Abstract
The protein phosphatase calcineurin is a key mediator of virulence and antifungal susceptibility of multiple fungal pathogens including Candida albicans, Cryptococcus neoformans, and Aspergillus fumigatus, and has clinical potential as a therapeutic target to increase the efficacy of the current antifungal armamentarium. Despite the importance of this signaling pathway, few components of the calcineurin-signaling pathway are known in C. albicans. Here we identified and analyzed additional components of the C. albicans calcineurin cascade, including the RCN1 (Regulator of Calcineurin1), MID1, and CCH1 genes, which mediate calcineurin functions in other species. When heterologous expressed in S. cerevisiae, C. albicans Rcn1 inhibited calcineurin function. Although rcn1/rcn1, mid1/mid1, and cch1/cch1 mutant strains share some phenotypes with calcineurin mutants, they do not completely recapitulate the phenotypes of a calcineurin mutant strain. These studies extend our understanding of the C. albicans calcineurin signaling cascade and its host-niche specific role in virulence.
Keywords: Candida albicans, calcineurin, Rcn1, Mid1, Cch1, fungal pathogenesis, fluconazole
Introduction
Calcineurin is a calcium, calmodulin-dependent serine-threonine specific protein phosphatase that is highly conserved from yeast to humans and mediates many important cellular processes (Hemenway and Heitman, 1999). In mammalian cells, calcineurin is involved in cardiac muscle differentiation (Chin et al., 1998; Kramer et al., 2003; Parsons et al., 2007), memory (Mansuy et al., 1998; Weitlauf and Winder, 2001), T-cell activation (Clipstone and Crabtree, 1992), and apoptosis (Krebs, 1998; Saito et al., 2000; Shibasaki and McKeon, 1995; Wang et al., 1999). The immunosuppressants FK506 and Cyclosporin A (CsA) exert their effect by entering cells and binding to an immunophilin protein partner (FKBP12 for FK506, and Cyclophilin A for CsA) (Cardenas et al., 1994; Cardenas et al., 1995; Clipstone et al., 1994; Ho et al., 1996). This protein-drug complex subsequently binds calcineurin and inhibits its activity and functions. In human T-cells, calcineurin activates a transcription factor (NF-AT), which promotes the expression of cytokines and T-cell proliferation (Crabtree, 1999). Due to the highly conserved nature of calcineurin, it was subsequently found that FK506 and CsA can inhibit not only mammalian calcineurin, but also fungal calcineurin (Blankenship et al., 2003a; Breuder et al., 1994; Foor et al., 1992; Nakamura et al., 1993; Steinbach et al., 2007).
Candida spp. are normal components of the human microbiota; however, under conditions of immunosuppression or altered host defenses these commensals have the ability to cause serious mucocutaneous and systemic disease (Odds, 1988). Diseases due to Candida spp. manifest a variety of clinical manifestations, ranging from mucocutaneous infections mouth (thrush), esophagus, and vagina to life-threatening systemic infections, where Candida spp. enter the bloodstream and disseminate throughout the body to infiltrat target organs (Edwards, 1991). Although C. albicans has historically accounted for the majority of candidal infections, following the introduction of the antifungal fluconazole numerous other species have increased in prevalence including C. glabrata, C. parapsilosis, C. tropicalis, and C. krusei (Hazen, 1995; Krcmery and Barnes, 2002; Merz et al., 1986; Nguyen et al., 1996; Pfaller et al., 1998). Despite intensive drug discovery efforts, there are still only 3 classes of antifungal drugs that are available to treat serious fungal infections, Amphotericin B, azoles and echinocandins. Additionally, despite their widespread usage the azoles are fungistatic rather than fungicidal. Thus, there is need for new strategies and therapeutics to combat fungal infections.
Interestingly, the combination of calcineurin inhibitors and the normally fungistatic antifungal fluconazole result in potent killing of C. albicans, as well as other more drug resistant species such as C. glabrata (Cruz et al., 2002; Marchetti et al., 2000; Onyewu et al., 2003). Additionally, C. albicans calcineurin mutants have attenuated virulence in murine models of systemic infection (Bader et al., 2003; Blankenship et al., 2003b), and have faster rates of disease resolution in murine keratitis models (Onyewu et al., 2006). The attenuated virulence of C. albicans calcineurin mutants in systemic disease is attributable to the inability of these strains to withstand the calcium stress imposed by serum and thus survive transit through the bloodstream (Blankenship and Heitman, 2005). However, the direct role of calcineurin in C. albicans virulence appears to be host niche-specific as there was no virulence defect seen in either a vaginal or a pulmonary model of infection (Bader et al., 2006). Thus, calcineurin inhibitors have two potential mechanisms of action in the clinic: 1) as single agents in cases of disseminated disease or ocular infections to directly impair survival of the yeasts, or 2) as combination therapy to enhance the efficacy of current antifungal therapies. However, the immunosuppressive nature of calcineurin inhibitors limits their use in systemic therapy. Therefore, we were interested in further characterizing the calcineurin signaling cascade to learn more about this important stress response pathway. We also wanted to elucidate other components that could serve as alternative drug targets that would circumvent the immunosuppressive effects of inhibiting calcineurin.
In C. albicans, calcineurin is required for cells to survive stressors such as high cations (Li+, Na+, and Ca2+), antifungal drug treatment (azoles), and the host bloodstream (Bader et al., 2003; Blankenship and Heitman, 2005; Blankenship et al., 2003b; Sanglard et al., 2003). Based upon homology with S. cerevisiae Crz1, the downstream transcription factor Crz1 (Cyert, 2003) was previously identified in C. albicans, and shown to shuttle into the nucleus in a calcineurin-dependent manner (Karababa et al., 2006). However, phenotypic analysis of crz1/crz1 strains only partially recapitulated a calcineurin mutant phenotype. Although crz1/crz1 mutant strains are sensitive to cations and membrane stresses, they exhibited an intermediate phenotype compared with calcineurin mutants (Karababa et al., 2006; Onyewu et al., 2004; Santos and de Larrinoa, 2005). Microarray studies have suggested that Crz1 is the primary mediator of the calcineurin-dependent transcriptional response (Karababa et al., 2006). As a first step towards elucidating other potential genes in the C. albicans calcineurin pathway, we took a candidate gene approach based on analogous signaling pathways in S. cerevisiae.
Few proteins are known that are direct binding partners of calcineurin; these include calmodulin, transcription factors (Crz1, C. albicans; Crz1/Tcn1 S. cerevisiae; NF-AT, mammalian cells), and the RCAN family of proteins (Beals et al., 1997; Cyert, 2003; Davies et al., 2007; Hilioti and Cunningham, 2003; Karababa et al., 2006; Klee et al., 1979; Matheos et al., 1997; Onyewu et al., 2004; Santos and de Larrinoa, 2005; Stathopoulos and Cyert, 1997). Members of the RCAN family have been identified in species including S. cerevisiae (Rcn1), C. neoformans (Cbp1), and humans (DSCR1/MCIP1) based upon a conserved FLSPPxSP domain (Davies et al., 2007; Gorlach et al., 2000; Hilioti and Cunningham, 2003; Strippoli et al., 2000a). The function of these proteins has been best explored in S. cerevisiae where they exert both positive and negative effects on calcineurin function. Rcn1 binds calcineurin and inhibits its function. However, upon phosphorylation by a GSK3 kinase Rcn1 is degraded thereby relieving calcineurin inhibition (Hilioti et al., 2004). In S. cerevisiae, Rcn1 expression is induced in a calcineurin-dependent manner, and the phosphorylated protein is itself a substrate for calcineurin (Gorlach et al., 2000; Hilioti and Cunningham, 2003; Hilioti et al., 2004; Kishi et al., 2007). Overexpression of RCAN family members (or their calcineurin binding domain) inhibits calcineurin function in both S. cerevisiae and in mammalian cells (Fuentes et al., 2000; Gorlach et al., 2000; Hilioti and Cunningham, 2003; Hilioti et al., 2004; Vega et al., 2002). Thus, RCANs serve as important control elements of the calcineurin cascade that could potentially be manipulated to therapeutically inhibit calcineurin function.
Another key aspect of calcineurin signaling is regulation of cellular calcium homeostasis and signaling. Direct targets of calcineurin include multiple calcium channels (Vcx1, Mid1/Cch1) (Bonilla et al., 2002; Cunningham and Fink, 1996). In S. cerevisiae, endoplasmic reticulum stress activates the Mpk1 pathway, which activates a plasma membrane calcium channel composed of Cch1 and Mid1 (Bonilla and Cunningham, 2003; Bonilla et al., 2002). Activation of the channel results in calcium influx and activation of calcineurin, which subsequently feedback inhibits the channel through dephosphorylation (Cunningham and Fink, 1994a). Thus, Mid1 and Cch1 control calcineurin activation. Previous studies in C. albicans characterized Mid1 and Cch1 roles in galvano- and thigmo-tropism (Brand et al., 2007). Deletion of either or both calcium channels significantly decreased calcium accumulation. However, the mutants differed in their response to various stimuli: Cch1 appears to play a greater role in hyphal orientation in response to electric fields, while loss of Mid1 had a more significant impact on hyphal tip reorientation in response to physical contract. Interestingly, calcineurin was required for the reorientation of hyphae in an electric field, but not involved in thigmotropism; however, Crz1 was required for both processes (Brand et al., 2007).
In this study, we used mutant analysis to investigate the roles C. albicans Rcn1, Mid1, and Cch1 homologs play in the response of C. albicans to stress. We found that while each of these proteins is required for the maximal resistance to some stressors, none of them is as important as calcineurin itself. Therefore, each of these proteins likely functions in only part of the calcineurin-signaling pathway.
Materials and methods
Strains and Media
All strains were routinely propagated on YPD medium (1% yeast extract, 2% bacto peptone, 2% dextrose, and 2% bacto agar (DIFCO)). YPD + 300 mM CaCl2 medium was made similarly to YPD except that the media was adjusted to pH 5 prior to autoclaving. The CaCl2 solution was sterilized separately and the two solutions were mixed after autoclaving. All strains used in this study are listed in Table 1.
Table 1.
