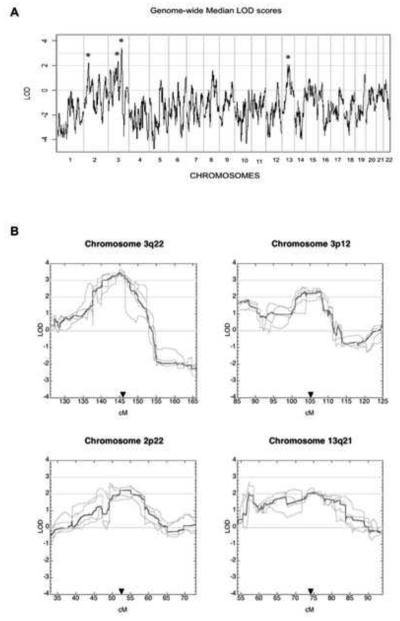
We observed linkage evidence in four regions: 3q22 (median LOD=3.43), 3p12 (median LOD = 2.35), 2p22 (median LOD = 2.21), and 13q21 (median LOD = 2.09). When used in conjunction with the nonparametric bootstrap, our approach yields high-resolution to identify candidate gene regions containing putative BMPR2-interacting genes. Imputation of the disease model by LOD score maximization indicates that the 3q22 locus alone predicts most FPAH cases in BMPR2-mutation carriers, providing strong evidence that BMPR2 and the 3q22 locus interact epistatically.
Conclusions
Our findings suggest that genotypes at loci in the newly-identified regions, especially at 3q22, could improve FPAH risk prediction in FPAH families and suggest other targets for therapeutic intervention.
Introduction
Familial pulmonary arterial hypertension (FPAH) (1) is an inherited disorder characterized by a sustained increase in mean pulmonary arterial pressure, a normal pulmonary capillary wedge pressure, and increased pulmonary vascular resistance. If unresponsive to therapeutic treatment, patients develop severe pulmonary dysfunction, which often leads to right heart failure, and eventually death (2). FPAH is a rare (incidence less than 1/10,000 per year) and genetically complex disease, segregating as an autosomal dominant trait, but displaying greatly reduced penetrance (3). Linkage analysis first mapped PPH1 to a 25-27 cM region of chromosome 2 and subsequent analysis identified loss-of-function mutations in Bone Morphogenetic Protein Receptor 2 (BMPR2) in the majority of FPAH families (3-8). While approximately 70% of FPAH patients carry a mutation in BMPR2, only 10-20% of BMPR2 mutation carriers in FPAH families develop symptoms. This suggests that additional genetic and/or environmental factors modify FPAH expression.
BMPR2 encodes the protein Bmpr-II, a subunit of the TGF-β receptor kinase superfamily. Highly conserved in vertebrates, the main ligands of Bmpr-II are the bone morphogenetic proteins Bmp 2, 4 and 7 (9-12). These growth factors are involved in the regulation of processes in many cell types including smooth muscle and endothelial cells (2). Loss of Bmpr-II function is thought to disrupt signaling pathways important in these cell types, leading to hypertrophy in the smooth muscles that surround pulmonary arterioles and to apoptosis-mediated loss of luminal integrity (13-15). An important line of evidence for the role of Bmpr-II in FPAH comes from Bmpr2+/− mice, and from transgenic mice in which Bmpr2 has been down-regulated in smooth muscle. In each case, the mice are predisposed to developing symptoms of pulmonary arterial hypertension (16, 17). Moreover, Bmpr-II and its ligands are involved in many basic developmental processes (11, 12, 18), and BMPR2 interactions with other genes involved in FPAH expression are unknown.
