Abstract
Pre-organization of enzyme active sites for substrate recognition typically comes at a cost to the stability of the folded form of the protein, and consequently enzymes can be dramatically stabilized by substitutions that attenuate the size and pre-organization “strain” of the active site. How this stability-activity trade-off constrains enzyme evolution has remained less certain, and it is unclear whether one should expect major stability insults as enzymes mutate towards new activities, or how these new activities manifest structurally. These questions are both germane and easy to study in β-lactamases, which are evolving on the timescale of years to confer resistance to an ever-broader spectrum of β-lactam antibiotics. To explore whether stability is a substantial constraint on this antibiotic resistance evolution, we investigated extended-spectrum mutants of class C β-lactamases which had evolved new activity versus third-generation cephalosporins. Five mutant enzymes had between 100- to 200-fold increased activity against the antibiotic cefotaxime in enzyme assays, and the mutant enzymes all lost thermodynamic stability – from 1.7 to 4.1 kcal/mol – consistent with the function-stability hypothesis. Intriguingly, several of the substitutions were 10 – 20 Å from the catalytic serine; the question arose how they conferred extended-spectrum activity. Eight structures, including complexes with inhibitors and extended-spectrum antibiotics, were determined by x-ray crystallography. Distinct mechanisms of action are revealed for each mutant, including changes in the flexibility and ground state structures of the enzyme. These results explain the structural bases for the antibiotic resistance conferred by these substitutions, and their corresponding decrease in protein stability, which will constrain the evolution of new antibiotic resistance.
Keywords: protein stability, AmpC beta-lactamase, antibiotic resistance, evolution, action-at-a-distance
Introduction
To engineer a protein for stability, among the surest places to make substitutions is the active site. This will appear paradoxical to many, because these active-site substitutions typically reduce the activity of the mutant protein, and an inactive-but-stable mutant enzyme is rarely sought. Still, this trading of activity for stability illustrates a point: active sites are typically those regions in proteins that manifest the most internal strain. In active sites, polar groups are sequestered from water 1; 2, like-charged residues cluster together, residues adopt strained conformations 3, and hydrophobic patches are exposed 4. As Warshel 1; 2 and others 5; 6 have suggested, this strain pre-organizes the active site for ligand recognition and catalysis, and is paid for at the time of folding by the net overall stability conferred by the rest of the protein. The conversion of active-site “strain” into increased stability was first tested in barnase 7, barstar 8, T4 lysozyme 9, and AmpC ß-lactamase 10, where point substitutions increased enzyme stability by up to 4 kcal/mol, or 30% of net overall stability. Subsequent studies are consistent with the idea that residues important for function may be suboptimal for stability 11; 12; 13; 14; 15; 16, and that enzymes may be substantially stabilized by active site substitutions that attenuate the activity 17; 18; 19.
If there is by now much support for the idea that enzymes can be stabilized by active site substitutions, the “stability-function hypothesis” 9, the logical corollary, that they will often lose stability as they gain activity, has been more controversial. In a study of the antibiotic resistance enzyme TEM-1 ß-lactamase, we found that clinically selected gain-of-activity mutants lost stability to the point where a secondary site substitution occurred, far from the active site, that restored stability and allowed further destabilizing gain-of-resistance mutants to evolve 20; 21; 22 Subsequently, Weinreich and colleagues showed that the pathways to stable and active TEM mutants was epistatic and cooperative, consistent with a stability constraint on enzyme evolution 23. On the other hand, Arnold has argued that the apparent stability-activity trade-offs may be unrelated, simply reflecting differing constraints: enzymes evolved for activity will give up stability because it is not being as strongly selected, whereas an enzyme being evolved for stability may give up activity for the same reason. Although there is agreement that enzymes that are stabilized 24; 25, or buffered against unfolding by chaperones 26, are more evolvable, consensus has not emerged around a correlation between stability and activity in enzyme evolution.
These two views may be reconcilable. At its heart, the stability-activity trade-off hypothesis is about competing physical interactions in the active site. For those enzymes that evolve new activity by increasing active site pre-organization, for instance by increasing active site size to accommodate larger substrates, trade-offs in stability seem physically necessary. However, not all gain-of-function mutants fit this pattern. For instance, a thermophilic enzyme selected for activity may give up its stability simply because this is not being selected for 27. Similarly, one can imagine a protein that evolves to recognize a simpler substrate, one that requires less pre-organization than its ancestral substrate; in this case, the mutant enzyme might well gain both activity and stability relative to the ancestral protein. But for situations where a new active site is being created, or an established one enlarged or further pre-organized, then by physical necessity we expect stability, unless otherwise compensated, to be sacrificed.
The experimental support for stability-activity trade-offs in enzyme evolution rests largely on a study in TEM-1 ß-lactamase 20, and it seemed worthwhile to explore generality in another enzyme. We turned to the class C β-lactamase AmpC which, though also a ß-lactamase, diverged from the class A enzymes hundreds of million years ago, with the two families sharing no measurable sequence identity and differing from each other in size, domain organization, and in mechanistic details 28; 29; 30. AmpC-family enzymes confer resistance to β-lactam antibiotics (Figure 1A and B), and are key resistance determinants in hospital acquired pathogens, especially to the cephalosporin ß-lactams. “β-lactamase stable” 3rd generation cephalosporins, such as ceftazidime and cefotaxime (Figure 1C), were introduced in the early 1980s to overcome resistance conferred largely by class A ß-lactamases, but they are also poor substrates for AmpC, whose active site is essentially too small to accommodate them. Thus, when these large 3rd generation cephalosporins form covalent adducts with AmpC they are forced into a conformation that is incompetent for catalysis, due to steric clashes between their bulky R1 side chain and residues Val211 and Tyr221 31. “Extended-spectrum” β-lactamase (ESBL) mutants with increased hydrolytic activity against third-generation cephalosporins have subsequently appeared, both in the clinic 32; 33; 34; 35 and as the product of in vitro evolution and mutagenesis 36; 37; 38; 39. Because these mutant enzymes were evolving to recognize substrates that were too large for the native active site, but in which catalysis would retain the same location and same chemistry, they seemed like good templates to explore for stability-activity trade-offs.
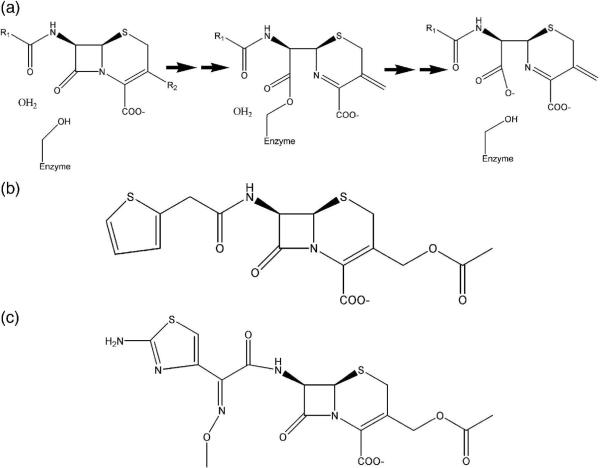
Figure 1.
Beta-lactamase reaction mechanism and representative antibiotic substrates. (a) Reaction mechanism of AmpC beta-lactamase. (b) The first-generation cephalosporin antibiotic cephalothin. (c) The third-generation cephalosporin antibiotic cefotaxime.
Here we investigate five extended-spectrum mutants of AmpC (Figure 2), asking the following questions. Are these mutants in fact better enzymes for the 3rd generation cephalosporins and are not simply, for instance, over-expressed in resistant cells? Does any gain of activity come with a concomitant loss of stability? Since stability-activity trade-offs are formulated as biophysical compensations, is the stability change thermodynamic, as reflected by reversible two-state thermal denaturation? The stability-function hypothesis applies to improved substrate complementarity arising from the introduction of structural defects or increasing residue pre-organization; to investigate this we determine x-ray crystal structures of the five mutant enzymes. These structures also reveal how substitutions that are distant from the active site, as occur with resistance mutations in many enzymes 40; 41; 42, can transduce their perturbations to the active site, and thereby increase catalytic activity.