Strains used in this study.
| Strain | Genotype | Background | Source |
|---|---|---|---|
| SC5314 | (Gillum et al., 1984) | ||
| DAY185 | URA3/ura3Δ::λimm434 HIS1/his1::hisG ARG4/arg4::hisG | BWP17 | (Davis et al., 2000) |
| JRB64 |
ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG::HIS1/his1::hisG arg4::hisG/arg4::hisG cnb1::UAU1/cnb1::URA3 |
BWP17 | (Blankenship et al., 2003b) |
| MCC85 |
ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG::CNB1-HIS1/his1::hisG arg4::hisG/arg4::hisG cnb1::UAU 1/cnb1::URA3 |
BWP17 | (Blankenship et al., 2003b) |
| SCCMP1M4 | cna1::frt/cna1::frt | SC5314 | (Bader, et al., 2006) |
| SCCMP1M2 | cna1::frt/cna1::frt + CNA1 | SC5314 | (Bader, et al., 2006) |
| OCC1.1 |
ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG::HIS1/his1::hisG arg4::hisG/arg4::hisG crz1::UAU1/crz1::URA3 |
BWP17 | (Onyewu, et al., 2004) |
| OCC7 |
ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG::CRZ1-HIS1/his1::hisG arg4::hisG/arg4::hisG crz1:: UA U1/crz1:: URA3 |
BWP17 | (Onyewu, et al., 2004) |
| JLR36.3 | rcn1::frt/rcn1::frt | SC5314 | this study |
| JLR37.1 | rcn1::frt/rcn1::frt | SC5314 | this study |
| JLR180 | rcn1::frt/rcn1::frt + RCN1 | JLR36.3 | this study |
| JLR323 | rcn1::frt/rcn1::frt + RCN1 | JLR37.1 | this study |
| JLR248 | mid1::frt/mid1::frt | SC5314 | this study |
| JLR253 | mid1::frt/mid1::frt | SC5314 | this study |
| JLR255 | mid1::frt/mid1::frt | SC5314 | this study |
| JLR284 | mid1::frt/mid1::frt + MID1 | JLR248 | this study |
| JLR301 | mid1::frt/mid1::frt + MID1 | JLR255 | this study |
| JLR48 | cch1::frt/cch1::frt | SC5314 | this study |
| JLR50 | cch1::frt/cch1::frt | SC5314 | this study |
| JLR265 | cch1::frt/cch1::frt | SC5314 | this study |
| JLR548 | crz1/crz1 rcn1/rcn1 | JLR36.3 | this study |
| JLR578 | crz1/crz1 rcn1/rcn1 | JLR37.1 | this study |
| JLR521 | mid1/mid1 cch1/cch1 | JLR248 | this study |
| JLR519 | mid1/mid1 cch1/cch1 | JLR255 | this study |
| K661 |
MATa ade2-1 can1-100 his3-11,15 leu2-3 112 trp1-1 ura3-1 vcx1D |
W303-1A | (Cunningham, et al, 1996) |
| K605 |
MATa ade2-1 can1-100 his3-11,15 leu2-3 112 trp1-1 ura3-1 pmc1::TRP1 |
W303-1A | (Cunningham, et al., 1994) |
| K665 |
MATa ade2-1 can1-100 his3-11,15 leu2-3 112 trp1-1 ura3-1 pmc1::TRP1 vcx1D |
W303-1A | (Cunningham, et al, 1996) |
Gene Disruptions
All deletion strains were generated in the SC5314 background. All primers used in strain construction are listed in Table 2. For disruption of the RCN1 gene, two ~500 bp regions with homology to the 5’ promoter and 3’ terminator region of RCN1 were PCR amplified, and cloned into plasmid pSFS2A (Reuss et al., 2004) with KpnI/XhoI, and NotI/SacI, respectively, generating plasmid pJLR1. Plasmid pJLR1 was digested with KpnI/SacI and the disruption cassette consisting of the SAT1 flipper cassette surrounded by ~500 bp of homology flanking RCN1 was gel purified. SC5314 was transformed with approximately 1 µg of DNA as previously described (Reuss et al., 2004). For all deletion strains, at least 3 independent transformations were performed at each step of disruption and an independent transformant was selected from each transformation for further analysis. Nourseothricin resistant isolates were selected on YPD + 200 µg/ml NAT (Werner). Correct integrants (RCN1/rcn1::SAT1) were confirmed by colony PCR and then by Southern blot. At least 3 independent strains that had correctly integrated the disruption cassette were grown overnight in YPM (1% yeast extract, 2% bacto peptone, 2% maltose, and 2% Bactoagar (Difco)) at 30°C and then plated onto YPD + 25 µg/ml NAT. Small colonies which represented those that had excised the SAT1 cassette (RCN1/rcn1::frt) were selected and confirmed by Southern blot. Three independent transformants were selected to undergo a second round of transformation to disrupt the remaining RCN1 allele.
Table 2.
Primers used in this study.
| Name | Sequence | Notes |
|---|---|---|
| JLR121 | AGTCAACGTAGGTACCATGAACCTTAGATAATTACG | RCN1 5′ flank KpnI |
| JLR120 | TCAACTTCTCGAGCATCAATATTGGTAATGATTAATG | RCN1 5′ flank XhoI |
| JLR118 | TCAATCATTGCGGCCGCAAGATCATCGATGTAATATGC | RCN1 3′ flank NotI |
| JLR119 | TACTTACAAGAGCTCGTCTTGGATAGAACATAAAATAC | RCN1 3′ flank SacI |
| JLR130 | TCAACTTCTCGAGGTCTTGGATAGAACATAAAATAC | RCN1 complementation 3' XhoI |
| JLR126 | AGTCAACGTAGGTACCCATACACTTGCTTCGCCTCTA | CCH1 5′ flank KpnI |
| JLR127 | TCAACTTCTCGAGGGTATTGCTGTTATCCATG | CCH1 5′ flank XhoI |
| JLR125 | TCAATCATTGCGGCCGCGAACTGTCATCTTGGAAAAATGC | CCH1 3′ flank NotI |
| JLR124 | TACTTACAAGAGCTCCTGATCCAGAAGATTTAGCT | CCH1 3′ flank SacI |
| JLR280 | ATGTGCTATTCATTAACCACTAGCCTTCATCATGATACCACCTTTT ATTCTACTACTATTTCTAATAATGAACTCATCATGGCCCCCCCTC GAGGAAGTT |
MID1 5′ deletion primer |
| JLR281 | AGAACATAAACAATCAATCAATCGATTCTTCTTCTTCTTTTCTATC TATTATATTCTATTTAAATGAACTATATGACAACGCTCTAGAACT AGTGGATCT |
MID1 3′ deletion primer |
| JLR163 | TCAACTTCTCGAGGAAGATTTGTCGAGTACTAG | MID1 complementation 3' XhoI |
| JLR133 | TCAATCATTGCGGCCGCCCTTTACTTTAATGGTTGTCA | MID1 3′ flank NotI |
| JLR132 | TACTTACAAGAGCTCGAGCTACATTAGTCATAGCAT | MID1 3′ flank SacI |
| JLR29 | ATTTCCCCTAACAGCATCTTTCCAAGTTCAAATATTTTCCCCTTTT TATATCTAAATTTCATAAATCCCAATCATGTCTGGCCCCCCCTCG AGGAAGTT |
CRZ1 5′ deletion primer |
| JLR295 | TAGAATAAAAAAACAACCAACCAACCAACCAACAGGAATAACT ATCGTGAATGACAACAACCTCAAAAAAAAACTAAGTGCTCTAGA ACTAGTGGATCT |
CRZ1 3′ deletion primer |
| JLR470 | TCAGGTACTAAGCTGATGCCAAGGAAACCAACAAAT | Ca RCN1 overexpression 5' PvuII |
| JLR473 | ACGTACTAGATCTAGAGTATGTTGACTTGGATAGAAC | Ca RCN1 overexpression 3' XbaI |
| JLR463 | TCAGGTACTACAGCTGATGGGTAATATTATAACGGAT | Sc RCN1 overexpression 5' PvuII |
| JLR466 | ACGTACTAGATCTAGAAAACAATTAACGAGCCATGG | Sc RCN1 overexpression 3' XbaI |
The RCN1 complementation cassette to reintroduce the RCN1 gene at the native locus was generated by PCR amplification of the full-length RCN1 gene plus ~500 bp of promoter sequence and ∼300 bp of terminator, cloned into plasmid pJLR-RCN3 (containing ~500 bp of homology to the 3’ terminator of RCN1 cloned into the NotI/SacI site of pSFS2A) and sequencing revealed no extraneous mutations were found. Because the two alleles of RCN1 differ in C. albicans, plasmids containing each allele were selected (pJLR35 and pJLR37) and used for complementations. Similarly to above, a KpnI/SacI gel purified fragment was used for transformation.
The rcn1/rcn1 crz1/crz1 strains were isolated by disrupting the CRZ1 gene in strains JLR36.3, JLR37.1, and JLR38.1. The CRZ1::SAT1 disruption cassette was obtained by amplifying the SAT1 flipper cassette using long primers with ~90 bp of homology to the region flanking CRZ1. PCRs were carried out using the following conditions: 94°C for 3 minutes, followed by 30 cycles of 94°C for 30 sec, 55°C for 30 sec, 68°C for 4 minutes, and a final extension of 68°C for 15 min. All PCRs were performed using exTAQ polymerase (Takara), and 5% DMSO. An ~4.2 kb PCR fragment was gel purified and 1 µg was used for transformation as discussed above.
The CCH1 disruption cassette was generated by PCR amplifying two ~500 bp fragments consisting of 5’ promoter and 3’ terminator regions of the CCH1 gene, and cloned into plasmid pSFS2A in two steps with KpnI/XhoI, and NotI/SacI respectively, yielding plasmid pJLR3. The procedure followed for disruption was the same as used for RCN1. Three independent transformants were confirmed by Southern blot.
The MID1 disruption cassette was produced by amplifying the SAT1 flipper cassette using long primers with ~90 bp of homology to the region flanking MID1 in a similar manner to the disruption of CRZ1. Three independent transformants were selected and confirmed by Southern blot. The MID1 complementation cassette was engineered by PCR amplifying the full MID1 ORF with ~500 bp of promoter and ~300 bp of terminator. This PCR fragment was cloned into plasmid pJLR-CCH3 (containing ~500 bp 3’ MID1 flanking homology cloned into the NotI/SacI site of pSFS2A) using KpnI/XhoI. Complementation was carried out as discussed above for RCN1. The mid1/mid1 cch1/cch1 strain was created by disrupting CCH1 in the mid1/mid1 strains JLR248, JLR253, and JLR255 using the disruption cassette for CCH1 described above.