Our goal is to identify genetic loci that interact with BMPR2 to influence FPAH expression. We conducted a high-resolution genome-wide linkage scan in 15 FPAH families containing 164 individuals, most of whom carry identified BMPR2 mutations (see methods). Using a high-resolution single nucleotide polymorphism (SNP) array, we sought to map modifier genes with high precision. However, there are computational challenges associated with these arrays due to linkage disequilibrium (LD). If LD is ignored or poorly modeled, linkage results can be biased (19-21). To make efficient use of dense SNP data without introducing bias, we combined linkage information from multiple, sparse subsamples (average spacing > 0.5 cM) using a novel, multi-scan approach. This approach avoids the need to model linkage disequilibrium, since the correlation between neighboring SNPs in each sparse subsample is negligible. Our novel multi-scan approach also allows us to construct narrow, accurate, intervals for linkage peaks, thus reducing the uncertainty in our estimates of the true trait locations. We used this method to identify and characterize four novel loci that potentially interact with BMPR2 mutations to influence FPAH expression. Our analyses, including penetrance estimation at these loci, suggest that much of the apparent reduced penetrance for BMPR2-mutation-carriers is explained by an interaction between BMPR2 and one or more of these loci, supporting an oligogenic disease model for FPAH.
Materials and Methods
We studied a cohort of patients referred to the Columbia University Pulmonary Hypertension Center between 1991 and 2007. Patients and their family members were diagnosed according to the Fourth World Symposium on Pulmonary Hypertension Dana Point 2008 consensus (REF Proceedings of the Fourth World Symposium on Pulmonary Hypertension). All patients diagnosis of FPAH was confirmed where other family members were clinically affected. Data from family members were collected, and medical records were reviewed to confirm the diagnosis of FPAH. Blood samples were obtained for all patients. All subjects provided written informed consent (and assent if indicated). The study was approved by the Columbia University Medical Center Institutional Review Board.
As part of our study design, we sequenced BMPR2 in the proband in each family for mutations. The 13 exons and adjacent intron/exon boundaries of BMPR2 were sequenced from genomic DNA using bidirectional dideoxy-sequencing performed on an ABI337 (Big DyeR; Applied Biosystems [ABI], Foster City, CA). Probands were also screened for BMPR2 deletions/duplications using multiplex ligation dependent probe amplification (MLPA) according to the manufacturer's instructions (MRC Holland: Amsterdam, Netherlands). Only families with an identified BMPR2 mutation were included. After identifying the proband's mutation, samples from all available members of the family were screened for the same mutation, and co-segregation with FPAH was confirmed.
With the exception of family members needed to connect pedigrees and marrying individuals, only BMPR2 mutation-carriers were included in our analysis. Thus, 70.1 % of all individuals carry a mutation at BMPR2 and 30.4 % of BMPR2 mutation carriers were clinically affected. Non-carrier family members were classified as “unknown”. This approach has been taken by other linkage studies, as in breast cancer (27), diabetes (37) and Huntington Disease (26). We used the GeneChip® Human Mapping 500K Array Set (Affymetrix, Inc, Santa Clara, CA) to produce ~500,000 genotypes on each subject. The mapping 500K Array set comprises two separate arrays, each capable of genotyping around 250,000 SNPs. One array uses the restriction enzyme, Nsp I (~262,000 SNPs), while the other uses the restriction enzyme, Sty I (~238,000 SNPs). Only arrays with >94% call rates were used for data analysis. Five percent of subjects were genotyped twice as internal measures of quality control. Replicate genotypes of these five subjects indicated at least 99.5% concordance of genotypes in all five subjects. Data were analyzed for potential genotyping errors by checking for Mendelian segregation within families and all genotypes at any SNP, yielding two or more Mendelian segregation errors, were excluded from the analysis. Similarly, if the genotypes at any SNP demonstrated a significant departure from Hardy-Weinberg proportions, those SNP data were also excluded.
We used Genehunter (2.0) (38, 39) to perform multipoint linkage analysis on the 15 families, which averaged 13 individuals per family, and ranged in size from 6 to 36. Two families were too large for linkage analysis with Genehunter (2.0), so a branch was pruned and analyzed as a separate family. One family had to be pruned twice, creating a total of 18 families with a total of 164 individuals in the analysis. We restricted attention to the 22 autosomes. For our initial analyses, we assumed 50% penetrance and a risk allele frequency of 0.006 when assuming dominant inheritance, and 0.010 when assuming recessive inheritance (22, 40-42).