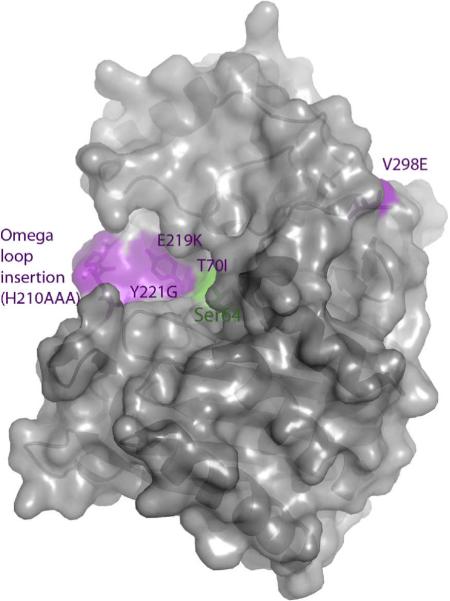
Figure 2.
ESBL substitutions under investigation in AmpC beta-lactamase, shown in purple. The catalytic serine is shown in green.
Results
Function-stability tradeoffs
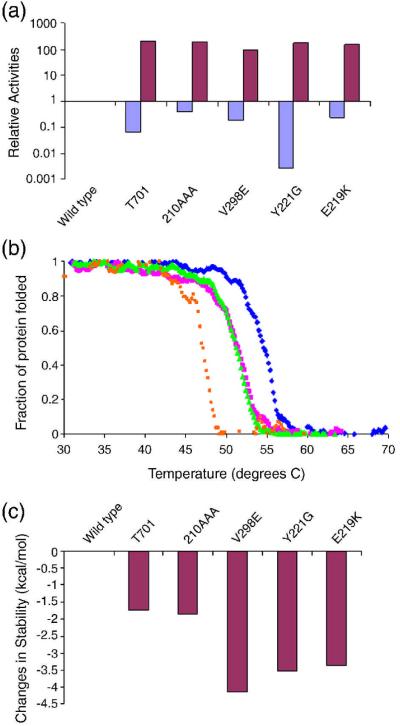
Five ESBL mutants – V298E 39, the “omega loop insertion” (H210AAA) 32; 33; 34, T70I 36, Y221G 37, and E219K 35; 38, were made by site-directed mutagenesis and purified to apparent homogeneity, and the kinetic constants kcat and Km against the first-generation cephalosporin cephalothin, and the third-generation cefotaxime, were determined. All of the ESBLs had increased activity against cefotaxime relative to wild-type AmpC, as measured by kcat, typically by 100- to 200-fold (Table 1, Figure 3A). Normally, one would use kcat/Km to compare the relative activities of the WT and mutant enzymes, but for class C β-lactamases this value is misleading. Since the acylation rate constant (k2) for ß-lactams is fast on the scale of the pre-covalent disassociation constant (k−1), the Km values for these enzymes does not reflect a disassociation constant, but rather the deacylation rate constant (k3 = kcat) divided by the acylation rate constant (k2). Thus kcat/Km collapses to simply k2, making what would ordinarily be considered the “specificity constant” misleading 43. Because kcat is slow for the WT enzyme, (kcat = 0.0448 s−1 for cefotaxime in wild-type AmpC, 44), Km values are typically too low to be measured, and IC50 is typically used as proxy for Km 44; 45. For these enzymes, kcat comparisons are more informative. This can be illustrated by example. For WT AmpC, the kcat for cefotaxime is 0.0448 s−1, and the IC50-derived Km is 0.8 μM, resulting in a kcat/Km of 5.6 × 104 M−1 s−1. For V298E, kcat is 4.4 sec −1 (Table 1), and the Km is 79 μM – resulting in the same kcat/Km value as the wild-type enzyme. However, 50 nM of V298 will hydrolyze 50 μM of ceftazidime, a physiologically relevant concentration, in minutes, while WT AmpC at the same concentration would take close to half a day. This gain in new activity against cefotaxime was accompanied by a corresponding loss of native activity against cephalothin, typically from 3- to 8-fold, but as high as 100-fold in the case of Y221G (Table 1, Figure 3A).
Table 1.
Kinetic and thermodynamic parameters of AmpC ESBL mutants.
| Mutation | Cα-Cα dist. to Ser64 (Å) | Melting temp (Tm) | ΔTm (°C) | ΔΔH (kcal/ mol) | ΔΔG (kcal/ mol) | Cephalothin | Cefotaxime | ||
|---|---|---|---|---|---|---|---|---|---|
| Km (μM) | kcat (s−1) | Km (μM) | kcat (s−1) | ||||||
| WT | NA | 54.6 | ----- | 182 | ----- | 27 | 420 | 0.8 | 0.0448 |
| T70I | 10.0 | 51.5 | −3.1 | 142 | −1.74 | 53 | 53.9 | 295 | 9.2 |
| V298E | 21.0 | 47.2 | −7.4 | 102 | −4.14 | 32 | 90.6 | 79 | 4.4 |
| E219K | 9.3 | 48.6 | −6.0 | 180 | −3.36 | 24 | 84.6 | 77 | 6.9 |
| Y221G | 4.6 | 50.8 | −3.8 | 199 | −2.13 | 99 | 4.2 | 337 | 8.0 |
| Ω loop insertion (res. 210) | 14.5 | 51.3 | −3.3 | 183 | −1.85 | 23 | 134 | 53 | 8.7 |
Figure 3.
Relative enzyme activities and thermodynamic stabilities of AmpC ESBL mutants. (a) Relative activities of AmpC ESBLs against the first-generation cephalosporin cephalothin (blue) and the third-generation cephalosporin cefotaxime (purple). (b) Characteristic thermal denaturation curves of selected AmpC ESBL mutants (V298E = orange, Omega loop insertion = green, T70I = magenta) relative to the wild-type enzyme (blue) (c) The differential stabilities of AmpC ESBL mutants relative to wild-type AmpC.
The stability of the ESBL mutants was determined by thermal denaturation, monitored by far-UV circular dichroism. To determine thermodynamic stability of these mutant enzymes, it is necessary to demonstrate their reversibility and two-state behavior upon thermal unfolding. Extensive previous work suggests that wild-type and mutant AmpC enzymes denature reversibly and in a two-state manner 46: they reversibly denature both by rapid cooling and by slow reannealing from the folded state in temperature melts, the Tm and ΔHu are very similar when measured by far-UV CD, near-UV CD, and by fluorescence, the Tm is unaffected by ramp rates between 0.5 and 2° C/min, and the transition itself is well-fit by a two-state model 46. Thermal denaturation of each extended-spectrum AmpC mutant monitored by far-UV was also consistent with a reversible, two-state model. Melting curves showed clear two-state behavior, with a clear and sharp transition at the melting temperature (Figure 3B). Denaturation was reversible, ranging from 90 to 100 percent of CD signal regained after quick cooling of the denatured enzymes. The thermal denaturation of a representative mutant, Y221G, monitored by fluorescence emission also showed a sharp transition with the same Tm of 50.8°C as when monitored by far-UV (Supplementary Material Figure S1A). A second ESBL mutant, the omega loop insertion, also showed a sharp transition with a Tm of 52.1°C (compared to a Tm of 51.3°C by CD) when monitored by far-UV (Supplementary Material Figure S1B). We therefore assume that these mutants, like the wild-type enzyme, denature in a reasonable approximation to reversible, two-state behavior, allowing us to determine true thermodynamic differences in enzyme stability relative to the wild-type enzyme.