In vitro stress testing
The minimum inhibitory concentrations 80 (MIC80) for all strains was determined using E-test strips (AB Biodisk). For the serial dilution spot assays, strains were grown overnight in YPD at 30°C, washed twice with PBS, and normalized to 1 × 107 cells/ml. 1:10 serial dilutions were spotted onto YPD, YPD containing fluconazole (Diflucan®, Pfizer), FK506 (Prograf, Astellas Pharma US, Inc) or cations. Cells were grown at 30°C and monitored for growth at 24 and 48 hours.
For liquid serum assays, strains were grown overnight in YPD at 30°C, washed twice with PBS, and inoculated into 100% FBS with or without 1 µg FK506 at 2000 cells/ml. Cultures were incubated at 30°C for 24 hours. This temperature was used because it does not affect the interaction between serum and FK506 (calcineurin mutants lose viability at both 30°C and 37°C), and cells incubated at 30°C do not form hyphae, so that the changes in fold population could be accurately measured. Appropriate serial dilutions of the cultures were plated onto YPD for CFU (colony forming unit) counts at 0 and 24 hours. The fold population change was determined by dividing the CFU’s at 24 hours by the CFU’s at 0 hours.
Heterologous complementation in S. cerevisiae
The C. albicans and S. cerevisiae RCN1 homologs were PCR amplified and subcloned into the pCR2.1 TOPO TA vector (Invitrogen) and sequenced. The RCN1 genes were released by cutting with PvuII and XbaI, gel purified, and cloned into the PvuII/XbaI site of plasmid pYES placing them under the control of the S. cerevisiae GAL1 promoter. Plasmids were confirmed by digestion and sequencing to have the desired structure. S. cerevisiae strains K661 (vcx1), K605 (pmc1), and K665 (vcx1 pmc1) (Cunningham and Fink, 1994b; Cunningham and Fink, 1996; Hilioti and Cunningham, 2003) were transformed with either pYES alone, or plasmids containing either the C. albicans or S. cerevisiae GAL1-RCN1. Cells were grown to an OD600=0.5 to 0.7, washed with 1 mL 0.1 M LiOAc TE, then resuspended in 0.1 M LiOAc TE. 50 µl of cells were mixed with 2.5 µl salmon sperm DNA (11 mg/ml) (Sigma), 1 µl plasmid, and 300 µl 40% PEG 0.1 M LiOAc The mixture was incubated at 30°C for 30 minutes, and then heat shocked at 42°C for 15 minutes. Cells were then washed with H2O and plated on SD-ura to select positive transformants. To test for complementation strains carrying pYES alone, GAL1-ScRCN1, or GAL1-CaRCN1 were streaked onto YPD pH 5, YP galactose pH 5, YPD pH 5 with 0.3 M CaCl2, and YP galactose pH 5 with 0.3 M CaCl2. The experiment was performed in duplicate with two different GAL1-RCN1 plasmids for each species.
Virulence Assays
For the disseminated candidiasis model, 5 × 106 cells of wild-type (SC5314), two independent rcn1/rcn1 (JLR36.3 and JLR37.1), and two rcn1/rcn1 + RCN1 strains (JLR180 and JLR323) were injected into the lateral tail vein. Five outbred ICR mice (NCI) were infected per group and monitored for survival. Log rank statistical analysis of the survival data was performed using the PRISM 4.02 program (GraphPad Software, San Diego, Calif.)
The oropharyngeal candidiasis model was performed using mice that were immunosuppressed with cortisone acetate and then orally inoculated as previously described (Park et al., 2005). Seven mice were infected per group. Both histology and colony forming units (CFUs) per gram of oral tissue after 5 days of infection were used to evaluate virulence. Differences in oral fungal burden among mice infected with the different strains were analyzed using the Wilcox Rank Sum test.
Results
Identification of the C. albicans RCN1 homolog
The RCAN (Regulator of Calcineurin) family of proteins are calcineurin binding proteins that modulate calcineurin activity in S. cerevisiae (RCN1), C. neoformans (CBP1), and mammalian cells (DSCR1/MCIP1) (Davies et al., 2007; Gorlach et al., 2000; Hilioti and Cunningham, 2003; Strippoli et al., 2000a; Strippoli et al., 2000b). In S. cerevisiae, Rcn1 was identified in a screen for proteins that inhibited calcineurin function when overexpressed (Hilioti and Cunningham, 2003). Subsequent studies have shown that the transcription of RCN1 in S. cerevisiae is induced by calcineurin and that Rcn1 has both positive and negative effects on calcineurin function depending on the cellular concentration and phosphorylation state of the protein (Hilioti et al., 2004; Kishi et al., 2007). The C. albicans Rcn1 homolog was identified based on the conserved KxFLSPPxSPP domain (Figure 1) (Hilioti and Cunningham, 2003). With the exception of this conserved domain the RCAN family members generally share little homology. Thus, it was expected that this would be the only conserved region of the C. albicans Rcn1 protein. The two alleles of RCN1 in C. albicans wild-type strain SC5314 were found to differ by ten amino acids, including 3 amino acid changes and an insert of 7 amino acids in Rcn1-1.
Figure 1. Structure of RCAN family members and C. albicans Rcn1.

A. Conserved domain of RCAN family members. ClustalW alignment of the conserved domain found in all RCAN family members (created with MacVector™). Shown are C. neoformans Cbp1, S. cerevisiae Rcn1, S. pombe SPAC13G6.15c, H. sapien DSCR1, and both C. albicans Rcn1 genes.
Three independent C. albicans rcn1/rcn1 disruption mutants were isolated using the SAT1 flipper cassette method described previously. For complementation experiments, a wild-type allele of RCN1 was reintroduced at the native locus in all three independent mutants. Two of the mutants were complemented with the RCN1-2 allele and one was complemented with the RCN1-1 allele. Both RCN1 alleles were capable of restoring wild-type phenotypes, and there was no observed functional difference between the two alleles.
rcn1/rcn1 strains are sensitive to LiCl, SDS, and fluconazole
Calcineurin mutants are exquisitely sensitive to cellular stress imposed by cell wall and cell membrane perturbing compounds, as well as various cations (Ca2+, Li+, and Na+) (Bader et al., 2003; Blankenship et al., 2003b; Sanglard et al., 2003). The susceptibility of calcineurin mutants is of particular interest in the clinical setting because calcineurin mutant strains, or wild-type strains in the presence of calcineurin inhibitory drugs (FK506 or Cyclosporin A), are killed by the normally fungistatic azole antifungal drugs (Cruz et al., 2002; Onyewu et al., 2003). Thus, we were interested in whether rcn1/rcn1 mutants would have similar sensitivities as calcineurin mutant strains. C. albicans rcn1/rcn1 deletion strains were more susceptible to LiCl, SDS, and fluconazole than the wild-type strain (SC5314), but less sensitive than cnb1/cnb1 or cna1/cna1 calcineurin mutant strains (Figure 2). In contrast to calcineurin mutant strains, rcn1/rcn1 mutants are not hypersensitive to calcium or serum. This suggests that the RCN1 gene may be involved in the execution of some, but not all calcineurin functions. Similar results have been found in studies of RCN1 homologs in both S. cerevisiae and C. neoformans. rcn1 deletion in S. cerevisiae resulted in a modest sensitivity to Li+ that was less severe than a calcineurin mutant phenotype (Gorlach et al., 2000; Hilioti and Cunningham, 2003). Likewise, C. neoformans cbp1 mutants were still able to grow at 37°C whereas calcineurin mutants are inviable at this temperature. However, deletion of cbp1 resulted in reduced virulence in a murine model of cryptococcal infection, albeit to a lesser degree than calcineurin mutants (Gorlach et al., 2000).
Figure 2. rcn1/rcn1 strains are sensitive to SDS, Li+, and fluconazole.
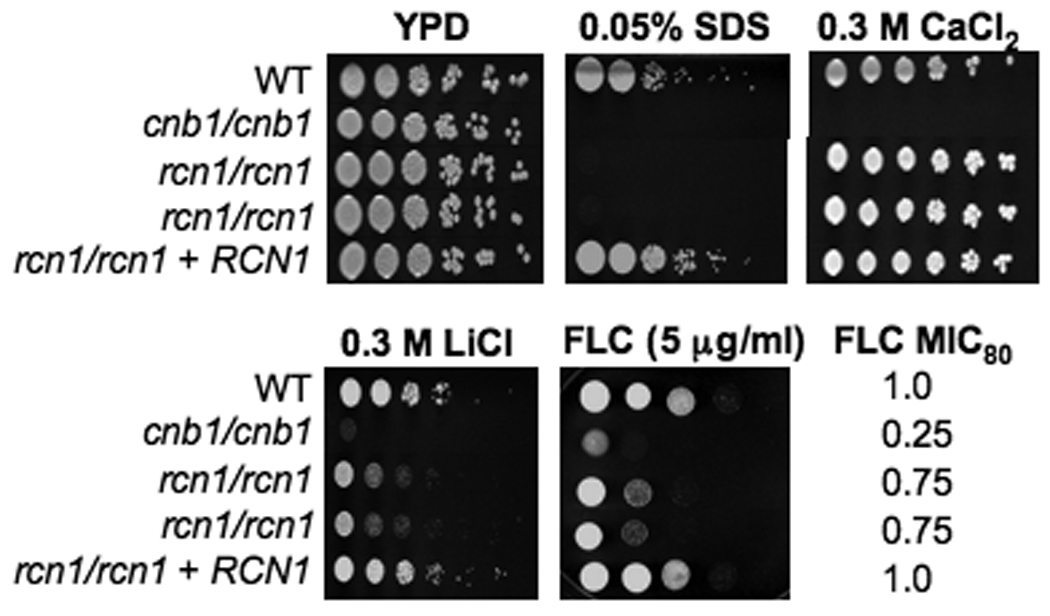
Serial dilutions of the wild-type (SC5314), cnb1/cnb1 (JRB64), rcn1/rcn1 (JLR36.3 and JLR37.1) and rcn1/rcn1 +RCN1 (JLR180) strains were spotted onto solid YPD media containing the designated salt or drug concentration and incubated at 30°C for 48 hours. Fluconazole MIC80 was determined by E-TEST.
