Abstract
Background
There is growing interest for identification of new targets for biomarker development in multiple sclerosis (MS). The goal of this study was to compare the concentration and the methylation patterns of cell-free plasma DNA (cfpDNA) in patients with relapsing-remitting multiple sclerosis (RRMS) and healthy individuals.
Methods
Three 30-patient cohorts were examined: patients with RRMS, in either remission or exacerbation, and healthy individuals as controls. Concentration of cfpDNA was determined using a standard fluorometric assay. Patterns of methylation in 56 gene promoters were determined by a microarray-based assay (MethDet-56). The data were analyzed to identify statistically relevant differences among the study groups.
Results
The concentration of cfpDNA in patients with RRMS was four to eight-fold higher compared to healthy controls. Significant differences in cfpDNA methylation patterns were detected in all three comparisons: RRMS patients in remission versus healthy controls were recognized with 79.2% sensitivity and 92.9% specificity; RRMS patients in exacerbation versus healthy controls were recognized with 75.9% sensitivity and 91.5% specificity; and RRMS patients in exacerbation versus those in remission were recognized with 70.8% sensitivity and 71.2% specificity.
Conclusion
Based on our findings, we conclude that patients with RRMS display unique disease- and state-specific changes of cfpDNA. Our findings are of clinical significance as they could be used in development of potentially new biomarkers for MS. This is the first report in our knowledge describing such changes of cfpDNA in patients with MS.
Keywords: multiple sclerosis, cell-free plasma DNA, DNA methylation, gene promoter, biomarker, microarray
Introduction
Multiple sclerosis (MS) is a chronic inflammatory disease of the central nervous system (CNS) of unknown etiology. The natural course of the disease exhibits several clinical patterns: relapsing-remitting, secondary progressive, primary progressive, and relapsing progressive [1, 2]. The etiologic ambiguity and the clinical variability of the disease create significant diagnostic and prognostic uncertainties and reduce the opportunities for early diagnosis and treatment. Such diagnostic limitations have stimulated growing interest for biomarker development in MS, which has evolved in multiple directions [3]. Protein-based approaches are focused on the detection of altered levels of inflammatory molecules (antibodies, cytokines, etc.) [4–6], or other cellular proteins found in the bodily fluids of MS patients [7, 8]. Genetic-based approaches concentrate on the identification of genes, related to disease susceptibility or severity (HLA genes, etc.) [9], or on disease-associated genetic variations (single nucleotide polymorphisms, etc.) that may predict risk for disease development and progression [10, 11]. New, high throughput technologies have also been used for biomarker research [12]. However, despite the current research, success at identification of a biomarker for MS remains elusive.
Disease-associated changes in DNA methylation have recently gained interest for biomarker development. DNA methylation is an epigenetic mechanism of long-term regulation of gene expression. It is a process of chemical modification of DNA that adds a methyl group to a cytosine nucleotide when it is located upstream from a guanosine nucleotide (CpG dinucleotides), frequently within and near gene promoters. Abnormal DNA methylation and gene expression is associated with various diseases such as cancers, lymphoproliferative disorders, rheumatoid arthritis, and systemic lupus erythematosus [13–16]. As for MS, putative epigenetic changes have been suggested to explain the gender bias, the low level concordance in homozygous twins, and the linkage to several genetic loci [17, 18].
Cell-free plasma DNA (cfpDNA) exists as heterogeneous polynucleotides in plasma of all humans. Lately, it has attracted attention as a disease biomarker because of its easy accessibility, established diagnostic value in genetic diseases, and utility for methylation analysis [19]. CfpDNA is believed to be released from proliferating, or dying cells, and to reflect the normal or abnormal turnover of cell populations, although its precise origins are still enigmatic [20]. CfpDNA concentration is found to be elevated in patients with trauma, cancer, inflammation, and stroke [21–23]. In MS, the utility of cfpDNA for biomarker development has not yet been evaluated. However, given the facts that MS is a disease of immune activation, central nervous system inflammation, oligodendrocyte and neuronal injury, and gliosis, one can hypothesize that the disease can induce changes in cfpDNA.
In this study, we analyzed cfpDNA in patients with relapsing-remitting MS (RRMS) and in healthy individuals. We found that cfpDNA in patients with RRMS displayed unique disease- and state specific characteristics. Our findings are clinically relevant and suggest that cfpDNA might be useful for biomarker development for MS.
Materials and methods
Patient enrollment and sample collection
The patients were prospectively enrolled in the study according to Institutional Review Board (IRB)-approved protocols and after obtaining a signed consent specifying the goals of the study. Patient confidentiality was ensured by using a coding system that was non-descriptive of patient personal information. The enrollment inclusion criteria involved: 1) presence of RRMS with documented clinical duration for at least 2 years but less than 15 years; 2) presence of some level of neurological impairment but ambulatory at baseline, 3) absence of any therapy for at least six months; 4) patient age between 20–70. Patients who had history of significant co-morbidity, substance abuse or polytherapy were excluded from the study. The diagnosis of RRMS was established based on the McDonalds criteria [3]. Disease remission [RRMS(r)] was defined as absence of any new or different from baseline clinical symptoms or neurological findings for at least 6 months. Disease exacerbation [RRMS(e)] was defined as presence of new or different from baseline clinical symptoms or neurological findings that lasted for at least 72 hours and required treatment. An outline of the patients’ demographics is provided in Table 1.
Table 1.