We employed a novel, multi-scan approach that uses subsamples of the original dense SNP data at sparsely spaced SNPs (average spacing > 0.5 cM). Each subsample retains all 164 individuals, but only keeps a small fraction (approximately 1%) of the original dense SNP genotypes for each individual. Since the linkage information in each subsample yields a genome-wide LOD, the information across multiple subsamples is easily summarized by the median LOD. By combining makes efficient use of all of the available dense SNP data. Moreover, since the SNPs within each subsample are sparsely spaced, the correlation, or LD, between neighboring SNPs is low and has little or no impact on the median LOD. This is also true for the standard approach, which bases inference on a single subsample. However, as noted above, a single subsample only retains a fraction of the original data, hence some information is always lost when using the standard approach. Therefore, to make efficient use of the original dense SNP data and to account for LD, we used our multi-scan approach to compute the genome-wide median LOD from five different, sparse subsamples. Each of our subsamples retained a random collection of roughly 4,400 equi-spaced SNPs (average spacing ~0.7 cM).
In the context of linkage analysis on affected sibpairs, Bacanu (2005) showed that, relative to the standard approach, the mean LOD has increased power to detect linkage. Due to software limitations, we chose to use the median LOD, but note that it has comparable performance to the mean LOD in the situations that we have examined (data not shown). Using both dominant and recessive disease models, we defined interesting regions of linkage as those regions where the median LOD consistently exceeded 2.0 across subsamples. Then, to estimate penetrance, we maximized the maximum LOD score (22).
In addition to using the median LOD, we improved the estimate of trait location for each of our linkage peaks by averaging the individual estimates of location obtained from each of the sparse subsamples. Then, we used our improved estimate in conjunction with a nonparametric bootstrap procedure (43) to construct approximate 95% confidence intervals for the true trait locations (Stewart et al., submitted). In the context of genetic map estimation, in which one seeks to estimate the locations of genetic markers (instead of disease genes), Stewart (44) showed that the bootstrap procedure accurately estimates the variance of each marker location estimate.
Results
We identified four novel regions linked to FPAH in BMPR2 mutation carriers: 3q22 (median LOD=3.43), 3p12 (median LOD=2.35), 2p22 (median LOD=2.21), and 13q21 (median LOD=2.09) (Table 1). Figure 1a shows the genome-wide median LOD assuming a dominant mode of inheritance. Figure 1b shows the median LOD and the five constituent LOD score curves used to compute the median LOD, for the four regions where the median LOD exceeds 2.0. As described previously (see Materials and Methods), each of the five curves is based on a randomly chosen subsample of the original dense SNP data, and each subsample contains roughly 4,400 equi-spaced SNPs. For each subsample and for a given chromosome, the maximum height of the median LOD and the locations where these maxima occur are similar (Figure 1b). Table 1 and Figure 1b show the trait location estimate derived from multiple scans (see methods). It can be seen that, for the locus at 13q21, the maximum median LOD occurs considerably distal to our final estimation of trait location derived from multiple scans. This also illustrates that a single scan alone can be misleading in some cases. In addition, Table 1 shows the trait location estimate and the approximate 95% confidence intervals for each of our novel trait loci. The confidence interval lengths, based on our multi-scan approach, are 9.6, 3.8, 9.3, and 18.7 cM for candidate regions 3q22, 3p12, 2p22, and 13q21, respectively. Moreover, our confidence intervals are shorter than those based on a single scan by 6.1, 3.3, 2.4, and 3.1 cM for the same candidate regions, respectively (Table 1).
Table 1.
Maximum median LOD scores for the loci that show linkage with FPAH, 95 % confidence intervals (CIs) and trait location estimates for each locus.
| Locus | Max median LOD score * |
95% CI (cM) (multiple subsample) |
Trait location estimate (cM) * |
95% CI (cM) (single subsample) |
|---|---|---|---|---|
| 3q22 | 3.43 | 141.760-151.348 | 145.99 | 137.316-153.036 |
| 3p12 | 2.35 | 102.139-105.948 | 105.06 | 101.000-108.060 |
| 2p22 | 2.21 | 49.464-58.710 | 52.63 | 47.33-58.96 |
| 13q21 | 2.09 | 58.930-77.596 | 74.01 | 57.160-78.974 |
Median LOD scores and trait location estimates were calculated based on five random subsamples of the dense SNP data.