Neither rcn1/rcn1 nor cnb1/cnb1 strains have a virulence defect in an oropharyngeal candidiasis model
Calcineurin mutant strains of C. albicans have attenuated virulence in murine models of disseminated disease (Bader et al., 2003; Blankenship et al., 2003b). This attenuation is due to the inability of C. albicans calcineurin mutants to withstand the calcium stress imposed by serum, and survive transit through the bloodstream to disseminate throughout the body (Blankenship and Heitman, 2005). Therefore, we tested the virulence of the rcn1/rcn1 mutant strains in this model. We found that the survival of mice infected with the rcn1/rcn1 mutant strains was similar to that of mice infected with wild-type C. albicans (Figure 3). Therefore, Rcn1 is not essential for maximal virulence during disseminated candidiasis.
Figure 3. Virulence of rcn1/rcn1 in a murine disseminated candidiasis model.
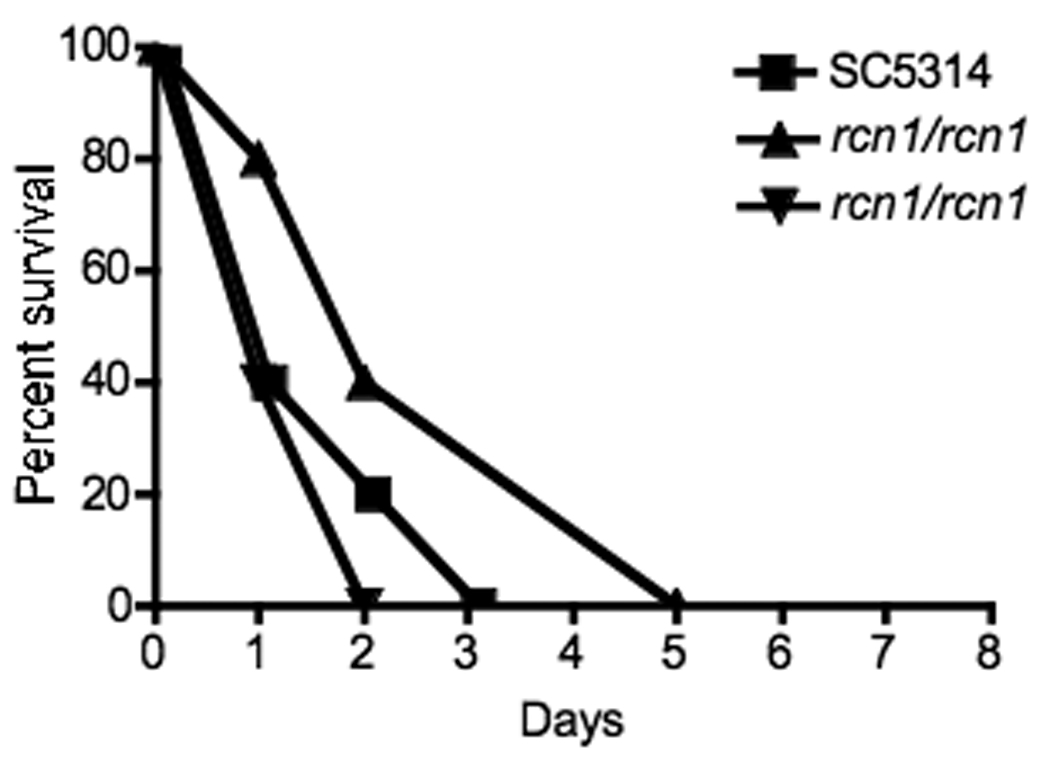
An inoculum of 5 × 106 cells of wild-type (SC5314), two independent rcn1/rcn1 (JLR36.3 and JLR37.1), and two rcn1/rcn1 + RCN1 (JLR180 and JLR323) strains were injected into the lateral tail vein. Five outbred ICR mice were infected per group. There was no significant difference in the survival between the wild-type and either the mutant or complemented strains (p = 0.06). Shown above are the wild-type and deletion mutant strains.
Interestingly, further studies have demonstrated that the virulence defect of calcineurin mutant strains is host niche-specific. In contrast to disseminated models of candidiasis, calcineurin mutants exhibit wild-type virulence in both pulmonary and vaginal models of infection (Bader et al., 2006). However, a murine keratitis model demonstrated faster resolution of infection in animals treated with the cnb1/cnb1 mutant strain or in those animals treated with both a calcineurin inhibitor and antifungal compared with either drug alone (Onyewu et al., 2006). Because oropharyngeal candidiasis is a major manifestation of Candida infection, particularly in HIV/AIDS patients (Fidel, 2006; Sweet, 1997), we tested the virulence of both cnb1/cnb1 and rcn1/rcn1 strains in this host-niche. After 5 days of infection, the oral fungal burden and histopathology of mice infected with these mutants was similar to mice infected with the wild-type strain (Figure 4 and data not shown). These results, indicate that neither calcineurin nor Rcn1 are required for virulence during oropharyngeal disease (Figure 4).
Figure 4. Virulence in a murine oropharyngeal candidiasis model.

The oropharyngeal candidiasis infections were performed according to Park, et al. 2005. Seven mice were infected per group. There was no significant difference between the wild-type SC5314 and rcn1/rcn1 mutant (p = 0.32) or between the wild-type DAY185 and cnb1/cnb1 mutant (p = 0.21).
Overexpression of C. albicans RCN1 can inhibit S. cerevisiae calcineurin function
To determine whether C. albicans Rcn1 is capable of interacting with calcineurin, the gene encoding it was expressed heterologously in S. cerevisiae. Our rationale for this experiment is as follows. In S. cerevisiae, RCN1 was originally identified in a screen for genes that, when overexpressed, inhibit calcineurin function and thus promote survival of a pmc1 strain in the presence of high Ca2+ (Hilioti and Cunningham, 2003). In S. cerevisiae, calcineurin plays a key role in regulating intracellular compartmentalization of Ca2+ (Cunningham and Fink, 1994a). Activated calcineurin induces the expression of the gene encoding Pmc1, a vacuolar Ca2+ pump, and thereby promotes influx of calcium into the vacuole. However, calcineurin also simultaneously inhibits the function of another vacuolar calcium pump, Vcx1, through a post-translational mechanism. The activity of either Pmc1 or Vcx1 is required to redistribute intracellular Ca2+ and enable the yeast to survive in high calcium conditions. In the presence of a functional calcineurin protein, deletion of PMC1 is lethal to cells in a high calcium environment; however, if calcineurin function is inhibited then Vcx1 is activated and cells survive calcium stress (Cunningham and Fink, 1994a; Hilioti and Cunningham, 2003). Thus, if overexpression of RCN1 inhibits calcineurin, pmc1 strains are rescued in the presence of calcium.
Plasmids were generated allowing expression of both the S. cerevisiae and C. albicans RCN1 genes under the control of the inducible S. cerevisiae GAL1 promoter. vcx1, pmc1, or pmc1 vcx1 S. cerevisiae strains were transformed with the plasmids. As expected, overexpression of S. cerevisiae RCN1 rescued pmc1 strains in the presence of 300 mM calcium (Figure 5). The C. albicans RCN1 homolog also rescued pmc1 strains in a high calcium environment, suggesting that C. albicans RCN1 is capable of interacting with and inhibiting S. cerevisiae calcineurin. Due to the highly conserved nature of calcineurin, CaRcn1 is likely to interact similarly with C. albicans calcineurin.
Figure 5. Overexpression of CaRCN1 rescues the viability of pmc1 strains.
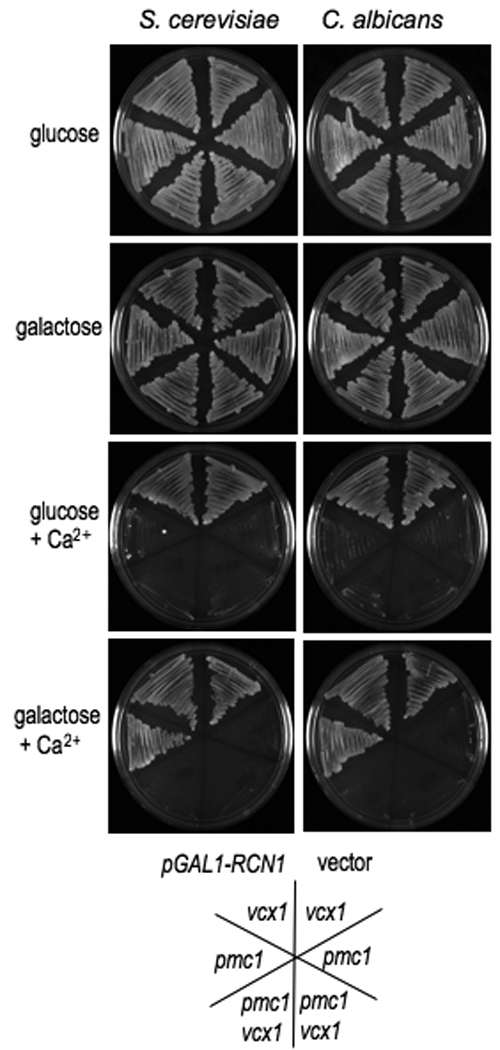
The C. albicans or S. cerevisiae RCN1 gene was cloned into plasmid pYES for expression in S. cerevisiae under the control of the GAL1 promoter. The plasmids were sequence confirmed and then strains pmc1::TRP1 vcx1 Δ (K665), pmc1::TRP1 (K605), and vcx1 (K661) were transformed. Under conditions of high calcium, calcineurin promotes the expression of PMC1 and inhibits Vcx1 function. pmc1 strains are unable to grow at high calcium levels unless calcineurin inhibition of Vcx1 is abolished. Overexpression of either the CaRCN1 or ScRCN1 gene allows growth of pmc1 strains in high calcium suggesting that calcineurin function is inhibited, whereas presence of the vector alone did not effect the growth of pmc1 strains. Inhibition of calcineurin also prevents the strong upregulation of PMC1 in response to high calcium, thus the high calcium conditions were chosen so that the basal expression of PMC1 was sufficient to support growth when calcineurin was inhibited in the vcx1 background. Strains were plated on solid YPD medium alone or containing 0.3 M CaCl2. The carbon source was either 2% glucose or 2% galactose, as indicated.