Patient demographics
| Gender | N | Age | Race* | ||||
|---|---|---|---|---|---|---|---|
| Ave | Median | Std Dev | Range | ||||
| Healthy Controls | Males | 12 | 47.4 | 46 | 8.6 | 30–59 | 10-2-0 |
| Females | 18 | 47.9 | 46 | 9.3 | 30–64 | 17-1-0 | |
| Total | 30 | 47.7 | 46 | 8.9 | 30–64 | 27-3-0 | |
| RRMS(r) | Males | 10 | 40.2 | 42 | 10.8 | 20–54 | 8-2-0 |
| Females | 20 | 47.2 | 46.5 | 10.6 | 26–67 | 18-1-1 | |
| Total | 30 | 44.8 | 45 | 11.0 | 20–67 | 26-3-1 | |
| RRMS(e) | Males | 5 | 36.6 | 34 | 6.6 | 29–44 | 4-1-0 |
| Females | 24 | 43 | 46 | 8.8 | 20–59 | 20-3-1 | |
| Total | 29 | 41.9 | 44 | 8.7 | 20–59 | 24-4-1 | |
RRMS(r) = RRMS in remission. RRMS(e) = RRMS in exacerbation. N = number of patients in each cohort.
C: Caucasian;
AA: African American; O: Other
Whole blood samples from RRMS patients were collected in EDTA-containing Vacutainer tubes during the clinical encounters and processed within 2 hours. The tubes were centrifuged at 2,600 g for 10 minutes at 4°C and the plasma-containing supernatants were collected, aliquoted and stored at −80°C. Similarly prepared plasma samples from healthy individuals were obtained from a commercially available source (Analytical Biological Services, Inc., Wilmington, DE, USA) and used as controls. The RRMS cohorts were similarly matched to the controls by race, gender, and age such that no demographic was statistically different between the cohorts (see Results).
DNA isolation and quantitation
DNA was isolated as previously described [24]. Briefly, 250μl of plasma were mixed with DNAzol BD (MRC, Cincinnati, OH, USA) and processed according to the manufacturer’s protocol. DNA was measured with Quant-it Picogreen (Molecular Probes; Eugene, OR; USA) according to the manufacturer’s protocol and analyzed using a BMG PolarStar reader (BMG Labtech, Offenburg, Germany). Background was subtracted and concentration determined using a standard curve. A two-tailed t-test with Satterthwaite adjustment was used to compare DNA concentration within cohorts.
DNA methylation assay
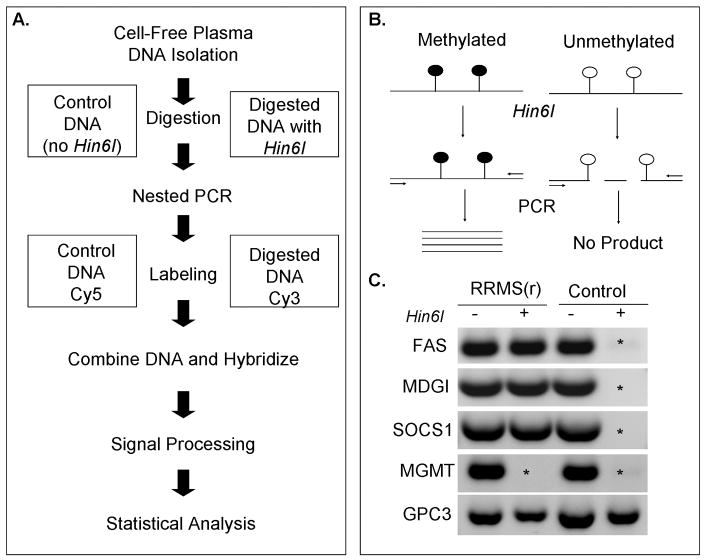
The DNA methylation (MethDet-56) assay was performed as previously described [24] (Fig. 1a). Each DNA sample (1ng) was divided into two equal aliquots. One of them was incubated with the methylation-sensitive restriction endonuclease Hin6I (Fermentas Inc, Glen Burnie, MD, USA), while the other was incubated without the enzyme (Fig. 1b). Following the digestion, nested PCR reactions were performed with primers that flanked selected Hin6I sites in each of the 56 gene promoters (Supplemental Table). Next, the PCR products were labeled with Cy3 or Cy5 (GE HealthCare, Piscataway, NJ, USA), mixed in 1:1 ratio, and hybridized to a custom designed MethDet-56 array (Microarrays, Inc., Huntsville, AL, USA) [25]. Each array contained three identical 8×8 sub-arrays (64 spots total) with three empty spots for background control (no DNA) and five control spots for non-specific control (probes for fragments that were not amplified). The arrays were scanned on a GenePix 4000B Microarray Scanner (Molecular Devices, Union City, CA, USA) with GenePix Pro 6.0 software. An example of the PCR products is shown (Fig. 1c).
Fig. 1. The DNA methylation assay.
A) General schema of the MethDet-56 analysis. B) Principle of the DNA methylation assay. Methylated DNA is resistant to Hin6I and can be amplified by PCR. C) Example of differential DNA methylation. DNA from RRMS(r) and healthy control plasma samples were treated with Hin6I (+) or left untreated (−). Note the differential amplification of FAS, MDGI and SOCS1 in the RRMS(r) sample compared to the control sample. In contrast, MGMT and GPC3 have identical patterns in both samples. Asterisk (*) marks the absence of a PCR product to indicate that the gene promoter is unmethylated.