The thermodynamic stability of all five ESBLs was decreased relative to the wild-type enzyme (Table 1, Figure 3C). Melting temperatures of these mutant enzymes diminished by between 3.1°C to 7.4°C, corresponding to a decrease in stability of 1.7 to 4.1 kcal/mol, a substantial change in stability for single substitutions, particularly since the overall stability of the wild-type enzyme is only 14 kcal/mol 46.
V298E apo structure
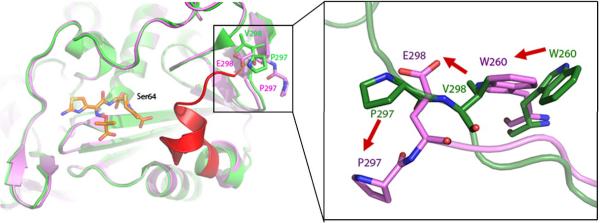
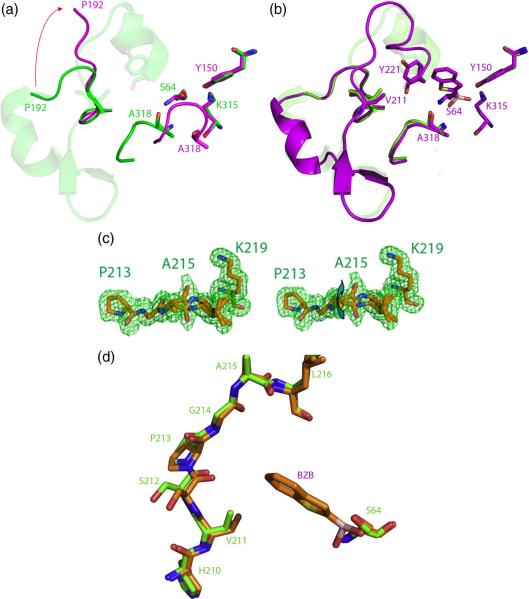
The structure of the V298E mutant enzyme was determined by xray crystallography to 2.64 Å resolution (Rwork=18.4%, Rfree=23.4%, Table 2). Well-defined 2Fo-Fc electron density at a contour level of 1σ was seen for most residues in the structure, with the exception of residues 285–296 (Figure 4, Supplementary Material Figure S2). The resulting structure reveals a “domino effect” of structural changes, starting at the point of substitution, and transmitted to the active site, 20 Å away (Figure 4). Val298 is found in a mini-hydrophobic core of the wild-type enzyme; when substituted to a glutamate, Glu298 moves to enable its interaction with solvent, opening up a cavity in the hydrophobic core (Figure 4). The sidechain of Trp260 adopts a conformation where it can fill this cavity. The new position of Glu298 clashes with the position of Pro297 in the wild-type enzyme; to avoid this clash, Pro297 swings out. This residue is on one end of a loop forming one wall of the active site; when it adopts this new conformation, it presumably takes the rest of the loop with it, as density is lost for residues 285–296 in this structure (Figure 4). The positions of key active site residues resemble that of the wild-type AmpC structure, with the exception of Tyr150, which adopts a new conformation where it hydrogen-bonds with Lys315. As an aside, we observed strong positive Fo-Fc density connecting atom Cε2 of Tyr150 and atom Nζ of Lys67. We also observed this feature in the omega loop insertion apo crystal structure (below), and a similar feature was observed in the crystal structure of the omega loop insertion GC1 from E. cloacae.32 In the GC1 structure this feature was left unmodeled but attributed to a reaction with sodium azide present in the crystallographic buffer; we have also chosen not to model this feature.
Table 2.
Crystallographic statistics
| Protein | V298E | Omega loop insertion | T70I/ BZB | Y221G | Y221G/ cefotaxime | E219K | E219K/ benzo(b) thiophene |
|---|---|---|---|---|---|---|---|
| Data collection | |||||||
| Space group | P21 | C2 | C2 | C2 | C2 | C2 | C2 |
| Cell dimensions | |||||||
| a, b, c (Å) | 81.271 | 117.582 | 114.162 | 117.717 | 117.80 | 118.247 | 117.06 |
| 75.852 | 77.926 | 77.361 | 76.789 | 77.58 | 76.876 | 77.41 | |
| 84.086 | 98.050 | 91.747 | 98.693 | 98.89 | 116.492 | 97.24 | |
| α, β, γ(°) | 90.00 | 90.00 | 90.00 | 90.00 | 90.00 | 90.00 | 90.0 |
| 115.96 | 115.96 | 122.12 | 116.62 | 118.60 | 130.89 | 115.58 | |
| 90.00 | 90.00 | 90.00 | 90.00 | 90.00 | 90.00 | 90.0 | |
| Resolution (Å) | 30.0–2.64 (2.70–2.64)* | 30.0–1.64 (1.68–1.64) | 30.0–2.64 (2.70–2.64) | 30.0–1.90 (1.95–1.90) | 60.02–2.30 (2.42–2.30) | 30.0–1.84 (1.88–1.84) | 30.0–1.63 (1.67–1.63) |
| Rmerge (%) | 8.9 | 6.1 | 7.1 | 6.8 | 10.6 | 9.4 | 6.1 |
| I / σI | 8.0 (2.0) | 8.7 (2.3) | 7.8 (2.1) | 7.7 (2.1) | 4.5 (2.3) | 5.1 (2.2) | 8.4 (2.0) |
| Completeness(%) | 100.0 (100.0) | 100.0 (99.9) | 98.4 (97.0) | 99.9 (99.9) | 96.9 (96.2) | 94.0 (93.2) | 99.9 (100.0) |
| Refinement | |||||||
| No. reflections | 28,233 (2080) | 92,466 (6818) | 34,977 (2548) | 58,776 (4295) | 32,017 (2363) | 60,887 (4418) | 92,583 (4868) |
| Rwork/Rfree (%) | 18.4 / 23.4 | 17.0 / 19.7 | 19.6 / 24.7 | 15.7 / 19.7 | 19.6 / 24.8 | 17.4 / 21.7 | 17.6 / 20.8 |
| No. atoms | |||||||
| Protein | 5577 | 6297 | 5316 | 5541 | 5610 | 5549 | 5565 |
| Ligand/ion | 6 | 0 | 12 | 74 | 28 | 10 | 44 |
| Water | 119 | 278 | 249 | 577 | 187 | 737 | 865 |
| B-factors (Å2) | |||||||
| Protein | 24.8 | 15.3 | 27.90 | 17.36 | 29.96 | 18.49 | 16.48 |
| Ligand/ion | 52.8 | 44.3 | 32.75 | 42.7 | 45.7 | 39.10 | 23.93 |
| Water | 37.358 | 36.9 | 30.81 | 30.71 | 30.79 | 27.65 | 28.41 |
| r.m.s. deviations | |||||||
| bond lengths (Å) | 0.013 | 0.016 | 0.011 | 0.015 | 0.013 | 0.015 | 0.010 |
| bond angles (°) | 1.494 | 1.486 | 1.414 | 1.460 | 1.437 | 1.465 | 1.383 |
Values in parenthesis represent highest resolution shells
Figure 4.
The x-ray crystal structure of V298E to 2.6 Å resolution, showing an overview of changes in the V298E mutant (purple) compared to the WT protein (green). Density is lost for the region of the loop shown in red, presumed to have flipped out. Structural changes are shown at the point of mutation (inset), with the mutant protein (purple) overlaid on the WT protein (green).