rcn1/rcn1 crz1/crz1 double mutants have modestly increased serum, Li+, and SDS sensitivity compared with either single mutant alone
Thus far, in C. albicans, only the Crz1 protein has been shown to act downstream of calcineurin (Karababa et al., 2006; Onyewu et al., 2004; Santos and de Larrinoa, 2005). Similar to rcn1/rcn1 strains, crz1/crz1 strains share a subset of cation and drug sensitivities with calcineurin mutant strains, but fail to entirely mimic the sensitivity profile of calcineurin deletion strains. Calcineurin likely has multiple downstream effectors and thus multiple deletion mutations may need to be combined to completely recapitulate calcineurin mutant phenotypes. Thus, we tested whether rcn1/rcn1 crz1/crz1 strains would be more sensitive to various stressors than either single mutant. Multiple independent double mutant strains were isolated and tested for sensitivity to a variety of stress conditions. The double mutants were found to be more sensitive to Li+ and SDS than either single mutant (Figure 6). Additionally, the double mutants had significantly reduced growth in serum compared with either single mutant; however, unlike calcineurin mutants, which are killed in serum, the rcn1/rcn1 crz1/crz1 mutants remained viable (Figure 7). Further experiments will determine whether the calcium stress of serum contributes to the decreased growth of the rcn1/rcn1 crz1/crz1 strains.
Figure 6. Phenotypic analysis of rcn1/rcn1 crz1/crz1 strains.
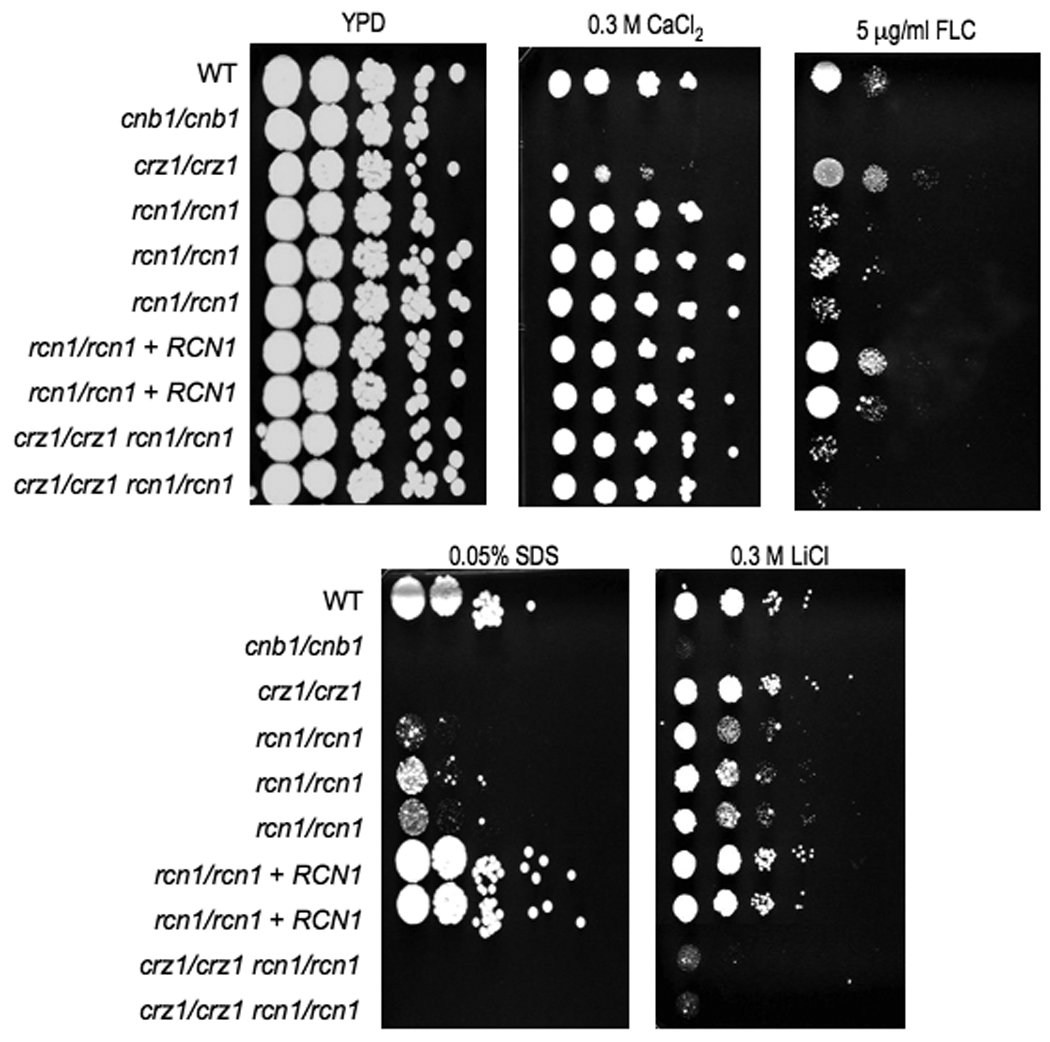
Wild-type strain (SC5314), cnb1/cnb1 (JRB64), rcn1/rcn1 mutants (JLR36.3, JLR37.1, and JLR38.1), rcn1/rcn1 +RCN1 complemented strain (JLR180, and JLR323), and rcn1/rcn1 crz1/crz1 double mutants (JLR548 and JLR578) were serially diluted and spotted onto solid YPD media containing the designated salt or drug concentration, and incubated at 30°C for 48 hours. The double mutants have a more severe phenotype than the single mutants on LiCl and fluconazole (FLU).
Figure 7. rcn1/rcn1 crz1/crz1 double mutants have increased serum sensitivity compared to either single mutant alone.
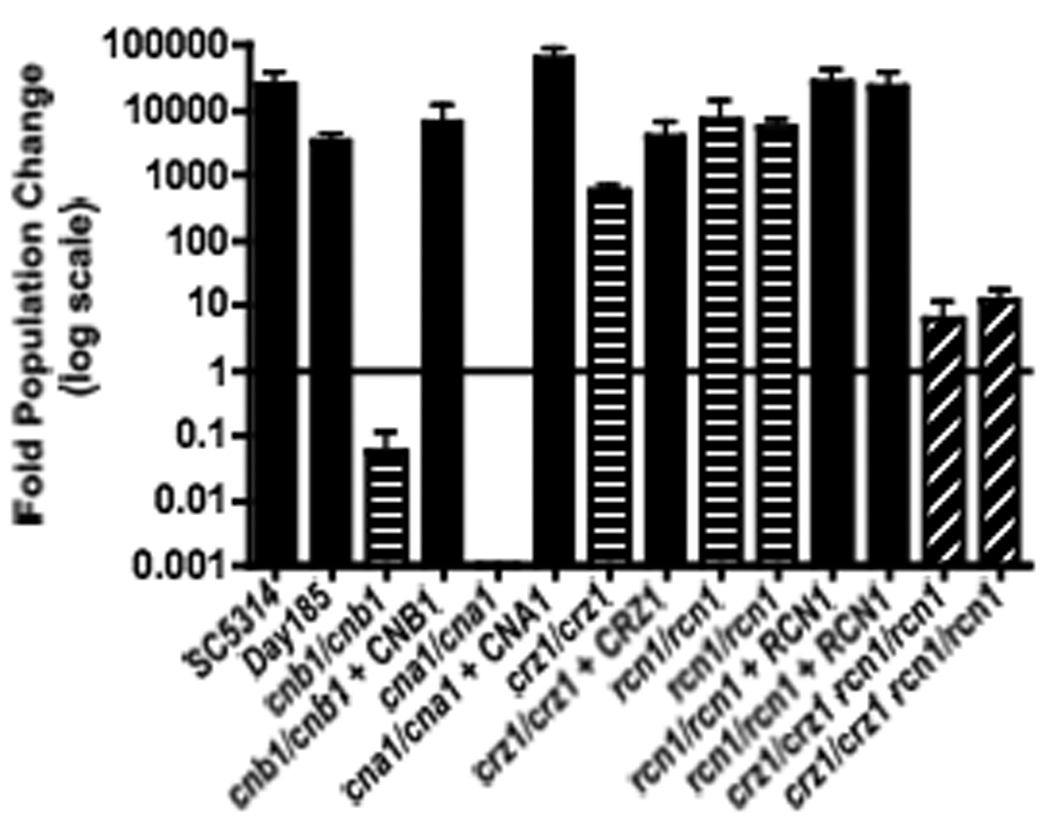
Strains were grown for 24 hours in 100% Fetal Bovine Serum at 30°C. Appropriate dilutions from each culture were plated onto YPD at 0 and 24 hours and then incubated at 30°C overnight. Fold population change was determined by dividing CFUs at 24 hours by CFUs at 0 hours. Error bars represent the standard error from two independent experiments. SC5314 and DAY185 served as the wild-type control strains. Strains tested were wild-type (SC5314 and DAY185), cnb1/cnb1 (JRB64), cnb1/cnb1 + CNB1 (MCC85), cna1/cna1 (SCCMP1M4), cna1/cna1 + CNA1 (SCCMP1M2), crz1/crz1 (OCC1.1), crz1/crz1 + CRZ1 (OCC7), rcn1/rcn1 (JLR36.3 and JLR37.1), rcn1/rcn1 + RCN1 (JLR180 and JLR323), and crz1/crz1 rcn1/rcn1 (JLR548 and JLR578)
Identification of C. albicans MID1 and CCH1 homologs; mid1/mid1 and cch1/cch1 strains are sensitive to LiCl and SDS
The C. albicans MID1 and CCH1 genes were identified based upon best hit reciprocal BLAST searches using the S. cerevisiae homologs. In S. cerevisiae, Mid1 and Cch1 form a calcium channel complex that promotes entry of calcium into cells in response to various stresses, including exposure to mating pheromone, endoplasmic reticulum stress, and cations (Bonilla and Cunningham, 2003; Bonilla et al., 2002; Fischer et al., 1997; Iida et al., 1994; Peiter et al., 2005). In response to ER stress, the calcium channels are activated by the Mpk1 pathway and are necessary for calcineurin activation and cell survival (Bonilla and Cunningham, 2003). Activated calcineurin dephosphorylates Mid1, resulting in feedback inhibition of the channel and preventing further influx of calcium (Bonilla et al., 2002). Therefore, the phenotypes of mid1 and cch1 cells in response to ER stress are similar to those of a calcineurin mutant. Consistent with the hypothesis that Mid1 and Cch1 form a complex, both the single and double mutants of S. cerevisiae have the same phenotype; however, in other species, such as C. neoformans, some CCH1-independent functions have been observed (Liu et al., 2006). Previous analysis in C. albicans on the role of calcium signaling in thigmotropism and galvanotropism reported that while Mid1 played a larger role in thigmotropism, Cch1 was more important for galvanotropism, suggesting that the proteins may have some differences in function (Brand et al., 2007). Recent studies examining the genes regulated in a calcineurin- and Crz1-dependent manner found that CCH1 is upregulated by activation of either calcineurin or Crz1 (Karababa et al., 2006).