Statistical analysis
Statistical analysis was done as previously described [24]. Briefly, background was subtracted from every spot on each sub-array. Spots with less than two-fold signal intensity of the non-specific binding controls were removed from the analysis. Methylation calls were determined for the remaining spots as: methylated, if the ratio of signals from the undigested and digested fragments (the Cy5/Cy3 ratio) was 4.0 or less; and unmethylated, if the ratio was greater than 4.0. Of the three subarrays, two calls had to be identical for each promoter, otherwise the gene promoter was removed from the next analysis. The final filter was used to remove gene promoters that had methylation calls for less than 75% of the samples in each cohort. Of the remaining set of genes, potentially informative genes were identified by Fisher’s Exact Test (p<0.1) and components of the most informative pattern were selected by the naïve Bayes algorithm. Twenty five rounds of five-fold cross-validation with independent selection of genes were performed to determine the sensitivity and specificity of each informative pattern. The sensitivity and specificity results for all rounds of cross-validations were averaged. The reported components were selected in more than 75% of cross-validation rounds.
Results
Patient demographics
The aim of the study was to analyze methylation in cfpDNA of patients with RRMS and healthy individuals and to identify disease-specific methylation patterns. Enrolled patients with RRMS were divided into two groups according to their disease state, remission, RRMS(r), and exacerbation, RRMS(e). Healthy individuals used as controls were chosen to match the gender, age and the ethnic background of the MS patients. The age, gender, and ethnic compositions of the study groups (healthy individuals, and the RRMS patients) were similar (p>0.18 for all comparisons) as determined by single-factor ANOVA for age and chi-square tests for gender and ethnic distribution (Table 1). Due to the difficulty in obtaining treatment naive RRMS patients in exacerbation, the RRMS(e) cohort contained fewer males and a decreased maximum age than the other two cohorts (5 vs. 10 and 12) and (59 vs. 64 and 67), respectively; although the median ages for all cohorts were within two years of each other.
Higher concentration of cfpDNA in blood of RRMS patients
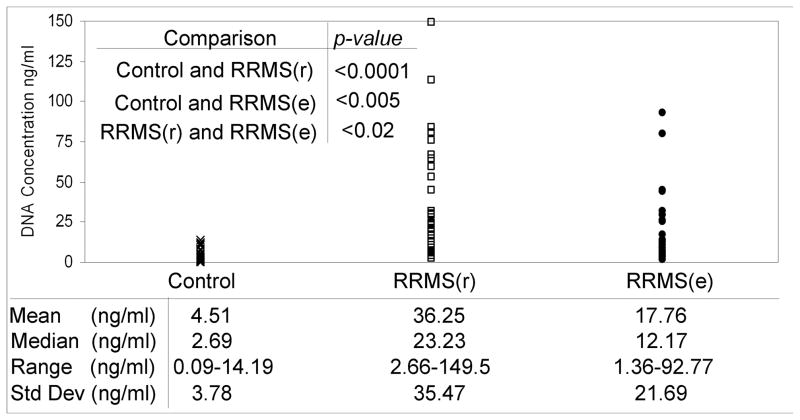
The concentration of cfpDNA was measured for all participants in the study. The results were then stratified according to individual’s clinical status and study group (Fig. 2). Group comparison of the cfpDNA concentrations was performed using a two-tailed t-test with Satterthwaite adjustment. Analysis of our data revealed that the cfpDNA concentration in the RRMS samples was significantly higher as compared to the healthy controls (mean concentration ± standard deviation): 36.25±35.37 vs. 4.51±3.36 for RRMS(r) vs. healthy controls, respectively. This represents an eight-fold increase in cfpDNA concentration over the control group (p<0.0001). Additionally, the RRMS(e) cohort had a 3.9-fold increase over the control group, 17.76±21.69 vs. 4.51±3.36 (p<0.005), respectively. The relative variabilities were comparable (standard deviation was close to the mean for all groups). While some differences were also observed in cfpDNA concentration between the RRMS(r) and RRMS(e) groups, their statistical significance was much smaller (p<0.02).
Fig. 2. Concentration of cfpDNA in RRMS.
The concentration of cfpDNA is significantly higher in both RRMS(r) and RRMS(e) groups as compared to the healthy controls.
Disease- and state-specific methylation patterns in cfpDNA of RRMS patients
CfpDNA methylation profiles of the three study groups were examined and compared. The goal of these experiments was to determine if MethDet-56 assay could identify a gene promoter methylation pattern that would detect and discriminate patients with RRMS from healthy controls as well as patients in disease remission from those in disease exacerbation. The cfpDNA methylation profile of all individual plasma samples was established using the MethDet-56 assay and the results were stratified according to group. For each comparison, differentially methylated genes were determined by the Fisher’s Exact test using p<0.1 as the cutoff. Then naïve Bayes algorithm was used in combination with five-fold cross-validation to identify genes repeatedly selected as informative by naïve Bayes algorithm. The frequency of methylation for each gene was determined by dividing the number of patients that had methylated promoters by the total number of patients within the cohort. As a first step, we examined and compared the methylation profiles of RRMS(r) and Control groups.
Fifteen genes (out of 56 or 26.8%) were identified as informative (Table 2A). The naïve Bayes algorithm and the cross validation analysis yielded a composite pattern, RRMS(r/c), that detected and discriminated RRMS(r) from Control samples with a sensitivity of 79.2% and a specificity of 92.9% (Table 3A). In contrast, the most differentially methylated gene (CDKN2B) had only a 71.0% sensitivity and 77.4% specificity for discrimination of RRMS(r) and Control samples. As a result, the pattern analysis approach was superior in discrimination accuracy compared to any single gene promoter, when tested individually.