Figure 1.
(A) Median LOD scores obtained from five genome-wide scans performed with five different subsamples of the data, plotted across all chromosomes. (B) Median LOD scores (black) for the chromosomes that show linkage to HPAH, along with the individual LOD scores from each scan (grey) each performed with different subsamples of the data. The arrows mark the estimate of trait location for each locus.
The signal on chromosome 3q22 revealed the highest LOD score and provided statistically significant evidence for linkage. Therefore, we estimated the penetrance for this locus (22) with a random subsample of our data in this region. A penetrance of 70% increased the evidence for linkage by 9.5% compared to the assumed penetrance of 50%, indicating that the locus at 3q22 has a strong genetic effect in BMPR2-mutation carriers. The 70% penetrance estimate suggests that genetic factors alone - in particular a locus on 3q22 - can account for most of the “reduced penetrance” of BMPR2 mutations in families in our study. We performed a similar analysis for 3p12, 2p22 and 13q21, but found no evidence to suggest a revision of the original trait model assumptions.
Examination of the LOD scores by family showed little evidence of locus heterogeneity, i.e., no families showed evidence against linkage at any of the four loci. However, with our limited number of FPAH families, we cannot conclude that all four loci are necessary for FPAH expression in BMPR2-mutation carriers. Similarly, the data were also too limited to identify a specific gene using family-based association testing. In addition, it is important to emphasize that, although all affected individuals carry BMPR2 mutations, our sample consists almost entirely of BMPR2-mutation carriers, affected and unaffected. An immediate consequence of this type of study design is the removal of any evidence of differential segregation at BMPR2 between affected and unaffected individuals, resulting in negative LOD scores at and around the BMPR2 locus (Figure 1a).
Discussion
We found evidence for four novel loci, at 3q22, 3p12, 2p22, and 13q21, which influence FPAH expression in BMPR2-mutation-carriers. Moreover, we minimized the estimated size of each candidate region by incorporating data from multiple subsamples of SNPs. Our linkage results strongly support an interaction between the locus on chromosome 3q22 and BMPR2 in FPAH expression. Three additional loci with suggestive linkage evidence, 3p12, 2p22 and 13q21, may also affect the risk for FPAH in the presence of BMPR2 mutations. Therefore, we propose a disease model in which the loci at 3q22, 3p12, 2p22 and 13q21 interact, singly or in some combination, with BMPR2 to produce an FPAH phenotype. Also, we do not rule out the possibility that some of these loci may have effects on FPAH expression independent of mutations in BMPR2, or may even contribute to idiopathic PAH, but we have not looked at such patients. With a sufficient number of idiopathic PAH patients and additional information (e.g., expression data and association analysis data), one could test whether the identified loci contribute to the non-BMPR2 PAH form.
Recently, much attention has been given to the search for “modifiers” of known so-called major-effect gene mutations in different diseases (23-26). Breast cancer is an example of a genetic disease in which a number of studies reported modifier genes related to BRCA1 and BRCA2 mutations. These studies, like the current one, have selected only mutation-carriers to include in the analysis, and also identified genetic factors that modify the risk conferred by BRCA1/2 mutations (27-29). Thus, using only mutation carriers in the analysis can identify loci that account for reduced penetrance.
Previous studies have proposed several modifiers of BMPR2 in FPAH, including members of the serotonin pathway (30, 31), the TGF-β pathway (32), the Golgi trafficking pathway (33) and potassium channel regulatory pathways (34, 35). Recently, studies examining gene expression differences in affected and unaffected BMPR2 mutation carriers revealed differential regulation of genes involved in estrogen signaling, cell proliferation, stress response and G-protein signaling. These results suggest that factors in these pathways might interact with BMPR2 to cause FPAH (36). Interestingly, the confidence intervals for our linkage peaks contain several genes involved in G-protein signaling, and other pathways implicated in FPAH expression, making it plausible that mutations in one or more of these candidate genes are responsible for our linkage signals (Table 2). However, association analyses on a larger sample and re-sequencing to identify genetic variants will be necessary to identify the genes and precise variants that interact with BMPR2 to cause disease.