Here both the MID1 and the CCH1 gene were deleted with the SAT1 flipper cassette in the wild-type SC5314 background, and mid1/mid1 cch1/cch1 double mutants were also generated. The susceptibility of mid1/mid1 and cch1/cch1 strains was tested in response to cations (Ca2+, Na+, and Li+), SDS, and fluconazole (Figure 8). Both single mutants were sensitive to LiCl and SDS, although not to the same extent as calcineurin mutants. Although neither mutant was sensitive to high Ca2+, the cch1/cch1 deletion strains were sensitive to low calcium environments. The fluconazole sensitivity of both single mutants was tested using E-test strips, and demonstrated modestly increased susceptibility (MIC80= 0.5 – 0.75) compared with wild-type (MIC80= 1.0 – 1.5). In all cases the mid1/mid1 cch1/cch1 double mutant behaved similarly to the single mutants, suggesting that at least for the phenotypes studied, neither protein has independent functions.
Figure 8. mid1/mid1 and cch1/cch1 are sensitive to Li+ cations and SDS.
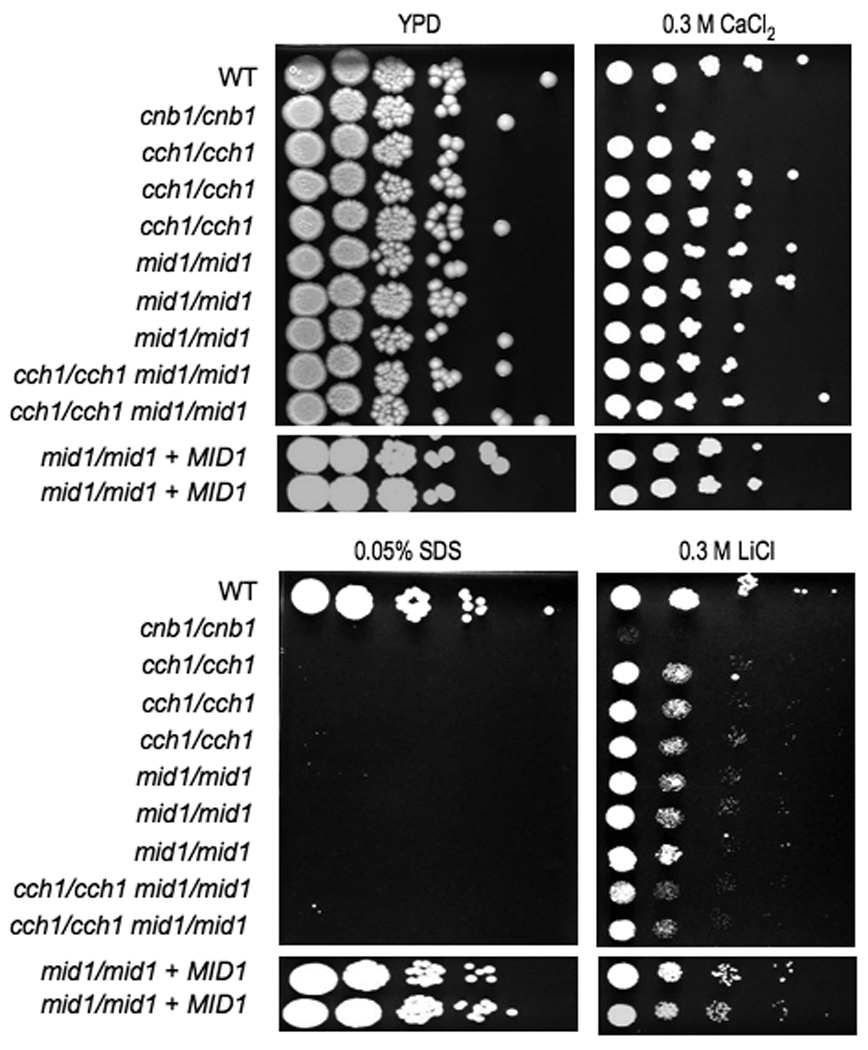
Serial dilutions of wild-type (SC5314), mid1/mid1 (JLR248, JLR253, and JLR255), cch1/cch1 (JLR48, JLR50, and JLR265), cnb1/cnb1 (JRB64), mid1/mid1 cch1/cch1 (JLR519 and JLR521), and mid1/mid1 + MID1 (JLR284 and JLR301) were spotted on solid YPD media containing the designated salt or drug concentration and incubated at 30°C for 48 hours.
Discussion
The calcineurin signaling pathway is a key mediator of stress responses in C. albicans and has clinical potential as a therapeutic target to enhance the efficacy of the current antifungal armamentarium. Here we have identified additional components of the calcineurin signaling pathway using a candidate gene approach based upon knowledge of the pathway in other organisms. Heterologous expression experiments provide evidence that Rcn1 is capable of inhibiting calcineurin function, similar to the S. cerevisiae homolog. Similarly to S. cerevisiae, an rcn1/rcn1 strain shares some similar sensitivities with calcineurin deletion strains; however, they exhibit a more intermediate phenotype and do not completely recapitulate the full severity of the calcineurin mutant phenotype. Microarray studies suggest that Crz1 may be solely responsible for the calcineurin-dependent transcriptional response (Karababa et al., 2006); however, calcineurin likely has other downstream effectors that are direct dephosphorylation targets regulated post-transcriptionally. Thus, to completely recapitulate a calcineurin mutant phenotype it is likely that multiple simultaneous gene deletions may be required. The double crz1/crz1 rcn1/rcn1 mutant had enhanced sensitivity to several stresses, suggesting that although both proteins interact with calcineurin, they may influence different downstream events. Further studies are needed to establish whether Rcn1 directly interacts with calcineurin in C. albicans in a manner similar to S. cerevisiae. The high degree of homology of calcineurin between these two fungi makes it likely that the relationship between Rcn1 and calcineurin may be similar in both species.
Several lines of evidence suggest that the Ca2+ channel subunits encoded by MID1 and CCH1 may play a role in calcineurin signaling. Studies in S. cerevisiae have shown that in response to endoplasmic reticulum stress the Mid1/Cch1 calcium channel is activated and the resulting influx of calcium activates calcineurin signaling (Bonilla and Cunningham, 2003; Bonilla et al., 2002). Studies on thigmotropism and galvanotropism showed that Mid1 and Cch1 are required for hyphal reorientation (respectively) and that calcineurin was required for the galvanotropic, but not the thigmotropic responses. However, Crz1 was required for both processes, suggesting that Crz1 could have calcineurin-independent functions (Brand et al., 2007). Additionally, microarray studies in C. albicans have shown that calcineurin regulates expression of the CCH1 gene (Karababa et al., 2006). Deletion analysis of CCH1 and MID1 suggests that these genes play a role in tolerance to membrane stress and tolerance to fluconazole, although they have an intermediate phenotype compared with calcineurin mutants. Further studies examining the role that these proteins play in activating calcineurin signaling will be required, including studying the activation of Crz1 in these mutants. For the phenotypes analyzed in this study, both single and double mutants had similar phenotypes suggesting that the two genes act as a complex (as in S. cerevisiae) or act in a linear pathway and thus there is no additive effect observed in the double mutant.
Interestingly, these studies also extend our understanding of the host-niche specific role of calcineurin in virulence. Previous studies demonstrated that calcineurin is required for virulence in murine models of systemic candidiasis (Bader et al., 2003; Blankenship et al., 2003b) and candidal keratitis (Onyewu et al., 2006), but is dispensable for virulence in murine vaginal and pulmonary infection models (Bader et al., 2006). Our results suggest that calcineurin is also not required for virulence in oropharyngeal infections. Thus, inhibition of calcineurin alone could be utilized as a novel mechanism of antifungal therapy in certain manifestation of candidal disease, such as bloodstream infections, or keratitis; however, in oropharyngeal, pulmonary, or vaginal candidal infections inhibition of calcineurin alone may not be sufficient. Combining calcineurin inhibition with azoles would likely provide enhanced antifungal activity in all host-niches.
The therapeutic potential of calcineurin inhibitors in combination therapy to enhance the efficacy of current antifungals makes furthering our understanding of this signaling pathway of considerable import. Prior to this study only one direct target of calcineurin, Crz1, had been extensively studied in C. albicans. The role of Rcn1 is of interest in C. albicans as peptides corresponding to the calcineurin binding segment of human RCAN family members have also been show to inhibit calcineurin function (Mulero et al., 2009), and could potentially be used as starting points to develop novel therapeutics.