Table 2.
Frequency of gene promoter methylation in cfpDNA
| A. RRMS(r/c) pattern | ||
|---|---|---|
| Gene | RRMS(r) | Controls |
| %Methylated | %Methylated | |
| CDH1# | 38.7 | 0.0 |
| CDKN2A* | 22.6 | 0.0 |
| CDKN2B# | 71.0 | 22.6 |
| FAS* | 61.3 | 25.8 |
| ICAM1 | 19.4 | 0.0 |
| MCJ* | 51.6 | 12.9 |
| MDGI* | 32.3 | 0.0 |
| MUC2 | 90.3 | 64.5 |
| MYF3 | 61.3 | 90.3 |
| PAX5 | 58.1 | 19.4 |
| PGK1* | 64.5 | 16.1 |
| RB1 | 38.7 | 67.7 |
| SOCS1 | 32.3 | 6.5 |
| SYK# | 77.4 | 22.6 |
| TP73* | 35.5 | 0.0 |
| B. RRMS(e/c) pattern | ||
|---|---|---|
| Gene | RRMS(e) | Controls |
| %Methylated | %Methylated | |
| BRCA1 | 55.2 | 16.7 |
| CCND2 | 24.1 | 0.0 |
| DAPK | 25.9 | 0.0 |
| FAS* | 69.0 | 22.2 |
| FHIT | 48.3 | 16.7 |
| MCT1 | 71.4 | 33.3 |
| MDGI* | 25.0 | 0.0 |
| MCJ* | 53.6 | 14.3 |
| CDKN2A* | 48.3 | 0.0 |
| TP73* | 27.6 | 0.0 |
| PGK1* | 58.6 | 17.9 |
| PR PROX** | 48.3 | 13.3 |
| C. RRMS(r/e) pattern | ||
|---|---|---|
| Gene | RRMS(e) | RRMS(r) |
| %Methylated | %Methylated | |
| CDH1# | 6.7 | 38.7 |
| CDKN2B# | 33.3 | 71.0 |
| HIC1 | 76.7 | 32.3 |
| PR PROX** | 46.7 | 12.9 |
| SYK# | 46.7 | 77.4 |
RRMS(r)= RRMS patients in remission; RRMS(e)=RRMS patients in exacerbation;
Gene promoters that were informative in both RRMS(r/c) and RRMS(e/c) patterns;
Gene promoters that were informative in both RRMS(r/c) and RRMS(r/e) patterns;
Gene promoters that were informative in both RRMS(e/c) and RRMS(r/e) patterns
Table 3.
Specificity and sensitivity of detection using patterns of methylation in gene promoters.
| A. RRMS(r) group vs. Healthy Controls | ||
|---|---|---|
| Predicted: | True: | |
| RRMS(r) | Controls | |
| RRMS(r) | 79.2 | 7.1 |
| Controls | 20.8 | 92.9 |
| B. RRMS(e) group vs. Healthy Controls | ||
|---|---|---|
| Predicted: | True: | |
| RRMS(e) | Controls | |
| RRMS(e) | 75.9 | 8.5 |
| Controls | 24.1 | 91.5 |
| C. RRMS(e) group vs. RRMS(r) group | ||
|---|---|---|
| Predicted: | True: | |
| RRMS(e) | RRMS(r) | |
| RRMS(e) | 70.8 | 28.8 |
| RRMS(r) | 29.2 | 71.2 |
Correctly identified RRMS and control samples (percent of correct identification) are shown in bold: specificity is the upper left hand box and sensitivity is the lower right hand box. RRMS(r) = RRMS patients, RRMS(e) = RRMS patients in exacerbation.
Next, using the same experimental approach we examined and compared the cfpDNA profile of RRMS(e) and Control groups. Fourteen differentially methylated gene promoters (or 25.0%) were identified as informative for the RRMS(e/c) methylation pattern (Table 2B). This composite pattern could detect and discriminate RRMS(e) from Control samples with a sensitivity of 75.9% and a specificity of 91.5% (Table 3B). As before, no single gene promoter could match this level of performance.
In the final comparison, made of RRMS(e) and RRMS(r) groups (Figure 2C), five differentially methylated gene promoters (or 8.9%) were identified as informative of RRMS(e/r) methylation pattern. This composite pattern detected and discriminated RRMS(e) from RRMS(r) samples with a sensitivity of 70.8% and a specificity of 71.2% (Table 3C).
A post hoc analysis of the three comparisons revealed several important characteristics of gene promoters within the methylation patterns. The patterns of RRMS, identified in comparisons to healthy controls, contained predominantly unique gene promoters: 9 of 15 selected were unique for the RRMS(r/c) pattern and 8 out of 14 were unique for the RRMS(e/c) pattern. The remaining gene promoters (CDKN2A, FAS, MCJ, MDGI, PGK1, and TP73) were informative for comparisons of both disease states with the controls (Table 4), indicating that RRMS in remission and exacerbation have common identifiable patterns. Additionally, six promoters were specific to only RRMS(r/c) pattern and five promoters specific to only RRMS(e/c) pattern (Table 4). These differences may be due to the clinical presentation of symptoms or to the location of the lesion.
Table 4.