Omega loop insertion apo structure
The three-residue omega loop insertion was originally identified as a clinical isolate in the class C P99 β-lactamase from E. cloacae and was subsequently characterized 32; 33; 34; 47. In addition, two structures of this extended-spectrum class C β-lactamase were determined, both the apo form 32, and in complex with a cefotaxime phosphonate transition-state analog 33. Inspired by this work, we decided to study the thermodynamic implications of this substitution, and for consistency, made the analogous mutation in AmpC. Since it was determined that the length, not the identity, of the insertion was crucial in imparting catalytic activity, and an insertion of three alanines had comparable extended-spectrum activity 34; 47; we used an insertion of three alanine residues after His210 to represent the omega loop insertion. The structure of the apo omega loop insertion structure was determined to 1.64 Å resolution by x-ray crystallography (Rwork=17.0%, Rfree=19.7%, Table 2). Overall, the omega loop insertion structure resembles the wild-type structure, the RMSD for Cα atoms are 0.16 Å. However, in the region of the omega loop, at the terminus of the active site, the changes are substantial. In the mutant, the insertion causes the omega loop to adopt a new conformation (Figures 5A and 5B); the Cα of Pro216 has moved by 8.9 Å, relative to the wild-type Pro213. Similarly, Val211 has moved by 8.5 Å, and has in essence been replaced by Ala211. In addition, the Tyr221 OH has moved by 1.1 Å, and the Tyr221 Cδ, the atom thought to sterically clash with a catalytically competent conformation of third-generation cephalosporins, has shifted by 0.6 Å. Well-defined 2Fo-Fc electron density at a contour level of 1 σ was seen for most residues in the structure, with the exception of residues 287–293 in the A monomer (the positions cannot be resolved for residues 285–290 in the A monomer of the wild-type enzyme). The key active site residues resemble that of the wild-type AmpC structure, with the exception of Tyr150, which adopts a new conformation where it hydrogen-bonds with Lys315.
Figure 5.
The x-ray structure of the “omega loop insertion” mutant (H210AAA) to 1.6 Å resolution. (a) The omega loop insertion structure (orange, 3 alanine insertion in purple) overlaid on the WT protein (green). (b) Fo-Fc omit density for residues 210–216 shown at 3σ.
T70I apo and holo structures
T70I was crystallized out of a buffer containing the transition-state analog benzo(b)thiophene-2-boronic acid (BZB, Ki 27 nM) 48, after repeated attempts to grow apo T70I crystals failed. The structure of T70I was determined by x-ray crystallography to 2.14 Å resolution (Rwork=19.6%, Rfree=24.7%, Table 2). Like all AmpC structures, the protein, though functionally a monomer, crystallized with two monomers in the asymmetric unit. Surprisingly, one monomer was an apo structure, and the other had the transition-state analog bound to the active site, and each monomer adopted different conformations (Figure 6). In the apo monomer no electron density was seen for residues 193–221, comprising the omega loop of the enzyme. In addition, the β-turn from residues 318–322 has flipped into the active site, disorganizing the region around the oxyanion hole formed by the backbone nitrogens of the catalytic Ser64 and Ala318 (Figure 6A). The active site of the T70I/benzo(b)thiophene structure (Figure 6B), conversely, resembles that of the WT AmpC/benzo(b)thiophene structure 48. In addition, unlike in the apo structure, strong electron density is seen for residues 193–221 at 1σ, allowing the positions for these residues to be determined (Figure 6B). This region now adopts a slightly different conformation than it does in the wild-type AmpC structure, predominately in the region from residues 211–216 (Figure 6B); it enlarges the active site in some regions, though is closer to the active site in others.
Figure 6.
Flexibility induced in extended-spectrum mutants T70I and E219K. (a) The T70I apo structure (purple) overlaid on the WT structure (green). Density is lost from residues 193–221 in the omega loop (shown in transparent green) in the T70I structure. (b) The T70I/benzo(b)thiophene (BZB) structure (purple) overlaid on the WT structure (green). Residues 193–221, lost in the T70I structure but present in the T70I/benzo(b)thiophene structure, are shown in cartoon. (c) Stereo view of the x-ray crystal structure of E219K/BZB to 1.63 Å resolution. Fo-Fc omit density is shown at 3σ. Arrow highlights the two conformations of residues 215–216, distinguished by a peptide flip. (d) The omega loop and active site of E219K/BZB (orange) overlaid with that of the wild-type AmpC structure (green), showing the conformational differences in the 211–213 region. The position of BZB is shown in the active site.
E219K apo and holo structures
The E219K apo structure was determined to 1.84 Å resolution (Rwork=17.4%, Rfree=21.7%, Table 2), and its complex with the transition-state analog inhibitor benzo(b)thiophene-2-boronic acid was determined to 1.63 Å resolution (Rwork=17.6%, Rfree=20.8%, Table 2). After an initial round of refinement using the wild-type enzyme as a model, negative Fo-Fc density was seen at 3σ for Glu219, with nearby positive Fo-Fc density at 3σ resembling a lysine. This residue was computationally mutated to a lysine and the structure was further refined. 2Fo-Fc density was unambiguous at the point of substitution for Lys219 in both the final structures, verifying the substitution (Supplementary Material figure S3A). The position of this side chain in both structures resembles that of Glu219 in the wild-type structure, pointing into solvent (Supplementary Material figure S3B). In the wild-type enzyme, the side chain of Glu219 hydrogen-bonds to its own backbone amide nitrogen. Substitution of this residue to a lysine removes this interaction, and the K219 backbone nitrogen now hydrogen-bonds with the backbone carbonyl of Leu216, which has moved closer to it (Figure 6C). This in turn affects the conformation of residues 215–216. In the WT structure, the backbone nitrogen of Asp217 hydrogen-bonds with the backbone carbonyl of Ala215. With the carbonyl of Leu216 now engaged with the backbone nitrogen of Lys219, residues 215–216 now adopt two conformations, with the peptide backbone flipped relative to each other (Figure 6C). Also, residues 211–213, also in the omega loop, now adopt a different conformation than in the wild-type AmpC structure (Figure 6D).
Y221G apo structure
The Y221G apo structure was determined to 1.90 Å resolution (Rwork=15.7%, Rfree=19.7%, Table 2). Few structural changes are seen in Y221G compared to the WT AmpC structure. Substitution of Tyr221 to a glycine creates a cavity, which alleviates the steric clash that occurs with the catalytically competent conformation of the third-generation cephalosporins (below). This opens up space in the active site that is only partially filled by Asp217 adopting a new conformation and moving into its place.
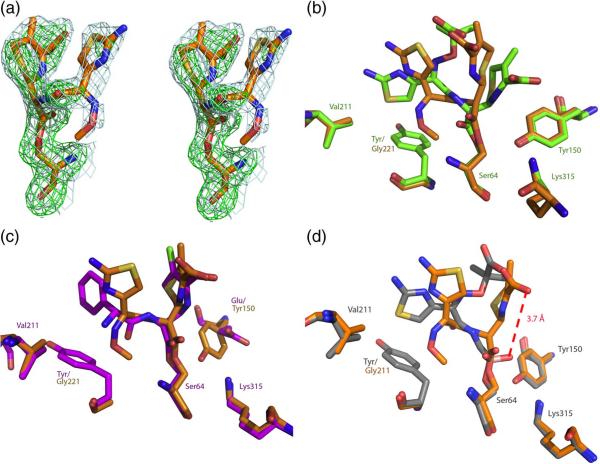
Y221G/cefotaxime structure
Y221G crystals were soaked into a 1.7 M KPi, pH 8.7, 50 mM cefotaxime, solution for one hour and then flash-frozen to obtain a structure of the Y221G/cefotaxime complex. The structure of the Y221G/cefotaxime complex was determined by x-ray crystallography to 2.3 Å resolution (Rwork=19.6%, Rfree=24.8%, Table 2). Strong 2Fo-Fc electron density was seen for the ligand at 1σ in the B monomer, and in an Fo-Fc omit map at 3σ (Figure 7A). We compared the Y221G/cefotaxime structure to the crystal structure of wild-type AmpC in complex with the closely related third-generation cephalosporin ceftazidime (Figure 7B, 31). Cefotaxime, which for the Y221G mutant enzyme has become a good substrate, adopts a different conformation than does ceftazidime in its complex with WT AmpC (Figure 7B), where the third generation cephalosporin is such a poor substrate that it functions as a covalent inhibitor. The new conformation of cefotaxime in the active site of Y221G resembles that of good substrates, such as loracarbef, in WT AmpC49 (Figure 7C). Cefotaxime's adoption of the loracarbef-like structure in the Y221G complex appears to make it competent for catalysis. In contrast with the AmpC/ceftazidime structure, where the lactam nitrogen of substrate is only 1.8 Å from where the attacking water is expected to be in the deacylation high-energy intermediate, in the Y221G/cefotaxime structure this same nitrogen has moved 3.7 Å from the putative water position, making it competent for catalysis (Figure 7D, 31). This movement is, in turn, allowed by the introduction of the cavity in Y221G, increasing the volume of the active site and its ability to accommodate 3rd generation cephalosporins.