Table 2.
Candidate genes within our 95 % confidence intervals (CIs) for the loci that show linkage.
| Locus | Candidate genes within our 95 % CIs. |
|---|---|
| 3q22 | PPP2P3A, NCK1*, MRAS*, PIK3CB, SPSB4, RASA2*, ATP1B3** |
| 3p12 | GBE1, |
| 2p22 | RAB19*, GPR113*, UCN, PLB1, PPP1CB, LBH, CAPN13, NLRC4, LTBP1***, RASGRP3* |
| 13q21 | PIBF1, KCTD12**, EDNRB*, SPRY2 |
G-protein signaling
Potassium transport
TGF-β signaling
Mouse models are commonly used to study the function of disease-related genes. However, locating interacting genes in inbred mice can be difficult because of greater gene sharing and increased locus homozygosity compared to humans. Furthermore, the mouse and human phenotypes differ and the contributing loci may be species, or even strain, specific. Nevertheless, when we locate the genes indicated by our linkage results, those genes' contribution to FPAH expression can be studied in the BMPR2 knockout mice.
The existence of strong linkage evidence means that the genes responsible for the linkage signal contribute substantially to FPAH susceptibility. We can identify such variants using association analysis. Identification of variants in these regions could improve disease prediction and genetic counseling for FPAH families and provide novel targets for therapeutic intervention. Because the chromosome 3 locus appears to strongly influence PAH expression, when identified, it may offer the best possibility for a therapeutic target.
Our analysis used a dense (~500 K genome-wide) SNP array. The utility of dense SNP data for linkage analysis is often compromised because most linkage analysis programs do not model the correlation between genotypes at nearby SNPs. As a result, most methods either analyze a single, sparse subsample of the original dense SNP data, or crudely approximate the correlation. The first approach is inefficient since information at many SNPs is ignored, and the utility of the second approach depends crucially on the fit between the crude approximation and the actual correlation. By contrast, our novel multi-scan approach makes efficient use of the original dense SNP data without introducing bias, and is robust to artifacts in the data such as genotyping errors, and errors in the relative ordering of SNPs. We have characterized this method and it compares favorably, in terms of power, accuracy, and computational speed to other methods currently in use (Stewart et al., submitted). Note that in some regions of the genome, random subsamples are virtually equally informative for linkage. In these regions the two approaches, multi-scan and single scan, will have similar performance. It is likely that the 13q21 linkage peak encompasses such a region.
Diseases such as FPAH that are caused by known mutations but have reduced penetrance provide a unique opportunity to answer questions concerning complex genetic interactions and disease modifiers. By reexamining BMPR2-mutation carriers in FPAH families, and by efficiently utilizing dense SNP data in our linkage analysis, we have identified new FPAH modifier loci that interact with BMPR2 to produce pulmonary arterial hypertension in these families. Moreover, our novel multi-scan approach uses all the available dense SNP data to map disease susceptibility genes with increased power and precision.
Acknowledgements
We would like to thank Adam Greenberg for his diligent assistance with regard to computer support; Anna Peljto for developing software to construct the confidence intervals; Helen Temple with patient collection; and Nicole Mallory for study coordination. This research was funded in part by grants NHLBI NLO60056, NIDDK DK31775, NINDS NS27941, NIMH MH48858, T32-MH65213 and the Columbia University Professional Schools Fund.