Differentially methylated gene promoters in RRMS patterns.
| Comparison | Promoters present in more than one pattern | Promoters specific to only one pattern | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RRMS(r/c) | CDKN2A | FAS | MCJ | MDGI | PGK1 | TP73 | CDH1 | CDKN2B | SYK | ICAM1 | MUC2 | MYF3 | PAX5 | RB1 | SOCS1 | |
| RRMS(e/c) | CDKN2A | FAS | MCJ | MDGI | PGK1 | TP73 | PR prox | BRCA1 | CCND2 | DAPK | FHIT | MCT1 | ||||
| RRMS(e/r) | CDH1 | CDKN2B | PR prox | SYK | HIC1 | |||||||||||
A composite of all three comparisons summarizing the gene promoters common to both remission and exacerbation and unique to disease status. RRMS (r/c) pattern = RRMS patients in remission vs. healthy controls. RRMS(e/c) pattern = RRMS patients in exacerbation vs. healthy controls, RRMS(e/r) pattern = RRMS patients in exacerbation vs. RRMS patients in remission.
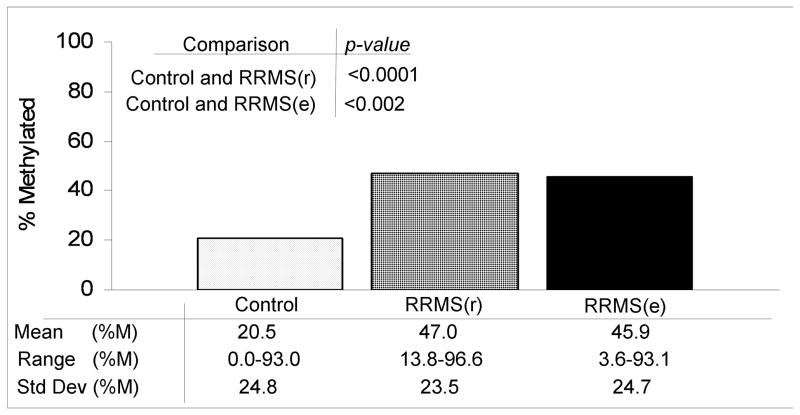
Overall, the informative gene promoters of the RRMS(r/c) and RRMS(e/c) patterns displayed increased frequency of methylation in the disease groups compared to the Control group. This frequency, reported as the percentage of methylated samples per promoter per group (Fig. 3), was 47.0% for RRMS(r), 45.9% for RRMS(e), and 20.5% for Control groups. Moreover, certain informative gene promoters demonstrated no methylation at all in any of the Control samples (Table 2A and B). In contrast, the informative gene promoters of the RRMS(r/e) pattern displayed a variation in the frequency of methylation, which did not result in any specific group patterning.
Fig. 3. Methylation of gene promoters in RRMS.
Mean percent methylation of gene promoters is higher in both RRMS(r) and RRMS(e) groups as compared to the healthy controls.
Discussion
Our experiments revealed that concentration of cfpDNA in RRMS patients was several-fold higher compared to healthy individuals and displayed unique disease- and state-specific gene promoter methylation patterns. Three patterns were identified two of them distinguished RRMS patients, either in remission or in exacerbation, from healthy controls with sensitivities near 80% and specificities above 90%. The third one distinguished RRMS patients in exacerbation from those in remission with greater than 70% sensitivity and specificity. Given that there were no specific patterns for gender, age, and race, the observed differences mostly, if not exclusively, correlated with the disease process. Therefore, we conclude that patients with RRMS display unique cfpDNA methylation profiles that may be used for developing of novel clinically important biomarkers for the disease. Our results suggest that cfpDNA methylation-based biomarkers are feasible and serve to establish a proof-of-principle for future studies with larger cohorts and patients with other neurological and inflammatory diseases.
Our study revealed two previously unknown attributes of cfpDNA in RRMS: increased concentrations and specific methylation profiles. CfpDNA is thought to originate from cells undergoing proliferation or death, so increased concentration may indicate homeostatic changes or presence of disease [20–23, 26]. Similarly, alterations in DNA methylation reflect the molecular features of pathological processes [13–15]. In this context, the elevated concentration and specific methylation patterns of cfpDNA in RRMS patients are probably related to the disease process and the associated immune activation, inflammation and cell death. Christensen et al. [27] have shown that the promoter methylation patterns in normal human tissues are different; therefore, the observed increase in cfpDNA concentration may be tissue specific. It is conceivable, that the similarities found in both RRMS(r/c) and RRMS(e/c) methylation patterns (CDKN2A, FAS, MCJ, MDGI, PGK1 and TP73) may be due to release of DNA from CNS tissues (Table 4). Likewise, the differences in the cfpDNA between RRMS patients in remission and in exacerbation may be reflective of different cellular events and dynamic shifts between inflammation, cell injury, or tissue repair. Several informative gene promoters were selected only for one pattern, either RRMS(r) or RRMS(e) (Table 4). At this moment we can only hypothesize that the selection may depend on the specific processes during exacerbation that cause the release of additional lesion-specific material. Further investigations are required to determine the cellular source(s) and the significance of cfpDNA in RRMS.
It is also possible, that the observed increase in cfpDNA in RRMS patients reflects the excessive activity of the immune system or is a compensatory mechanism that modulates its additional activation [28–30]. Two toll-like receptors (TLR) bind DNA - TLR3 binds to double-stranded DNA and TLR9 – to single-stranded CpG DNA [29, 31, 32]. These DNA are involved in systemic autoimmune diseases, including SLE and RA [33–36]. In experimental autoimmune encephalitis, the mouse model of MS, both TLR9 and MyD88 modulate the autoimmune process [37], while inhibitors to TLR9 prevent activation of TLR9-positive cells [38]. Increased cfpDNA may also stimulate a defective immune system [30, 39–41].