Acknowledgements
We thank Joachim Morschhauser for the generous gift of the pSFS2A SAT1 flipper cassette for gene disruptions as well as the C. albicans calcineurin A deletion and complemented strains, and Kyle Cunningham for the S. cerevisiae strains used in the heterologous expression experiments. We also thank Norma Solis and Trang Phan for assistance with the oropharyngeal candidiasis experiments. This work was supported by NIAID R01 grants AI42159 and AI50438, and by NIDCR R01 grant DE017088.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bader T, Bodendorfer B, Schroppel K, Morschhauser J. Calcineurin is essential for virulence in Candida albicans. Infect Immun. 2003;71:5344–5354. doi: 10.1128/IAI.71.9.5344-5354.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader T, Schroppel K, Bentink S, Agabian N, Kohler G, Morschhauser J. Role of calcineurin in stress resistance, morphogenesis, and virulence of a Candida albicans wild-type strain. Infect Immun. 2006;74:4366–4369. doi: 10.1128/IAI.00142-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beals CR, Clipstone NA, Ho SN, Crabtree GR. Nuclear localization of NF-ATc by a calcineurin-dependent, cyclosporin-sensitive intramolecular interaction. Genes Dev. 1997;11:824–834. doi: 10.1101/gad.11.7.824. [DOI] [PubMed] [Google Scholar]
- Blankenship JR, Heitman J. Calcineurin is required for Candida albicans to survive calcium stress in serum. Infect Immun. 2005;73:5767–5774. doi: 10.1128/IAI.73.9.5767-5774.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenship JR, Steinbach WJ, Perfect JR, Heitman J. Teaching old drugs new tricks: reincarnating immunosuppressants as antifungal drugs. Curr Opin Investig Drugs. 2003a;4:192–199. [PubMed] [Google Scholar]
- Blankenship JR, Wormley FL, Boyce MK, Schell WA, Filler SG, Perfect J, Heitman J. Calcineurin is essential for Candida albicans survival in serum and virulence. Eukaryotic Cell. 2003b;2:422–430. doi: 10.1128/EC.2.3.422-430.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonilla M, Cunningham KW. Mitogen-activated protein kinase stimulation of Ca(2+) signaling is required for survival of endoplasmic reticulum stress in yeast. Mol Biol Cell. 2003;14:4296–4305. doi: 10.1091/mbc.E03-02-0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonilla M, Nastase KK, Cunningham KW. Essential role of calcineurin in response to endoplasmic reticulum stress. EMBO J. 2002;21:2343–2353. doi: 10.1093/emboj/21.10.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand A, Shanks S, Duncan VM, Yang M, Mackenzie K, Gow NA. Hyphal orientation of Candida albicans is regulated by a calcium-dependent mechanism. Curr Biol. 2007;17:347–352. doi: 10.1016/j.cub.2006.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuder T, Hemenway CS, Movva NR, Cardenas ME, Heitman J. Calcineurin is essential in cyclosporin A- and FK506-sensitive yeast strains. Proc Natl Acad Sci U S A. 1994;91:5372–5376. doi: 10.1073/pnas.91.12.5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas ME, Hemenway C, Muir RS, Ye R, Fiorentino D, Heitman J. Immunophilins interact with calcineurin in the absence of exogenous immunosuppressive ligands. Embo J. 1994;13:5944–5957. doi: 10.1002/j.1460-2075.1994.tb06940.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas ME, Muir RS, Breuder T, Heitman J. Targets of immunophilin-immunosuppressant complexes are distinct highly conserved regions of calcineurin A. Embo J. 1995;14:2772–2783. doi: 10.1002/j.1460-2075.1995.tb07277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin ER, Olson EN, Richardson JA, Yang Q, Humphries C, Shelton JM, Wu H, Zhu W, Bassel-Duby R, Williams RS. A calcineurin-dependent transcriptional pathway controls skeletal muscle fiber type. Genes Dev. 1998;12:2499–2509. doi: 10.1101/gad.12.16.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clipstone NA, Crabtree GR. Identification of calcineurin as a key signalling enzyme in T-lymphocyte activation. Nature. 1992;357:695–697. doi: 10.1038/357695a0. [DOI] [PubMed] [Google Scholar]
- Clipstone NA, Fiorentino DF, Crabtree GR. Molecular analysis of the interaction of calcineurin with drug-immunophilin complexes. J Biol Chem. 1994;269:26431–26437. [PubMed] [Google Scholar]
- Crabtree GR. Generic signals and specific outcomes: signaling through Ca2+, calcineurin, and NF-AT. Cell. 1999;96:611–614. doi: 10.1016/s0092-8674(00)80571-1. [DOI] [PubMed] [Google Scholar]
- Cruz MC, Goldstein AL, Blankenship JR, Del Poeta M, Davis D, Cardenas ME, Perfect J, McCusker JH, Heitman J. Calcineurin is essential for survival during membrane stress in Candida albicans. EMBO J. 2002;21:546–559. doi: 10.1093/emboj/21.4.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham KW, Fink GR. Ca2+ transport in Saccharomyces cerevisiae. J Exp Biol. 1994a;196:157–166. doi: 10.1242/jeb.196.1.157. [DOI] [PubMed] [Google Scholar]
- Cunningham KW, Fink GR. Calcineurin-dependent growth control in Saccharomyces cerevisiae mutants lacking PMC1, a homolog of plasma membrane Ca2+ ATPases. J Cell Biol. 1994b;124:351–363. doi: 10.1083/jcb.124.3.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham KW, Fink GR. Calcineurin inhibits VCX1-dependent H+/Ca2+ exchange and induces Ca2+ ATPases in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2226–2237. doi: 10.1128/mcb.16.5.2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyert MS. Calcineurin signaling in Saccharomyces cerevisiae: how yeast go crazy in response to stress. Biochem Biophys Res Commun. 2003;311:1143–1150. doi: 10.1016/s0006-291x(03)01552-3. [DOI] [PubMed] [Google Scholar]
- Davies KJ, Ermak G, Rothermel BA, Pritchard M, Heitman J, Ahnn J, Henrique-Silva F, Crawford D, Canaider S, Strippoli P, et al. Renaming the DSCR1/Adapt78 gene family as RCAN: regulators of calcineurin. Faseb J. 2007;21:3023–3028. doi: 10.1096/fj.06-7246com. [DOI] [PubMed] [Google Scholar]
- Davis D, Edwards JE, Jr, Mitchell AP, Ibrahim AS. Candida albicans RIM101 pH response pathway is required for host-pathogen interactions. Infect Immun. 2000;68:5953–5959. doi: 10.1128/iai.68.10.5953-5959.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards JE., Jr Invasive candida infections--evolution of a fungal pathogen. N Engl J Med. 1991;324:1060–1062. doi: 10.1056/NEJM199104113241511. [DOI] [PubMed] [Google Scholar]
- Fidel PL., Jr Candida-host interactions in HIV disease: relationships in oropharyngeal candidiasis. Adv Dent Res. 2006;19:80–84. doi: 10.1177/154407370601900116. [DOI] [PubMed] [Google Scholar]
- Fischer M, Schnell N, Chattaway J, Davies P, Dixon G, Sanders D. The Saccharomyces cerevisiae CCH1 gene is involved in calcium influx and mating. FEBS Lett. 1997;419:259–262. doi: 10.1016/s0014-5793(97)01466-x. [DOI] [PubMed] [Google Scholar]
- Foor F, Parent SA, Morin N, Dahl AM, Ramadan N, Chrebet G, Bostian KA, Nielsen JB. Calcineurin mediates inhibition by FK506 and cyclosporin of recovery from α-factor arrest in yeast. Nature. 1992;360:682–684. doi: 10.1038/360682a0. [DOI] [PubMed] [Google Scholar]
- Fuentes JJ, Genesca L, Kingsbury TJ, Cunningham KW, Perez-Riba M, Estivill X, de la Luna S. DSCR1, overexpressed in Down syndrome, is an inhibitor of calcineurin-mediated signaling pathways. Hum Mol Genet. 2000;9:1681–1690. doi: 10.1093/hmg/9.11.1681. [DOI] [PubMed] [Google Scholar]
- Gillum AM, Tsay EY, Kirsch DR. Isolation of the Candida albicans gene for orotidine-5'-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol Gen Genet. 1984;198:179–182. doi: 10.1007/BF00328721. [DOI] [PubMed] [Google Scholar]
- Gorlach J, Fox DS, Cutler NS, Cox GM, Perfect JR, Heitman J. Identification and characterization of a highly conserved calcineurin binding protein, CBP1/calcipressin, in Cryptococcus neoformans. EMBO J. 2000;19:3618–3629. doi: 10.1093/emboj/19.14.3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazen KC. New and emerging yeast pathogens. Clin Microbiol Rev. 1995;8:462–478. doi: 10.1128/cmr.8.4.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemenway CS, Heitman J. Calcineurin. Structure, function, and inhibition. Cell Biochem Biophys. 1999;30:115–151. doi: 10.1007/BF02737887. [DOI] [PubMed] [Google Scholar]
- Hilioti Z, Cunningham KW. The RCN family of calcineurin regulators. Biochemical and Biophysical Research Communications. 2003;311:1089–1093. doi: 10.1016/s0006-291x(03)01515-8. [DOI] [PubMed] [Google Scholar]
- Hilioti Z, Gallagher DA, Low-Nam ST, Ramaswamy P, Gajer P, Kingsbury TJ, Birchwood CJ, Levchenko A, Cunningham KW. GSK-3 kinases enhance calcineurin signaling by phosphorylation of RCNs. Genes and Development. 2004;18:35–47. doi: 10.1101/gad.1159204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho S, Clipstone N, Timmermann L, Northrop J, Graef I, Fiorentino D, Nourse J, Crabtree GR. The mechanism of action of cyclosporin A and FK506. Clin Immunol Immunopathol. 1996;80:S40–S45. doi: 10.1006/clin.1996.0140. [DOI] [PubMed] [Google Scholar]
- Iida H, Nakamura H, Ono T, Okumura MS, Anraku Y. MID1, a novel Saccharomyces cerevisiae gene encoding a plasma membrane protein, is required for Ca2+ influx and mating. Molecular and Cellular Biology. 1994;14:8259–8271. doi: 10.1128/mcb.14.12.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karababa M, Valentino E, Pardini G, Coste AT, Bille J, Sanglard D. CRZ1, a target of the calcineurin pathway in Candida albicans. Mol Microbiol. 2006;59:1429–1451. doi: 10.1111/j.1365-2958.2005.05037.x. [DOI] [PubMed] [Google Scholar]