We also fondly acknowledge the memory of Dr. Jane Morse who dedicated her life to finding a cure for FPAH and who worked with the families in this study.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Simonneau G, Robbins IM, Beghetti M, et al. Updated clinical classification of pulmonary hypertension. J Am Coll Cardiol. 2009;54:S43–54. doi: 10.1016/j.jacc.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 2.Sztrymf B, Coulet F, Girerd B, et al. Clinical outcomes of pulmonary arterial hypertension in carriers of BMPR2 mutation. Am J Respir Crit Care Med. 2008;177:1377–83. doi: 10.1164/rccm.200712-1807OC. [DOI] [PubMed] [Google Scholar]
- 3.Morse JH, Jones AC, Barst RJ, Hodge SE, Wilhelmsen KC, Nygaard TG. Mapping of familial primary pulmonary hypertension locus (PPH1) to chromosome 2q31-q32. Circulation. 1997;95:2603–6. doi: 10.1161/01.cir.95.12.2603. [DOI] [PubMed] [Google Scholar]
- 4.Lane KB, Machado RD, Pauciulo MW, et al. Heterozygous germline mutations in BMPR2, encoding a TGF-beta receptor, cause familial primary pulmonary hypertension. The International PPH Consortium. Nat Genet. 2000;26:81–4. doi: 10.1038/79226. [DOI] [PubMed] [Google Scholar]
- 5.Deng Z, Haghighi F, Helleby L, et al. Fine mapping of PPH1, a gene for familial primary pulmonary hypertension, to a 3-cM region on chromosome 2q33. Am J Respir Crit Care Med. 2000;161:1055–9. doi: 10.1164/ajrccm.161.3.9906051. [DOI] [PubMed] [Google Scholar]
- 6.Thomson JR, Machado RD, Pauciulo MW, et al. Sporadic primary pulmonary hypertension is associated with germline mutations of the gene encoding BMPR-II, a receptor member of the TGF-beta family. J Med Genet. 2000;37:741–5. doi: 10.1136/jmg.37.10.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Machado RD, Pauciulo MW, Thomson JR, et al. BMPR2 haploinsufficiency as the inherited molecular mechanism for primary pulmonary hypertension. Am J Hum Genet. 2001;68:92–102. doi: 10.1086/316947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Machado RD, Aldred MA, James V, et al. Mutations of the TGF-beta type II receptor BMPR2 in pulmonary arterial hypertension. Hum Mutat. 2006;27:121–32. doi: 10.1002/humu.20285. [DOI] [PubMed] [Google Scholar]
- 9.Massague J. How cells read TGF-beta signals. Nat Rev Mol Cell Biol. 2000;1:169–78. doi: 10.1038/35043051. [DOI] [PubMed] [Google Scholar]
- 10.Chen D, Zhao M, Mundy GR. Bone morphogenetic proteins. Growth Factors. 2004;22:233–41. doi: 10.1080/08977190412331279890. [DOI] [PubMed] [Google Scholar]
- 11.Liu F, Ventura F, Doody J, Massague J. Human type II receptor for bone morphogenic proteins (BMPs): extension of the two-kinase receptor model to the BMPs. Mol Cell Biol. 1995;15:3479–86. doi: 10.1128/mcb.15.7.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Newman JH, Phillips JA, 3rd, Loyd JE. Narrative review: the enigma of pulmonary arterial hypertension: new insights from genetic studies. Ann Intern Med. 2008;148:278–83. doi: 10.7326/0003-4819-148-4-200802190-00006. [DOI] [PubMed] [Google Scholar]
- 13.Morrell NW, Yang X, Upton PD, et al. Altered growth responses of pulmonary artery smooth muscle cells from patients with primary pulmonary hypertension to transforming growth factor-beta(1) and bone morphogenetic proteins. Circulation. 2001;104:790–5. doi: 10.1161/hc3201.094152. [DOI] [PubMed] [Google Scholar]
- 14.Mandegar M, Fung YC, Huang W, Remillard CV, Rubin LJ, Yuan JX. Cellular and molecular mechanisms of pulmonary vascular remodeling: role in the development of pulmonary hypertension. Microvasc Res. 2004;68:75–103. doi: 10.1016/j.mvr.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 15.Davies RJ, Morrell NW. Molecular Mechanisms of Pulmonary Arterial Hypertension: Role of Mutations in the Bone Morphogenetic Protein Type II Receptor. Chest. 2008;134:1271–7. doi: 10.1378/chest.08-1341. [DOI] [PubMed] [Google Scholar]