We and others have previously identified specific cfpDNA methylation patterns for breast, ovarian, and pancreatic cancers [24, 25, 42]. We have also shown that inflammatory and neoplastic diseases of the same organ can be differentially detected using the same analytical approach [43], thus supporting our current interpretations for RRMS. Our studies further suggest that methylation analysis in cfpDNA can be used not only for disease detection (separation of sick and healthy individuals) but also for diagnosis (defining identity of the disease). In addition, the observed differences in methylation patterns of RRMS patients in exacerbation and in remission indicate that such analysis can identify two different states of a single disease. As such, methylation patterns of multiple genes, compared to a single molecular marker, can better identify a complex disease such as MS, and utilization of a composite methylation pattern yields a higher accuracy. Exploratory work in this direction, as well as validation of discovered patterns in blinded samples, is currently in progress.
Our results have potentially significant clinical implications. The methylation patterns that differentiate RRMS patients from healthy controls could be expanded into biomarkers for detection and diagnosis of the disease. Differences in the methylation patterns between RRMS patients in exacerbation and remission can also be developed into a predictive biomarker for exacerbation. Testing for cfpDNA during clinic visits is relatively uncomplicated, as it requires a small volume of plasma (250μl) and standard laboratory equipment. Such approach may be particularly useful in individuals with vague clinical presentation, limited access to MRI, or following a clinically isolated syndrome. Finally, cfpDNA methylation patterns might become useful for assessing the efficacy of MS therapies and individualization of patient management. These lines of investigating work will undoubtedly require large longitudinal studies with various patient cohorts and control groups.
In conclusion, our study demonstrates differences in cfpDNA concentration and patterns of methylation between RRMS patients and healthy individuals. Our findings suggest that cfpDNA might be useful in biomarker development for RRMS and support transitioning to larger studies with a wider assortment of potentially methylated promoters. Besides implications in disease diagnosis and management, such studies may eventually provide insights into the origin and natural history of RRMS.
Supplementary Material
Acknowledgments
The project was supported by the NS060311 grant from NINDS (VL) and philanthropy to the Rush University MS Center (RB, DS). Authors are grateful to Melinda Kopec, R.N., Cynthia Dendrinos, R.N., Carmen Petrizzo, R.N. and Raquel Brillante, N.P., for their invaluable help with collection of blood samples and clinical data. VL is grateful to Dr. John Flax (PrecisionMed, Inc.) for a subset of plasma samples, and to Dr. Joel Peek (Microarrays, Inc.) for printing microarrays and technical support with microarray experiments.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Compston A, Coles A. Multiple sclerosis. Lancet. 2008;372(9648):1502–1517. doi: 10.1016/S0140-6736(08)61620-7. [DOI] [PubMed] [Google Scholar]
- 2.Polman CH, Reingold SC, Edan G, Filippi M, Hartung HP, Kappos L, Lublin FD, Metz LM, McFarland HF, O’Connor PW, et al. Diagnostic criteria for multiple sclerosis: 2005 revisions to the “McDonald Criteria”. Ann Neurol. 2005;58(6):840–846. doi: 10.1002/ana.20703. [DOI] [PubMed] [Google Scholar]
- 3.Bielekova B, Martin R. Development of biomarkers in multiple sclerosis. Brain. 2004;127(Pt 7):1463–1478. doi: 10.1093/brain/awh176. [DOI] [PubMed] [Google Scholar]
- 4.Berger T, Rubner P, Schautzer F, Egg R, Ulmer H, Mayringer I, Dilitz E, Deisenhammer F, Reindl M. Antimyelin antibodies as a predictor of clinically definite multiple sclerosis after a first demyelinating event. N Engl J Med. 2003;349(2):139–145. doi: 10.1056/NEJMoa022328. [DOI] [PubMed] [Google Scholar]
- 5.Dore-Duffy P, Newman W, Balabanov R, Lisak RP, Mainolfi E, Rothlein R, Peterson M. Circulating, soluble adhesion proteins in cerebrospinal fluid and serum of patients with multiple sclerosis: correlation with clinical activity. Ann Neurol. 1995;37(1):55–62. doi: 10.1002/ana.410370111. [DOI] [PubMed] [Google Scholar]
- 6.Freedman MS, Thompson EJ, Deisenhammer F, Giovannoni G, Grimsley G, Keir G, Ohman S, Racke MK, Sharief M, Sindic CJ, et al. Recommended standard of cerebrospinal fluid analysis in the diagnosis of multiple sclerosis: a consensus statement. Arch Neurol. 2005;62(6):865–870. doi: 10.1001/archneur.62.6.865. [DOI] [PubMed] [Google Scholar]