Figure 7.
X-ray structure of Y221G/cefotaxime (2.3 Å). (a) Stereo view of quality of density of cefotaxime in the active site of Y221G. 2Fo-Fc density (blue) is shown at a contour level of 1σ, and Fo-Fc.omit density (green) is shown at a contour level of 3σ. (b) The Y221G/cefotaxime structure (orange) overlaid with the WT/ceftazidime structure (green). (c) The Y221G/cefotaxime structure (orange) overlaid with the WT/loracarbef structure (purple). (d) The Y221G/cefotaxime structure (orange) overlaid with the WT/ceftazidime deacylation transition state analog (gray).
Discussion
Two key observations emerge from this study. First, each of the five mutant enzymes has achieved its two-orders of magnitude activity gains at the cost of substantial stability loss. The minimum stability loss is 1.7 kcal/mol (3.3°C), for T70I, whereas V298E loses 4.1 kcal/mol (7.4°C), fully 30% of the net stability of the native protein. Second, the crystal structures reveal that the substitutions, though distant from one another by up to 30 Å, have the same overall effect, enlarging the active site by introducing physical defects, allowing the mutant enzymes to accommodate the large, third-generation cephalosporins. We consider these points in turn.
Destabilization of the mutant enzymes
The stability insults conferred by these gain-of-function substitutions are easily appreciated. In V298E, substituting a charged residue into a well-packed hydrophobic core causes a cascade of conformational changes that are revealed in the x-ray structure of the mutant enzyme (Figure 4). Similarly, in Y221G, the substitution introduces a straight-forward structural defect, creating a hole in the protein structure and loss of van der Waals interactions that are only partially fulfilled by movement of Asp217 into this region. In the H210AAA insertion mutant, the expanded omega loop conformation reduces packing, van der Waals interactions, hydrogen bonding, and hydrophobic surface area burial (Figure 5). Finally, both T70I and E219K lose hydrogen bonds that ordinarily stabilize this same omega-loop region of the enzyme. In WT AmpC, the hydroxyl of Thr70 hydrogen bonds with the backbone carbonyl of Glu219 in this loop, while in the structure of T70I this interaction has been lost and not replaced, leading to substantial conformational change in the mutant protein (Figure 6). In E219K, the substitution of the lysine disrupts a hydrogen bond that, in the WT enzyme, the native glutamate side chain made to its own backbone nitrogen, which otherwise has no partner in the WT structure. This internal hydrogen-bond stabilizes the loop conformation adopted in this region of the structure and its loss is likely responsible for the destabilization of E219K. Thus, all five mutant enzymes are textbook illustrations of how to disrupt protein structure and stability, should that ever be one's goal.
Translating stability loss into activity gain
The activity that these mutants need to gain is the ability to hydrolyze ß-lactam substrates that are too large for the native active site, the 3rd generation cephalosporins. Each of the mutants accomplishes this by introducing structural insults to the integrity of the protein. Three mechanisms may be considered through which these stability defects are translated into increased activity: by changing the ground state of the enzyme, by changing enzyme flexibility, or by changing its dynamics. Here, we will not further distinguish between flexibility and dynamics, as our structural and thermodynamic results are insufficiently resolved to do so, but will simply refer to both under the rubric of “flexibility”, understanding that dynamics may play a role.
Perhaps the simplest mutant to understand is Y221G, which changes the ground state, apo conformation of the enzyme to accommodate 3rd generation cephalosporins. Tyr221 is a highly conserved residue whose presence forces 3rd generation cephalosporins into a catalytically incompetent conformation, owing to a steric clash with their large R1 side chains (above). Substitution of Tyr221 by a glycine allows the ß-lactam to relax into a catalytically competent conformation. Thus, in the Y221G/cefotaxime complex (Figure 7), this third-generation cephalosporin adopts a conformation resembling that adopted by good substrates in WT x-ray structures (Figure 7C). Similarly, the x-ray structure of H210AAA reveals a well-resolved, larger conformation for the omega loop, indicating a change in the ground state for this mutant enzyme. This insertion effectively replaces Val211 with an alanine; Val211 is the second residue, in addition to Tyr221, that in WT AmpC clashes with the R1 side chain of 3rd generation cephalosporins. This structure is largely consistent with that of the enzyme that inspiring our work on this mutant, the clinically isolated GC1 ESBL from E. cloacae 32; 33; 34; 47. One difference between the two structures is that the GC1 apo structure revealed local disorder in residues 213–215 32, which includes one residue of the insertion, suggesting increased flexibility relative to that of the P99 enzyme. Conversely, in the AmpC apo structure the omega loop adopts a new, well-ordered conformation that expands active site volume (Figure 5).
The V298E mutant falls into the same category of affecting the ground state conformation of the enzyme—enlarging it—though here flexibility may also play a role. The introduction of a glutamate into the hydrophobic core of the protein is a gross structural insult, and to relieve this the enzyme undergoes substantial reorganization, flipping the now Glu298 into bulk solvent. This perturbation, 21 Å from the catalytic serine, begins a cascade of structural changes, starting with a flip of Trp260 into the hole created by the movement of Glu298. What used to be the loop defined by residues 285 to 296, which in the WT structure formed a bounding wall of the active site, is both moved outwards and disordered—it has largely disappeared from the electron density maps—reflecting both increased active site volume and, potentially, flexibility. The net effect is to increase the volume of the active site, improving its ability to accommodate the catalytically competent conformation of the bulky 3rd generation cephalosporins. This, then, provides a relatively rare structural view into a phenomenon common to resistance enzymes as diverse as dihydrofolate reductase 42, HIV protease 40, HIV reverse transcriptase 50, and BCR-ABL kinase 41 —the occurrence of resistance substitutions far from the active site.
The mutant enzymes T70I and E219K appear to gain activity against 3rd generation cephalosporins via increased flexibility. The T70I apo structure has two notable conformational changes relative to WT: notwithstanding the 2.14 Å resolution to which it was determined, density is missing for residues 193–221 in the active site “omega loop”, and an active site ß turn (residues 318–322) has flipped conformations to disrupt the catalytically critical oxyanion hole, formed by the backbone nitrogens of Ser64 and Ala318. Since this mutant enzyme is catalytically active, the x-ray structure can only represent one conformation accessible to the protein, suggesting that it is more flexible. Indeed, in the complex with the transition-state analog BZB, the active site of T70I resolves into a conformation that is again catalytically competent, resembling the native active site in conformation and overall volume. These observations are consistent with activity being gained by increased flexibility in T70I, which appears to be much more plastic on ligand binding than does the WT enzyme51. The situation with E219K is more subtle. We observe little substantial conformational change in either the E219K apo or inhibitor-bound structure relative to WT AmpC, though the mutant is 150-fold more active against cefotaxime. The loss of the internal hydrogen bond between the side chain and backbone nitrogen of residue 219 appears to loosen the structure, with the 215–216 peptide bond adopting a second conformation in the holo structure, while residues 211–213 also undergo a conformational change (Figure 6D). Neither of these conformations are seen in a 1.07 Å structure of WT AmpC 52 that revealed other inherent conformational states, suggesting increased flexibility in the E219K enzyme, again in the omega loop region.