- 16.Tada Y, Majka S, Carr M, et al. Molecular effects of loss of BMPR2 signaling in smooth muscle in a transgenic mouse model of PAH. Am J Physiol Lung Cell Mol Physiol. 2007;292:L1556–63. doi: 10.1152/ajplung.00305.2006. [DOI] [PubMed] [Google Scholar]
- 17.Hong KH, Lee YJ, Lee E, et al. Genetic ablation of the BMPR2 gene in pulmonary endothelium is sufficient to predispose to pulmonary arterial hypertension. Circulation. 2008;118:722–30. doi: 10.1161/CIRCULATIONAHA.107.736801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Said SI. Mediators and modulators of pulmonary arterial hypertension. Am J Physiol Lung Cell Mol Physiol. 2006;291:L547–58. doi: 10.1152/ajplung.00546.2005. [DOI] [PubMed] [Google Scholar]
- 19.Huang Q, Shete S, Amos CI. Ignoring linkage disequilibrium among tightly linked markers induces false-positive evidence of linkage for affected sib pair analysis. Am J Hum Genet. 2004;75:1106–12. doi: 10.1086/426000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boyles AL, Scott WK, Martin ER, et al. Linkage disequilibrium inflates type I error rates in multipoint linkage analysis when parental genotypes are missing. Hum Hered. 2005;59:220–7. doi: 10.1159/000087122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li B, Leal SM. Ignoring intermarker linkage disequilibrium induces false-positive evidence of linkage for consanguineous pedigrees when genotype data is missing for any pedigree member. Hum Hered. 2008;65:199–208. doi: 10.1159/000112367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greenberg DA, Abreu P, Hodge SE. The power to detect linkage in complex disease by means of simple LOD-score analyses. Am J Hum Genet. 1998;63:870–9. doi: 10.1086/301997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dipple KM, McCabe ER. Modifier genes convert "simple" Mendelian disorders to complex traits. Mol Genet Metab. 2000;71:43–50. doi: 10.1006/mgme.2000.3052. [DOI] [PubMed] [Google Scholar]
- 24.Daw EW, Chen SN, Czernuszewicz G, et al. Genome-wide mapping of modifier chromosomal loci for human hypertrophic cardiomyopathy. Hum Mol Genet. 2007;16:2463–71. doi: 10.1093/hmg/ddm202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Daw EW, Lu Y, Marian AJ, Shete S. Identifying modifier loci in existing genome scan data. Ann Hum Genet. 2008;72:670–5. doi: 10.1111/j.1469-1809.2008.00449.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gayan J, Brocklebank D, Andresen JM, et al. Genomewide linkage scan reveals novel loci modifying age of onset of Huntington's disease in the Venezuelan HD kindreds. Genet Epidemiol. 2008;32:445–53. doi: 10.1002/gepi.20317. [DOI] [PubMed] [Google Scholar]
- 27.Nathanson KL, Shugart YY, Omaruddin R, et al. CGH-targeted linkage analysis reveals a possible BRCA1 modifier locus on chromosome 5q. Hum Mol Genet. 2002;11:1327–32. doi: 10.1093/hmg/11.11.1327. [DOI] [PubMed] [Google Scholar]
- 28.Chenevix-Trench G, Milne RL, Antoniou AC, Couch FJ, Easton DF, Goldgar DE. An international initiative to identify genetic modifiers of cancer risk in BRCA1 and BRCA2 mutation carriers: the Consortium of Investigators of Modifiers of BRCA1 and BRCA2 (CIMBA) Breast Cancer Res. 2007;9:104. doi: 10.1186/bcr1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ripperger T, Gadzicki D, Meindl A, Schlegelberger B. Breast cancer susceptibility: current knowledge and implications for genetic counselling. Eur J Hum Genet. 2008 doi: 10.1038/ejhg.2008.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marcos E, Fadel E, Sanchez O, et al. Serotonin-induced smooth muscle hyperplasia in various forms of human pulmonary hypertension. Circ Res. 2004;94:1263–70. doi: 10.1161/01.RES.0000126847.27660.69. [DOI] [PubMed] [Google Scholar]