- 7.Rejdak K, Petzold A, Stelmasiak Z, Giovannoni G. Cerebrospinal fluid brain specific proteins in relation to nitric oxide metabolites during relapse of multiple sclerosis. Mult Scler. 2008;14(1):59–66. doi: 10.1177/1352458507082061. [DOI] [PubMed] [Google Scholar]
- 8.Whitaker JN. Myelin basic protein in cerebrospinal fluid and other body fluids. Mult Scler. 1998;4(1):16–21. doi: 10.1177/135245859800400105. [DOI] [PubMed] [Google Scholar]
- 9.Barcellos LF, Sawcer S, Ramsay PP, Baranzini SE, Thomson G, Briggs F, Cree BC, Begovich AB, Villoslada P, Montalban X, et al. Heterogeneity at the HLA-DRB1 locus and risk for multiple sclerosis. Hum Mol Genet. 2006;15(18):2813–2824. doi: 10.1093/hmg/ddl223. [DOI] [PubMed] [Google Scholar]
- 10.Gregory SG, Schmidt S, Seth P, Oksenberg JR, Hart J, Prokop A, Caillier SJ, Ban M, Goris A, Barcellos LF, et al. Interleukin 7 receptor alpha chain (IL7R) shows allelic and functional association with multiple sclerosis. Nat Genet. 2007;39(9):1083–1091. doi: 10.1038/ng2103. [DOI] [PubMed] [Google Scholar]
- 11.Hafler DA, Compston A, Sawcer S, Lander ES, Daly MJ, De Jager PL, de Bakker PI, Gabriel SB, Mirel DB, Ivinson AJ, et al. Risk alleles for multiple sclerosis identified by a genomewide study. N Engl J Med. 2007;357(9):851–862. doi: 10.1056/NEJMoa073493. [DOI] [PubMed] [Google Scholar]
- 12.Oksenberg JR, Baranzini SE, Sawcer S, Hauser SL. The genetics of multiple sclerosis: SNPs to pathways to pathogenesis. Nat Rev Genet. 2008;9(7):516–526. doi: 10.1038/nrg2395. [DOI] [PubMed] [Google Scholar]
- 13.Baylin SB. DNA methylation and gene silencing in cancer. Nat Clin Pract Oncol. 2005;2 (Suppl 1):S4–11. doi: 10.1038/ncponc0354. [DOI] [PubMed] [Google Scholar]
- 14.Delgado J. Novel epigenetic targets in lymphoproliferative disorders. Curr Cancer Drug Targets. 2008;8(5):378–391. doi: 10.2174/156800908785133222. [DOI] [PubMed] [Google Scholar]
- 15.Sanchez-Pernaute O, Ospelt C, Neidhart M, Gay S. Epigenetic clues to rheumatoid arthritis. J Autoimmun. 2008;30(1–2):12–20. doi: 10.1016/j.jaut.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 16.Strickland FM, Richardson BC. Epigenetics in human autoimmunity. Epigenetics in autoimmunity - DNA methylation in systemic lupus erythematosus and beyond. Autoimmunity. 2008;41(4):278–286. doi: 10.1080/08916930802024616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Casaccia-Bonnefil P, Pandozy G, Mastronardi F. Evaluating epigenetic landmarks in the brain of multiple sclerosis patients: a contribution to the current debate on disease pathogenesis. Prog Neurobiol. 2008;86(4):368–378. doi: 10.1016/j.pneurobio.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chao MJ, Ramagopalan SV, Herrera BM, Lincoln MR, Dyment DA, Sadovnick AD, Ebers GC. Epigenetics in multiple sclerosis susceptibility: difference in transgenerational risk localizes to the major histocompatibility complex. Hum Mol Genet. 2009;18(2):261–266. doi: 10.1093/hmg/ddn353. [DOI] [PubMed] [Google Scholar]
- 19.Bearzatto A, Conte D, Frattini M, Zaffaroni N, Andriani F, Balestra D, Tavecchio L, Daidone MG, Sozzi G. p16(INK4A) Hypermethylation detected by fluorescent methylation-specific PCR in plasmas from non-small cell lung cancer. Clin Cancer Res. 2002;8(12):3782–3787. [PubMed] [Google Scholar]
- 20.Anker P, Stroun M, Maurice PA. Spontaneous release of DNA by human blood lymphocytes as shown in an in vitro system. Cancer Res. 1975;35(9):2375–2382. [PubMed] [Google Scholar]
- 21.Leon SA, Shapiro B, Sklaroff DM, Yaros MJ. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977;37(3):646–650. [PubMed] [Google Scholar]
- 22.Lo YM, Rainer TH, Chan LY, Hjelm NM, Cocks RA. Plasma DNA as a prognostic marker in trauma patients. Clin Chem. 2000;46(3):319–323. [PubMed] [Google Scholar]
- 23.Rainer TH, Wong LK, Lam W, Yuen E, Lam NY, Metreweli C, Lo YM. Prognostic use of circulating plasma nucleic acid concentrations in patients with acute stroke. Clin Chem. 2003;49(4):562–569. doi: 10.1373/49.4.562. [DOI] [PubMed] [Google Scholar]
- 24.Melnikov A, Scholtens D, Godwin A, Levenson V. Differential methylation profile of ovarian cancer in tissues and plasma. J Mol Diagn. 2009;11(1):60–65. doi: 10.2353/jmoldx.2009.080072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Melnikov AA, Scholtens D, Talamonti MS, Bentrem DJ, Levenson VV. Methylation profile of circulating plasma DNA in patients with pancreatic cancer. J Surg Oncol. 2009;99(2):119–122. doi: 10.1002/jso.21208. [DOI] [PubMed] [Google Scholar]
- 26.Giacona MB, Ruben GC, Iczkowski KA, Roos TB, Porter DM, Sorenson GD. Cell-free DNA in human blood plasma: length measurements in patients with pancreatic cancer and healthy controls. Pancreas. 1998;17(1):89–97. doi: 10.1097/00006676-199807000-00012. [DOI] [PubMed] [Google Scholar]