How do these results fit with the stability-activity trade-off model? At the simplest level, the observation that the AmpC ESBLs have in every case lost stability is consistent with the model. At atomic resolution, too, the structural underpinnings of the “stability-function” hypothesis are confirmed—larger, more difficult substrates are recognized by the introduction of stability defects that increase either the ground-state size of the AmpC active site, or its ability to flex when confronted by the larger substrates.
It is this physical basis of the model that is key to understanding where the “stability-function” hypothesis is relevant, and how it may be reconciled with the observations of other groups. Clearly, stability-activity trade-offs are not linked by physical law; one can imagine substitutions that increase stability without effect on activity—indeed, the stability restoring mutant in TEM-1 ESBLs, M182T, is an example of such, 20; 22 and the evolution of thermophilic mutants that do not sacrifice activity is another 27. Nor does the evolution of new function necessarily require stability sacrifices. Indeed, results from this study suggest that an enzyme could mutate to a new substrate profile and gain stability. Thus, for all five mutant enzymes studied here, the kcat values for smaller 1st generation substrates, such as cephalothin, is actually reduced, typically by 3- to 8-fold, and by 100-fold for Y221G, for the destabilized mutants (Table 1). The same increase in active site volume and flexibility that allows the larger 3rd generation cephalosporins to be accommodated by the AmpC ESBL mutants decreases complementarity for smaller substrates, which fit the native active site snugly. If one inverts the evolution of AmpC, and imagines that Y221G is the native enzyme, then one would be able to evolve a “mutant” that has the WT sequence with 100-fold greater activity for the smaller cephalothin, and substantially improved stability. This would be a case where the active site has shrunk and new interactions with the rest of the folded protein have been (re)introduced, consistent with the physical model underlying the “stability-function” hypothesis.
Still, this case remains the exception that proves the rule. In enzymes evolving under the pressure of antimicrobial or antineoplastic chemotherapy, such as dihydrofolate reductase 42, HIV protease 40, BCR-ABL tyrosine kinase 41, HIV reverse transcriptase 50, and ß-lactamases, gain-of-function and many inhibitor-resistant mutants are likely to sacrifice stability, not because stability is a necessary correlate to activity, by some implied law, but rather because introduction of structural defects or further pre-organization into active sites typically reduce stability. It is a testament to the importance of stability in enzyme evolution that “restabilizing” and “recatalyzing” substitutions have been found in vivo to compensate for activity and stability losses occurring as a side effect of primary drug-resistance substitutions 20; 21; 22; 53. The role of stability as a constraint in enzyme evolution 23; 26 may well have implications for strategies to reduce resistance evolution under chemotherapeutic pressure 26.
Materials and Methods
Construction and purification of AmpC β-lactamases
Mutants of AmpC were created using the overlap extension polymerase chain reaction 54. Both WT and mutant enzymes were expressed and purified as described 49; 55, except Y221G, where the cells were lysed using a Microfluidizer M-110 at 18,000 psi in 50 mM Tris-HCl, pH 7, and the lysate was purified. The mutant enzymes were purified from an m-aminophenylboronic acid affinity column 55.
Kinetic measurements
The activity of each mutant enzyme was determined by its hydrolysis of the β-lactam substrate cephalothin (Sigma, St Louis, MO) in a 50 mM Tris-HCl buffer containing 0.01% Triton-X at pH 7.0. Reaction rates were measured in a Hewlett–Packard HP-8453 spectrometer. Values of kcat and Km were determined from Michaelis-Menten plots, and parameters were fit using Kaleidagraph (Synergy, Reading, PA). The extinction coefficients used were: AmpC: 2.45 OD/mg/mL/cm; cephalothin: Δε265 = −8790 M−1 cm−1; cefotaxime: Δε260 = −6710 M−1 cm−1. At high substrate concentrations, 1 mm pathlength cuvettes were used to obtain kinetic data.
Crystallization and structure determination
AmpC extended-spectrum mutants were crystallized in the following conditions: V298E: 30% PEG 8000, 0.1M sodium cacodylate, 0.2 M ammonium sulfate, pH 6.5; omega loop insertion: 1.7M KPi, pH 8.0; T70I/benzo(b)thiophene 2-boronic acid (BZB) and E219K/BZB: 1.7M KPi, pH 8.7, 360 μM BZB; Y221G and E219K: 1.7M KPi, pH 8.7. Y221G was also soaked in 50mM cefotaxime, 1.7 M KPi, pH 8.7, for 1 hour. Diffraction was measured at beamline 8.3.1 at the Advanced Light Source (Lawrence Berkeley National Labs, Berkeley, CA). Reflections were indexed, integrated and scaled using MOSFLM 56 and SCALA in CCP4 57. Molecular replacement was accomplished with Molrep in CCP4 57, using the appropriate apo structure, typically the wild-type structure (1KE4) as a search model. Model building and refinement was completed with Coot 58 and REFMAC5 59 in the CCP4 suite 57.
Thermal Denaturation
Enzymes were denatured by raising the temperature in 0.1 °C increments at a ramp rate of 2 °C/min in 50 mM potassium phosphate (pH 6.8), 50 mM potassium chloride, 38% (v/v) ethylene glycol buffer, using a Jasco 715 spectropolarimeter with a Peltier-effect temperature controller and an in-cell temperature monitor 46. Denaturation was marked by an obvious transition in the far-UV CD (223 nm) signal. Consistent with previous work with AmpC enzymes, which demonstrated reversible, two-state behavior (see Results) 10; 46, all mutant enzyme melts were reversible and apparently two-state, as judged by >90% return of CD signal upon quick cooling following denaturation, and a clear, sharp transition in the denaturation curve. Denaturation of two representative mutant enzyme (Y221G and H210AAA) was also measured by the intensity of the integrated fluorescence emission for all wavelengths above 300 nm, exciting at 280 nm, using a fluorescence detector on the Jasco instrument, and compared to thermal denaturation monitored by far-UV to investigate two-state behavior. Temperature of melting (Tm) and van't Hoff enthalpy of unfolding (ΔHvh) values were calculated using EXAM 60. The free energy of unfolding relative to WT was calculated using the method of Becktel and Schellman: ΔΔGu = ΔTmΔSu WT 61. A negative value of ΔΔGu indicates a stability loss. The ΔSu WTwas 0.56 kcal mol−1 K−1 46.
PDB Accession codes
X-ray crystal structure coordinates have been deposited in the Protein Data Bank with the following accession codes: V298E = 3IXD; H210AAA = 3IWI; T70I/BZB = 3IXG; Y221G = 3IWO; Y221G/CTX = 3IXH; E219K = 3IWQ; E219K/BZB = 3IXB.