- 31.Long L, MacLean MR, Jeffery TK, et al. Serotonin increases susceptibility to pulmonary hypertension in BMPR2-deficient mice. Circ Res. 2006;98:818–27. doi: 10.1161/01.RES.0000215809.47923.fd. [DOI] [PubMed] [Google Scholar]
- 32.Phillips JA, 3rd, Poling JS, Phillips CA, et al. Synergistic heterozygosity for TGFbeta1 SNPs and BMPR2 mutations modulates the age at diagnosis and penetrance of familial pulmonary arterial hypertension. Genet Med. 2008;10:359–65. doi: 10.1097/GIM.0b013e318172dcdf. [DOI] [PubMed] [Google Scholar]
- 33.Sehgal PB, Mukhopadhyay S. Pulmonary arterial hypertension: a disease of tethers, SNAREs and SNAPs? Am J Physiol Heart Circ Physiol. 2007;293:H77–85. doi: 10.1152/ajpheart.01386.2006. [DOI] [PubMed] [Google Scholar]
- 34.Young KA, Ivester C, West J, Carr M, Rodman DM. BMP signaling controls PASMC KV channel expression in vitro and in vivo. Am J Physiol Lung Cell Mol Physiol. 2006;290:L841–8. doi: 10.1152/ajplung.00158.2005. [DOI] [PubMed] [Google Scholar]
- 35.Burg ED, Remillard CV, Yuan JX. Potassium channels in the regulation of pulmonary artery smooth muscle cell proliferation and apoptosis: pharmacotherapeutic implications. Br J Pharmacol. 2008;153(Suppl 1):S99–S111. doi: 10.1038/sj.bjp.0707635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.West J, Cogan J, Geraci M, et al. Gene expression in BMPR2 mutation carriers with and without evidence of Pulmonary Arterial Hypertension suggests pathways relevant to disease penetrance. BMC Med Genomics. 2008;1:45. doi: 10.1186/1755-8794-1-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim SH, Ma X, Klupa T, et al. Genetic modifiers of the age at diagnosis of diabetes (MODY3) in carriers of hepatocyte nuclear factor-1alpha mutations map to chromosomes 5p15, 9q22, and 14q24. Diabetes. 2003;52:2182–6. doi: 10.2337/diabetes.52.8.2182. [DOI] [PubMed] [Google Scholar]
- 38.Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES. Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet. 1996;58:1347–63. [PMC free article] [PubMed] [Google Scholar]
- 39.Kruglyak L, Lander ES. Faster multipoint linkage analysis using Fourier transforms. J Comput Biol. 1998;5:1–7. doi: 10.1089/cmb.1998.5.1. [DOI] [PubMed] [Google Scholar]
- 40.Hodge SE, Abreu PC, Greenberg DA. Magnitude of type I error when single-locus linkage analysis is maximized over models: a simulation study. Am J Hum Genet. 1997;60:217–27. [PMC free article] [PubMed] [Google Scholar]
- 41.Pal DK, Durner M, Greenberg DA. Effect of misspecification of gene frequency on the two-point LOD score. Eur J Hum Genet. 2001;9:855–9. doi: 10.1038/sj.ejhg.5200724. [DOI] [PubMed] [Google Scholar]
- 42.Greenberg DA, Abreu PC. Determining trait locus position from multipoint analysis: accuracy and power of three different statistics. Genet Epidemiol. 2001;21:299–314. doi: 10.1002/gepi.1036. [DOI] [PubMed] [Google Scholar]
- 43.Efron BaT RJ. An Introduction to the Bootstrap. Chapman & Hall; New York: 1993. [Google Scholar]
- 44.Stewart WC. Improving estimates of genetic maps: a meta-analysis-based approach. Genet Epidemiol. 2007;31:408–16. doi: 10.1002/gepi.20221. [DOI] [PubMed] [Google Scholar]
- Bacanu S. Multipoint linkage analysis for a very dense set of markers. Bioinformatics. 2005;29:195–203. doi: 10.1002/gepi.20089. [DOI] [PubMed] [Google Scholar]