- 27.Christensen BC, Houseman EA, Marsit CJ, Zheng S, Wrensch MR, Wiemels JL, Nelson HH, Karagas MR, Padbury JF, Bueno R, et al. Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS Genet. 2009;5(8):e1000602. doi: 10.1371/journal.pgen.1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gelman AE, Zhang J, Choi Y, Turka LA. Toll-like receptor ligands directly promote activated CD4+ T cell survival. J Immunol. 2004;172(10):6065–6073. doi: 10.4049/jimmunol.172.10.6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kabelitz D. Expression and function of Toll-like receptors in T lymphocytes. Curr Opin Immunol. 2007;19(1):39–45. doi: 10.1016/j.coi.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 30.Vollmer J, Tluk S, Schmitz C, Hamm S, Jurk M, Forsbach A, Akira S, Kelly KM, Reeves WH, Bauer S, et al. Immune stimulation mediated by autoantigen binding sites within small nuclear RNAs involves Toll-like receptors 7 and 8. J Exp Med. 2005;202(11):1575–1585. doi: 10.1084/jem.20051696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leadbetter EA, Rifkin IR, Hohlbaum AM, Beaudette BC, Shlomchik MJ, Marshak-Rothstein A. Chromatin-IgG complexes activate B cells by dual engagement of IgM and Toll-like receptors. Nature. 2002;416(6881):603–607. doi: 10.1038/416603a. [DOI] [PubMed] [Google Scholar]
- 32.Pietschmann K, Beetz S, Welte S, Martens I, Gruen J, Oberg HH, Wesch D, Kabelitz D. Toll-like receptor expression and function in subsets of human gammadelta T lymphocytes. Scand J Immunol. 2009;70(3):245–255. doi: 10.1111/j.1365-3083.2009.02290.x. [DOI] [PubMed] [Google Scholar]
- 33.Brentano F, Schorr O, Gay RE, Gay S, Kyburz D. RNA released from necrotic synovial fluid cells activates rheumatoid arthritis synovial fibroblasts via Toll-like receptor 3. Arthritis Rheum. 2005;52(9):2656–2665. doi: 10.1002/art.21273. [DOI] [PubMed] [Google Scholar]
- 34.Holm CK, Petersen CC, Hvid M, Petersen L, Paludan SR, Deleuran B, Hokland M. TLR3 ligand polyinosinic:polycytidylic acid induces IL-17A and IL-21 synthesis in human Th cells. J Immunol. 2009;183(7):4422–4431. doi: 10.4049/jimmunol.0804318. [DOI] [PubMed] [Google Scholar]
- 35.Komatsuda A, Wakui H, Iwamoto K, Ozawa M, Togashi M, Masai R, Maki N, Hatakeyama T, Sawada K. Up-regulated expression of Toll-like receptors mRNAs in peripheral blood mononuclear cells from patients with systemic lupus erythematosus. Clin Exp Immunol. 2008;152(3):482–487. doi: 10.1111/j.1365-2249.2008.03646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakano S, Morimoto S, Suzuki J, Nozawa K, Amano H, Tokano Y, Takasaki Y. Role of pathogenic auto-antibody production by Toll-like receptor 9 of B cells in active systemic lupus erythematosus. Rheumatology (Oxford) 2008;47(2):145–149. doi: 10.1093/rheumatology/kem327. [DOI] [PubMed] [Google Scholar]
- 37.Prinz M, Garbe F, Schmidt H, Mildner A, Gutcher I, Wolter K, Piesche M, Schroers R, Weiss E, Kirschning CJ, et al. Innate immunity mediated by TLR9 modulates pathogenicity in an animal model of multiple sclerosis. J Clin Invest. 2006;116(2):456–464. doi: 10.1172/JCI26078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duramad O, Fearon KL, Chang B, Chan JH, Gregorio J, Coffman RL, Barrat FJ. Inhibitors of TLR-9 act on multiple cell subsets in mouse and man in vitro and prevent death in vivo from systemic inflammation. J Immunol. 2005;174(9):5193–5200. doi: 10.4049/jimmunol.174.9.5193. [DOI] [PubMed] [Google Scholar]
- 39.Fischer M, Ehlers M. Toll-like receptors in autoimmunity. Ann N Y Acad Sci. 2008;1143:21–34. doi: 10.1196/annals.1443.012. [DOI] [PubMed] [Google Scholar]
- 40.Hansen BS, Hussain RZ, Lovett-Racke AE, Thomas JA, Racke MK. Multiple toll-like receptor agonists act as potent adjuvants in the induction of autoimmunity. J Neuroimmunol. 2006;172(1–2):94–103. doi: 10.1016/j.jneuroim.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 41.Krieg AM, Vollmer J. Toll-like receptors 7, 8, and 9: linking innate immunity to autoimmunity. Immunol Rev. 2007;220:251–269. doi: 10.1111/j.1600-065X.2007.00572.x. [DOI] [PubMed] [Google Scholar]
- 42.Melnikov AA, Scholtens DM, Wiley EL, Khan SA, Levenson VV. Array-based multiplex analysis of DNA methylation in breast cancer tissues. J Mol Diagn. 2008;10(1):93–101. doi: 10.2353/jmoldx.2008.070077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liggett T, Melnikov A, Yi Q, Replogle C, Brand R, Kaul K, Talamonti M, Abrams R, Levenson V. Differential methylation of cell-free circulating DNA in patients with pancreatic cancer and chronic pancreatitis. Cancer. doi: 10.1002/cncr.24893. In Press. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.