Supplementary Material
Acknowledgements
This work was supported by NIH grants GM63813 (to BKS). VLT was partly supported by a National Science Foundation Predoctoral Fellowship. We thank Sandri Soelaiman and Ray Nagatani for some mutagenesis, protein purification and setting crystal trays, Kerim Babaoglu for crystallographic assistance, and Sarah Boyce and Jens Carlsson for reading this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Warshel A. Energetics of Enzyme Catalysis. Proc. Natl. Acad. Sci. USA. 1978;75:5250–5254. doi: 10.1073/pnas.75.11.5250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Warshel A, Sussman F, Hwang J-K. Evaluation of Catalytic Free Energies in Genetically Modified Proteins. J. Mol. Biol. 1988;201:139–159. doi: 10.1016/0022-2836(88)90445-7. [DOI] [PubMed] [Google Scholar]
- 3.Herzberg O, Moult J. Analysis of the Steric Strain in the Polypeptide Backbone of Protein Molecules. Proteins. 1991;11:223–229. doi: 10.1002/prot.340110307. [DOI] [PubMed] [Google Scholar]
- 4.Clackson T, Wells JA. A hot spot of binding energy in a hormone-receptor interface. Science. 1995;267:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 5.Richards FM. The interpretation of protein structures: total volume, group volume distributions and packing density. J. Mol. Biol. 1974;82:1–14. doi: 10.1016/0022-2836(74)90570-1. [DOI] [PubMed] [Google Scholar]
- 6.Williams RJ. The Entatic State. Cold Spring Harbor Symposia on Quantitative Biology. 1972;36:53–62. doi: 10.1101/sqb.1972.036.01.010. [DOI] [PubMed] [Google Scholar]
- 7.Meiering EM, Serrano L, Fersht AR. Effect of active site residues in barnase on activity and stability. J. Mol. Biol. 1992;225:585–9. doi: 10.1016/0022-2836(92)90387-y. [DOI] [PubMed] [Google Scholar]
- 8.Schreiber G, Buckle AM, Fersht AR. Stability and function: two constraints in the evolution of barstar and other proteins. Structure. 1994;2:945–51. doi: 10.1016/s0969-2126(94)00096-4. [DOI] [PubMed] [Google Scholar]
- 9.Shoichet BK, Baase WA, Kuroki R, Matthews BW. A Relationship Between Protein Stability and Protein Function. Proc. Nat. Acad. Sci USA. 1995;92:452–456. doi: 10.1073/pnas.92.2.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beadle BM, Shoichet BK. Structural bases of stability-function trade-offs in enzymes. J. Mol. Biol. 2002;321:285–296. doi: 10.1016/s0022-2836(02)00599-5. [DOI] [PubMed] [Google Scholar]
- 11.Ota M, Isogai Y, Nishikawa K. Structural requirement of highly-conserved residues in globins. FEBS Lett. 1997;415:129–33. doi: 10.1016/s0014-5793(97)01110-1. [DOI] [PubMed] [Google Scholar]
- 12.Mukaiyama A, Haruki M, Ota M, Koga Y, Takano K, Kanaya S. A hyperthermophilic protein acquires function at the cost of stability. Biochemistry. 2006;45:12673–9. doi: 10.1021/bi060907v. [DOI] [PubMed] [Google Scholar]
- 13.Hargrove MS, Olson JS. The stability of holomyoglobin is determined by heme affinity. Biochemistry. 1996;35:11310–8. doi: 10.1021/bi9603736. [DOI] [PubMed] [Google Scholar]
- 14.Chen YC, Lim C. Common physical basis of macromolecule-binding sites in proteins. Nucleic Acids Res. 2008;36:7078–87. doi: 10.1093/nar/gkn868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liang S, Zhang J, Zhang S, Guo H. Prediction of the interaction site on the surface of an isolated protein structure by analysis of side chain energy scores. Proteins. 2004;57:548–57. doi: 10.1002/prot.20238. [DOI] [PubMed] [Google Scholar]
- 16.Ota M, Kinoshita K, Nishikawa K. Prediction of catalytic residues in enzymes based on known tertiary structure, stability profile, and sequence conservation. J Mol Biol. 2003;327:1053–64. doi: 10.1016/s0022-2836(03)00207-9. [DOI] [PubMed] [Google Scholar]
- 17.Kidokoro S, Miki Y, Endo K, Wada A, Nagao H, Miyake T, Aoyama A, Yoneya T, Kai K, Ooe S. Remarkable activity enhancement of thermolysin mutants. FEBS Lett. 1995;367:73–6. doi: 10.1016/0014-5793(95)00537-j. [DOI] [PubMed] [Google Scholar]
- 18.Zhi W, Srere PA, Evans CT. Conformational stability of pig citrate synthase and some active-site mutants. Biochemistry. 1991;30:9281–9286. doi: 10.1021/bi00102a021. [DOI] [PubMed] [Google Scholar]
- 19.Garcia C, Nishimura C, Cavagnero S, Dyson HJ, Wright PE. Changes in the apomyoglobin folding pathway caused by mutation of the distal histidine residue. Biochemistry. 2000;39:11227–37. doi: 10.1021/bi0010266. [DOI] [PubMed] [Google Scholar]
- 20.Wang X, Minasov G, Shoichet BK. Evolution of an Antibiotic Resistance Enzyme Constrained by Stability and Activity Trade-Offs. J. Mol. Biol. 2002;320:85–95. doi: 10.1016/S0022-2836(02)00400-X. [DOI] [PubMed] [Google Scholar]
- 21.Huang W, Palzkill T. A natural polymorphism in beta-lactamase is a global suppressor. Proc Natl Acad Sci U S A. 1997;94:8801–6. doi: 10.1073/pnas.94.16.8801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sideraki V, Huang W, Palzkill T, Gilbert HF. A secondary drug resistance mutation of TEM-1 beta-lactamase that suppresses misfolding and aggregation. Proc Natl Acad Sci U S A. 2001;98:283–8. doi: 10.1073/pnas.011454198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weinreich DM, Delaney NF, Depristo MA, Hartl DL. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science. 2006;312:111–4. doi: 10.1126/science.1123539. [DOI] [PubMed] [Google Scholar]
- 24.Bloom JD, Labthavikul ST, Otey CR, Arnold FH. Protein stability promotes evolvability. Proc Natl Acad Sci U S A. 2006;103:5869–74. doi: 10.1073/pnas.0510098103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bloom JD, Arnold FH. In the light of directed evolution: pathways of adaptive protein evolution. Proc Natl Acad Sci U S A. 2009;106(Suppl 1):9995–10000. doi: 10.1073/pnas.0901522106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tokuriki N, Tawfik DS. Chaperonin overexpression promotes genetic variation and enzyme evolution. Nature. 2009;459:668–73. doi: 10.1038/nature08009. [DOI] [PubMed] [Google Scholar]
- 27.Giver L, Gershenson A, Freskgard PO, Arnold FH. Directed evolution of a thermostable esterase. Proc Natl Acad Sci U S A. 1998;95:12809–13. doi: 10.1073/pnas.95.22.12809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Massova I, Mobashery S. Kinship and Diversification of Bacterial Penicillin-Binding Proteins and Beta-Lactamases. Antimicrob. Agents Chem. 1998;42:1–17. doi: 10.1128/aac.42.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Galleni M, Lamotte-Brassear J, Raquet X, Dubus A, Monnaie D, Knox JR, Frere J-M. The Enigmatic Catalytic Mechanism of Active-Site Serine Beta-lactamases. Biochemical Pharmacology. 1995;49:1171–1178. doi: 10.1016/0006-2952(94)00502-d. [DOI] [PubMed] [Google Scholar]
- 30.Matagne A, Misselyn-Bauduin AM, Joris B, Erpicum T, Granier B, Frere JM. The diversity of the catalytic properties of class A beta-lactamases. Biochem J. 1990;265:131–46. doi: 10.1042/bj2650131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Powers RA, Caselli E, Focia PJ, Prati F, Shoichet BK. Structures of ceftazidime and its transition-state analogue in complex with AmpC beta-lactamase: implications for resistance mutations and inhibitor design. Biochemistry. 2001;40:9207–14. doi: 10.1021/bi0109358. [DOI] [PubMed] [Google Scholar]