- Kishi T, Ikeda A, Nagao R, Koyama N. The SCFCdc4 ubiquitin ligase regulates calcineurin signaling through degradation of phosphorylated Rcn1, an inhibitor of calcineurin. Proc Natl Acad Sci U S A. 2007;104:17418–17423. doi: 10.1073/pnas.0704951104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klee CB, Crouch TH, Krinks MH. Calcineurin: a calcium- and calmodulin-binding protein of the nervous system. Proc Natl Acad Sci U S A. 1979;76:6270–6273. doi: 10.1073/pnas.76.12.6270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer D, Fresu L, Ashby DS, Freeman TC, Genazzani AA. Calcineurin controls the expression of numerous genes in cerebellar granule cells. Mol Cell Neurosci. 2003;23:325–330. doi: 10.1016/s1044-7431(03)00057-5. [DOI] [PubMed] [Google Scholar]
- Krcmery V, Barnes AJ. Non-albicans Candida spp. causing fungaemia: pathogenicity and antifungal resistance. J Hosp Infect. 2002;50:243–260. doi: 10.1053/jhin.2001.1151. [DOI] [PubMed] [Google Scholar]
- Krebs J. The role of calcium in apoptosis. Biometals. 1998;11:375–382. doi: 10.1023/a:1009226316146. [DOI] [PubMed] [Google Scholar]
- Liu M, Du P, Heinrich G, Cox GM, Gelli A. Cch1 mediates calcium entry in Cryptococcus neoformans and is essential in low-calcium environments. Eukaryot Cell. 2006;5:1788–1796. doi: 10.1128/EC.00158-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansuy IM, Mayford M, Jacob B, Kandel ER, Bach ME. Restricted and regulated overexpression reveals calcineurin as a key component in the transition from short-term to long-term memory. Cell. 1998;92:39–49. doi: 10.1016/s0092-8674(00)80897-1. [DOI] [PubMed] [Google Scholar]
- Marchetti O, Entenza JM, Sanglard D, Bille J, Glauser MP, Moreillon P. Fluconazole plus cyclosporine: a fungicidal combination effective against experimental endocarditis due to Candida albicans. Antimicrob Agents Chemother. 2000;44:2932–2938. doi: 10.1128/aac.44.11.2932-2938.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheos DP, Kingsbury TJ, Ahsan US, Cunningham KW. Tcn1p/Crz1p, a calcineurin-dependent transcription factor that differentially regulates gene expression in Saccharomyces cerevisiae. Genes Dev. 1997;11:3445–3458. doi: 10.1101/gad.11.24.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merz WG, Karp JE, Schron D, Saral R. Increased incidence of fungemia caused by Candida krusei. J Clin Microbiol. 1986;24:581–584. doi: 10.1128/jcm.24.4.581-584.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulero MC, Aubareda A, Orzaez M, Messeguer J, Serrano-Candelas E, Martinez-Hoyer S, Messeguer A, Perez-Paya E, Perez-Riba M. Inhibiting the Calcineurin-NFAT (Nuclear Factor of Activated T Cells) Signaling Pathway with a Regulator of Calcineurin-derived Peptide without Affecting General Calcineurin Phosphatase Activity. J Biol Chem. 2009;284:9394–9401. doi: 10.1074/jbc.M805889200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Liu Y, Hirata D, Namba H, Harada S-i, Hirokawa T, Miyakawa T. Protein phosphatase type 2B (calcineurin)-mediated, FK506-sensitive regulation of intracellular ions in yeast is an important determinant for adaptation to high salt stress conditions. EMBO J. 1993;12:4063–4071. doi: 10.1002/j.1460-2075.1993.tb06090.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen MH, Peacock JE, Jr, Morris AJ, Tanner DC, Nguyen ML, Snydman DR, Wagener MM, Rinaldi MG, Yu VL. The changing face of candidemia: emergence of non-Candida albicans species and antifungal resistance. Am J Med. 1996;100:617–623. doi: 10.1016/s0002-9343(95)00010-0. [DOI] [PubMed] [Google Scholar]
- Odds FC. Candida and candidosis. 2nd edn. London; Philadelphia: Baillière Tindall; 1988. [Google Scholar]
- Onyewu C, Afshari NA, Heitman J. Calcineurin promotes infection of the cornea by Candida albicans and can be targeted to enhance fluconazole therapy. Antimicrob Agents Chemother. 2006;50:3963–3965. doi: 10.1128/AAC.00393-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onyewu C, Blankenship JR, Del Poeta M, Heitman J. Ergosterol biosynthesis inhibitors become fungicidal when combined with calcineurin inhibitors against Candida albicans, Candida glabrata, and Candida krusei. Antimicrob Agents Chemother. 2003;47:956–964. doi: 10.1128/AAC.47.3.956-964.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onyewu C, Wormley FL, Perfect J, Heitman J. The calcineurin target, Crz1, functions in azole tolerance but is not required for virulence of Candida albicans. Infection and immunity. 2004;72:7330–7333. doi: 10.1128/IAI.72.12.7330-7333.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H, Myers CL, Sheppard DC, Phan QT, Sanchez AA, J EE, Filler SG. Role of the fungal Ras-protein kinase A pathway in governing epithelial cell interactions during oropharyngeal candidiasis. Cell Microbiol. 2005;7:499–510. doi: 10.1111/j.1462-5822.2004.00476.x. [DOI] [PubMed] [Google Scholar]
- Parsons SA, Millay DP, Sargent MA, Naya FJ, McNally EM, Sweeney HL, Molkentin JD. Genetic disruption of calcineurin improves skeletal muscle pathology and cardiac disease in a mouse model of limb-girdle muscular dystrophy. J Biol Chem. 2007;282:10068–10078. doi: 10.1074/jbc.M609368200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiter E, Fischer M, Sidaway K, Roberts SK, Sanders D. The Saccharomyces cerevisiae Ca2+ channel Cch1pMid1p is essential for tolerance to cold stress and iron toxicity. FEBS Lett. 2005;579:5697–5703. doi: 10.1016/j.febslet.2005.09.058. [DOI] [PubMed] [Google Scholar]
- Pfaller MA, Jones RN, Doern GV, Sader HS, Hollis RJ, Messer SA. International surveillance of bloodstream infections due to Candida species: frequency of occurrence and antifungal susceptibilities of isolates collected in 1997 in the United States, Canada, and South America for the SENTRY Program. The SENTRY Participant Group. J Clin Microbiol. 1998;36:1886–1889. doi: 10.1128/jcm.36.7.1886-1889.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuss O, Vik A, Kolter R, Morschhauser J. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene. 2004;341:119–127. doi: 10.1016/j.gene.2004.06.021. [DOI] [PubMed] [Google Scholar]
- Saito S, Hiroi Y, Zou Y, Aikawa R, Toko H, Shibasaki F, Yazaki Y, Nagai R, Komuro I. beta-Adrenergic pathway induces apoptosis through calcineurin activation in cardiac myocytes. J Biol Chem. 2000;275:34528–34533. doi: 10.1074/jbc.M002844200. [DOI] [PubMed] [Google Scholar]
- Sanglard D, Ischer F, Marchetti O, Entenza J, Bille J. Calcineurin A of Candida albicans: involvement in antifungal tolerance, cell morphogenesis and virulence. Mol Microbiol. 2003;48:959–976. doi: 10.1046/j.1365-2958.2003.03495.x. [DOI] [PubMed] [Google Scholar]
- Santos M, de Larrinoa IF. Functional characterization of the Candida albicans CRZ1 gene encoding a calcineurin-regulated transcription factor. Curr Genet. 2005;48:88–100. doi: 10.1007/s00294-005-0003-8. [DOI] [PubMed] [Google Scholar]
- Shibasaki F, McKeon F. Calcineurin functions in Ca(2+)-activated cell death in mammalian cells. J Cell Biol. 1995;131:735–743. doi: 10.1083/jcb.131.3.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stathopoulos AM, Cyert MS. Calcineurin acts through the CRZ1/TCN1-encoded transcription factor to regulate gene expression in yeast. Genes Dev. 1997;11:3432–3444. doi: 10.1101/gad.11.24.3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbach WJ, Reedy JL, Cramer RA, Jr, Perfect JR, Heitman J. Harnessing calcineurin as a novel anti-infective agent against invasive fungal infections. Nat Rev Microbiol. 2007;5:418–430. doi: 10.1038/nrmicro1680. [DOI] [PubMed] [Google Scholar]
- Strippoli P, Lenzi L, Petrini M, Carinci P, Zannotti M. A new gene family including DSCR1 (Down Syndrome Candidate Region 1) and ZAKI-4: characterization from yeast to human and identification of DSCR1-like 2, a novel human member (DSCR1L2) Genomics. 2000a;64:252–263. doi: 10.1006/geno.2000.6127. [DOI] [PubMed] [Google Scholar]
- Strippoli P, Lenzi L, Petrini M, Carinci P, Zannotti M. A new gene family including DSCR1 (Down Syndrome Candidate Region 1) and ZAKI-4: Characterization from yeast to human and identification of DSCR1-like 2, a novel human member ( DSCR1L2) Genomics. 2000b;64:252–263. doi: 10.1006/geno.2000.6127. [DOI] [PubMed] [Google Scholar]
- Sweet SP. Selection and pathogenicity of Candida albicans in HIV infection. Oral Dis. 1997;3 Suppl 1:S88–S95. doi: 10.1111/j.1601-0825.1997.tb00383.x. [DOI] [PubMed] [Google Scholar]
- Vega RB, Yang J, Rothermel BA, Bassel-Duby R, Williams RS. Multiple domains of MCIP1 contribute to inhibition of calcineurin activity. J Biol Chem. 2002;277:30401–30407. doi: 10.1074/jbc.M200123200. [DOI] [PubMed] [Google Scholar]
- Wang HG, Pathan N, Ethell IM, Krajewski S, Yamaguchi Y, Shibasaki F, McKeon F, Bobo T, Franke TF, Reed JC. Ca2+-induced apoptosis through calcineurin dephosphorylation of BAD. Science. 1999;284:339–343. doi: 10.1126/science.284.5412.339. [DOI] [PubMed] [Google Scholar]
- Weitlauf C, Winder D. Calcineurin, synaptic plasticity, and memory. Scientific World Journal. 2001;1:530–533. doi: 10.1100/tsw.2001.259. [DOI] [PMC free article] [PubMed] [Google Scholar]