- 32.Crichlow GV, Kuzin AP, Nukaga M, Mayama K, Sawai T, Knox JR. Structure of the extended-spectrum class C beta-lactamase of Enterobacter cloacae GC1, a natural mutant with a tandem tripeptide insertion. Biochemistry. 1999;38:10256–61. doi: 10.1021/bi9908787. [DOI] [PubMed] [Google Scholar]
- 33.Nukaga M, Kumar S, Nukaga K, Pratt RF, Knox JR. Hydrolysis of third-generation cephalosporins by class C beta-lactamases. Structures of a transition state analog of cefotoxamine in wild-type and extended spectrum enzymes. J Biol Chem. 2004;279:9344–52. doi: 10.1074/jbc.M312356200. [DOI] [PubMed] [Google Scholar]
- 34.Nukaga M, Taniguchi K, Washio Y, Sawai T. Effect of an amino acid insertion into the omega loop region of a class C beta-lactamase on its substrate specificity. Biochemistry. 1998;37:10461–8. doi: 10.1021/bi980184i. [DOI] [PubMed] [Google Scholar]
- 35.Matsumura N, Minami S, Mitsuhashi S. Sequences of homologous beta-lactamases from clinical isolates of Serratia marcescens with different substrate specificities. Antimicrob Agents Chemother. 1998;42:176–9. doi: 10.1128/aac.42.1.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raimondi A, Sisto F, Nikaido H. Mutation in Serratia marcescens AmpC beta-lactamase producing high- level resistance to ceftazidime and cefpirome. Antimicrob Agents Chemother. 2001;45:2331–9. doi: 10.1128/AAC.45.8.2331-2339.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Z, Yu Y, Musser JM, Palzkill T. Amino acid sequence determinants of extended spectrum cephalosporin hydrolysis by the class C P99 beta-lactamase. J Biol Chem. 2001;276:46568–74. doi: 10.1074/jbc.M102757200. [DOI] [PubMed] [Google Scholar]
- 38.Tsukamoto K, Ohno R, Nukaga M, Sawai T. The effect of amino acid substitution at position 219 of Citrobacter freundii cephalosporinase on extension of its substrate spectrum. Eur J Biochem. 1992;207:1123–7. doi: 10.1111/j.1432-1033.1992.tb17150.x. [DOI] [PubMed] [Google Scholar]
- 39.Morosini MI, Negri MC, Shoichet B, Baquero MR, Baquero F, Blazquez J. An extended-spectrum AmpC-type beta-lactamase obtained by in vitro antibiotic selection. FEMS Microbiol Lett. 1998;165:85–90. doi: 10.1111/j.1574-6968.1998.tb13131.x. [DOI] [PubMed] [Google Scholar]
- 40.Muzammil S, Ross P, Freire E. A major role for a set of non-active site mutations in the development of HIV-1 protease drug resistance. Biochemistry. 2003;42:631–8. doi: 10.1021/bi027019u. [DOI] [PubMed] [Google Scholar]
- 41.Shah NP, Nicoll JM, Nagar B, Gorre ME, Paquette RL, Kuriyan J, Sawyers CL. Multiple BCR-ABL kinase domain mutations confer polyclonal resistance to the tyrosine kinase inhibitor imatinib (STI571) in chronic phase and blast crisis chronic myeloid leukemia. Cancer Cell. 2002;2:117–25. doi: 10.1016/s1535-6108(02)00096-x. [DOI] [PubMed] [Google Scholar]
- 42.Volpato JP, Pelletier JN. Mutational `hot-spots' in mammalian, bacterial and protozoal dihydrofolate reductases associated with antifolate resistance: sequence and structural comparison. Drug Resist Updat. 2009;12:28–41. doi: 10.1016/j.drup.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 43.Dubus A, Wilkin JM, Raquet X, Normark S, Frere JM. Catalytic mechanism of active-site serine beta-lactamases: role of the conserved hydroxy group of the Lys-Thr(Ser)-Gly triad. Biochem J. 1994;301:485–94. doi: 10.1042/bj3010485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trehan I, Morandi F, Blaszczak LC, Shoichet BK. Using steric hindrance to design new inhibitors of class C beta-lactamases. Chem Biol. 2002;9:971–80. doi: 10.1016/s1074-5521(02)00211-9. [DOI] [PubMed] [Google Scholar]
- 45.Mazzella LJ, Pratt RF. Effect of the 3'-leaving group on turnover of cephem antibiotics by a class C beta-lactamase. Biochem J. 1989;259:255–60. doi: 10.1042/bj2590255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beadle BM, McGovern SL, Patera A, Shoichet BK. Functional analyses of AmpC beta-lactamase through differential stability. Protein Sci. 1999;8:1816–24. doi: 10.1110/ps.8.9.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nukaga M, Haruta S, Tanimoto K, Kogure K, Taniguchi K, Tamaki M, Sawai T. Molecular evolution of a class C beta-lactamase extending its substrate specificity. J Biol Chem. 1995;270:5729–35. doi: 10.1074/jbc.270.11.5729. [DOI] [PubMed] [Google Scholar]
- 48.Powers RA, Blazquez J, Weston GS, Morosini MI, Baquero F, Shoichet BK. The complexed structure and antimicrobial activity of a non-beta-lactam inhibitor of AmpC beta-lactamase. Protein Sci. 1999;8:2330–7. doi: 10.1110/ps.8.11.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Patera A, Blaszczak LC, Shoichet BK. Crystal structures of substrate and inhibitor complexes with AmpC -lactamase: Possible implications for substrate-assisted catalysis. J. Am. Chem. Soc. 2000;122:10504–10512. [Google Scholar]
- 50.Ghosn J, Chaix ML, Delaugerre C. HIV-1 resistance to first- and second-generation non-nucleoside reverse transcriptase inhibitors. AIDS Rev. 2009;11:165–73. [PubMed] [Google Scholar]
- 51.Beadle BM, Trehan I, Focia P, Shoichet BK. Structural Milestones in the Pathway of an Amide Hydrolase: substrate, acyl, and product complexes of cephalothin with AmpC b-lactamase. Structure. 2002;10:413–424. doi: 10.1016/s0969-2126(02)00725-6. [DOI] [PubMed] [Google Scholar]
- 52.Chen Y, Minasov G, Roth TA, Prati F, Shoichet BK. The deacylation mechanism of AmpC beta-lactamase at ultrahigh resolution. J Am Chem Soc. 2006;128:2970–6. doi: 10.1021/ja056806m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schock HB, Garsky VM, Kuo LC. Mutational anatomy of an HIV-1 protease variant conferring cross-resistance to protease inhibitors in clinical trials. Compensatory modulations of binding and activity. J Biol Chem. 1996;271:31957–63. doi: 10.1074/jbc.271.50.31957. [DOI] [PubMed] [Google Scholar]
- 54.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–9. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 55.Usher KC, Blaszczak LC, Weston GS, Shoichet BK, Remington SJ. Three-dimensional structure of AmpC beta-lactamase from Escherichia coli bound to a transition-state analogue: possible implications for the oxyanion hypothesis and for inhibitor design. Biochemistry. 1998;37:16082–92. doi: 10.1021/bi981210f. [DOI] [PubMed] [Google Scholar]
- 56.Leslie AGW. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography. 1992;26 [Google Scholar]
- 57.The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–3. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 58.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 59.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–55. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 60.Kirchoff W. EXAM: A two-state thermodynamic analysis program. Gaithersburg; Maryland: 1993. [Google Scholar]
- 61.Becktel WJ, Schellman JA. Protein stability curves. Biopolymers. 1987;26:1859–77. doi: 10.1002/bip.360261104. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.